Abstract
During the development of multicellular organisms, cellular diversity is often achieved through asymmetric cell divisions that produce two daughter cells having different developmental potentials. Prior to an asymmetric cell division, cellular components segregate to opposite ends of the cell defining an axis of polarity. The mitotic spindle rotationally aligns along this axis of polarity, thereby ensuring that the cleavage plane is positioned such that segregated components end up in individual daughter cells. Here we report our characterization of a novel gene required for spindle alignment in Caenorhabditis elegans. During the first mitosis in spd-3(oj35) embryos the spindle failed to align along the anterior/posterior axis, leading to abnormal cleavage configurations. spd-3(oj35) embryos had additional defects reminiscent of dynein/dynactin loss-of-function possibly caused by the mislocalization of dynactin. Surprisingly, we found that SPD-3∷GFP localized to mitochondria. Consistent with this localization, spd-3(oj35) worms exhibited slow growth and increased ATP concentrations, which are phenotypes similar to those described for other mitochondrial mutants in C. elegans. To our knowledge, SPD-3 is the first example of a link between mitochondria and spindle alignment in C. elegans.
ASYMMETRIC cell divisions are an essential feature of metazoan development as they provide cell diversity within a multicellular organism. During determinative asymmetric cell divisions, specific cellular components are first polarized to opposite ends of the cell. The cleavage plane is then positioned such that it bisects the polarization axis, thereby giving rise to daughter cells with different developmental potentials (Hyman and White 1987; White and Strome 1996). These actions ensure that the polarized components are segregated to different daughter cells, where they act to determine the differentiated state.
The single-cell embryo of Caenorhabditis elegans undergoes an asymmetric cell division. Upon fertilization, a centrosome enters the oocyte along with the paternal pronucleus and splits into two. Initially, the daughter centrosomes are aligned perpendicular to the long axis of the embryo. The axis of polarity is determined by the site of fertilization: the sperm entry site designates the posterior of the embryo. This is achieved by an interaction of the sperm centrosomes with the cell cortex, which causes cortical material to flow away from the centrosomes (Hird and White 1993; Goldstein and Hird 1996; Golden 2000; Munro et al. 2004). As polarity is being established, the pronuclei migrate toward each other, meeting at ∼70% egg length (slightly toward the posterior). When the pronuclei meet, the entire pronuclear/centrosome complex rotates 90°, placing the centrosomes, and consequently the developing mitotic spindle, along the anterior/posterior (A/P) axis. Because the cleavage plane is always set up to bisect the mitotic spindle (Rappaport 1986), the process of rotational alignment ensures that polarized components are segregated to unique daughter cells, thereby making the division determinative.
Microtubules are required for rotational alignment. Exposure to the microtubule depolymerizing drug nocodazole or mutations in genes such as zyg-9 and mbk-2 result in short astral microtubules that are unable to reach the cortex (Hyman and White 1987; Matthews et al. 1998; Pang et al. 2004). Consequently, the pronuclear/centrosome complex fails to rotate, resulting in an improperly aligned mitotic spindle. Because the resulting cleavage plane does not coordinate with the polarity axis, cell division with a misaligned mitotic spindle often does not segregate polarized determinants into appropriate daughter cells and the embryos exhibit gross developmental defects.
The microtubule-associated minus end-directed motor dynein, together with its activator dynactin, is required for rotational alignment (Skop and White 1998; Gonczy et al. 1999). Cells deficient in dynein or dynactin function are defective in several additional early developmental processes including meiosis, pronuclear migration, centrosome separation, and chromosome segregation (Skop and White 1998; Gonczy et al. 1999; Yoder and Han 2001). Generally, it is believed that spindle alignment occurs by the capture of astral microtubule plus ends at the cell cortex by localized or locally activated dynein/dynactin complexes, followed by shortening of the captured microtubules (White and Strome 1996; Tsou et al. 2002). Spindle rotation is thought to occur as a result of shortening more captured microtubules from one centrosome over the other. Several models exist that predict mechanisms for this break in the symmetry of pulling forces (Cowan and Hyman 2004).
While the search and capture model is attractive, the actual mechanism that determines spindle orientation remains poorly understood. Recent evidence has implicated several “housekeeping” genes as having specific and essential roles during spindle alignment. Proteins involved in diverse processes such as secretion and nuclear pore formation are required for proper spindle orientation (H. Zhang and J. G. White, personal communication; Skop et al. 2001; Schetter et al. 2006). Here we present evidence of another unexpected player in the process of spindle rotational alignment: mitochondria.
MATERIALS AND METHODS
C.elegans strains and alleles:
The Bristol strain N2 was used as the standard wild-type strain. Culturing, handling, and genetic manipulation of C. elegans were performed using standard procedures (Brenner 1974). Temperature-sensitive strains were maintained at 16° and L4 hermaphrodites were shifted to 25° for 24 hr prior to analysis. The following strains were used: WH113 (spd-3(oj35)), MT5734 (nDf41 IV/nT1[unc(n754), let](IV;V)), MT5241 (unc-5(e53), unc-44(e362)), DR1213 (unc-44(e362), unc-24(e138)), CB4856 (Hawaiian mapping strain), WH204 (unc-119(ed3);ojIs1[β-tubulin∷GFP unc-119(+)]), WH258 (unc-119(ed3); ojIs5[dnc-2∷GFP unc-119(+)]), WH400 (spd-3(oj35); ojIs5[dnc-2∷GFP unc-119(+)]), WH342 (unc-119(ed3); ojIs31[spd-3∷GFP unc-119(+)]), MQ130 (clk-1(qm30)), TR1450 (unc-54(r293)), and VC1332 (H34C03.1(ok1817) IV/nT1[qIs51](IV;V)) (International C. elegans Gene Knockout Consortium).
RNA-mediated interference:
For spd-3, H34C03.2, Y57A10A.26, and Y48C3A.3 RNA-mediated interference (RNAi) by injection, dsRNA was synthesized in vitro using T3 and T7 (or just T7) in vitro transcription kits (Ambion, Austin, TX) with cDNA clones (yk262b3 or yk1669c08 for spd-3, yk1310d6 for H34C03.2, yk111g2 for Y57A10A.26, and OST104E5-1 for Y48C3A.3) or genomic DNA [H34C03.2 primers (T7 site in boldface type)]: TAATACGACTCACTATACGCGTCTATAAATCCCCGAAAGT and TAATACGACTCACTATATGTCACCAGGCCGCATCAT.
All dsRNA was injected at final concentrations of 1–4 μg/μl into young hermaphrodite adults (Fire et al. 1998). Worms were allowed to recover for 24 hr before embryos were collected. dnc-1, smg-1, smg-2, atp-2, cco-1, and cyc-1 RNAi were administered by feeding. Feeding vectors were obtained from the Ahringer library (Kamath et al. 2003). The dnc-1 RNAi feeding vector used to test the DNC-1 antibody specificity was constructed by cloning the full-length yk11c8 cDNA into the L4440 feeding vector, followed by transformation into Escherichia coli HT115(DE3) bacteria. The E. coli HT115(DE3) bacteria containing the feeding vectors were cultured and used to seed RNAi plates (NGM + 1 m IPTG + 25 μg/ml carbenicillin). L4 hermaphrodites were allowed to feed for ≥24 hr before analysis (Timmons and Fire 1998). As a control for smg-2 RNAi efficiency, unc-54(r293) worms were fed RNAi in parallel with spd-3(oj35) worms. smg-2 RNAi has been previously reported to suppress the paralysis phenotype seen in unc-54(r293), which was consistent with our studies (Hodgkin et al. 1989).
Mapping/cloning:
Three-point mapping was done at 16° by crossing spd-3(oj35) males to either MT5241 or DR1213 hermaphrodites. Several heterozygous F1 progeny were singled and allowed to self-fertilize. The resulting F2 progeny were screened for recombinants containing one marker but losing the other. Recombinant worms were singled and homozygosed. After lines were established, several worms from each line were tested for the spd-3(oj35) allele by shifting to the restrictive temperature and screening for dead embryos.
SNP mapping was done by crossing CB4856 Hawaiian males to either unc-5(e53), spd-3(oj35) hermaphrodites or spd-3(oj35),unc-24(e138) hermaphrodites (Jakubowski and Kornfeld 1999). Several heterozygous F1 progeny were singled and allowed to self-fertilize. The resulting F2 progeny were screened for unc worms that had lost the spd-3(oj35) allele as distinguished by fertility at 25°. Worms were then genotyped at the appropriate SNPs (Table 1) to determine the region of recombination.
TABLE 1.
spd-3 SNP map
| Cosmid | Primers | Enzyme |
|---|---|---|
| C31H1 | AAAACTGGTCGAGGCACAC | HpyCH4IV |
| AGCCACGTAGCCACTGTAAC | ||
| C49A9 | TTTTGTTGTCGCATCACTTCTG | SspI |
| AACTTCTAAATGCTGCCCTTCC | ||
| T13A10 | GCTTGAGTGCCAGTTGTTATGTG | — |
| CTGCGGAAAAGGGTATGAGAA | ||
| Y73B6Bl | CCTGCAGGTGCGGATTGAG | — |
| CTAACCGATGCGCTGTGAACG | ||
| F38A5 | GCACCATCTTCTGCTCCAAC | XbaI |
| TTAACCTTTGGAGTGACTGCG | ||
| Y73B6A | TTATAGCTTGAACAACGGGACATC | — |
| CTCATTCGTTCGTGCTATTTCTCT | ||
| H34C03 | TCCGCCAATTTCCAC | — |
| CATCATGCGCCCACTTCT | ||
| R13H7 | ACAAGATCGTTTTCAGGGTCAAT | — |
| AGGGGGAGAATAAAACATCGTAA | ||
| B0478 | AAGAAGTGCCACCCAAAAAGAG | — |
| AAACCTAGCCGCCAAATGAC | ||
| C25A8 | CCCTTATGCCATTACTTATTCGTG | — |
| GTCGCCGGCGTTTTTACTTT | ||
| Y43B11AR | TTCGCAGTAGTAAAAGGTAAACAA | BsrSI |
| CGGCGACGGCGAGAAGT | ||
| D2024 | TAACCGCCACGAAAAAGATAGGAT | — |
| GGGACTTAGAATTACTGCGTTTGA | ||
| C48A7 | TGGTGCACGAAGAAGGAAGAG | DraI |
| GCCCCCGGAAATCAGAAATG | ||
| R05G6 | GGTGTTCAAACATGCGACG | PstI |
| TTTGGACGGATAGCTACATACG |
A single-nucleotide polymorphism (SNP) map was designed for spd-3 mapping. Primers were designed using DNAStar Lasergene PrimerSelect and used to amplify ∼1 kb of DNA containing a SNP between the Bristol and the Hawaiian strain. Several of these SNPs were contained in restriction enzyme cut sites in one strain but not the other, in which case the appropriate restriction enzymes were used to determine the genotype of the recombinant strain. In other cases, strains were genotyped by sequencing (no enzyme listed).
For rescue experiments, cosmid DNA containing the putative wild-type spd-3 gene was injected individually into spd-3(oj35) young adult worms at a concentration of 100 μg/ml along with pTG96, a GFP coinjection marker (Yochem et al. 1998) at a concentration of 1 mg/ml (Figure 1A). Progeny were screened for the uptake of a GFP-expressing transgene using a fluorescence dissecting microscope; green worms were singled and allowed to self-fertilize. Once lines were established, L1 worms were shifted to 25° to test for rescue of the spd-3(oj35) postembryonic phenotype of sterility.
Figure 1.—
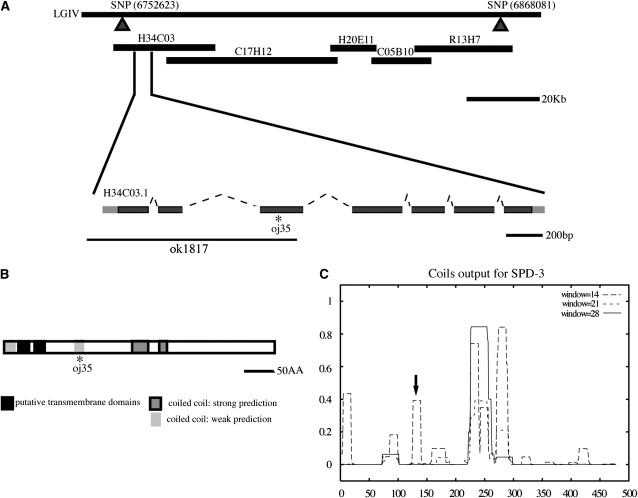
spd-3 is a novel gene with putative transmembrane domains and predicted coiled coils. (A) spd-3 is equivalent to the novel gene H34C03.1. The oj35 allele contains a single-base-pair change in the third exon. The ok1817 deletion allele removes the 5′-UTR through exon 3 of the spd-3 gene. (B) The predicted gene product contains two putative transmembrane domains and predicted coiled coils. (C) The y-axis represents the probability of coiled-coil conformations in the SPD-3 protein. A weak coil prediction is destroyed in the oj35 allele (arrow).
The PCR product used for rescue was generated using the primers 5′ TCACATGAAAGCAATAAAACCAAT 3′ and 5′ TCGGAAAATAAATTGGAAAAGGAG 3′.
The spd-3 complementation test was carried out by crossing spd-3(oj35) to VC1332 (H34C03.1(ok1817) IV/nT1[qIs51](IV;V)), which carries a deletion including the first three exons of the gene (WormBase). The spd-3(oj35)/H34C03.1(ok1817) L4 worms were shifted to 25° overnight to screen for embryonic lethality.
SPD-3 domain searches were done using the following websites: http://smart.embl-heidelberg.de/, http://www.ch.embnet.org/software/COILS_form.html, and http://www.ch.embnet.org/software/TMPRED_form.html.
Measuring embryonic lethality:
Dead egg counts were done by placing an L4 worm at restrictive temperature and shifting to a new plate every 24 hr. Dead eggs and larvae were scored 24 hr after the shift.
Live imaging:
For colocalization experiments, NGM agar plates were seeded with OP50 culture containing 2 μm MitoTracker CMXRos (Invitrogen, San Diego) (Chen et al. 2000). SPD-3∷GFP (or wild-type control) L4 worms were allowed to feed on the plates for 24 hr prior to imaging. Mitochondria in oj35 embryos were visualized with rhodamine 6G (Sigma, St. Louis) as described (Badrinath and White 2003).
All slides for live imaging were prepared by placing three to five young adult worms into a drop of M9 buffer on a glass coverslip. The worms were cut open using a scalpel to release the embryos. A glass slide containing a 2% agarose pad was lowered onto the coverslip and sealed with Vaseline.
Nomarski imaging was done using either a Nikon diaphot 300 inverted microscope or a Nikon optiphot-2 upright microscope. Data were collected using 4-D Grabber software as described previously (Skop and White 1998). Fluorescent imaging was done using either a multiphoton microscope as described previously (Wokosin et al. 2003; M. Z. Nazir, K. W. Eliceiri, A. Ahmed, E. Hathaway, A. Hashmi, V. Agarwal, Y. Rao, S. Kumar, T. Lukas, K. M. Riching, C. T. Rueden, Y. Wang and J. G. White, unpublished results) or a spinning-disk confocal (QLC100; Visitech International) and collected with Open Lab 4.0.3. Images were analyzed using ImageJ software.
Quantification of the average fluorescence intensity in DNC-2∷GFP and DNC-2∷GFP; spd-3(oj35) embryos:
Images of DNC-2∷GFP and DNC-2∷GFP; spd-3(oj35) embryos were taken in parallel under the same conditions using a multiphoton microscope. A single focal plane (adjusted manually) was observed over time. We used the elliptical tool in ImageJ to select the embryonic region during metaphase and then calculated the average intensity of this region. Student's t-test with two-tailed equal variance was used to determine if the difference between DNC-2∷GFP and DNC-2∷GFP; spd-3(oj35) was statistically significant.
Immunohistochemistry:
The following peptide sequence was used to generate rabbit polyclonal DNC-1 antibody: Ac-DPNEPQFTAPDPRRQSLC-amide. Protein production, antibody production, and affinity purification were performed by Quality Controlled Biochemicals (Hopkinton, MA). Slides for indirect immunofluorescence were prepared by placing 30–40 adult worms in M9 buffer on a subbed slide and slicing the worms open to release the embryos. After placing a coverslip over the embryos, excess M9 was wicked out using filter paper. The slides were then placed on a metal block on dry ice for 30 min, after which the coverslip was cracked off using a flat-edge razor to crack the eggshells of the embryos. The embryos were then fixed in 100% methanol for 10 min at room temperature and blocked with PBS + 0.5% BSA + 0.5% Tween 20 (PBSBT) for 30 min at room temperature. The slides were incubated with primary antibodies at 4° overnight. The slides were washed with PBS + 0.5% Tween 20 (PBST) 3× 10 min. The slides were then incubated with Alexa [488]-conjugated secondary antibodies (Molecular Probes, Eugene, OR) at 1:200 for 1–2 hr at room temperature. After washing with PBST, the slides were mounted with 8 μl VectaShield and a glass coverslip and sealed with nail polish. Slides were viewed using a Bio-Rad (Hercules, CA) 1024 confocal microscope. The following primary antibodies were used: mouse α-tubulin (n357) at 1:100 (Amersham Pharmacia Biotech, Piscataway, NJ) and rabbit α-DNC-1 at 1:400.
SPD-3∷GFP construct:
SPD-3∷GFP vector was constructed using full-length genomic spd-3 DNA that was cloned into pFJ1 vector. pFJ1 contains an N-terminal GFP tag and pie-1 regulatory elements, as well as unc-119(+). An altered version of pFJ1, which contains a GFP tag at the C terminus of the insert, was also used. The GFP vectors were introduced into unc-119(ed3) worms by biolistic bombardment to create integrated lines (Praitis et al. 2001).
Suppressor screen:
Two L4 spd-3(oj35) worms were placed in each well of a 12-well plate at 25° and fed E. coli HT115(DE3) bacteria containing dsRNA from the Ahringer library (Kamath et al. 2003). After 3 days, plates were screened for larvae (significantly higher in number than controls). Candidates were retested two times for consistency.
Pull-down assay:
Worm lysate was obtained by passing wild-type worms grown in liquid culture through a French press two times and removing cellular debris by high-speed centrifugation. SPD-3∷GST was constructed by cloning an 882-bp fragment of spd-3 cDNA (encoding the last 294 amino acids of the predicted SPD-3 protein) into the pGEX3X vector. SPD-3∷GST was produced by transforming the vector into E. coli BL21 Rosetta cells and inducing protein production with IPTG. Cells were lysed by sonication, and protein was purified with glutathione Sepharose 4B beads (Amersham). Purified protein was concentrated using centriprep columns (Amicon, Danvers, MA) and dialyzed into 1× PBS using PD-10 desalting columns (Amersham). Final concentrations were estimated by comparison to BSA standards on a Coomassie-stained SDS–PAGE gel.
Approximately 500 μg of purified protein were coupled to 500 μl of glutathione Sepharose 4B beads with 1.5 mm dithiobis(succinimidyl)propionate (Pierce, Rockford, IL). Worm lysate was precleared by incubating with beads bound with GST alone for 1 hr at 4°. Precleared lysate was then transferred to beads bound with SPD-3∷GST and incubated for 1 hr at 4°. All beads were washed three times with 5% 1 m Tris, pH 8.0 + 3% 5 m NaCl + 1% NP40 (IP buffer) and then three times with IP buffer minus NP40. Binding partners were eluted with 0.5% SDS in 1× PBS (SDS elution buffer), and proteins were precipitated by adding 3–4% trichloroacetic acid and leaving them on ice in a 4° incubator overnight. Tubes were centrifuged at maximum speed for 30 min at 4°. The protein pellet was washed with acetone two times, dried completely, resuspended in SDS sample buffer (Bio-Rad), and run on a 10% SDS polyacrylamide gel.
Coomassie bands exclusive to SPD-3∷GST were excised, trypsin digested, and analyzed at the University of Wisconsin Molecular Interaction Facility using Mascot after MALDI-Mass Spectrometry. The reported C. elegans homolog to MTX2 is ZC97.1 (Armstrong et al. 1999). However, Y57A10A.26 has stronger sequence conservation than ZC97.1 (e-value: 6.9e-12 over 90% of the protein sequence vs. 9.1e-4 over 85.5% of sequence).
Growth rate experiments:
Growth rates were measured by placing a single [wild type, clk-1(qm30), or spd-3(oj35)] L4 worm on a 35-mm plate at 16° and recording the number of days until the appearance of F1 L4 worms.
Mitochondrial inhibitor drugs:
Fifty microliters of a 40-μm antimycin A (Sigma) solution in ethanol were spread on a 35-mm NGM plate seeded with OP50. Fifty microliters of a 20-mm sodium azide (Sigma) solution in water were spread on a 35-mm NGM plate seeded with OP50. Water and ethanol alone were used for controls. Plates were allowed to dry overnight, and growth-rate experiments were performed as described above.
ATP concentration measurements:
Mixed-stage wild-type and spd-3(oj35) hermaphrodites were shifted to 25° for 24 hr and then harvested, washed twice with M9 buffer, resuspended in cell lysis buffer, and frozen in liquid nitrogen. After samples were thawed, ATP concentration was determined using a bioluminescence assay kit (Roche, Indianapolis) and total protein concentration was measured using a BCA protein assay kit (Pierce).
RESULTS
Cloning spd-3:
spd-3(oj35) was isolated in an EMS-mutagenesis screen in search of cell division mutants (O'Connell et al. 1998). It is a temperature-sensitive, maternal-effect mutant (O'Connell et al. 1998). Homozygous mothers shifted to the restrictive temperature at the fourth larval stage produce embryos manifesting the spd-3(oj35) mutant phenotype (hereafter referred to as oj35 embryos). spd-3 animals shifted to the restrictive temperature just prior to hatching develop into sterile adults (O'Connell et al. 1998). spd-3 was initially mapped to the left arm of chromosome IV between 0 and 6.3 MU using linkage analysis, three-point mapping, and deficiency mapping (O'Connell et al. 1998).
To further narrow the spd-3 region, we used a combination of classic three-point mapping and SNP mapping. We mapped spd-3 to a small region covered by five overlapping cosmids (Figure 1A). We next showed that cosmid H34C03 partially rescues the oj35 phenotype. This cosmid contains six predicted open reading frames. Subsequently, we narrowed the rescuing fragment to a PCR product containing only two genes that compose the H34C03 operon. Operons are defined as a set of two or more genes sharing a single promoter; genes in C. elegans operons may or may not be functionally related to each other (Blumenthal and Gleason 2003). The H34C03 operon consists of only two genes: a ubiquitin carboxy-terminal hydrolase and a novel gene, neither of which show any obvious phenotypes with RNAi (data not shown). Upon sequencing the entire operon in the oj35 mutant, we found a single mutation in the downstream novel gene H34C03.1 (Figure 1A). The oj35 allele contained a cytosine-to-thymine transition resulting in a leucine-to-phenylalanine change at amino acid 130. We subsequently showed that an H34C03.1∷GFP transgene under pie-1 control elements rescued defects in the oj35 embryos (Table 2). In addition, an amino-terminal deletion (ok1817) in H34C03.1 failed to complement the oj35 allele, suggesting that this is a loss-of-function mutation (Figure 1A). Taken together, we concluded that spd-3 is equivalent to H34C03.1.
TABLE 2.
SPD-3:GFP rescues oj35 embryonic defects
| Genotype | % dead eggs | % transverse spindle in P0 |
|---|---|---|
| SPD-3∷GFP | 1.0 ± 0.89 (n = 6) | 0 (n = 4) |
| unc-5(e53), spd-3(oj35) | 99.3 ± 1.7 (n = 6) | 100 (n = 4) |
| SPD-3∷GFP; unc-5(e53), spd-3(oj35) | 4.0 ± 3.2 (n = 6) | 0 (n = 6) |
All experiments were done at restrictive temperature.
The predicted SPD-3 protein does not contain any recognizable functional domains. However, there are two putative transmembrane domains in the amino terminus and several predicted coiled coils (Figure 1B). The oj35 mutation destroys a weakly predicted coil (Figure 1C, arrow). While there are unambiguous orthologs in both C. briggsae and C. remanei, there are no obvious homologs outside of nematodes.
spd-3(oj35) is defective in meiosis, pronuclear migration, and mitotic spindle alignment:
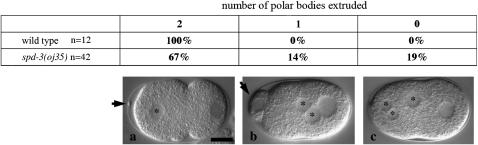
Multiple defects in oj35 early embryos were observed using noninvasive, live Nomarski imaging. The earliest defect that we observed in oj35 embryos was the failure to faithfully extrude polar bodies. C. elegans embryos normally undergo two rounds of meiosis shortly after fertilization. The meiotic spindle aligns perpendicular with the anterior cortex, resulting in the extrusion of two compact polar bodies (Figure 2a, arrow). In oj35 embryos, the number of polar bodies was variable, with some embryos completely failing to extrude extra maternal pronuclei (Figure 2c), some having abnormally large polar bodies (Figure 2b, arrow), and some with normal extrusion. The failure to extrude polar bodies, as well as the presence of large polar bodies, suggests possible defects in the alignment of the meiotic spindle.
Figure 2.—
spd-3(oj35) is defective in meiosis. In wild-type embryos, two female pronuclei are faithfully extruded as polar bodies (a, arrow), leaving a single female pronucleus in the zygote (a, asterisk). oj35 embryos are defective in meiosis, often failing to extrude polar bodies resulting in extra maternal pronuclei in the zygote (b and c, asterisks). Often, the polar bodies that are extruded are very large (b, arrow) compared to wild type. Bar, 10 μm.
Pronuclear migration was often delayed in oj35 embryos. In wild-type embryos, the pronuclei form shortly after the completion of meiosis. The paternal pronucleus is located in the posterior, while the maternal pronucleus is most often located in the anterior. Shortly following the formation of the pronuclear envelopes, the maternal pronucleus in wild-type embryos migrated toward the paternal pronucleus at an average rate of 0.126 ± 0.039 μm/sec (n = 10). However, in oj35 embryos, the maternal pronuclear migration rate was 0.0674 ± 0.0371 μm/sec (n = 12), almost 50% slower than that in wild type. Often, the paternal pronuclear envelope broke down near the posterior cortex before the pronuclei met.
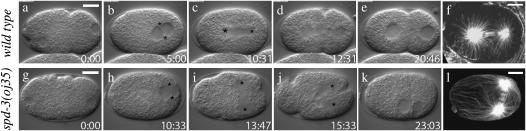
The most apparent and detrimental defect observed in oj35 embryos was the failure to properly orient the first mitotic spindle. Following fertilization in a wild-type embryo, the pronuclear/centrosomal complex rotates 90° to align the mitotic spindle along the A/P axis, i.e., the axis of polarity (Figure 3, a–e). This rotation failed in oj35 embryos, resulting in a mitotic spindle aligned transversely to the A/P axis (Figure 3, g–k). The misaligned spindle gave rise to a mispositioned cleavage plane that did not coordinate with the polarity axis, resulting in a failure of the asymmetric division. It is worth noting that even in instances when the pronuclei did meet prior to nuclear envelope breakdown, the spindle was still misaligned, suggesting that the rotation failure was not simply a result of delayed pronuclear migration.
Figure 3.—
spd-3(oj35) embryos are defective in mitotic spindle alignment. In the single-cell embryo, the centrosomes are associated with the paternal pronucleus designating the posterior of the embryo, while the maternal pronucleus is usually located in the anterior (a and g). The pronuclei migrate toward each other, meeting near the posterior (b and h, centrosomes are marked by asterisks). In wild-type embryos, the pronuclear/centrosome complex then rotates 90°, aligning the mitotic spindle along the anterior/posterior axis to coordinate with the axis of polarity (c, centrosomes marked by asterisks). This rotation fails in the oj35 embryo, resulting in a mitotic spindle aligned transverse to the anterior/posterior axis (i, centrosomes marked by asterisks). The cleavage plane bisects the spindle (d and j), resulting in a proper asymmetric division in wild type (e) but not in oj35 embryos (k). In wild-type embryos, astral microtubules interact with the cell cortex and appear to be captured and stabilized (f). In oj35 embryos, astral microtubules appear to grow along the cortex (l) and are excessive in length. Bar, 10 μm. Tubulin is visualized with α-tubulin n357 (Amersham Pharmacia Biotech).
It has been shown that astral microtubule interactions with the cell cortex are imperative for spindle rotation to occur. Loss-of-function alleles of genes required for microtubule growth such as zyg-9 and mbk-2 result in short spindles that fail to rotate, presumably due to the inability of the short astral microtubules to interact with the cortex (Matthews et al. 1998; Pang et al. 2004). Exposure to microtubule depolymerizing drugs such as nocodazole also results in this phenotype (Hyman and White 1987). Given these observations, we examined the microtubules in fixed oj35 embryos by immunofluorescent staining. Surprisingly, we found that astral microtubules in oj35 embryos were robust and sufficient in length to interact with the cortex (Figure 3l). Indeed, the microtubules in the mutant embryos appeared excessive in length, compared to wild type. In wild-type embryos, several microtubules appear to have been captured and presumably stabilized upon reaching the cortex (Figure 3f). However, in oj35 embryos, the microtubules appear to have contacted the cortex and then continued to grow along it (Figure 3l).
spd-3(oj35) embryos exhibit defects in dynactin localization:
Interestingly, a similar spindle alignment defect is observed with loss-of-function of dynactin components (Skop and White 1998; Gonczy et al. 1999). The dynein/dynactin complex has been well characterized for its role in spindle alignment in various systems, including the C. elegans early embryo (Carminati and Stearns 1997; Mcgrail and Hays 1997; Skop and White 1998; Gonczy et al. 1999; O'Connell and Wang 2000). Therefore, we next examined whether the dynactin complex was localized properly in oj35 embryos.
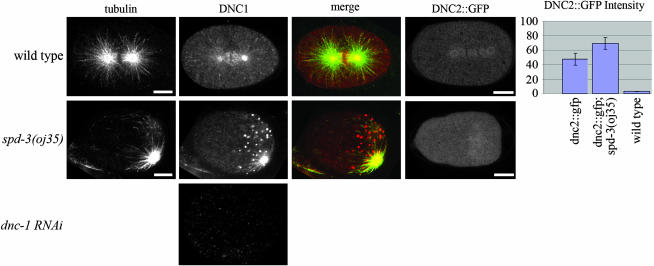
We examined the localization of DNC-1 (p150glued) in oj35 embryos using an antibody to DNC-1. In wild-type embryos, DNC-1 localizes to the centrosomes, along spindle microtubules, and to the plus ends of astral microtubules (Figure 4). Surprisingly, we found that although the microtubule localization of DNC-1 appeared normal in oj35 embryos, the majority of the protein appeared to localize into posterior structures resembling P granules. In addition, the centrosome labeling appeared brighter than that in wild type, a phenotype reminiscent of defects in dynein heavy chain function (Schmidt et al. 2005).
Figure 4.—
spd-3(oj35) embryos have defects in dynactin localization. In wild-type embryos, DNC-1 localizes to the centrosomes, the mitotic spindle, and the plus ends of astral microtubules (n = 12). oj35 embryos exhibit an additional, anomalous localization of DNC-1 to posterior structures resembling P granules (n = 12). DNC-1 signal is absent in dnc-1 RNAi embryos. DNC-2∷GFP in wild-type embryos is present throughout the cytoplasm, with enrichments in the pericentriolar region and along the mitotic spindle (n = 5). This enrichment is decreased in oj35 embryos, while the cytoplasmic signal is significantly increased (n = 6). DNC-2∷GFP images portrayed are snapshots taken from live imaging, while tubulin (n357) and DNC-1 antibodies were used with fixed embryos. Bar, 10 μm.
During mitosis, DNC-2∷GFP (p50, dynamitin) normally localizes cytoplasmically with an enrichment in the pericentriolar region and along microtubules (Figure 4). This microtubule localization is particularly apparent on the mitotic spindle where there is a distinct absence of signal on the chromosomes. In oj35 embryos, DNC-2∷GFP exhibited a slight enrichment in the pericentriolar region and the mitotic spindle and also a higher, delocalized cytoplasmic signal (Figure 4). Quantification of DNC2∷GFP fluorescence intensity showed a consistent increase in the oj35 background (Figure 4 graph). In addition, the contrasting lack of staining at the centrioles and the chromosomes is not as obvious in the oj35 embryos compared to wild type.
SPD-3 localizes to mitochondria:
To determine the subcellular localization of SPD-3, we generated transgenic worms expressing SPD-3∷GFP under the control of the pie-1 promoter and the 3′-UTR to enhance expression in the early embryo. The transgene consists of full-length genomic spd-3 DNA tagged with GFP at the N or the C terminus and rescued the embryonic lethality and P0 spindle orientation defects in the oj35 mutants (Table 2). Several lines were produced with a C-terminal GFP tag and one with an N-terminal GFP tag, which all showed identical localization patterns.
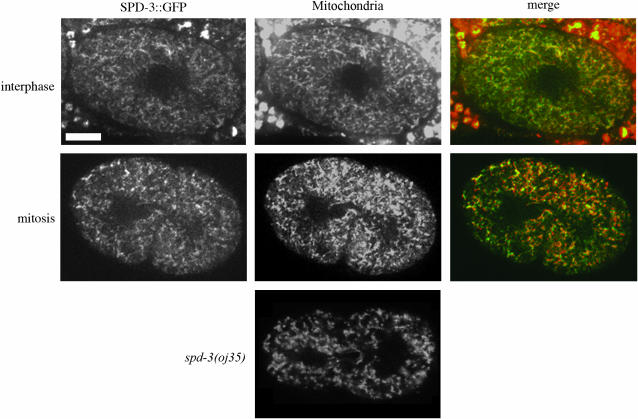
Surprisingly, we found that SPD-3∷GFP localized to mitochondria (Figure 5). In the early C. elegans embryo, mitochondria are diffusely distributed throughout the cytoplasm with no distinct localization pattern. In most organisms during interphase, mitochondria are localized throughout the cytoplasm. However, in some organisms such as yeast and the nematode Acrobeloides, mitochondria cluster around the centrosomes during mitosis, presumably to ensure proper segregation into separate daughter cells (Badrinath and White 2003; Yaffe et al. 2003). This centrosome clustering is consistent with a possible dependence on dynein. However, in C. elegans embryos, mitochondria do not undergo any obvious rearrangements during mitosis (Figure 5) (Badrinath and White 2003), making SPD-3∷GFP localization even more surprising. It is worth noting that mitochondrial morphology and localization as assessed with mitochondrial vital dyes were not affected in oj35 embryos, at least at levels that we could detect in these studies (Figure 5).
Figure 5.—
SPD-3∷GFP colocalizes with mitochondria. SPD-3∷GFP has a similar localization pattern throughout the cell cycle (top, interphase; bottom, mitosis) and colocalizes with MitoTracker CMXRosetta. Bar, 10 μm. There are no gross abnormalities in mitochondrial morphology or localization in oj35 embryos as visualized with Rhodamine 6G (Sigma) (n = 3).
We identified the protein product of C. elegans gene Y57A10A.26 as a potential interactor with SPD-3 in a GST pull-down assay. This gene has strong homology to murine metaxin 2, which has been shown in mice to localize to the mitochondrial outer membrane and is involved in transporting proteins into the mitochondrion (Armstrong et al. 1999). Although Y57A10A.26 alone did not exhibit any RNAi phenotype (data not shown), knocking down both Y57A10A.26 and its homolog Y48C3A.3 (36% identities, 55% positives) by RNAi gave 20% embryonic lethality after 31 hr, quickly followed by sterility (n = 30). Because the worms exhibit sterility so soon after the RNAi takes effect, we were unable to thoroughly analyze embryonic defects. However, it is worth noting that in one instance we did witness an embryo phenocopy of the oj35 spindle alignment defect.
The identification of the mitochondrial protein metaxin as a potential binding partner for SPD-3 and the suggestion that loss-of-function of the two metaxin genes leads to sterility, a postembryonic defect seen in oj35 worms (O'Connell et al. 1998), provide further evidence that SPD-3 does localize to mitochondria and suggest that SPD-3 either may be part of a protein import complex along with the metaxin 2-like protein or may itself be imported by this complex.
spd-3(oj35) worms exhibit metabolic defects:
A common phenotype in C. elegans mutants with perturbations in metabolism is reduced growth rates. We therefore measured the growth rates of oj35 worms at permissive temperature by recording the generation time of the worms. We found that oj35 worms had reduced growth rates compared to wild type, similar to that of strains with disruption in mitochondrial function such as clk-1(qm30) (Table 3) (Lakowski and Hekimi 1996; Dillin et al. 2002).
TABLE 3.
Generation rates
| Genotype | Temperature | Generation rate (days) |
|---|---|---|
| Wild type (n = 30) | 16° | 5.03 ± 0.18 |
| spd-3(oj35) (n = 29) | 16° | 7.62 ± 0.68 |
| clk-1(qm30) (n = 19) | 16° | 8.63 ± 0.68 |
| Drug exposure | Temperature | Generation rate (days) |
| Water (control) (n = 5) | 20° | 4.0 ± 0 |
| Sodium azide (20 mm) (n = 3) | 20° | 12.67 ± 2.3 |
| Ethanol (control) (n = 5) | 20° | 4.0 ± 0 |
| Antimycin A (40 μm) (n = 4) | 20° | 5.5 ± 0.58 |
Generation rates were measured as the number of days it took a larval-4-stage parent to produce larval-4-stage progeny.
We wondered whether metabolic defects in worms might affect dynein motor function due to alterations in energy production. We addressed this possibility by comparing the concentration of free ATP between wild-type and oj35 worms. Surprisingly, oj35 worms exhibited almost 40% greater ATP concentration compared to wild type. spd-3(oj35) worms contained 0.148 ± 0.016 μm ATP/μg total protein (n = 3) while wild-type worms contained 0.093 ± 0.003 μm ATP/μg total protein (n = 3). Functional disruptions in other proteins required for normal growth patterns including CLK-1 (required for ubiquinone biosynthesis) and DAF-2 (involved in insulin growth factor signaling) also result in increased ATP concentrations, suggesting that SPD-3 may be involved in these pathways (Braeckman et al. 1999; Dillin et al. 2002).
To address whether disruptions in metabolism could lead to dynein loss-of-function phenotypes, we disrupted the electron transport chain using various mitochondrial inhibitor drugs including antimycin A, a complex III inhibitor, and sodium azide, a complex IV inhibitor commonly used as a chemical surrogate for hypoxia (Scott et al. 2002). While exposure to these drugs led to slow growth (Table 3), there were no obvious defects in the early embryos analyzed after 24 hr of drug exposure in the mother. In addition, disruption of various other mitochondrial genes through mutations [clk-1(qm30)] (n = 23) or RNAi [atp-2 (ATP synthase) (n = 13), cyc-1 (cytochrome C) (n = 19), and cco-1 (cytochrome C oxidase) (n = 20)] did not resemble phenotypes seen in embryos with dynein loss-of-function (Sonnichsen et al. 2005), suggesting that the purported dynein loss-of-function is specific to the loss of SPD-3 function.
spd-3 genetically interacts with smg-1:
In an attempt to identify genetic interactors, we performed a suppressor screen using RNAi. RNAi has proven to be a convenient method to significantly knock down gene expression in C. elegans and can be administered to worms through feeding (Timmons and Fire 1998). Using a feeding library designed in the Ahringer lab (Kamath et al. 2003), we screened for genes on chromosomes I and II that suppressed oj35 phenotypes when depleted by RNAi. Normally, oj35 larvae placed at the restrictive temperature produce almost all dead eggs. We searched for genetic suppressors by screening for decreased levels of lethality after RNAi treatment.
We identified smg-1 as a suppressor of oj35. oj35 worms exposed to smg-1 RNAi at restrictive temperature produce an average of 9.83 ± 4.25 (n = 9) larvae per plate compared to untreated mutant worms with an average of 0.5 ± 0.84 (n = 8) larvae per plate. Further, we showed that smg-1 RNAi rescued the spindle rotation defect in six of seven oj35 single-cell embryos observed.
SMG-1 is a kinase that has been well studied in worms for its role in nonsense-mediated mRNA decay (NMD) (Grimson et al. 2004). However, there is evidence that smg gene products may also be involved in other types of regulation of mRNA expression levels (Mango 2001; Mitrovich and Anderson 2005). The oj35 allele contains a missense, rather than a nonsense, mutation. Therefore, if smg-1 RNAi is suppressing the oj35 phenotype through regulation of gene expression levels, it must be through a mechanism other than NMD. Supporting the hypothesis that spd-3 expression is regulated by the smg gene products, gene chip data suggest that spd-3(+) RNA levels are elevated 2.27 ± 0.64-fold in smg mutant backgrounds compared to wild type (D. Markwardt and P. Anderson, personal communication).
We reasoned that if suppression of the oj35 phenotype was the result of changes in expression levels by the disruption of the SMG complex, then loss-of-function of other smg genes would lead to the same suppression. However, smg-2 RNAi does not suppress the oj35 phenotype (n = 10), but does suppress the paralysis phenotype seen in unc-54(r293) worms (n = 10) done in parallel as an RNAi control. This suggests that smg-1 RNAi suppression of the oj35 phenotype may not be through regulation of gene expression levels by the SMG complex, but rather through an alternative, specific function of SMG-1.
DISCUSSION
SPD-3 and dynein:
We have shown that oj35 embryos have defects in meiosis, maternal pronuclear migration, and mitotic spindle alignment leading to embryonic lethality. Interestingly, these defects are similar to those seen in weak loss-of-function of the dynein motor protein or components of its activating protein complex, dynactin (Skop and White 1998; Gonczy et al. 1999; Yoder and Han 2001; Schmidt et al. 2005). In addition, we showed that SPD-3 is required for proper localization or expression of the dynactin components DNC-2 (p50 dynamitin) and DNC-1 (p150glued). Interestingly, while dynein and dynactin localize to microtubules, the nuclear envelope, the cell cortex, and centrosomes (Skop and White 1998; Gonczy et al. 1999), SPD-3 was found to localize to mitochondria. Furthermore, oj35 worms have additional metabolic defects including altered ATP levels and slow growth, which are common phenotypes seen in C. elegans worms with disruptions in mitochondrial function (Lakowski and Hekimi 1996; Dillin et al. 2002), making oj35 worms distinct from typical dynein/dynactin loss-of-function mutants.
SPD-3 and the cytoskeleton:
It has been shown in several systems that dynein/dynactin is required for proper subcellular localization of several organelles including mitochondria (Burkhardt et al. 1997; Helfand et al. 2002; Varadi et al. 2004). While there appears to be no specific localization of mitochondria in the C. elegans early embryo, it is possible that localization in the early embryo is not random, but too subtle to detect in these studies. Therefore, SPD-3 may be required for proper dynein function in spindle alignment as well as proper subcellular localization of organelles such as mitochondria. While we cannot rule out this possibility, we deem this unlikely as mitochondrial localization and morphology do not appear to be perturbed in the oj35 mutant embryos.
SPD-3 and mitochondria:
Surprisingly, we found SPD-3∷GFP localized at mitochondria. Several pieces of evidence shed clues as to a potential function of SPD-3 in relation to metabolism: the slow growth exhibited by oj35 worms introduces the possibility that SPD-3 may be required for optimal mitochondrial function, as this is a common phenotype seen when the electron transport chain is compromised (Lakowski and Hekimi 1996; Dillin et al. 2002).
In addition, we have identified a potential interaction between SPD-3 and Y57A10A.26, a protein with homology to murine metaxin 2. Metaxin 2 has been shown in mice to localize to the mitochondrial outer membrane and is involved in mitochondrial protein import (Armstrong et al. 1999). Decreased function of Y57A10A.26 and a similar, potentially redundant gene product (Y48C3A.3) via RNAi in C. elegans results in embryonic lethality followed by sterility, phenotypes also observed in spd-3(oj35) worms. This raises the possibility that SPD-3 is involved in mitochondrial protein import.
Furthermore, we have shown that ATP levels are increased in oj35 worms compared to wild type. Mutations in clk-1 or daf-2 also exhibit alterations in growth patterns and increased ATP levels (Braeckman et al. 1999; Dillin et al. 2002). CLK-1 is required for the biosynthesis of ubiquinone, a carrier in the electron transport chain (Jonassen et al. 1998), while DAF-2 is an insulin growth factor receptor that is thought to affect longevity by ultimately regulating gene expression through a phosphatidylinositol 3-kinase signaling pathway (Kenyon et al. 1993; Kimura et al. 1997). This suggests the possibility that SPD-3 may also be involved in biosynthesis or signaling pathways.
Interestingly, it has been shown that disrupting the function of the mitochondrial ATP synthase, stunted, in Drosophila embryos results in defects in spindle alignment in the syncytial blastoderm (Kidd et al. 2005). The authors show that ATP levels in the cortices of stunted mutant embryos are decreased compared to wild type. They suggest a model in which the reduced energy level is selectively affecting molecular motors such as dynein. While we have shown that ATP levels in oj35 worms are higher than those in wild type, it remains possible that elevated ATP levels are affecting dynein regulation, as it has been shown previously that dynein activity can be dependent on ATP and ADP concentrations (Shiroguchi and Toyoshima 2001).
However, the fact that we are unable to mimic the oj35 phenotype with mitochondrial inhibitor drugs, or by the disruption of other mitochondrial genes, suggests that the metabolic perturbation in oj35 worms is not directly leading to the embryonic defects reminiscent of dynein loss-of-function. In fact, disrupting mitochondrial function with drugs or RNAi of various genes required in the electron transport chain resulted in almost no observable defects in early embryonic development, implying that the early embryo is surprisingly tolerant to perturbations in the electron transport chain. This observation is consistent with previous studies that suggest that the embryo does not require large amounts of ATP for early development and the small percentage of ATP produced by mitochondria-independent processes such as glycolysis may be sufficient for early developmental progression (Feng et al. 2001).
Partial suppression of the oj35 phenotype by smg-1 RNAi:
smg-1 loss-of-function via RNAi partially suppresses the embryonic lethality and spindle alignment defects caused by the oj35 mutant allele. Because oj35 does not contain a nonsense mutation, the suppression must be through a mechanism other than NMD. Gene-chip data provide evidence that the SMG protein complex is regulating spd-3(+) expression levels. When the SMG complex is compromised through mutations, spd-3(+) expression is increased more than twofold.
However, the fact that depletion of another member of the SMG complex by smg-2 RNAi does not suppress the oj35 phenotype suggests that smg-1 RNAi suppression of the oj35 phenotype may not be through regulation of gene expression levels by the SMG complex, but through an alternative, specific function of the SMG-1 kinase. In addition to its role in NMD, SMG-1 orthologs in other systems have been implicated in signaling pathways regulating rearrangements of the actin cytoskeleton and stress response (Schmidt et al. 1996; Brumbaugh et al. 2004). Specifically, the human SMG-1 ortholog has recently been identified as a phosphoinositide 3-kinase, which is activated by genotoxic stress (Abraham 2004; Brumbaugh et al. 2004). Given that SPD-3∷GFP localizes to mitochondria, it is quite possible that SMG-1 is necessary to regulate expression levels of genes such as spd-3 in response to oxidative stress. However, an alternative explanation is that SMG-1 and SPD-3 are both involved in a stress-response signaling pathway, and it is the depletion of SMG-1 kinase activity in relation to this process that is suppressing the oj35 phenotype. Given that phosphorylation of various subunits of the dynein complex has been shown to regulate motor activity as well as membrane localization (Dillman and Pfister 1994; Lin et al. 1994; Pfister et al. 1996), SPD-3 may play an additional role in regulating dynein activity, expression levels, and localization. The fact that dynactin components are mislocalized in oj35 embryos is consistent with this hypothesis. These observations lead us to consider the intriguing possibility that SPD-3 may be acting in multiple signaling cascades required for C. elegans early embryonic development.
While the specific function of SPD-3 remains elusive, we believe SPD-3 is the first example of a mitochondrial localized gene product required for spindle alignment in the C. elegans embryo. Further studies are needed to clarify the precise mechanism through which SPD-3 is affecting both dynactin localization and metabolism and elucidate the role mitochondria are playing in early C. elegans development.
Acknowledgments
We are grateful to Dave Markwardt and Phil Anderson for communications and sharing unpublished data regarding smg-1 regulation, as well as Grzegorz Sabat at the University of Wisconsin Molecular Interaction Facility for analysis of the pull-down assay. We thank the labs of Judith Kimble, Sean Carroll, and Ann Palmenburg for equipment use. This work was supported by grants from the National Institutes of Health (R01 GM052454-09)
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession no. NM-068635.
References
- Abraham, R. T., 2004. The ATM-related kinase, hSMG-1, bridges genome and RNA surveillance pathways. DNA Repair 3: 919–925. [DOI] [PubMed] [Google Scholar]
- Armstrong, L. C., A. J. Saenz and P. Bornstein, 1999. Metaxin 1 interacts with metaxin 2, a novel related protein associated with the mammalian mitochondrial outer membrane. J. Cell Biochem. 74: 11–22. [PubMed] [Google Scholar]
- Badrinath, A. S., and J. G. White, 2003. Contrasting patterns of mitochondrial redistribution in the early lineages of Caenorhabditis elegans and Acrobeloides sp. PS1146. Dev. Biol. 258: 70–75. [DOI] [PubMed] [Google Scholar]
- Blumenthal, T., and K. S. Gleason, 2003. Caenorhabditis elegans operons: form and function. Nat. Rev. Genet. 4: 112–120. [DOI] [PubMed] [Google Scholar]
- Braeckman, B. P., K. Houthoofd, A. De Vreese and J. R. Vanfleteren, 1999. Apparent uncoupling of energy production and consumption in long-lived Clk mutants of Caenorhabditis elegans. Curr. Biol. 9: 493–496. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh, K. M., D. M. Otterness, C. Geisen, V. Oliveira, J. Brognard et al., 2004. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol. Cell 14: 585–598. [DOI] [PubMed] [Google Scholar]
- Burkhardt, J. K., C. J. Echeverri, T. Nilsson and R. B. Vallee, 1997. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol. 139: 469–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati, J. L., and T. Stearns, 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138: 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F., B. M. Hersh, B. Conradt, Z. Zhou, D. Riemer et al., 2000. Translocation of C. elegans CED-4 to nuclear membranes during cell death. Science 287: 1485–1489. [DOI] [PubMed] [Google Scholar]
- Cowan, C. R., and A. A. Hyman, 2004. Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu. Rev. Cell. Dev. Biol. 20: 427–453. [DOI] [PubMed] [Google Scholar]
- Dillin, A., A. L. Hsu, N. Arantes-Oliveira, J. Lehrer-Graiwer, H. Hsin et al., 2002. Rates of behavior and aging specified by mitochondrial function during development. Science 298: 2398–2401. [DOI] [PubMed] [Google Scholar]
- Dillman, 3rd, J. F., and K. K. Pfister, 1994. Differential phosphorylation in vivo of cytoplasmic dynein associated with anterogradely moving organelles. J. Cell Biol. 127: 1671–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, J., F. Bussiere and S. Hekimi, 2001. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell 1: 633–644. [DOI] [PubMed] [Google Scholar]
- Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver et al., 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Golden, A., 2000. Cytoplasmic flow and the establishment of polarity in C. elegans 1-cell embryos. Curr. Opin. Genet. Dev. 10: 414–420. [DOI] [PubMed] [Google Scholar]
- Goldstein, B., and S. N. Hird, 1996. Specification of the anteroposterior axis in Caenorhabditis elegans. Development 122: 1467–1474. [DOI] [PubMed] [Google Scholar]
- Gonczy, P., S. Pichler, M. Kirkham and A. A. Hyman, 1999. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 147: 135–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson, A., S. O'Connor, C. L. Newman and P. Anderson, 2004. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans. Mol. Cell. Biol. 24: 7483–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand, B. T., A. Mikami, R. B. Vallee and R. D. Goldman, 2002. A requirement for cytoplasmic dynein and dynactin in intermediate filament network assembly and organization. J. Cell Biol. 157: 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird, S. N., and J. G. White, 1993. Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans. J. Cell Biol. 121: 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., A. Papp, R. Pulak, V. Ambros and P. Anderson, 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman, A. A., and J. G. White, 1987. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. J. Cell Biol. 105: 2123–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski, J., and K. Kornfeld, 1999. A local, high-density, single-nucleotide polymorphism map used to clone Caenorhabditis elegans cdf-1. Genetics 153: 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen, T., M. Proft, F. Randez-Gil, J. R. Schultz, B. N. Marbois et al., 1998. Yeast Clk-1 homologue (Coq7/Cat5) is a mitochondrial protein in coenzyme Q synthesis. J. Biol. Chem. 273: 3351–3357. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin et al., 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kenyon, C., J. Chang, E. Gensch, A. Rudner and R. Tabtiang, 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. [DOI] [PubMed] [Google Scholar]
- Kidd, T., R. Abu-Shumays, A. Katzen, J. C. Sisson, G. Jimenez et al., 2005. The epsilon-subunit of mitochondrial ATP synthase is required for normal spindle orientation during the Drosophila embryonic divisions. Genetics 170: 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, K. D., H. A. Tissenbaum, Y. Liu and G. Ruvkun, 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277: 942–946. [DOI] [PubMed] [Google Scholar]
- Lakowski, B., and S. Hekimi, 1996. Determination of life-span in Caenorhabditis elegans by four clock genes. Science 272: 1010–1013. [DOI] [PubMed] [Google Scholar]
- Lin, S. X., K. L. Ferro and C. A. Collins, 1994. Cytoplasmic dynein undergoes intracellular redistribution concomitant with phosphorylation of the heavy chain in response to serum starvation and okadaic acid. J. Cell Biol. 127: 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango, S. E., 2001. Stop making nonSense: the C. elegans smg genes. Trends Genet. 17: 646–653. [DOI] [PubMed] [Google Scholar]
- Matthews, L. R., P. Carter, D. Thierry-Mieg and K. Kemphues, 1998. ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. J. Cell Biol. 141: 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrail, M., and T. S. Hays, 1997. The microtubule motor cytoplasmic dynein is required for spindle orientation during germline cell divisions and oocyte differentiation in Drosophila. Development 124: 2409–2419. [DOI] [PubMed] [Google Scholar]
- Mitrovich, Q. M., and P. Anderson, 2005. mRNA surveillance of expressed pseudogenes in C. elegans. Curr. Biol. 15: 963–967. [DOI] [PubMed] [Google Scholar]
- Munro, E., J. Nance and J. R. Priess, 2004. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev. Cell 7: 413–424. [DOI] [PubMed] [Google Scholar]
- O'Connell, C. B., and Y. L. Wang, 2000. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol. Biol. Cell 11: 1765–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, K. F., C. M. Leys and J. G. White, 1998. A genetic screen for temperature-sensitive cell-division mutants of Caenorhabditis elegans. Genetics 149: 1303–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, K. M., T. Ishidate, K. Nakamura, M. Shirayama, C. Trzepacz et al., 2004. The minibrain kinase homolog, mbk-2, is required for spindle positioning and asymmetric cell division in early C. elegans embryos. Dev. Biol. 265: 127–139. [DOI] [PubMed] [Google Scholar]
- Pfister, K. K., M. W. Salata, J. F. Dillman, 3rd, K. T. Vaughan, R. B. Vallee et al., 1996. Differential expression and phosphorylation of the 74-kDa intermediate chains of cytoplasmic dynein in cultured neurons and glia. J. Biol. Chem. 271: 1687–1694. [DOI] [PubMed] [Google Scholar]
- Praitis, V., E. Casey, D. Collar and J. Austin, 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport, R., 1986. Establishment of the mechanism of cytokinesis in animal cells. Int. Rev. Cytol. 105: 245–281. [DOI] [PubMed] [Google Scholar]
- Schetter, A., P. Askjaer, F. Piano, I. Mattaj and K. Kemphues, 2006. Nucleoporins NPP-1, NPP-3, NPP-4, NPP-11 and NPP-13 are required for proper spindle orientation in C. elegans. Dev. Biol. 289: 360–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, A., J. Kunz and M. N. Hall, 1996. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc. Natl. Acad. Sci. USA 93: 13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, D. J., D. J. Rose, W. M. Saxton and S. Strome, 2005. Functional analysis of cytoplasmic dynein heavy chain in Caenorhabditis elegans with fast-acting temperature-sensitive mutations. Mol. Biol. Cell 16: 1200–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, B. A., M. S. Avidan and C. M. Crowder, 2002. Regulation of hypoxic death in C. elegans by the insulin/IGF receptor homolog DAF-2. Science 296: 2388–2391. [DOI] [PubMed] [Google Scholar]
- Shiroguchi, K., and Y. Y. Toyoshima, 2001. Regulation of monomeric dynein activity by ATP and ADP concentrations. Cell Motil. Cytoskeleton 49: 189–199. [DOI] [PubMed] [Google Scholar]
- Skop, A. R., and J. G. White, 1998. The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Curr. Biol. 8: 1110–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop, A. R., D. Bergmann, W. A. Mohler and J. G. White, 2001. Completion of cytokinesis in C. elegans requires a brefeldin A-sensitive membrane accumulation at the cleavage furrow apex. Curr. Biol. 11: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen, B., L. B. Koski, A. Walsh, P. Marschall, B. Neumann et al., 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434: 462–469. [DOI] [PubMed] [Google Scholar]
- Timmons, L., and A. Fire, 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Tsou, M. F., A. Hayashi, L. R. DeBella, G. McGrath and L. S. Rose, 2002. LET-99 determines spindle position and is asymmetrically enriched in response to PAR polarity cues in C. elegans embryos. Development 129: 4469–4481. [DOI] [PubMed] [Google Scholar]
- Varadi, A., L. I. Johnson-Cadwell, V. Cirulli, Y. Yoon, V. J. Allan et al., 2004. Cytoplasmic dynein regulates the subcellular distribution of mitochondria by controlling the recruitment of the fission factor dynamin-related protein-1. J. Cell Sci. 117: 4389–4400. [DOI] [PubMed] [Google Scholar]
- White, J., and S. Strome, 1996. Cleavage plane specification in C. elegans: how to divide the spoils. Cell 84: 195–198. [DOI] [PubMed] [Google Scholar]
- Wokosin, D. L., J. M. Squirrell, K. W. Eliceiri and J. G. White, 2003. An optical workstation with concurrent, independent multiphoton imaging and experimental laser microbeam capabilities. Rev. Sci. Instrum. 74: 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, M. P., N. Stuurman and R. D. Vale, 2003. Mitochondrial positioning in fission yeast is driven by association with dynamic microtubules and mitotic spindle poles. Proc. Natl. Acad. Sci. USA 100: 11424–11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem, J., T. Gu and M. Han, 1998. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics 149: 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder, J. H., and M. Han, 2001. Cytoplasmic dynein light intermediate chain is required for discrete aspects of mitosis in Caenorhabditis elegans. Mol. Biol. Cell 12: 2921–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]