Abstract
As part of the Saccharomyces Genome Deletion Project, sets of presumably isogenic haploid and diploid strains that differed only by single gene deletions were constructed. We found that one set of 96 strains (containing deletions of ORFs located between YOR097C and YOR192C) in the collection, which was derived from the haploid BY4741, has an additional mutation in the MSH3 mismatch repair gene.
GROUPS of researchers involved in the Saccharomyces Genome Deletion Project generated sets of haploid and diploid strains containing individual deletions in nearly all nonessential genes in the yeast genome (Winzeler et al. 1999; Giaever et al. 2002). In these strains, each individual open reading frame (ORF) was replaced by the KanMX drug-resistance gene. This collection has been screened for a variety of interesting phenotypes, including drug sensitivities, growth in various types of media, spontaneous mutation rates, ability to respond to osmotic stress, and sporulation proficiency (Winzeler et al. 1999; Deutschbauer et al. 2002; Giaever et al. 2002; Huang et al. 2003). The assumption in this type of analysis is that the strains are identical except for the single mutation introduced by transformation. Although, in general, this assumption is likely to be correct, it has been noted that some mutations result in genomic instability that leads to additional changes. For example, mec1 haploid strains often become disomic for chromosome IV (Gasch et al. 2001). In this report, we describe another, and more subtle, problem: the de novo acquisition of a mutation in one of the progenitor strains used to construct a subset of the deletions.
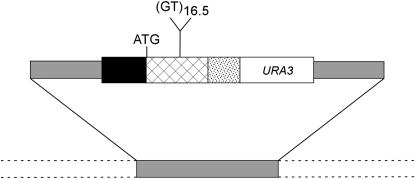
Our discovery of this problem was in conjunction with our analysis of the effect of chromosome context on microsatellite instability. In a previous study (Hawk et al. 2005), we showed that the stability of a microsatellite varied by ∼16-fold, depending on genome location, and that most of this variation reflected context-specific variation in the rate of DNA mismatch repair (MMR). This study was based on the analysis of 10 genomic locations. To extend this study, we constructed a plasmid (pKRL1, Figure 1) that contained a galactose-inducible URA3-GT fusion gene (Wierdl et al. 1996) inserted into the KanMX-coding region of the pFA6-KanMX4 plasmid (Wach et al. 1994). The URA3-GT fusion gene has an in-frame insertion [a poly(GT) sequence 33 bp in length] in a nonessential region of the fusion gene. A NotI fragment derived from this plasmid, containing this fusion gene and flanking sequences from the KanMX gene, was transformed into derivatives of the haploid strain BY4741 in which different genes had been deleted using the KanMX cassette. Transformants in which the reporter gene was inserted within the KanMX cassette were phenotypically Ura+ and G418 sensitive (Figure 1). Using this method, we could insert the same reporter sequence into any ORF of the genome that had been previously tagged with the KanMX cassette.
Figure 1.—
Construction of derivatives of the BY4741 collection with insertions of the URA3-GT reporter gene. In a previous study (Wierdl et al. 1996), we constructed a fusion gene with a GAL1,10 promoter regulating transcription of a Ura3 fusion protein. Into a region of the gene that did not affect gene function, we inserted an in-frame (33 bp) poly(GT) tract. A plasmid with this gene (pMBW1) was treated with KpnI and XhoI, and the fragment containing the fusion gene was treated with the Klenow fragment to create blunt ends, which were then inserted into the NruI site of pFA6-kanMX4. In the resulting plasmid (pKRL1), the URA3-GT fusion gene is inserted into the middle of the KanMX gene; the fusion gene and KanMX are transcribed in opposite directions. To construct strains with the fusion gene inserted in different positions, we treated pKRL1 with NotI and transformed strains with single KanMX insertions replacing individual ORFs. We selected Ura+ transformants and screened for those transformants that become G418 sensitive. Shaded boxes indicate KanMX; solid box, GAL1,10 promoter; crosshatched box, LYS2 sequences; stippled box, HIS4 sequences; and open box, URA3. The dotted lines indicate the chromosomal sequences flanking the KanMX gene.
Strains with the URA3-GT fusion gene are sensitive to 5-fluoroorotic acid (5-FOA), a compound that kills strains with a wild-type URA3 gene (Boeke et al. 1984). Since alterations in the length of the microsatellite that result in loss of the normal reading frame (for example, a deletion of 2 bp) cause the cells to be 5-FOAR, the rate of appearance of 5-FOAR derivatives is a measurement of the rate of microsatellite instability (Hawk et al. 2005). Rates are determined by measurement of the frequencies of 5-FOAR derivatives in multiple independent cultures and conversion of these frequencies to rates using the method of the median (Lea and Coulson 1949).
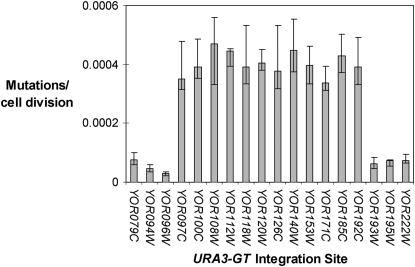
We inserted the URA3-GT construct into ∼50 derivatives of BY4741, representing 50 random genomic locations. The rates of instability observed in different genomic locations varied from 1 × 10−5 to 5 × 10−4. Two of the insertions with the highest rates [YOR108W (5 × 10−4) and YOR185C (4 × 10−4)] were located close together on chromosome XV. To determine whether there was a region on chromosome XV that had elevated microsatellite instability, we constructed 18 derivatives of BY4741 with insertions of the reporter gene within ORFs on chromosome XV and measured the rates of instability in these strains. As shown in Figure 2, all 12 insertions in the region located between YOR097C and YOR192C showed significantly elevated rates of microsatellite instability relative to the 6 insertions outside of this region. The transition between the rate of high instability and low instability was abrupt, occurring between ORFs YOR096W and YOR097C at one junction and between ORFs YOR192C and YOR193W at the other.
Figure 2.—
Microsatellite mutation rates in deletion strains. Haploid strains with the URA3-GT reporter at different genomic locations were constructed as described above. Mutation rates were calculated by fluctuation analysis as described previously (Hawk et al. 2005). Error bars indicate 95% confidence limits.
The 96 strains with disruptions of the ORFs between YOR097C and YOR192C were produced by a single lab as part of the Saccharomyces Deletion Project (A. Chu, personal communication). This same lab also produced two additional 96-strain deletion sets. We measured mutation rates of our reporter integrated in strains from these other sets and found no significant increase in rates (data not shown). These data suggest that the progenitor “wild-type” strain used by this lab to generate the YOR097C–YOR192C set, but not the additional two sets, had acquired a mutation that resulted in elevated levels of microsatellite instability. The gene disruptions made for ORFs in the two regions flanking the YOR097C–YOR192C chromosome segment were made by other labs (A. Chu, personal communication), presumably using a different isolate of BY4741.
In yeast, loss of the capacity to repair mismatches greatly elevates microsatellite instability (Strand et al. 1993). In most eukaryotes, there are two systems that recognize mismatches resulting from misincorporation errors or DNA polymerase slippage events (reviewed by Harfe and Jinks-Robertson 2000). A complex of Msh2p/Msh6p/Mlh1p/Pms1p recognizes base–base mismatches and insertions and deletions of one base, whereas a complex of Msh2p/Msh3p/Mlh1p/Pms1p recognizes insertions and deletions of 1–14 bases (Harfe and Jinks-Robertson 2000). The stability of dinucleotide microsatellites is affected much more strongly by an msh3 mutation than by an msh6 mutation (Sia et al. 1997). Previously, Huang et al. (2003) screened the BY4741 deletion collection for mutants that had elevated rates of forward mutation at the CAN1 locus. These mutators did not map in the YOR097C–YOR192C region. Since we have previously shown that msh3 strains have very elevated instability for dinucleotide microsatellites, but a wild-type rate of mutation at the CAN1 locus (Sia et al. 1997), these results suggested that the progenitor strain used to make the deletions might have an msh3 mutation.
We sequenced the MSH3 gene in five of the strains containing deletions of genes located on the right arm of chromosome XV. We found a single base-pair deletion (loss of an A located at position 1551) in MSH3 in the three strains containing deletions of genes between YOR097C and YOR192C (YOR097C, YOR153W, YOR192C), but not in those containing deletions of genes outside of this region (YOR096W, YOR193W). The observed frameshift mutation results in a truncated Msh3 protein, composed of 521 instead of 1047 amino acids. To verify that this msh3 mutation was responsible for the elevated microsatellite instability, we inserted the wild-type MSH3 gene into strains that had the URA3-GT reporter at YOR097C and YOR192C. In these derivatives, the rates of microsatellite instability were substantially reduced (Figure 3), demonstrating that the msh3 mutation is responsible for the elevated rates of microsatellite instability.
Figure 3.—
Microsatellite mutation rates in deletion strains with msh3 replaced with MSH3. Strains with integrated URA3-GT at two different sites (YOR97C and YOR192C) were transformed with the plasmid pEAI218 (provided by E. Alani, Cornell University) digested with KpnI and PvuII. The resulting DNA fragment contains a wild-type MSH3 gene with an adjacent LEU2 gene. Transformants were selected on media lacking leucine. Correct integration at the site of the endogenous MSH3 gene was confirmed by sequencing. Mutation rates in the original strains with the URA3-GT construct and in the MSH3 derivatives are shown by shaded bars and solid bars, respectively. Error bars indicate 95% confidence limits.
Our results illustrate the difficulties of constructing an isogenic collection of yeast strains, as well as the difficulties of detecting departures from non-isogenicity. It is likely that, when equivalent isogenic knockout collections are constructed in higher eukaryotes, these collections will have similar problems. The appearance of a de novo spontaneous mutation in the progenitor strains used to construct a collection of deletions may seem improbable; however, because the strains in the YOR097C–YOR192C region share the same four auxotrophic mutations as BY4741, it is unlikely that these strains were constructed with a contaminating unrelated yeast strain that had an msh3 mutation. An alternative possibility is that some genes, such as MSH3, might be more susceptible to mutations than the “standard” genes (CAN1 and URA3) usually used to measure forward mutation rates. It is also possible that this mutation arose in BY4741 cells that were kept in a mutagenic environmental condition (high temperature, low temperature, stationary phase, etc.). Finally, we point out that the observations reported in this article do not influence our earlier conclusion that the efficiency of MMR varies in different regions of the yeast genome (Hawk et al. 2005), since this previous study did not utilize the knockout collection.
Acknowledgments
We thank J. Lieb and E. Alani for providing deletion strains and the MSH3-replacement plasmid, respectively. We thank Sue Jinks-Robertson for suggesting that we investigate the construction of the deletion collection and A. Chu and G. Volckaert for providing details about the construction of deletions in BY4741. The research was supported by National Institutes of Health grants GM24110 (T.D.P.) and CA63264 (R.A.F.).
References
- Boeke, J., F. Lacrute and G. R. Fink, 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoroorotic resistance. Mol. Gen. Genet. 197: 345–346. [DOI] [PubMed] [Google Scholar]
- Deutschbauer, A. M., R. Williams, A. M. Chu and R. W. Davis, 2002. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99: 15530–15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch, A. P., M. Huang, S. Metzner, D. Botstein, S. J. Elledge et al., 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell 12: 2987–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles et al., 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387. [DOI] [PubMed] [Google Scholar]
- Harfe, B. D., and S. Jinks-Robertson, 2000. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34: 359–399. [DOI] [PubMed] [Google Scholar]
- Hawk, J. D., L. Stefanovic, J. C. Boyer, T. D. Petes and R. A. Farber, 2005. Variation in efficiency of DNA mismatch repair at different sites in the yeast genome. Proc. Natl. Acad. Sci. USA 102: 8639–8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, M.-E., A.-G. Rio, A. Nicolas and R. D. Kolodner, 2003. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. USA 100: 11529–11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea, D. E., and C. A. Coulson, 1949. The distribution of the number of mutants in a bacterial population. J. Genet. 49: 264–285. [DOI] [PubMed] [Google Scholar]
- Sia, E. A., R. J. Kokoska, M. Dominska, P. Greenwell and T. D. Petes, 1997. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 17: 2851–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand, M., T. A. Prolla, R. M. Liskay and T. D. Petes, 1993. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365: 274–276. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wierdl, M., C. N. Greene, A. Datta, S. Jinks-Robertson and T. D. Petes, 1996. Destabilization of simple repetitive DNA sequences by transcription in yeast. Genetics 143: 713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]