Abstract
The male-specific lethal (MSL) complex, which includes two noncoding RNA on X (roX)1 and roX2 RNAs, induces histone H4-Lys16 acetylation for twofold hypertranscription of the male X chromosome in Drosophila melanogaster. To characterize the role of roX RNAs in this process, we have identified evolutionarily conserved functional domains of roX RNAs in several Drosophila species (eight for roX1 and nine for roX2). Despite low homology between them, male-specific expression and X chromosome-specific binding are conserved. Within roX RNAs of all Drosophila species, we found conserved primary sequences, such as GUUNUACG, in the 3′ end of both roX1 (three repeats) and roX2 (two repeats). A predicted stem–loop structure of roX2 RNA contains this sequence in the 3′ stem region. Six tandem repeats of this stem–loop region (72 nt) of roX2 were enough for targeting the MSL complex and inducing H4-Lys16 acetylation on the X chromosome without other parts of roX2 RNA, suggesting that roX RNAs might play important roles in regulating enzymatic activity of the MSL complex.
BOTH RNA on X (roX)1 and roX2 play an essential role in equalizing the level of transcription on the X chromosome in Drosophila males (XY) to that of females (XX) (Park and Kuroda 2001). The male-specific lethal (MSL) complex is composed of roX1 and/or roX2 RNAs with MSL1, MSL2, MSL3, male absent on the first (MOF) (histone acetyltransferase), and maleless (MLE) (RNA helicase) (Meller et al. 2000). This complex has been shown to bind to hundreds of sites on the male X chromosome (Kelley et al. 1999; Meller et al. 2000) and increase its gene expression by approximately twofold through specific histone H4-Lys16 acetylation (Smith et al. 2000; Hamada et al. 2005; Straub et al. 2005). roX1 (3700 nt) and roX2 (500 nt) RNAs (Amrein and Axel 1997; Meller et al. 1997; Park et al. 2005) are functionally redundant despite their differences in size and primary sequence (Franke and Baker 1999; Meller and Rattner 2002). Previously, a small 25/30-nt identity and a MSL binding site (a male-specific DNase I hypersensitive site, DHS) were common motifs found in both roX genes (Franke and Baker 1999; Kageyama et al. 2001; Park et al. 2003), but subsequent experiments showed that they were not necessary for the function of roX RNA (Park et al. 2003; Stuckenholz et al. 2003). Given that there is no apparent sequence homology within the two roX RNAs, secondary or tertiary structures shared between them are likely to be crucial for the function in the MSL complex as manifested in other known noncoding RNAs (Ban et al. 2000; Dror et al. 2005). However, the large size of roX1 (3700 nt) and roX2 (500 nt) RNAs has made it difficult to predict functional secondary structures. Several Drosophila species have shown male-specific MSL proteins binding to the X chromosome (Bone and Kuroda 1996; Marin et al. 1996), raising a possibility that roX RNAs also exist in the other Drosophila species and have similar functions. Therefore, we hypothesized that the functional domains (primary sequences and/or secondary structures) of roX RNAs are evolutionarily conserved, as is the case in ribosomal RNAs (Ban et al. 2000). In this study, we cloned roX RNAs from numerous Drosophila species, identified evolutionarily conserved domains within the roX RNAs, and performed functional analysis of each roX RNA and a stem–loop structure.
MATERIALS AND METHODS
Fly genotypes and immunostaining:
Wild-type Drosophila simulans, D. yakuba, D. erecta, D. ananassae, D. pseudoobscura, D. willistoni, D. mojavensis, and D. virilis were acquired from the Tucson stock center at the University of Arizona. The D. melanogaster genotypes used in this study were: wild type, y w and roX−, y w roX1ex6 Df(1)roX252 P{w+ 4Δ4.3} (Park et al. 2002). The P{w+ 4Δ4.3} element is required to supply essential genes lost in Df(1)roX252 (Meller and Rattner 2002).
After crossing y w/Y; [roX2 transgene] males to y w roX1ex6 Df(1)roX252 P{w+ 4Δ4.3} females, male rescue frequencies of the roX2 transgenes (Figure 4F) were calculated by ratio of the male (y w roX1ex6 Df(1)roX252 P{w+ 4Δ4.3}/Y; [roX2 transgene]/+)/female (y w roX1ex6 Df(1)roX252 P{w+ 4Δ4.3}/y w; [roX2 transgene]/+) progeny from adult flies collected during 10 days after the first day of adult eclosion. For immunostaining of Figure 4, D and E, polytene chromosomes of salivary glands were used from y w roX1ex6 Df(1)roX252 P{w+ 4Δ4.3}/Y; [roX2 transgene]/+ male larvae acquired from the cross described above. Immunostaining of MSL proteins and RNA in situ hybridization of roX RNAs were performed as previously described (Kelley et al. 1999).
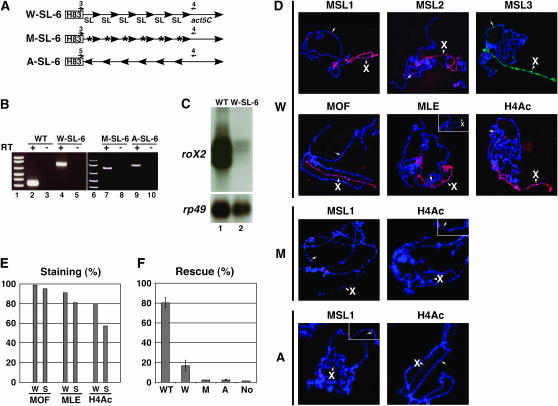
Figure 4.—
A conserved stem–loop of roX2 can assemble and localize the MSL complex to the X chromosome. (A) The construct of six tandem repeats of the stem–loop (72 bp/repeat) in roX2 RNA (SL-6). Act5C is used for poly(A) adenylation of the construct. W-SL-6, wild type; M-SL-6, mutant in the G1 region (blue characters in Figure 3F); A-SL-6, antisense of wild type. (B) RT–PCR analysis showing expression of the transcripts from total RNAs purified from male transgenic adult flies containing wild-type roX2 (WT) or W-, M-, and A-SL-6 RNAs in a roX− mutant. Primers 1 and 2 (Figure 3B) and primers 3 or 5 and 4 (A) were used in WT and W-, M-, and A-SL-6 for PCR, respectively. (C) Northern analysis of WT and W-SL-6 using total RNAs of B, with rp49 as a loading control. (D) Polytene chromosome immunostainings of MSL proteins and H4-Lys16 acetylation showing the MSL complex binding on the W-SL-6 transgene (arrow, 49A5) and the entire X chromosome (arrowhead, X). Another nucleus is represented to show heterogeneous staining of MLE (small rectangle, top right). W, W-SL-6; M, M-SL-6 (transgene, 82A5); A, A-SL-6 (transgene, 90C5). (E) Comparison of X-chromosomal immunostaining percentages (%) of MOF, MLE, and H4-Lys16 acetylation colocalized with MSL3 between wild-type roX2 (W) and W-SL-6 (S) transgenic flies in a roX− mutant. The number of counted nuclei was 323 (minimum)–463 (maximum). (F) Male rescue frequency (male/female) by WT roX2 or W-, M-, or A-SL-6 RNA in a roX− mutant. Full genotypes of crosses are described in materials and methods. Averages of male viability (%) are represented with standard deviations for three WT roX2 or four W-, two M-, and three A-SL-6 transgenic lines tested. No, no transgene.
Transgene construction and transformation:
To create W-SL-6 and M-SL-6 transgenes (Figure 4A), a monomer of a stem–loop region (72 bp) of the roX2 gene was amplified during PCR with a 5′ primer (5′-CTCGGGAAAAGACGTGTAAAATGTTGC-3′) and a 3′ primer (wild type, WT, 5′-CCCGAGTTAAGGCGCGTAAAACGTT-3′; M, 5′-CCCGAGTTTTCGCGACATAAAACAA-3′) containing an AvaI site (underline) and cloned into the pCRII-TOPO vector (Invitrogen, Carlsbad, CA). After sequencing, this monomer was excised by AvaI digestion, self-ligated, and subcloned into an AvaI-digested pCRII-TOPO vector. After a trimer was cloned, a plasmid containing six tandem repeats was constructed by blunt end ligation between two trimers. This hexamer was subcloned into a NotI/BamHI-digested pCaSpeR Hsp83-act plasmid (containing an act5C gene fragment to provide a 3′ poly(A) site). To create an A-SL-6 transgene (Figure 4A), a wild-type hexamer was subcloned into a BglII/NotI-digested pCaSpeR Hsp83-act plasmid. Transgenic flies were made by P-element-mediated transformation at Model System Genomics of Duke University.
RT–PCR analysis:
To check the male-specific expression of roX1 and roX2 genes in other Drosophila species (Figure 1B), oligo(dT)-primed cDNAs were made from 5 μg total RNAs of male and female adults. For roX1, the 5′ primers (D. melanogaster, 5′-ACCAGCAGTTGATTTGCG-3′; D. simulans and D. erecta, 5′-TCTATTGGCCTTGATTATTAAC-3′; D. yakuba, 5′-ACTGGGCGCCTACAATGCG-3′; D. ananassae, 5′-CGAGCCGCTCATGTTCGCA-3′; D. pseudoobscura, 5′-CCCTCTGTTGGTCAATCGTTC-3′; D. mojavensis, 5′-GAGGGCACTTAGAGTGTCAAC-3′; D. virilis, 5′-ACCTGCTGCGTCCCTCTGC-3′) and the 3′ primers (D. melanogaster, 5′-ATTTCGATTTTCTTTTTATAGTTTGGG-3′; D. simulans and D. erecta, 5′-CGGCTCAGGCGTATAACGAT-3′; D. yakuba, 5′-CGGCTCAAGCGTATAACGATT-3′; D. ananassae, 5′-CGGCACAGGCGTATAACGG-3′; D. pseudoobscura, 5′-GCTCAGACGTATAACGTTTCC-3′; D. mojavensis, 5′-CGGCTCAGACGTATAACAGTT-3′; D. virilis, 5′-CGGCTCGGACGTATAACGTT-3′) were used for PCR. For roX2, the 5′ primers (D. melanogaster, 5′-TATATCATAAGTCGAGCGTTTAG-3′; D. yakuba, 5′-CGGCCTGGTCACACTGAGCT-3′; D. ananassae, 5′-ACCCTCTCTAGATCTTACGAC-3′; D. pseudoobscura, 5′-CTTTTCCCGCTAAAAATAATTCAG-3′; D. mojavensis, 5′-GTTCTTGCATCAGATAGTTAGG-3′; D. virilis, 5′-GTTCATCATCAGACAGCTAGG-3′) and the 3′ primers (D. melanogaster and D. yakuba, 5′-ACTGGTTAAGGCGCGTAAAAC-3′; D. ananassae, 5′-CTGGTTAAGGCGCGTAAAAC-3′; D. pseudoobscura, 5′-GGCTCGTAAAACGTTACCATTG-3′; D. mojavensis, 5′-ATTGTTAAGGCGCGTATAACGT-3′; D. virilis, 5′-GTTAAGGCACGTATAACGTTAC-3′) were used during PCR.
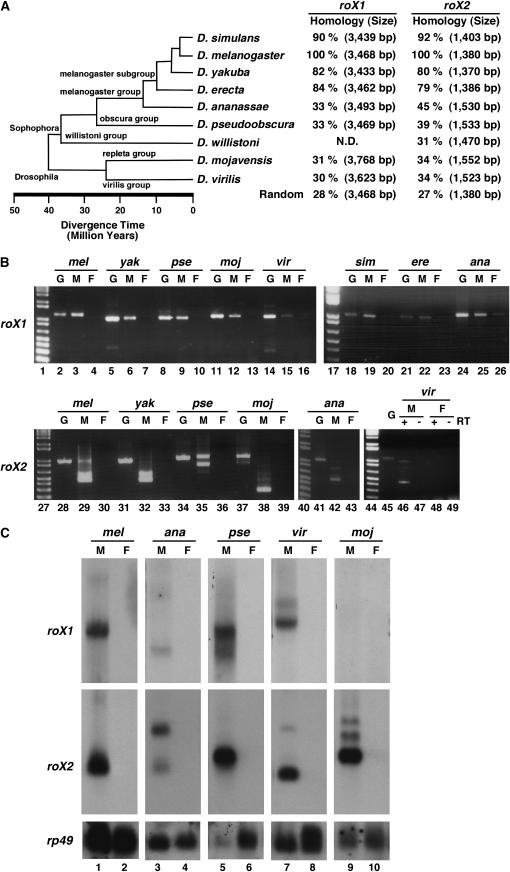
Figure 1.—
The evolutionary conservation of roX RNAs in other Drosophila species. (A) The estimated time distance of divergence of several Drosophila species (http://www.flybase.org/blast/). According to the 5′ and 3′ end sequences of c20 roX1 cDNA and W-H83roX2 cDNA, which can rescue the roX− mutant (Meller and Rattner 2002; Stuckenholz et al. 2003; Park et al. 2005), the genomic sequences of roX1 (3468 bp) and roX2 (1380 bp) of D. melanogaster were used to find roX1 and roX2 genomic sequences of other Drosophila species. By a pairwise alignment of the ClustalW program (http://www.ebi.ac.uk/clustalw) between roX genomic sequences of D. melanogaster and other Drosophila species, we predicted roX gene sizes of other Drosophila species and obtained the homology percentage (%) comparing the alignment scores between them. ND, not determined; Random, no homology control (mof and pka genes of D. melanogaster). (B) RT–PCR analysis of roX1 and roX2 RNAs in several Drosophila species. In roX1, primers from the 3′ region containing the evolutionarily conserved sequences (Figure 3A) were used for PCR (no RT control, data not shown). In roX2, primers from each end of the whole gene except DHS were used for PCR. G, genomic DNA; M, male; F, female; lane 1, a 1-kb plus DNA ladder (Invitrogen). (C) Northern analysis of roX1 and roX2 RNAs in distantly related Drosophila species. Using total RNA (20 μg) of adult flies, one membrane was made, cut, and then hybridized with species-specific roX1 or roX2 probes. Membranes were stripped off and reprobed with rp49 of D. melanogaster as a loading control. M, male; F, female.
To check the expression of roX2 clones from WT and W-, M-, and A-SL-6 transgenics in the roX− mutant (Figure 4B), oligo(dT)-primed cDNAs were made from 5 μg total RNAs of male adults, and the 5′ primers [WT, 5′-GCCATCGAAAGGGTAAATTGG-3′ (primer 1); W-, M-SL-6, 5′-CAGTGTGATGGATAATTCGCC-3′ (3); A-SL-6, 5′-CGGTACATCGAATTCGTTAAC-3′ (5)] and the 3′ primers [WT, 5′-ATTGCGACTTGTACAATGTTGCGTT-3′ (2); W-, M-, A-SL-6, 5′-GCGATCCTTCTTAGAAGCACT-3′ (4)] were used in PCR.
Northern analysis:
For Northern analysis (Figures 1C and 4C), total RNAs from adult flies were prepared using TRIzol Reagent (GIBCO-BRL, Carlsbad, CA) and 20 μg of total RNAs were loaded in each lane. In Figure 1C, specific probes for several Drosophila species were prepared by random priming (Invitrogen), using PCR products purified from Figure 1B. In Figure 4C, roX2 whole genomic sequence (1380 bp) was used to make the probe. Following overnight hybridization at 42° in hybridization solution (30% formamide, 1 m NaCl, 100 mm NaPO4 pH 7.0, 7% SDS, 10× Denhardt's, 100 μg/ml ssDNA-fish), the membrane was washed two times in 2× SSC, 0.1% SDS at 42°.
RESULTS AND DISCUSSION
Male-specific expression and X chromosome-specific binding of roX RNAs in other Drosophila species:
Using the full sequence or evolutionarily conserved partial sequence of roX1 and roX2 genes of D. melanogaster (http://www.flybase.org/blast/), we have identified eight roX1 and nine roX2 genes in different Drosophila species (Figure 1A). In distantly related Drosophila species ranging from D. ananassae to D. virilis, homology percentages were low enough to be similar to unrelated controls (Figure 1A), indicating that roX1 and roX2 sequences highly diverged as noncoding RNA during evolution. In all Drosophila species tested by RT–PCR, roX RNAs were expressed only in males (Figure 1B), suggesting that roX1 and roX2 RNAs of other Drosophila species have similar expression patterns to those of D. melanogaster. Interestingly, the results demonstrate a presence of alternative splicing patterns in the roX2 RNA species investigated (Figure 1B), which is important for the function of roX2 RNA (Park et al. 2005). In Northern analysis, male-specific transcripts of roX1 and roX2 were detected in D. simulans, D. yakuba, and D. erecta with similar sizes to the roX RNAs of D. melanogaster (data not shown; Park et al. 2003), when radio-labeled roX probes of D. melanogaster were used with the low-stringency hybridization method (see materials and methods). However, roX RNAs from more distantly related Drosophila species showed no cross-hybridization with roX probes of D. melanogaster, even though protein-coding rp49 probes of D. melanogaster were hybridized with rp49 RNA in every Drosophila species we tested (Figure 1C). This result suggests that the nucleotide sequences of roX RNAs of these more distantly related species differ significantly from those of D. melanogaster, which is consistent with the low homology percentage shown in Figure 1A. Using species-specific probes of roX1 (∼1 kb of evolutionarily conserved 3′ end regions) and roX2 (∼1 kb of the entire region except DHS) in distantly related fly species, we detected male-specific transcripts similar in size to roX1 (∼3700 nt) and roX2 (∼500 nt) of D. melanogaster (Figure 1C). One exception was roX1 RNA in D. mojavensis. Even though we used an additional roX1 probe from the 5′ end of the sequence (2.5 kb), a roX1 transcript was not detected in Northern blot analysis (Figure 1C, top section, lane 9), suggesting that roX1 RNA of D. mojavensis might not be functional in the adult male.
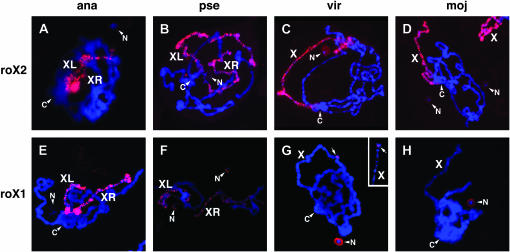
In RNA in situ hybridization performed using species-specific roX2 (Figure 2, A–D) and roX1 (Figure 2, E–H) probes in polytene chromosomes, D. ananassae and D. pseudoobscura showed metacentric X chromosomes with two arms (XL and XR; Bone and Kuroda 1996) painted by roX1 and roX2 RNAs. However, in D. pseudoobscura, the roX1 signal on the X chromosome (Figure 2F) was weaker than the roX2 signal (Figure 2B). At this point it is not known if this is caused by a dominant function of roX2 RNA in the salivary gland or a low hybridization efficiency of the roX1 probe. Unexpectedly a weak roX1 signal was detected in the nucleolus of D. pseudoobscura (Figure 2F), but no roX2 signal was detected in the nucleolus (Figure 2B). In D. virilis, a single band of roX1 signal was detected on the X chromosome (Figure 2G), contrary to the roX2 signal that was found along the entire X chromosome (Figure 2C). Through cytological mapping of polytene chromosomes, we found that the single band of roX1 signal in D. virilis was localized to the tip of the X chromosome in several nuclei. Considering that the roX1 gene of D. melanogaster is located in the distal region of the X chromosome (3F), this signal could be derived from a nascent transcript of the D. virilis roX1 gene. In D. virilis, we also observed a weak roX2 and a strong roX1 signal in the nucleolus (Figure 2, C and G). In D. mojavensis, no roX1 signal was detected on the X chromosome (Figure 2H) in contrast to the strong roX2 signal on the whole X chromosome (Figure 2D), which is consistent with Northern blot analysis of adult flies (Figure 1C, lane 9). Considering functional redundancy between roX1 and roX2 in D. melanogaster (Franke and Baker 1999; Meller and Rattner 2002), it is possible that roX2 might play a major role during dosage compensation of the X chromosome in D. mojavensis. roX1 signal was also detected in the nucleolus of D. mojavensis (Figure 2H), similar to D. pseudoobscura and D. virilis (Figure 2, F and G), which suggests that roX1 RNA might have different function(s) in the nucleolus instead of the X chromosome in other distantly related Drosophila species.
Figure 2.—
X chromosome-specific binding of roX RNAs in other Drosophila species. (A–H) RNA in situ hybridization in male polytene chromosomes of salivary glands of several Drosophila species using species-specific roX2 (A–D) and roX1 (E–H) probes. In D. virilis (G), arrows indicate roX1 signal from the X chromosome. Another roX1 signal from a different X chromosome in another nucleus is shown to verify its distal location (small rectangle, top right). XL and XR, metacentric X chromosomes; C, chromocenter; N, nucleolus; X, X chromosome.
Evolutionarily conserved primary sequences and stem–loop structures in roX RNAs:
By alignment of the nucleotide sequence of roX RNAs cloned from several Drosophila species (supplemental Figure S1 at http://www.genetics.org/supplemental/), we identified several evolutionarily conserved regions within the roX1 and roX2 genes despite the low homology scores of the whole region (Figure 1A). The already known roX-consensus sequence for MSL binding, so-called DHS (Kageyama et al. 2001; Park et al. 2003), is well retained in similar regions (middle of roX1 and 3′ end of roX2) even in distantly related Drosophila species (Figure 3, A–D). In particular, CTCTC and GAGA (red characters in top line of Figure 3, C and D), which showed strong defects in MSL binding when mutated (Park et al. 2003), are highly conserved even though the number of nucleotides flanking CTCTC and GAGA is variable among different Drosophila species.
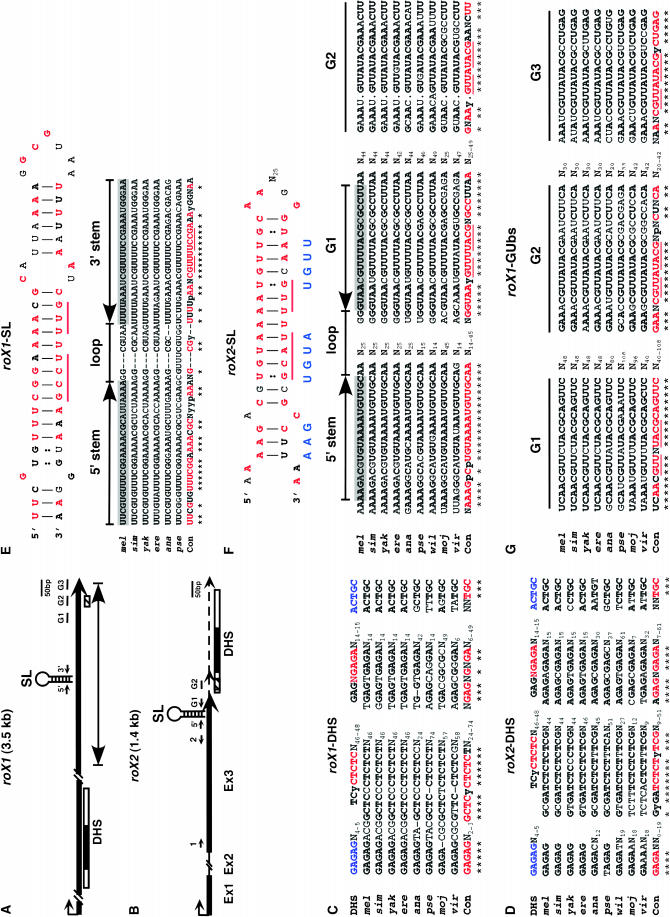
Figure 3.—
Evolutionarily conserved primary sequences and stem–loop structures of roX RNAs. (A and B) Gene structure of roX1 (A) and roX2 (B) RNA showing location of stem–loop region (SL), 5′ stem (5′), 3′ stem (3′), and GUUNUACG boxes (G1, G2, and G3). Thick line, the major transcript; dotted line, 3′ minor transcript in roX2; black box in DHS, 110-bp segment containing conserved sequences for MSL binding located in both roX1 and roX2 genes (Park et al. 2003); hatched box, a small 25/30-nt identity found from initial sequence comparison between roX genes (Franke and Baker 1999); line with arrowheads at both ends (A), an essential domain (∼600 bp) found by serial deletion (each ∼300 bp) analysis of the roX1 gene (Stuckenholz et al. 2003). (C and D) Alignment of consensus sequences within the DHS of roX1 (C) and roX2 (D). In the first line (DHS), blue and red characters represent the sites that showed mild and strong defects in MSL binding, respectively, when mutagenized previously (Park et al. 2003). (E) A stem–loop structure previously predicted in roX1 RNA (Stuckenholz et al. 2003) and an alignment of stem–loop (SL) sequences of roX1 RNA. (F and G) A predicted stem–loop structure in roX2 RNA from the mfold program (F) and alignments of 5′ stem and GUUNUACG box sequences in roX2 RNAs (F) and roX1 RNAs (G). All alignments were performed using the ClustalW program and manually from nine (roX2) and eight (roX1) Drosophila species except those in E (six species). Within the consensus sequence (Con), red characters in boldface type represent perfect matches (no mismatch or a nucleotide mismatch in only one species aligned) and black characters in boldface type represent less perfect matches (a nucleotide mismatch in two species aligned) except those in E (red boldface type, no mismatch; black boldface type, a nucleotide mismatch). Red and black characters in boldface type in stem–loop structures (E and F) represent the consensus sequences from alignments. N, random; p, purine; y, pyrimidine base; *, consensus sequence.
Previously, an ∼600-bp region in the 3′ end of roX1 RNA was identified as an essential domain from serial deletion analysis of the roX1 gene (∼300 bp each) (Figure 3A) (Stuckenholz et al. 2003). This region includes a stem–loop structure, which was shown to be critical for proper RNA function. Upon comparison of this stem–loop region found from different Drosophila species, we were able to identify conserved primary sequences and secondary structures (Figure 3E), suggesting that this region may be evolutionarily important for the function of roX1 RNA. So far we have not been able to identify this conserved region in more distantly related Drosophila species, D. virilis and D. mojavensis, both of which lost their roX1 RNA-binding activity along the entire X chromosome in polytene chromosomes as shown in Figure 2, G and H, respectively. It is possible that the lack of localization of roX1 RNA on their X chromosomes might be a result of the loss of this conserved stem–loop structure through evolution.
Alignment of nine roX2 RNAs from different Drosophila species revealed a stretch of conserved sequence (GUUNUACG box-1, GUb-1) in the 3′ end of roX2 RNA, which is located at the 3′ stem region of the putative stem–loop structure predicted using the mfold program (Figure 3, B and F). Further inspection of the alignment of the roX2 sequence allowed us to identify another conserved GUUNUACG sequence (GUb-2) downstream of GUb-1, which is located at the 3′ minor transcript of roX2 RNA (Figure 3, B and F). The GUb-2 region was previously found as a small 25/30-nucleotide identity in both roX genes (Franke and Baker 1999), but it was shown that the deletion of this region containing GUb-2 did not affect the function of roX2 RNA (Park et al. 2003). This raises a possibility that multiple GUb's in the roX2 RNA might be functionally redundant, considering its conservation through evolution and multiple occurrences within roX2 (and roX1, see below). In D. pseudoobscura, D. mojavensis, and D. virilis we found an additional GUb (GUUNUACG) sequence upstream of GUb-1 (supplemental Figure S1 at http://www.genetics.org/supplemental/). Interestingly, the 3′ stem region of the predicted stem–loop structure in roX1 RNA contains a GUUNUCCG sequence (Figure 3E), which is similar to the GUb (GUUNUACG) sequence of roX2 RNA. Although more experiments are required to test if these two stem–loop structures found in roX1 and roX2 RNAs have a similar function, it is possible that those two stem loops with similar nucleotide motifs (e.g., GUb at 3′ stem) might explain the functional redundancy between roX RNAs despite no apparent resemblance otherwise.
In search of more clues for functional redundancy between roX RNAs, we attempted to find GUUNUACG motifs in the roX1 RNA. Upon detailed analysis of several roX1 RNA sequences from different fly species, we identified three GUb's in the 3′ end of roX1 RNA in eight Drosophila species (Figure 3, A and G). The second GUUNUACG (GUb-2) found in roX1 RNA is a previously identified 25/30-nt region (Franke and Baker 1999). Similar to GUb-2 of roX2 RNA, deletion of this GUb-2 did not affect function of roX1 RNA (Stuckenholz et al. 2003), which might also be attributed to the presence of other GUb's in the roX1 RNA. D. ananassae and D. virilis contain another GUb sequence (total of four GUb's) and D. mojavensis contains two more GUb sequences (total of five GUb's) in the more upstream region of roX1 RNA (supplemental Figure S1 at http://www.genetics.org/supplemental/). However, an evolutionarily conserved 5′ stem sequence around the GUb regions of roX1 RNA has not yet been identified in eight Drosophila species. This suggests several possibilities. Alignment imperfection may have not enabled us to detect the conserved 5′ stem sequence. It is also possible that this region functions as a sequence without secondary structure or that other distantly related Drosophila species (for example, D. mojavensis and D. virilis) have lost their secondary structure around the GUb regions during evolution, such as the stem–loop structure identified previously (Figure 3E) (Stuckenholz et al. 2003). At this point we are not certain if these GUUNUACG sequences are necessary for the function of roX1 and roX2 RNAs. However, it is interesting that they are evolutionarily conserved in the 3′ ends both of roX1 and of roX2 RNAs, which are functionally redundant in spite of low similarity between total sequences and a low homology between several Drosophila species.
A stem–loop region of roX2 RNA alone can induce the X chromosome-specific binding of the MSL complex and H4-Lys16 acetylation:
To determine the functional importance of the putative stem–loop region of roX2 including a GUb sequence at its 3′ stem, we made tandem repeats of the stem–loop region of roX2 RNA (W-SL-6) under the control of constitutive promoter (hsp 83) (Figure 4A). The size of W-SL-6 RNA is 432 nt (72 nt × 6), similar to the major isoform of roX2 RNA (∼500 nt) (Park et al. 2005). RT–PCR analysis confirmed that the W-SL-6 construct expresses a transcript including SL-6 RNA (Figure 4B, lane 4) in roX− mutant flies (see materials and methods). However, the steady-state level of W-SL-6 RNA in the W-SL-6 transgenic flies was much lower than that of roX2 RNA from wild-type roX2 transgenic flies even though both transcripts are expressed from the hsp83 promoters (Figure 4C), suggesting that the other parts of roX2 might be required for the stability of the RNA.
Interestingly, in the polytene chromosome of W-SL-6 transgenic flies we found that all five MSL proteins were detected not only on the autosomal transgenic location (three different locations tested, arrow in Figure 4D), but also on the X chromosome, indicating that SL-6 itself is sufficient to attract MSL proteins to the site of its own transcription and then target the MSL complex to the X chromosome. Unlike the other MSL proteins (MSL1, MSL2, MSL3, and MOF), which showed strong and consistent signals on the X chromosome in all nuclei (Figure 4D), MLE showed heterogeneous staining with variable degrees of intensity (Figure 4D). To compare binding efficiency of W-SL-6 and wild-type roX2 RNAs with MSL proteins, we performed double staining of MSL3 with either MOF or MLE protein in the polytene chromosome of each transgenic fly (Figure 4E). First, the numbers of nuclei showing MSL3 staining were counted and next, double staining (MSL3 + MOF or MSL3 + MLE) from them was counted. The percentage of double staining was calculated [(MSL3 + MOF)/MSL3 or (MSL3 + MLE)/MSL3]. Even though it is slightly lower in W-SL-6 transgenic lines, the percentages of double staining of MSL3 + MOF and MSL3 + MLE were comparable in W-SL-6 and wild-type roX2 RNAs (Figure 4E), suggesting that all five MSL proteins are assembled with the W-SL-6 RNA. As shown in individual staining (Figure 4D), MLE binding to the X chromosome in W-SL-6 transgenic flies was heterogeneous with variable intensity unlike consistent binding of MSL3 within every nucleus in double staining (data not shown), which implies that weaker staining of MLE in some nuclei is not due to the weak binding of other MSL proteins. At this point we do not know yet whether a more efficient interaction between MLE (RNA helicase) and other MSL proteins may require other regions of roX2 RNA outside of the stem loop and then the instability of W-SL-6 RNA (Figure 4C) may be caused by heterogeneous binding of MLE. Interestingly, the MSL complex assembled with the SL-6 RNA was able to induce H4-Lys16 acetylation on the X chromosome and the autosomal transgene (Figure 4, D and E), although the percentage of double staining of MSL3 and H4-Lys16 acetylation [(MSL3 + H4lys16Ac)/MSL3] in W-SL-6 RNA (57%) was a little lower than that of wild-type roX2 RNA (80%).
To analyze the function of W-SL-6 RNA in flies, a rescue assay was performed by expressing W-SL-6 transgenes (four independent lines) in the roX− mutant and counting survival of male flies (Figure 4F). In contrast to the immunostaining results including positive histone H4 lysine 16 acetylation on the X chromosome (Figure 4, D and E), a rescue frequency of SL-6 was low (17%) compared to that of the wild-type roX2 transgene (80%). One explanation for this partial rescue efficiency of the W-SL-6 transgene is that it might be caused by low stability of W-SL-6 RNA (Figure 4C) due to the absence of other parts of roX2 RNA in SL-6, which are necessary for better interaction with MSL proteins.
To confirm the specific interaction between the stem–loop of roX2 RNA and MSL proteins, we tested other tandem repeats (hexamers) that have mutations in a GUb sequence (Figure 3F) of the 3′ stem region (M-SL-6) or an antisense transcript of stem–loop (A-SL-6) (Figure 4A). These hexamers did not either attract MSL proteins or induce H4-Lys16 acetylation (Figure 4D). In addition, they showed low frequency for rescue (∼2.2%) of the roX− deficiency male, which is similar to no transgenic control (1.5%). Considering the moderate rescue frequency of wild-type hexamer (W-SL-6, 17%), this result suggests that a GUb sequence or stem–loop structure (or both) within roX RNA plays an important role in the interaction with MSL proteins.
Although roX1 (3700 nt) and roX2 (500 nt) RNAs are apparently different in size and primary sequence, they function redundantly in dosage compensation on the Drosophila X chromosome (Franke and Baker 1999; Meller and Rattner 2002), suggesting that they share common functional domains. In several Drosophila species, male-specific binding to the X chromosome by roX RNAs is evolutionarily conserved (Figure 2), indicating that roX RNAs keep common functional domains despite evolutional change as noncoding RNA (∼40 million years apart, Figure 1A). Considering low homology in total sequences and no cross-hybridization between D. melanogaster and other distant Drosophila species (Figure 1), functional domains could be short primary sequences and/or the secondary structures. Using a comparative evolutionary approach, we successfully found stretches of conserved motifs (GUb) and putative stem–loop structures within the roX RNAs from several different Drosophila species (Figure 3).
roX1 and roX2 double-mutant males die from failure of dosage compensation on the X chromosome in contrast to females that suffer no harmful effects. In the roX-deficient male fly, MSL proteins show little to no ability to localize to the X chromosome and mostly mislocalize to the heterochromatic chromocenter (Meller and Rattner 2002). These observations suggest that roX RNAs are important for accurate targeting of the MSL complex to the X chromosome. It is unknown how MSL proteins interact with roX RNAs to make the MSL complex functional or if roX RNAs regulate enzymatic activity of MSL proteins. However, our data showed that the conserved stem–loop region of roX2 is a core functional domain sufficient to attract MSL proteins, assemble MSL complexes, and target them to the X chromosome, followed by subsequent acetylation of histone H4 lys16 on the X chromosome (Figure 4). This suggests a possibility that roX RNA is required not only to assemble the MSL complex, but also to regulate enzymatic activity of MSL proteins by the conserved stem–loop region. A more detailed study about the conserved functional domains of roX RNAs will reveal how noncoding RNA regulates protein components in a ribonucleoprotein complex for chromatin organization.
Acknowledgments
We thank T. Chan and R. Snepar for technical support. We are grateful to M. Kuroda for anti-MSL antibodies. This work was supported by grants from the American Heart Association (0535548T), the New Jersey Commission on Cancer Research (05-1963-CCR), and the University of Medicine and Dentistry of New Jersey Foundation.
References
- Amrein, H., and R. Axel, 1997. Genes expressed in neurons of adult male Drosophila. Cell 88: 459–469. [DOI] [PubMed] [Google Scholar]
- Ban, N., P. Nissen, J. Hansen, P. B. Moore and T. A. Steitz, 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Bone, J. R., and M. I. Kuroda, 1996. Dosage compensation regulatory proteins and the evolution of sex chromosomes in Drosophila. Genetics 144: 705–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror, O., R. Nussinov and H. Wolfson, 2005. ARTS: alignment of RNA tertiary structures. Bioinformatics 21: ii47–ii53. [DOI] [PubMed] [Google Scholar]
- Franke, A., and B. S. Baker, 1999. The roX1 and roX2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol. Cell 4: 117–122. [DOI] [PubMed] [Google Scholar]
- Hamada, F. N., P. J. Park, P. R. Gordadze and M. I. Kuroda, 2005. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 19: 2289–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama, Y., G. Mengus, G. Gilfillan, H. G. Kennedy, C. Stuckenholz et al., 2001. Association and spreading of the Drosophila dosage compensation complex from a discrete roX1 chromatin entry site. EMBO J. 20: 2236–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, R. L., V. H. Meller, P. R. Gordadze, G. Roman, R. L. Davis et al., 1999. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98: 513–522. [DOI] [PubMed] [Google Scholar]
- Marin, I., A. Franke, G. J. Bashwa and B. S. Baker, 1996. The dosage compensation system of Drosophila is co-opted by newly evolved X chromosomes. Nature 383: 160–163. [DOI] [PubMed] [Google Scholar]
- Meller, V. H., and B. P. Rattner, 2002. The roX RNAs encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 21: 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller, V. H., K. H. Wu, G. Roman, M. I. Kuroda and R. L. Davis, 1997. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell 88: 445–457. [DOI] [PubMed] [Google Scholar]
- Meller, V. H., P. R. Gordadze, Y. Park, X. Chu, C. Stuckenholz et al., 2000. Ordered assembly of roX RNAs into MSL complexes on the dosage compensated X chromosome in Drosophila. Curr. Biol. 10: 136–143. [DOI] [PubMed] [Google Scholar]
- Park, Y., and M. I. Kuroda, 2001. Epigenetic aspects of X-chromosome dosage compensation. Science 293: 1083–1085. [DOI] [PubMed] [Google Scholar]
- Park, Y., R. L. Kelley, H. Oh, M. I. Kuroda and V. H. Meller, 2002. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science 298: 1620–1623. [DOI] [PubMed] [Google Scholar]
- Park, Y., G. Megnus, X. Bai, Y. Kageyama, V. H. Meller et al., 2003. Sequence-specific targeting of Drosophila roX genes by the MSL dosage compensation complex. Mol. Cell 11: 977–986. [DOI] [PubMed] [Google Scholar]
- Park, Y., H. Oh, V. H. Meller and M. I. Kuroda, 2005. Variable splicing of non-coding roX2 RNAs influences targeting of MSL dosage compensation complexes in Drosophila. RNA Biol. 2: 157–164. [DOI] [PubMed] [Google Scholar]
- Smith, E. R., A. Pannuit, W. Gu, A. Steurnagel, R. G. Cook et al., 2000. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 20: 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub, T., G. D. Gilfillan, V. K. Maier and P. B. Becker, 2005. The Drosophila MSL complex activates the transcription of target genes. Genes Dev. 19: 2284–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckenholz, C., V. H. Mrller and M. I. Kuroda, 2003. Functional redundancy within roX1, a noncoding RNA involved in dosage compensation in Drosophila melanogaster. Genetics 164: 1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]