Abstract
The circadian mechanism appears remarkably conserved between Drosophila and mammals, with basic underlying negative and positive feedback loops, cycling gene products, and temporally regulated nuclear transport involving a few key proteins. One of these negative regulators is PERIOD, which in Drosophila shows very similar temporal and spatial regulation to TIMELESS. Surprisingly, we observe that in the housefly, Musca domestica, PER does not cycle in Western blots of head extracts, in contrast to the TIM protein. Furthermore, immunocytochemical (ICC) localization using enzymatic staining procedures reveals that PER is not localized to the nucleus of any neurons within the brain at any circadian time, as recently observed for several nondipteran insects. However, with confocal analysis, immunofluorescence reveals a very different picture and provides an initial comparison of PER/TIM-containing cells in Musca and Drosophila, which shows some significant differences, but many similarities. Thus, even in closely related Diptera, there is considerable evolutionary flexibility in the number and spatial organization of clock cells and, indeed, in the expression patterns of clock products in these cells, although the underlying framework is similar.
CIRCADIAN rhythms are generated by a system of interlocked autoregulatory feedback loops in which both negative and positive feedback play prominent roles. The insect model has been developed most forcefully in Drosophila melanogaster, where two cycling proteins, PERIOD (PER) and TIMELESS (TIM), act as negative regulators of their own transcription through the positively acting bHLH-PAS transcription factors CLOCK (CLK) and CYCLE (CYC, also known as BMAL1) (Hall 2003; Collins and Blau 2007). The CLK protein also cycles and its regulation is interlocked with that of PER, in that CLK/CYC also activate PDP1ɛ and VRILLE (VRI), with the latter modulating expression of Clk (Cyran et al. 2003; Benito et al. 2007).
One of the most compelling features of per and tim regulation is that the mRNAs cycle with a peak a few hours in advance of the rhythm in their protein products in the fly's head (Hall 2003). The protein cycles have been visualized in a small subset of neurons within the CNS, termed the lateral and the dorsal neurons. The best studied are the ventral lateral neurons (LNvs) because they can also be counterstained for the neuropeptide pigment-dispersing factor PDF (Helfrich-Forster and Homberg 1993; Nassel et al. 1993; Helfrich-Forster 1995). Within these neurons, PER and TIM proteins can be seen to translocate to the nucleus late at night, when their proteins are at their peak (Shafer et al. 2002). During the day, both proteins show dramatic reductions in their abundance that correlates with their hyperphosphorylation and subsequent degradation (Edery et al. 1994; Zeng et al. 1996; Naidoo et al. 1999). PER appears to require TIM for its stability, in part to protect it from phosphorylation by the DOUBLETIME kinase (DBT), so once TIM levels start to fall, PER levels also become compromised (Kloss et al. 1998; Price et al. 1998; Hall 2003). However, post-transcriptional control means that, even in constant darkness, PER and TIM levels ebb and flow with similar patterns that are seen under light–dark (LD) cycles (Hall 2003). A number of other kinases (Hall 2003) and phosphatases (Sathyanarayanan et al. 2004; Fang et al. 2007) have also been shown to modulate the stabilities of PER and TIM.
Comparative analysis of this insect model of circadian gene regulation has been most comprehensively studied in the giant silkmoth, Antheraea pernyii. While the photoreceptors in the moth appear to show cycles of PER antigenicity similar to that seen in the fly (Reppert et al. 1994), in the central brain, a very small number of neurons co-express cycling PER and TIM, yet the two proteins remain stubbornly cytoplasmic (Sauman and Reppert 1996). This observation might initially appear to preclude a canonical autoregulatory role for these two proteins. However, when transformed into per-null flies—although behavioral rhythmicity is markedly attenuated in the transgenic compared to control transformant flies—Antheraea pernyii PER(ApPER) does appear to locate to the nucleus of lateral neurons and photorecepeptors during the night phase (Levine et al. 1995). In addition, when ApPER is used in Drosophila cell lines to reconstitute a circadian pacemaker, it does appear to act as a negative regulator of ApCLK/ApBMAL1-mediated transactivation, with the added bonus of A. pernyii TIM acting to enhance this negative regulation (Chang et al. 2003). In Drosophila, TIM can shuttle in and out of the nucleus, so it may be that although TIM (or PER) cannot be seen in the nucleus at particular time points in the silkmoth, it is nevertheless present (Ashmore et al. 2003; Nawathean and Rosbash 2004). Thus perhaps even in Antheraea, the canonical model holds. Furthermore, a recent study has revealed that double-stranded RNA interference knockdown of per in another silkmoth, Bombyx mori, generates a modest disruption in the circadian larval eclosion gate, consistent with the view that per in Lepidoptera plays a similar biological role to Drosophila, irrespective of any differences in temporal expression patterns (Sandrelli et al. 2007).
However, a survey of PER-like immunoreactivity in a number of insect orders once again reveals a recalcitrant PER antigen that is exclusively confined to the cytoplasm in the various neuronal cell types where it is found (Zavodska et al. 2003b). We have therefore studied the regulation of the per and tim genes within the circadian clock of the housefly Musca domestica, which had a common ancestor with Drosophila ∼100 MYA (Hennig 1981). The Musca per ortholog can rescue per-null arrhythmia in Drosophila hosts to a surprisingly robust degree compared to the per transgene from a more closely related species such as D. pseudoobscura (Piccin et al. 2000). These results might suggest that Musca per might not seem to provide a very promising avenue for further comparative work, particularly as a related muscid, Lucilia cuprina, shows an expression pattern for per gene products very similar to that of Drosophila (Warman et al. 2000). However, we will see that the study of clock gene regulation in Musca provides some interesting twists and turns that should be considered carefully when analyzing the results of other comparative circadian studies.
MATERIALS AND METHODS
Fly maintenance:
Musca embryos and larvae were raised on a medium made of bran (55 g), heat-inactivated yeast (3 g), milk (150 ml), and the antimycotic nipagin (0.35 g) until pupariation. After eclosion, adult flies were fed on water, sugar, and dried milk. Flies were maintained at 25° under 12 hr light/12 hr dark cycles (LD12:12). In our studies, we used both a wild type (gift from A. Malacrida and G. Gasperi, University of Pavia) and a white strain (gift from Daniel Bopp, University of Zurich) without noting any significant difference (Hediger et al. 2001). However, locomotor activity and confocal microscopy were performed on the white strain only, whereas pupal eclosion was tested on flies carrying both white and apterous mutations (again from Daniel Bopp).
Gene expression analyses:
Sequences for the Musca domestica circadian genes vri, cyc, Clk, and “insect” cryptochrome (cry) were obtained using a degenerate PCR approach with primers designed according to conserved protein regions from Drosophila and mouse clock gene orthologs. Short fragments (60–250 bp) were then amplified from Musca head cDNA, cloned, and sequenced. Gene sequence was then extended by primer walking or 3′-RACE. Musca tim sequences were obtained by screening a housefly genomic library with D. melanogaster tim as a probe (E. Rosato, J. Clayton, B. Collins and S. Campesan, unpublished results). These clock gene sequences will constitute the subject of a separate publication. The Musca per sequence has been described previously (Piccin et al. 2000). Real-time PCR quantification of reverse-transcribed mRNA from whole-head extracts was carried out for Musca per, tim, vri, cyc, Clk, and cry under LD, constant darkness (DD), and constant light (LL) conditions. The constitutively expressed Musca rp49 gene was used as an amplification standard. Primers used are listed in Table 1. In one experimental design, fly cultures (from the final larval stage) were entrained to LD12:12 and collected at 4-hr intervals during one final LD cycle and the following day in DD. Similarly, flies exposed to LL from the late larval stage were used to assess the expression of clock genes in LL. For both experimental designs, at least three independent animal groups were reared and sacrificed. For each experiment, 20 3- to 5-day-old males were collected at the appropriate time point, and the heads stored at −80° until RNA extraction.
TABLE 1.
Gene-specific primers used in real-time PCR
| Transcript | Primer | Primer sequencea | Product size (bp) (cDNA) |
|---|---|---|---|
| rp49 | rp49 fw | 5′-GTTATGCCAAATTGTCG^CACA | 123 |
| rp49 rev | 5′-GGCGGGTACGTTTGTTGG | ||
| period | per fw | 5′-ACGAAAACACTCTTAAG^CCCAA | 140 |
| per rev | 5′-TTTGCTGTTGTCGTTCTCCTG | ||
| timelessb | tim fw | 5′-TGTTGCTCTTGATACTGGATAGTG | 118 |
| tim rev | 5′-AGCAGCATGCCATAGAAGTG | ||
| cryptochrome | cry fw | 5′-TGGGTTAGTTCATCGGCATTTG | 157 |
| cry rev | 5′-GCCAGGGTTCATGG^ATAAACAG | ||
| vrille | vri fw | 5′-AATGAGGCCACAAATG^TTCAC | 156 |
| vri rev | 5′-GGCGCTGACCTGCTGTTT | ||
| cycle | cyc fw | 5′-CCTGTTCATAGATCAAAG^AGCC | 111 |
| cyc rev | 5′-TCAGCCAAGGCGGGTAT | ||
| Clock | clk fw | 5′-TTATATCACCCCTACCG^CATC | 100 |
| clk rev | 5′-TTACGCTGAAGCTGTTCCT |
Intron position is marked with a caret (^).
timeless primers were designed to anneal to different exons separated by an ∼2.5-kb intron.
Total RNA was isolated with Trizol (Sigma, St. Louis) according to the manufacturer's instructions. One microgram of total RNA was reverse transcribed using oligo(dT) (24mer) and SuperscriptII reverse transcriptase (Invitrogen, San Diego). To safeguard against amplification of possible contaminant genomic DNA, the primers were designed either to anneal only to a template corresponding to the spliced transcript or (in the case of timeless amplification) to include a large (2.5-kb) intron in the genomic template. In the latter case, PCR conditions were optimized, so that only the short (cDNA) product would amplify. This “cDNA specificity” was confirmed in pilot real-time PCRs with pure genomic DNA as a template (resulting in no product).
A total of 5 μl of diluted cDNA were used for a 20-μl PCR reaction (Hot start Taq polymerase (Takara) 0.32 unit, Taq buffer 1×, dNTPs 200 μm each, Syber green 1:25,000, primers 400 nm each). Amplifications were carried out on a Rotor-Gene 3000 (Corbet Research) for 40 cycles (94° for 20 sec; 60° for 30 sec, 72° for 30 sec) following an initial denaturation/Taq activation step (95° for 5 min). Clock genes and rp49 were amplified in separate tubes in duplicates or triplicates for each primer combination (Table 1) and each cDNA sample was amplified in parallel to negative controls. Product size was confirmed by melting analysis and 2.5% agarose gel electrophoresis. Data were analyzed and quantified with the Rotor-Gene6 analysis software. Relative values were standardized to rp49 and normalized to the sample with highest expression. Values represent the mean of three independent experiments plus or minus standard deviation. For in situ hybridization to Mdper, a 482-bp probe was generated that spanned the MdPER C-domain (Piccin et al. 2000).
Western blots:
Fly heads and thoraxes were collected separately in LD12:12 and DD and Western blots were run essentially as described previously (Edery et al. 1994; Peixoto et al. 1998). Primary antibodies used to assess PER and TIM levels were rabbit αDmPER-I (diluted 1:10,000, a gift from Jeff Hall, Brandeis University), rabbit αDmPER-II and rat αDmTIM (both 1:1000 and gifts from Michael Young, Rockefeller University), and rabbit αMdPER 774 (1:500, generated by Seth Racey, Leicester University). An α-rabbit HRP conjugate (Sigma; 1:6000) was employed as a secondary antibody and the reaction was developed by ECL. Nonspecific bands labeled by the primary antibodies were used to correct for loading inaccuracies. Autoradiographs were quantified with Scion Image software.
Immunohistochemistry and immunofluorescence:
Immunohistochemistry was performed on 6-μm vertical sections of paraffin-embedded Musca heads, initially using αDmPER-II (1:1000); experiments were then repeated using αDmPER-I (1:1000) and αMdPER 774 (1:500). An α-rabbit HRP conjugate (Sigma; 1:2000) was employed as a secondary antibody and the reaction was developed enzymatically with DAB. Samples were dehydrated and mounted with DPX (Fisher). Preparations were viewed with a Nikon Eclipse TE 200 inverted microscope and images were taken with a color CCD camera (JVC Ky F50). Immunofluorescence was performed using αDmPER-I (1:10,000), a rabbit α-crab pigment dispersing hormone (PDH) (1:2500, gift from Simon Webster, University of Bangor: the same antibody was also used at the same dilution for DAB staining), and αDmTIM (1:1000) in conjunction with α-rabbit Cy2 (1:400, Jackson ImmunoResearch Laboratories) and α-rat Cy3 (1:400, Jackson ImmunoResearch Laboratories) fluorescent secondary antibodies. Whole-mount confocal microscopy was performed on a Zeiss LSM 510 (objective ×63 oil NA 1.4, lasers Argon 488 and Hene 543, pin hole 1) and a Leica SP5 (objective ×40 oil NA1.25, lasers Argon 488 and DPSS 561, pin hole 1) system. The mounting medium was 3% (w/v) propylgallate in 20% PBS and 80% glycerol. Pictures were adjusted for contrast and brightness in Adobe Photoshop. The confocal results described are based on >30 brains analyzed in three independent experiments with the Zeiss system and an additional 20 brains analyzed with the Leica system.
Behavior:
Locomotor behavior of Musca individuals carrying the white eye mutation was recorded for 3-day-old adults by an infra-red detector that was attached to a petri dish 9 cm in diameter. Data were collected for at least 5 days and activity counts were collated in 30-min time bins. Analysis of the period of locomotor activity was carried out using chi-square periodogram analysis (Refinetti 2000). Pupal adult eclosion was monitored by placing individual pupae homozygous for white and apterous into Eppendorf tubes, and adult emergees were counted every 2 hr.
RESULTS
Rhythmic behavior:
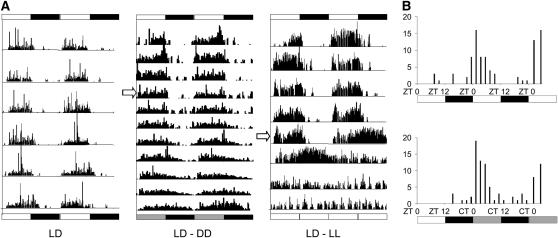
We examined locomotor activity rhythms in adult M. domestica in LD, DD, and LL at 25° after prior entrainment to LD12:12 for at least 7 days. Figure 1A illustrates examples of individual locomotor patterns in each of the three conditions. Under entrainment, Musca showed rhythmic behavior with almost all of the locomotor activity restricted to the day phase (Helfrich et al. 1985). Under free-running conditions, the flies showed an average period of 24.4 ± 0.1 hr (N = 27). In DD, however, locomotor activity began to impinge into the subjective night from the first day in free-running conditions. In LL, rhythmicity was soon lost (Figure 1A). Pupal/adult eclosion also showed a clear circadian rhythm in LD and DD with peaks of emergence at Zeitgeber time (ZT)2 and circadian time 2. Exhaustion of the observer prevented a prolonged observation period, but a second peak of emergence occurred ∼24 hr after the first in both LD and DD (Figure 1B).
Figure 1.—
Rhythmic phenotypes in M. domestica. (A) Locomotor activity of individual flies is double plotted so that day 1 and day 2 activity is placed on the first horizontal line, day 2 and day 3 on the second line, etc. (Left) LD12:12. (Middle) Four days in LD12:12 followed (open arrow) by 8 days in DD. (Right) Five days in LD12:12 after which (open arrow) the flies were maintained in LL for 4 days. (B) Circadian pupal–adult eclosion. Newly emerging flies were observed in dim red light after previous entrainment in LD12:12. Eclosions were followed every 2 hr.
mRNA cycles:
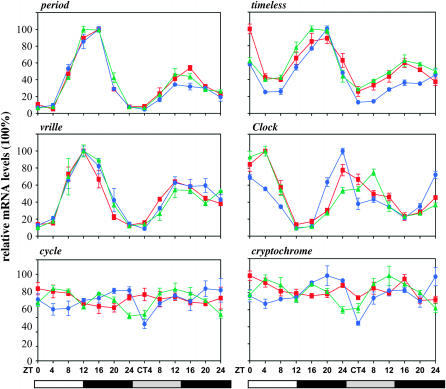
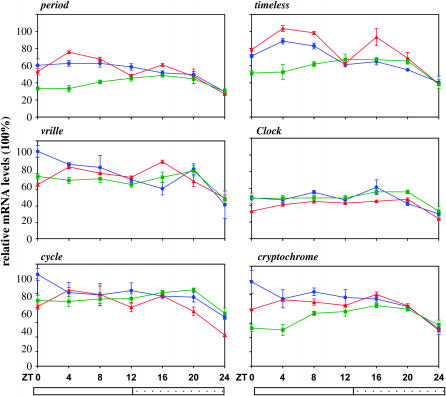
Quantitative real-time PCR was performed from total RNA extracts derived from male Musca heads collected every 4 hr both under LD and during the first day of DD in three independent experiments. Peaks of mRNA were observed at ZT12–16 for per, tim, and vri and this pattern was continued into the first cycle of DD. As in Drosophila, MdClk mRNA peaked with an opposite phase to Mdper, namely at the end of both the actual (LD) and the subjective (DD) night (Figure 2). The arrhythmic expression of Mdcyc was also consistent with that reported for the fruit fly, but the absence of robust cycling for Mdcry was surprising (Figure 2). As expected, exposure to constant light resulted in loss of cycling for all the clock genes (Figure 3), which is in agreement with the behavioral arrhythmia observed under the same conditions (Figure 1A).
Figure 2.—
Clock gene expression in Musca heads in LD and DD. Real-time PCR was performed for the six clock genes in LD followed by DD. Light regime appears below each column of figures. Each color represents a different biological replicate.
Figure 3.—
Clock gene expression in Musca heads in LL. Real-time PCR was performed for the six clock genes as in Figure 2. Each color represents a different biological replicate.
MdPER and MdTIM protein cycles:
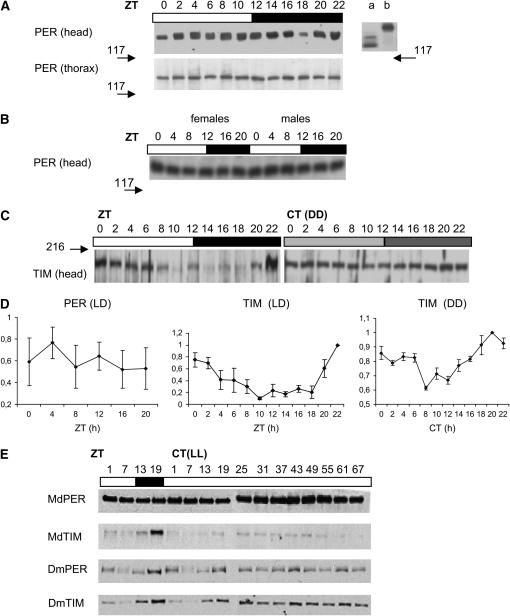
Figure 4, A and B, shows typical Western blots using αDmPER-I, but the results were essentially identical using all three anti-PER antibodies. The MdPER band has a size of ∼130–140 kDa, consistent with its predicted molecular weight based on primary sequence data (Piccin et al. 2000), and shows no evidence for any cycling in LD12:12. We repeated the blot several times, also with other PER antisera and separately for head and thorax (Figure 4A) or male and female (Figure 4B) extracts, but have never observed cycling (Figure 4D). In all blots examined, no obvious temporal changes in electrophoretic mobility that could be attributed to circadian modulation of phosphorylation were detected (Edery et al. 1994).
Figure 4.—
PER and TIM expression in Musca. (A) Separate Western blots in LD12:12 of heads and thoraxes, showing that MdPER does not cycle in either tissue although it is more highly expressed in the former than in the latter. The numbers 117 and 216 and their associated arrows represent kilodaltons and show the positions of the molecular-weight markers. (Right) A Western blot of the Drosophila MM1 transformant heads that carry the null mutation per01, plus the Mdper transgene under the control of Drosophila per 5′ sequences (Piccin et al. 2000). Lane a is the MM1 transformant and lane b represents wild-type Drosophila heads harvested at ZT8. (B) Western blot of male and female heads taken in 4-hr intervals in LD12:12. No evidence for cycling in the abundance of MdPER is observed, suggesting that there are no sex-specific differences in this trait. Molecular-weight markers in kilodaltons are arrowed. (C) Representative MdTIM head blots in LD12:12 and DD. The arrow represents a molecular-weight marker in kilodaltons. (D) Mean (±SEM) levels of MdPER (LD12:12) and MdTIM (LD12:12 and DD) in Musca heads. Loading differences were corrected using constitutive nonspecific cross-reacting bands as a comparison. Data were normalized to the highest level of clock protein on each Western blot. Several replicates were performed for each experiment. (E) Musca PER is stable in constant light. Western blots are shown for Musca and Drosophila PER and TIM in 67 hr of constant bright light after previous entrainment in LD12:12. Note the stability of MdPER compared to MdTIM and to dPER and dTIM, which show levels slightly above the minimum levels in LD (see text).
We also examined MdTIM cycling in Musca heads using a rat anti-TIM antibody (αDmTIM 1:1000) raised against Drosophila TIM. Although weak, we detected a band of approximately the correct size that appears to cycle with a peak at the end of the night phase in LD12:12 and, dampened, also in DD (Figure 4C). We confirmed these findings over three separate full replicates (Figure 4D). We next investigated expression of MdPER in LL and found it to be similarly stable over more than two cycles, with no evidence for light-induced degradation (Figure 4E). In contrast, both Drosophila PER and TIM cycle robustly in LD and this cycle dampens in LL, eventually stabilizing at just above minimal LD levels by the third day in LL (Figure 4E and see Marrus et al. 1996). MdTIM, however, immediately degrades in response to light, implying that MdPER does not rely on MdTIM for stability (Figure 4E).
Spatial localization of clock proteins:
The failure of MdPER to cycle in head extracts does not necessarily imply that MdPER does not cycle. It is possible that, in a small subset of neurons that might be acting as pacemaker cells, rhythms in MdPER expression may be present, but masked by noncycling MdPER in other cell types. We therefore performed both in situ hybridizations and antibody stainings on Musca heads.
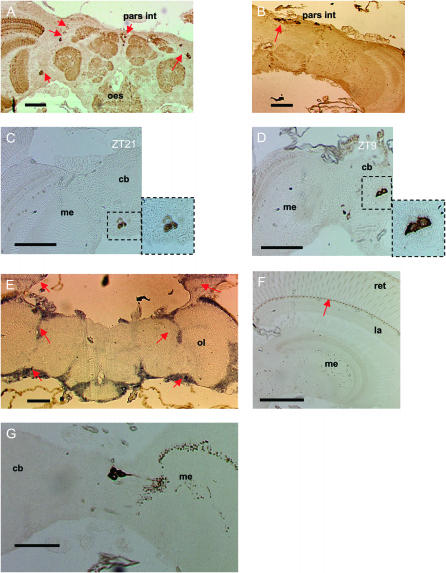
Initial immunohistochemistry (IHC) using αDmPER-II (1:1000) on paraffin sections revealed results similar to those also obtained with αDmPER-I (1:1000). Anti-rabbit HRP-conjugated secondary antibody and DAB were used for detection of the signal. Many groups of cells were observed to stain during day (ZT9) and night (ZT21) and several were localized in the area between the optic lobes and the central brain, both dorsally and ventrally (Figure 5A). Intense staining was observed in the pars intercerebralis (Figure 5, A and B). However, labeling of these cells in control experiments with nonimmune rabbit serum suggested that staining of the pars intercerebralis likely represented an artifact (Figure 5B). In all the immunopositive cells, the staining was exclusively cytoplasmic at both time points (ZT9 and ZT21) with characteristic “doughnut” patterns (Figure 5, C and D). To validate these results, we performed in situ hybridization on paraffin sections at ZT16 using a digoxigenin-labeled Mdper probe in an attempt to localize Mdper expression in the same area identified by IHC. In situ hybridization of Mdper closely resembles the pattern of per expression in D. melanogaster (Kloss et al. 1998). We detected staining in the photoreceptor cells and in a broad region between the optic lobes and the central brain (Figure 5E) in general agreement with the localization of the lateral PER immunoreactive cells (Figure 5A). However, we did not detect staining within photoreceptor cells by IHC, but a structure at the base of the photoreceptors was strongly labeled at all times (Figure 5F).
Figure 5.—
Clock protein expression in head sections of Musca. (A) A section at ZT21 labeled with αDmPER-I and represents a general staining pattern obtained with all available anti-PER antibodies. Several groups of cells are labeled (arrows), including the pars intercerebralis and a dorsal and ventral group of neurons that are lateral to the central brain. (B) Replacing the αDmPER-I antibody with rabbit normal serum results in staining of pars intercerebralis cells (arrow), suggesting that these are nonspecific for PER staining. (C and D) During both the night (ZT21) and the day (ZT9) anti-PER immunoreactivity was exclusively cytoplasmic with cells showing a characteristic “doughnut” shape. (E) In situ hybridization to Mdper. Arrows denote hybridization to regions where the dorsal and lateral PER-positive neurons are located and the photoreceptors. (F) Although no staining is observed in photoreceptor nuclei at night, a structure at the base of the photoreceptor can be seen to stain strongly at all times (arrow). (G) PDF-expressing cells and their projections. pars int, pars intercerebralis; oes, esophagus; me, medulla; la, lamina; ret, retina; ol, optic lobe. Bars, 100 μm.
We also used a rabbit anti-crab PDH antibody that in Drosophila recognizes PDF-expressing cells and identified some immunoreactive neurons between the central brain and the optic lobes (Figure 5G). As we could not perform MdPER-PDH double staining, we can neither confirm nor exclude the possibility that these PDH immunoreactive cells correspond to any of the lateral cells that stain for PER. No staining was achieved for MdTIM, precluding any further investigation using these methods.
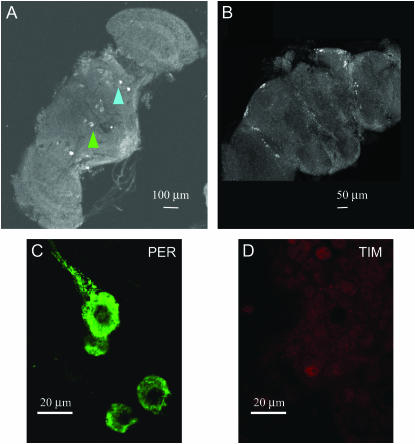
In the experiments described above, visualization of the signal is based upon an enzymatic reaction that is stopped before saturation is achieved. Hence the possibility exists that the intense cytoplasmic staining observed could mask a small amount of nuclear PER. Moreover, paraffin embedding obliged us to use a high concentration of the anti-PER antibodies, raising questions about the specificity of the signal. To investigate these issues, whole-mount brains were labeled using fluorescent secondary antibodies and examined with confocal microscopy. Using αDmPER-I (1:10,000), at low magnification we detected only two groups of neurons in Musca in medial and medio-lateral regions (Figure 6A, ZT24) compared to the six clusters that we found in Drosophila with the same reagent and method (Figure 6B, ZT24). These two groups, each consisting of two neurons, show strong PER cytoplasmic staining which, for one group, also extends into the axons (Figure 6C). These cells are strongly labeled at every time point (ZT8, ZT19, and ZT24), but they do not stain for TIM (Figure 6D, ZT24). Because of the lack of cell-specific markers, we cannot conclude definitively that these neurons are localized among the large number of PER immunoreactive cells previously visualized with DAB staining, although some of these also showed labeling of axonal projections.
Figure 6.—
Confocal microscopy of a whole-mount Musca brain. (A) PER within the Musca brain stained with αDm PER-I antibody at ZT24. Two medial neurons (green arrowhead) and two medio-lateral neurons (blue arrowhead) show intense staining at this magnification. (B) D. melanogaster shows six clusters of PER immunoreactive neurons (αDm PER-I) in lateral and dorsal regions of the brain at ZT24 (this is the same as Figure 2 in Rosato et al. 2006). (C and D) Musca medial and medio-lateral neurons show intense PER cytoplasmic immunoreactivity but do not show staining with αDmTIM antibodies at ZT24.
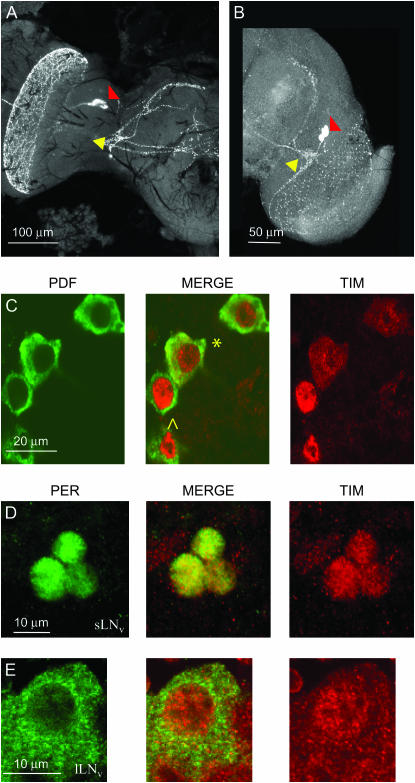
We then used the α-crab PDH antibody to identify the PDF-expressing cells (Figure 7A). In Musca, as in Drosophila (Figure 7B), they consist of two clusters of cells with large (∼20 μm in diameter in Musca) and small (∼10 μm in diameter in Musca) somata; thus we refer to them as large and small ventral lateral neurons (s-, l-LNvs) as in the fruit fly. In Drosophila, both s- and l-LNvs express PER and TIM in a coordinated fashion (Shafer et al. 2002). Because of their similar organization, we asked whether Musca LNvs also express clock proteins. As PDH and PER antisera are made from the same host (rabbit), we could use only anti-PDH and anti-DmTIM (rat) for colocalization studies. We observed that, in Musca, both types of lateral ventral neurons co-express MdTIM. However, although the small lateral neurons ventral (s-LNvs) show nuclear staining at ZT24, as in Drosophila, the l-LNvs show both cytoplasmic and nuclear subcellular distribution of MdTIM. It is also evident that the labeling is far more intense for the small than the large LNvs, which is at odds with the situation described for the fruit fly (Figure 7C; see also Figure 5 in Dissel et al. 2004).
Figure 7.—
Confocal images of PDF-expressing neurons within Drosophila and Musca brains. (A) Musca adult brain at ZT24. s-LNv and l-LNv ventral lateral neurons are marked with yellow and red arrowheads, respectively. (B) Drosophila adult brain at ZT24 with s-LNv (yellow) and l-LNv neurons (red) expressing PDF. (C) Two s-LNvs (^) and two l-LNvs (*) neurons in Musca co-expressing PDF and TIM at ZT24. TIM is predominantly nuclear at this time, although cytoplasmic staining is also observed in the l-LNvs. (D) s-LNv neurons in Musca co-expressing PER and TIM at ZT24. Both antigens are nuclear. (E) l-LNv neurons in Musca co-expressing PER and TIM at ZT24. TIM is found both in the nucleus and in the cytoplasm, whereas PER is only cytoplasmic.
We then investigated the colocalization of MdPER and MdTIM in the LNvs by knowing the approximate position of the LNvs in the Musca brain and by using the different cell sizes and the unequal subcellular distribution of MdTIM to distinguish between the two types of neurons. Indeed, MdPER and MdTIM colocalize to the LNvs. In the s-LNvs, both proteins are nuclear at ZT24, but in the l-LNvs, MdTIM is nuclear and cytoplasmic at the same time whereas MdPER, also expressed at very low levels, is present only in the cytoplasm (Figure 7, D and E), showing that the subcellular distribution of these two proteins is not coordinated between the two neuronal groups.
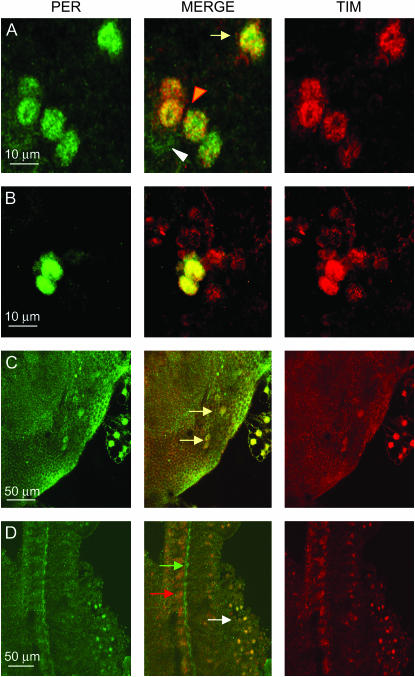
In Drosophila, the ventral lateral clusters also compose a single neuron that is very similar to the s-LNvs in size and timing of development, but is found in a position closer to the l-LNvs than to the s-LNvs (Helfrich-Forster 2003). It is also the only LNv that does not express PDF, and as such we refer to it as PDF-null LNv (Pn-LNv). In Musca, this single neuron seems to have expanded into a cluster of about four cells (Figure 8A). Unfortunately, the signal/background ratio is always quite low, making the identification and precise counting of every neuron very difficult. The Pn-LNvs are located more anteriorly than the lateral neurons dorsal (LNds; see below) and always show nuclear colocalization of MdPER and MdTIM at the end of the night.
Figure 8.—
Confocal images of PER- and TIM-expressing neurons in Musca brain at ZT24. (A) Four PDF null LNvs (Pn-LNvs, orange arrowhead) are interspersed among the l-LNvs (white arrowhead; note that only PER staining is visible). Two LNds are also visible (yellow arrow). As in the sLNvs, the staining intensity for LNds and Pn-LNvs is higher than for the l-LNvs. (B) Nuclear staining for PER and TIM in a six- to eight-cell cluster equivalent to the Drosophila LNds. Only two neurons are in the same focal plane. (C) Dorsal neurons expressing PER and TIM (arrows). The staining is nuclear for both proteins. (D) Photoreceptor staining in Musca. MdPER and MdTIM colocalize within the photoreceptor nuclei (white arrow). PER staining can once again be seen in a structure at the base of the photoreceptors (green arrow; see also Figure 5F); TIM does not stain in this structure although strong staining for TIM can be seen in cells lying underneath (red arrow).
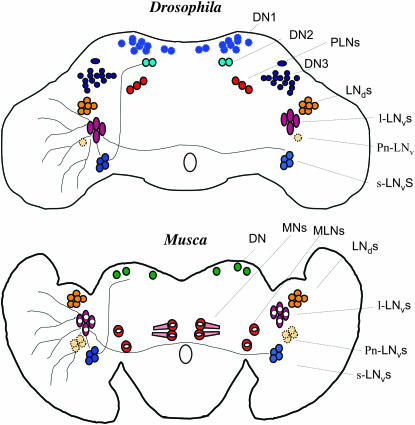
In Drosophila, the LNds are located dorsal and posterior to the l-LNvs and appear as a cluster of usually six neurons, similar in size to the s-LNvs (Helfrich-Forster 2003). These cells also appear to be present in Musca and, like the s-LNvs, both MdPER and MdTIM are fully nuclear at ZT24 (Figure 8B). In addition, we have identified in the dorsal brain of the housefly a few neurons expressing both MdPER and MdTIM in the nucleus, but we are not able to relate any of these cells to the dorsal clusters described for Drosophila (Figure 8C). Finally, we have also observed nuclear staining in photoreceptors at this time for both MdPER and MdTIM (Figure 8D). In another set of experiments, we analyzed the expression of MdPER and MdTIM at ZT24, -18, and -6. As before, we could identify PER/TIM-expressing neurons and staining in photoreceptor cells only at ZT24 but not at earlier time points, which confirms the cycling of both proteins in important central and peripheral clock cells of Musca (Figure 9). Figure 10 illustrates all the relevant neuronal clusters that we have identified in Musca and compares them to those described in Drosophila.
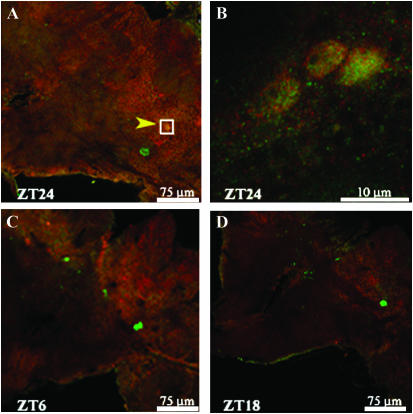
Figure 9.—
Confocal images of PER- and TIM-expressing neurons at different time points in Musca. (A) The PER-expressing medial lateral neurons are in the proximity of the PER- and TIM-expressing s-LNvs, here shown arrowed and boxed. (B) Larger magnification of the boxed region of A showing nuclear colocalization of PER and TIM in sLNvs at ZT24. (C and D) At ZT6 and -18, the same region shows the brightly stained medial lateral neurons but indicates the absence of PER and TIM staining in the sLNvs.
Figure 10.—
Neurons expressing PER in Musca and Drosophila brains. DN1, DN2, and DN3, dorsal neurons; PLNs, posterior lateral neurons; MNs, medial neurons; MLNs, medio-lateral neurons.
DISCUSSION
Drosophila has contributed an enormous wealth of experimental data and insight into the molecular dissection of the circadian clock. The negative feedback model was developed in the fly with PER (Siwicki et al. 1988; Hardin et al. 1990), and the fact that the murine clock is also built around the negative regulation of PER proteins further supports the generality of the higher eukaryote model (Shearman et al. 2000). One of the anchors for this model is that the per-encoded mRNAs and proteins cycle in pacemaker cells and that PER also enters the nucleus in a temporally regulated manner. Exactly what causes this temporal change in the relative abundance of PER between different cellular compartments is currently under debate (Nawathean and Rosbash 2004; Meyer et al. 2006), but the general observation that, in several insect orders, PER can be found to be exclusively cytoplasmic in its neuronal expression is inconsistent with its “dedicated” role as a circadian transcriptional regulator. One group of “neuronal” cells that do show nuclear expression of PER are the photoreceptors of both the giant silkmoth A. pernyii and the hawkmoth Manduca sexta where cycles in PER immunoreactivity have been documented in the former species, but not in the latter (Sauman and Reppert 1996; Wise et al. 2002). However, in the hawkmoth, nuclear staining of PER was consistently observed in four neurons within each hemisphere in the pars lateralis, a neurosecretory region, although no circadian cycling of PER abundance was noted (Wise et al. 2002). In contrast, in A. pernyii, a similar group of four cells exclusively express cytoplasmic PER, which cycles in concert with TIM (Sauman et al. 1996).
While it can be argued that in some of these studies perhaps the antigenicity did not reflect PER in these insects, or that PER does indeed enter the nucleus at low undetectable levels to engage the negative feedback loop, it is odd that PER antigenicity, whether cycling in the firebrat (Zavodska et al. 2003a) or not in the hawkmoth (Wise et al. 2002) or the beetle (Frisch et al. 1996), or “not known” in a variety of other insects (Zavodska et al. 2003b), does not usually appear to be nuclear. In fact, on the basis of our initial results with Musca, we might have been tempted to add the housefly to the list of species with “noncanonical” patterns of PER regulation. This would have been based on the failure of MdPER (1) to cycle in Musca heads in Western blots and its associated inability to “degrade” in response to constant light, (2) to show the extensive phosphorylation patterns that are observed in Drosophila, and (3) to translocate into the nucleus with its apparently exclusive cytoplasmic localization in several groups of neurons on the basis of enzymatic staining methods. Such a conclusion would have been surprising, given that MdPER strongly rescues circadian locomotor rhythms in Drosophila per01 transformants (Piccin et al. 2000) and that Mdper, Mdtim, Mdvri, and MdClk mRNAs cycle quite robustly, with phases similar to those in the fruit fly in LD and DD, and become arrhythmic in LL, further revealing their clock-like nature. Of interest was that, in contrast to Drosophila, Musca cry unexpectedly did not cycle robustly. The cry gene plays a pivotal switch role in the evolution of the clock because in the central clock mechanism it can act predominantly either as a photoreceptor, as in Drosophila, or as a negative regulator, as in the mouse (Shearman et al. 2000). However, cry in the fly can also act as a negative regulator in peripheral neuronal tissue, indicating that cell-specific trans-acting factors can modulate the light-sensitive function of this molecule (Collins et al. 2006). The two types of CRYs can be clustered phylogenetically, and the Musca cry identified by our screen is a Drosophila-like molecule. The distribution of the two types of cry's do not follow simple phylogenetic rules in that mouse-like cry is found exclusively in some insects such as bees and Tribolium, whereas in others, such as the dipteran Anopheles gambiae or in the monarch butterfly Danaus plexippus, both types of cry have been identified (Zhu et al. 2005; Rubin et al. 2006; Yuan et al. 2007). At present, we cannot state conclusively whether a mouse-like cry is present in the Musca genome. Nevertheless, it is intriguing that Musca cry does not appear to cycle at the mRNA level, at least at the level of whole-head preparations, but it is too early to say whether this may imply a different function compared to the Drosophila cry.
As mentioned above, the absence of MdPER cycling in Western blots further contradicts the Drosophila model. Although the formal possibility exists that the band that we are detecting represents a cross-reacting protein, we are confident that we are reliably observing MdPER in Western blots for the following reasons. We identified the same band with several antibodies and separately in heads and thoraxes of both males and females. In per01 D. melanogaster transformants carrying the Mdper transgene (Piccin et al. 2000), the same antibodies identify a band of similar size (Figure 4A). αDmPER-II is able to recognized the cycling PER of the blowfly L. cuprina (Warman et al. 2000), further confirming that the polyclonal antibodies that we used recognize regions of PER that are highly conserved in Diptera.
That our initial conclusion (which we held for several years, namely that Musca represented a noncanonical type of circadian clock) was premature and incorrect came to light once we applied immunofluorescence with confocal microscopy. We observed medial and medio-lateral neurons that expressed high levels of cytoplasmic MdPER but not MdTIM, in night and day, as we had with the enzymatic method. On closer scrutiny, less dramatic PER and TIM colabeling of groups of neurons became apparent, even though the staining was much fainter. This might have been due to a general low level of expression of MdPER and MdTIM in these neurons, to a low affinity for Musca proteins of our anti-Drosophila antibodies, to an increased difficulty for the antibodies to penetrate a much bigger brain, or, most likely, to a combination of all these reasons. Whatever the cause, the immunostaining was very faint with both antisera but especially with anti-TIM, making the identification of Musca “clock neurons” technically very challenging. Nevertheless, we identified many cells showing immunoreactivity for either anti-PER or anti-TIM or both. However, in this study we have considered as putative clock neurons only those cells where colocalization of the two immunosignals was clearly evident, as we do not have any other reliable criterion to judge the specificity of the labeling produced by each of the antibodies. We could not detect any reliable TIM and PER double staining at times earlier than ZT24 both in central clock neurons and in photoreceptor cells. Therefore, we suggest that in clock cells there is a cycle in the abundance of the two proteins that might be mirrored by a cycle in their subcellular localization.
Thus, in contrast to our initial conclusions, neuronal clusters in Musca largely correspond to those in Drosophila, suggesting that we have identified homologous structures (see Figures 7–9). It is interesting to note that in Musca at ZT24 the l-LNvs are the only group of putative clock cells to show cytoplasmic PER and nuclear-cytoplasmic TIM and that both types of reagents give much weaker signals. This might suggest that this neuronal cluster might have a special function in the circadian network, as suggested for Drosophila (Collins et al. 2005). Furthermore, in Drosophila the l-LNvs are the only group of neurons where, under particular environmental conditions, for example, in constant conditions, PER and TIM nuclear accumulation can be decoupled (Yang and Sehgal 2001; Shafer et al. 2002; Rieger et al. 2006). These cells possibly represent a strategic point in the neuronal network of Diptera where a physiological response to a combination of environmental variables might be amplified for entrainment (Collins et al. 2005). As for the more nuclear subcellular distribution of MdTIM in the l-LNvs of Musca compared to MdPER, this could reflect a more prominent role for MdTIM as a negative regulator in the housefly, as has been suggested for A. pernyii (Chang et al. 2003).
As in Drosophila, PDF localizes with the two groups of lateral neurons in Musca, providing a helpful additional marker for these putative clock neurons. In both Drosophila and Musca, these cells are located within the accessory medulla (also termed the anterior base of the medulla; Figure 7, A and B), and their patterns of projections from the LNvs are also similar, as has been previously reported (Helfrich-Forster 1995; Pyza and Meinertzhagen 1997; Miskiewicz et al. 2004). Our results with anti-PER and TIM reagents therefore support the long-standing suspicions of Pyza and Meinertzhagen (1997) that the PDF-expressing neurons in Musca are in fact “clock” cells (Pyza and Meinertzhagen 1997; Miskiewicz et al. 2004).
In other insect orders, PDF does not colocalize with PER or TIM antigens (Zavodska et al. 2003b), although some colocalization of PER and PDF may be present in the beetle Pachymorpha sexguttata (Frisch et al. 1996). It therefore may be that the Diptera compared with these other insects have a fundamental difference in this aspect of clock neuronal biology. Even so, in Antheraea, for example, although the PDH- and PER/TIM-expressing neurons are not the same, in terms of their relative anatomical positions, they could be functionally related (Sauman and Reppert 1996). We did, however, find a number of PER- and TIM-expressing neurons that were localized in the region of the Musca l-LNvs that were PDF null. In Drosophila, one such neuron has been identified (Helfrich-Forster 1995; Kaneko et al. 1997), and its role in the circadian mechanism is being clarified (Rieger et al. 2006).
One outstanding issue involves why the MdPER protein does not cycle in whole-head Western blots? In the muscid L. cuprina (sheep blow fly), cycles in both gene products are found (Warman et al. 2000), yet in the medfly, Ceratitis capitata, a similar situation to Musca is observed, with cycles in per mRNA expression, but no PER cycling in Westerns (Mazzotta et al. 2005). We suspect that the Musca PER protein that is highly and apparently constitutively expressed in the medio- and medio-lateral neurons, if indeed it is PER, plays a role different from that found in the lateral neurons. Microarray studies in Drosophila have revealed that per does more than simply control cycling output transcripts and that a large number of mRNAs that do not cycle are either up- or downregulated in per-null mutants (Claridge-Chang et al. 2001; Y. Lin et al. 2002). This, in turn, would suggest that there are downstream functions for PER that do not require cycling per products. Consequently, the apparently stable PER in these neurons may play a role different from the familiar negative regulator theme, particularly given the apparent lack of TIM in these cells. Given PER's intimate association with the DBT kinase, which earmarks it for degradation (Kloss et al. 1998; Price et al. 1998), it would be interesting to see whether DBT, or indeed other kinases such as casein kinase 2, which have been implicated with PER stability (J. M. Lin et al. 2002; Akten et al. 2003), are also localized in these nonrhythmic MdPER-expressing neurons.
Future work will be aimed at elucidating the roles of the various clock molecules in Musca. Obviously, transgenic Musca, in which misexpression of MdPER, targeted or not, would be very helpful for functional studies (Hediger et al. 2001), and the development of such techniques is underway in our laboratories. Musca therefore will prove to be a useful model for studying the evolution of the circadian system as it is phylogenetically far enough away from Drosophila to be interesting, yet close enough to have at least the possibility of being studied by using some of the techniques that are available in the fruit fly. One final thought relates to the findings in other insect orders, namely that PER is observed to be cytoplasmic only in brain neurons (Zavodska et al. 2003b). All of these studies have used enzymatic IHC reactions except one, which also used confocal microscopy, but only for analysis of PDF-expressing neurons. Had we concluded our analyses without confocal microscopy, we would have come to conclusions similar to those of other laboratories, even to the point of suggesting that PDF-expressing cells did not colocalize with PER (compare Figure 5D with 5G). However, we observe that in fact Musca appears to have anatomical substrates for the PER/TIM and PDF molecules similar to those of Drosophila, albeit with some intriguing differences in the way in which MdPER behaves in Western blots and in the numbers of neurons that express these clock proteins. Perhaps a more detailed reanalysis of spatial and temporal clock gene product expression in other insect brains might be timely.
Acknowledgments
We thank the Biotechnology and Biological Sciences Research Council for a David Philips Fellowship to E.R. and a committee Ph.D. studentship to V.C. We also thank the European Community for grants to C.P.K. and R.C. and the Wellcome Trust for a grant to C.P.K. and E.R. and for a program grant to E.L. C.P.K. acknowledges the Royal Society for a Wolfson Research Merit Fellowship. This work was supported in part by the Grant Agency of the Czech Republic, grant 204/04/0862 (I.S.), and the Ministry of Education of the Czech Republic, project 2B06129 (I.S.)
References
- Akten, B., E. Jauch, G. K. Genova, E. Y. Kim, I. Edery et al., 2003. A role for CK2 in the Drosophila circadian oscillator. Nat. Neurosci. 6: 251–257. [DOI] [PubMed] [Google Scholar]
- Ashmore, L. J., S. Sathyanarayanan, D. W. Silvestre, M. M. Emerson, P. Schotland et al., 2003. Novel insights into the regulation of the timeless protein. J. Neurosci. 23: 7810–7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito, J., H. Zheng and P. E. Hardin, 2007. PDP1epsilon functions downstream of the circadian oscillator to mediate behavioral rhythms. J. Neurosci. 27: 2539–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, D. C., H. G. McWatters, J. A. Williams, A. L. Gotter, J. D. Levine et al., 2003. Constructing a feedback loop with circadian clock molecules from the silkmoth, Antheraea pernyi. J. Biol. Chem. 278: 38149–38158. [DOI] [PubMed] [Google Scholar]
- Claridge-Chang, A., H. Wijnen, F. Naef, C. Boothroyd, N. Rajewsky et al., 2001. Circadian regulation of gene expression systems in the Drosophila head. Neuron 32: 657–671. [DOI] [PubMed] [Google Scholar]
- Collins, B. H., and J. Blau, 2007. Even a stopped clock tells the right time twice a day: circadian timekeeping in Drosophila. Pflugers Arch. Eur. J. Physiol. 454: 857–867. [DOI] [PubMed] [Google Scholar]
- Collins, B. H., S. Dissel, E. Gaten, E. Rosato and C. P. Kyriacou, 2005. Disruption of Cryptochrome partially restores circadian rhythmicity to the arrhythmic period mutant of Drosophila. Proc. Natl. Acad. Sci. USA 102: 19021–19026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, B. H., E. O. Mazzoni, R. Stanewsky and J. Blau, 2006. Drosophila CRYPTOCHROME is a circadian transcriptional repressor. Curr. Biol. 16: 441–449. [DOI] [PubMed] [Google Scholar]
- Cyran, S. A., A. M. Buchsbaum, K. L. Reddy, M. C. Lin, N. R. Glossop et al., 2003. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112: 329–341. [DOI] [PubMed] [Google Scholar]
- Dissel, S., V. Codd, R. Fedic, K. J. Garner, R. Costa et al., 2004. A constitutively active cryptochrome in Drosophila melanogaster. Nat. Neurosci. 7: 834–840. [DOI] [PubMed] [Google Scholar]
- Edery, I., L. J. Zwiebel, M. E. Dembinska and M. Rosbash, 1994. Temporal phosphorylation of the Drosophila period protein. Proc. Natl. Acad. Sci. USA 91: 2260–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Y., S. Sathyanarayanan and A. Sehgal, 2007. Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1). Genes Dev. 15: 1506–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch, B., G. Fleissner, C. Brandes and J. C. Hall, 1996. Staining in the brain of Pachymorpha sexguttata mediated by an antibody against a Drosophila clock-gene product: labeling of cells with possible importance for the beetle's circadian rhythms. Cell Tissue Res. 286: 411–429. [DOI] [PubMed] [Google Scholar]
- Hall, J. C., 2003. Genetics and molecular biology of rhythms in Drosophila and other insects. Adv. Genet. 48: 1–280. [DOI] [PubMed] [Google Scholar]
- Hardin, P. E., J. C. Hall and M. Rosbash, 1990. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343: 536–540. [DOI] [PubMed] [Google Scholar]
- Hediger, M., M. Niessen, E. A. Wimmer, A. Dubendorfer and D. Bopp, 2001. Genetic transformation of the housefly Musca domestica with the lepidopteran derived transposon piggyBac. Insect Mol. Biol. 10: 113–119. [DOI] [PubMed] [Google Scholar]
- Helfrich, C., B. Cymborowski and W. Engelmann, 1985. Circadian activity rhythm of the house fly continues after optic tract severance and lobectomy. Chronobiol. Int. 2: 19–32. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster, C., 1995. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 92: 612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster, C., 2003. The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microsc. Res. Tech. 62: 94–102. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster, C., and U. Homberg, 1993. Pigment-dispersing hormone-immunoreactive neurons in the nervous system of wild-type Drosophila melanogaster and of several mutants with altered circadian rhythmicity. J. Comp. Neurol. 337: 177–190. [DOI] [PubMed] [Google Scholar]
- Hennig, W., 1981. Insect Phylogeny. Piman Press, Bath, England.
- Kaneko, M., C. Helfrich-Forster and J. C. Hall, 1997. Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: newly identified pacemaker candidates and novel features of clock gene product cycling. J. Neurosci. 17: 6745–6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss, B., J. L. Price, L. Saez, J. Blau, A. Rothenfluh et al., 1998. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell 94: 97–107. [DOI] [PubMed] [Google Scholar]
- Levine, J. D., I. Sauman, M. Imbalzano, S. M. Reppert and F. R. Jackson, 1995. Period protein from the giant silkmoth Antheraea pernyii functions as a circadian clock element in Drosophila melanogaster. Neuron 15: 147–157. [DOI] [PubMed] [Google Scholar]
- Lin, J. M., V. L. Kilman, K. Keegan, B. Paddock, M. Emery-Le et al., 2002. A role for casein kinase 2α in the Drosophila circadian clock. Nature 420: 816–820. [DOI] [PubMed] [Google Scholar]
- Lin, Y., M. Han, B. Shimada, L. Wang, T. M. Gibler et al., 2002. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 99: 9562–9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrus, S. B., H. Zeng and M. Rosbash, 1996. Effect of constant light and circadian entrainment of perS flies: evidence for light-mediated delay of the negative feedback loop in Drosophila. EMBO J. 15: 6877–6886. [PMC free article] [PubMed] [Google Scholar]
- Mazzotta, G. M., F. Sandrelli, M. A. Zordan, M. Mason, C. Benna et al., 2005. The clock gene period in the medfly Ceratitis capitata. Genet. Res. 86: 13–30. [DOI] [PubMed] [Google Scholar]
- Meyer, P., L. Saez and M. W. Young, 2006. PER-TIM interactions in living Drosophila cells: an interval timer for the circadian clock. Science 311: 226–229. [DOI] [PubMed] [Google Scholar]
- Miskiewicz, K., E. Pyza and F. W. Schurmann, 2004. Ultrastructural characteristics of circadian pacemaker neurones, immunoreactive to an antibody against a pigment-dispersing hormone in the fly's brain. Neurosci. Lett. 363: 73–77. [DOI] [PubMed] [Google Scholar]
- Naidoo, N., W. Song, M. Hunter-Ensor and A. Sehgal, 1999. A role for the proteasome in the light response of the timeless clock protein. Science 285: 1737–1741. [DOI] [PubMed] [Google Scholar]
- Nassel, D. R., S. Shiga, C. J. Mohrherr and K. R. Rao, 1993. Pigment-dispersing hormone-like peptide in the nervous system of the flies Phormia and Drosophila: immunocytochemistry and partial characterization. J. Comp. Neurol. 331: 183–198. [DOI] [PubMed] [Google Scholar]
- Nawathean, P., and M. Rosbash, 2004. The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol. Cell 13: 213–223. [DOI] [PubMed] [Google Scholar]
- Peixoto, A. A., J. M. Hennessy, I. Townson, G. Hasan, M. Rosbash et al., 1998. Molecular coevolution within a Drosophila clock gene. Proc. Natl. Acad. Sci. USA 95: 4475–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccin, A., M. Couchman, J. D. Clayton, D. Chalmers, R. Costa et al., 2000. The clock gene period of the housefly, Musca domestica, rescues behavioral rhythmicity in Drosophila melanogaster. Evidence for intermolecular coevolution? Genetics 154: 747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, J. L., J. Blau, A. Rothenfluh, M. Abodeely, B. Kloss et al., 1998. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94: 83–95. [DOI] [PubMed] [Google Scholar]
- Pyza, E., and I. A. Meinertzhagen, 1997. Neurites of period-expressing PDH cells in the fly's optic lobe exhibit circadian oscillations in morphology. Eur. J. Neurosci. 9: 1784–1788. [DOI] [PubMed] [Google Scholar]
- Refinetti, R., 2000. Circadian Physiology, CRC Press, Boca Raton, FL.
- Reppert, S. M., T. Tsai, A. L. Roca and I. Sauman, 1994. Cloning of a structural and functional homolog of the circadian clock gene period from the giant silkmoth Antheraea pernyi. Neuron 13: 1167–1176. [DOI] [PubMed] [Google Scholar]
- Rieger, D., O. T. Shafer, K. Tomioka and C. Helfrich-Forster, 2006. Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J. Neurosci. 26: 2531–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato, E., E. Tauber and C. P. Kyriacou, 2006. Molecular genetics of the fruit-fly circadian clock. Eur. J. Hum. Genet. 14: 729–738. [DOI] [PubMed] [Google Scholar]
- Rubin, E. B., Y. Shemesh, M. Cohen, S. Elgavish, H. M. Robertson et al., 2006. Molecular and phylogenetic analyses reveal mammalian-like clockwork in the honey bee (Apis mellifera) and shed new light on the molecular evolution of the circadian clock. Genome Res. 16: 1352–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrelli, F., S. Cappellozza, C. Benna, A. Saviane, A. Mastella et al., 2007. Phenotypic effects induced by knock-down of the period clock gene in Bombyx mori. Genet. Res. 89: 73–84. [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan, S., X. Zheng, R. Xiao and A. Sehgal, 2004. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell 116: 603–615. [DOI] [PubMed] [Google Scholar]
- Sauman, I., and S. M. Reppert, 1996. Circadian clock neurons in the silkmoth Antheraea pernyi: novel mechanisms of Period protein regulation. Neuron 17: 889–900. [DOI] [PubMed] [Google Scholar]
- Sauman, I., T. Tsai, A. L. Roca and S. M. Reppert, 1996. Period protein is necessary for circadian control of egg hatching behavior in the silkmoth Antheraea pernyi. Neuron 17: 901–909. [DOI] [PubMed] [Google Scholar]
- Shafer, O. T., M. Rosbash and J. W. Truman, 2002. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J. Neurosci. 22: 5946–5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman, L. P., S. Sriram, D. R. Weaver, E. S. Maywood, I. Chaves et al., 2000. Interacting molecular loops in the mammalian circadian clock. Science 288: 1013–1019. [DOI] [PubMed] [Google Scholar]
- Siwicki, K. K., C. Eastman, G. Petersen, M. Rosbash and J. C. Hall, 1988. Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron 1: 141–150. [DOI] [PubMed] [Google Scholar]
- Warman, G. R., R. D. Newcomb, R. D. Lewis and C. W. Evans, 2000. Analysis of the circadian clock gene period in the sheep blow fly Lucilia cuprina. Genet. Res. 75: 257–267. [DOI] [PubMed] [Google Scholar]
- Wise, S., N. T. Davis, E. Tyndale, J. Noveral, M. G. Folwell et al., 2002. Neuroanatomical studies of period gene expression in the hawkmoth, Manduca sexta. J. Comp. Neurol. 447: 366–380. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and A. Sehgal, 2001. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron 29: 453–467. [DOI] [PubMed] [Google Scholar]
- Yuan, Q., D. Metterville, A. D. Briscoe and S. M. Reppert, 2007. Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol. Biol. Evol. 24: 948–955. [DOI] [PubMed] [Google Scholar]
- Zavodska, R., I. Sauman and F. Sehnal, 2003. a The cycling and distribution of PER-like antigen in relation to neurons recognized by the antisera to PTTH and EH in Thermobia domestica. Insect Biochem. Mol. Biol. 33: 1227–1238. [DOI] [PubMed] [Google Scholar]
- Zavodska, R., I. Sauman and F. Sehnal, 2003. b Distribution of PER protein, pigment-dispersing hormone, prothoracicotropic hormone, and eclosion hormone in the cephalic nervous system of insects. J. Biol. Rhythms 18: 106–122. [DOI] [PubMed] [Google Scholar]
- Zeng, H., Z. Qian, M. P. Myers and M. Rosbash, 1996. A light-entrainment mechanism for the Drosophila circadian clock. Nature 380: 129–135. [DOI] [PubMed] [Google Scholar]
- Zhu, H, Q. Yuan, A. D. Briscoe, O. Froy, A. Casselman et al., 2005. The two CRYs of the butterfly. Curr. Biol. 15: R953–R954. [DOI] [PubMed] [Google Scholar]