Abstract
During Drosophila development Suppressor of Hairless [Su(H)]-dependent Notch activation upregulates transcription of the Enhancer of split-Complex [E(spl)-C] genes. Drosophila melanogaster E(spl) genes share common transcription regulators including binding sites for Su(H), proneural, and E(spl) basic-helix-loop-helix (bHLH) proteins. However, the expression patterns of E(spl) genes during development suggest that additional factors are involved. To better understand regulators responsible for these expression patterns, recently available sequence and annotation data for multiple Drosophila genomes were used to compare the E(spl) upstream regulatory regions from more than nine Drosophila species. The mγ and mβ regulatory regions are the most conserved of the bHLH genes. Fine analysis of Su(H) sites showed that high-affinity Su(H) paired sites and the Su(H) paired site plus proneural site (SPS + A) architecture are completely conserved in a subset of Drosophila E(spl) genes. The SPS + A module is also present in the upstream regulatory regions of the more ancient mosquito and honeybee E(spl) bHLH genes. Additional transcription factor binding sites were identified upstream of the E(spl) genes and compared between species of Drosophila. Conserved sites provide new understandings about E(spl) regulation during development. Conserved novel sequences found upstream of multiple E(spl) genes may play a role in the expression of these genes.
REGULATION of gene expression is central to the process of cell differentiation during development. Multiple levels of regulation allow tight control of gene activation that results in specific patterns of expression and cell-type specification. At one level, gene expression patterns are controlled by proteins that bind to specific DNA sequences in the regulatory regions of genes and either activate or repress transcription. It is becoming increasingly clear that transcriptional regulation often depends on the interaction between multiple proteins and that the cellular context determines which players will be available to bind and affect transcription (Levine and Tjian 2003). Transcription factor binding sites clustered together in specific orientations are termed modules. These modules coordinate protein–protein interactions that result in activation or repression of gene expression (Halfon et al. 2002).
The Drosophila Enhancer of split [E(spl)] genes present an interesting study of complex modular transcriptional regulation. The E(spl) locus contains 13 different genes: HLHm3, HLHm5, HLHm7, HLHm8, HLHmβ, HLHmγ, and HLHmδ encode a set of basic-helix-loop-helix (bHLH) transcriptional repressors; mα, m2, m4, and m6 code for proteins that are members of the Bearded family; m1 codes for a putative protease inhibitor; and groucho codes for a transcriptional corepressor (Delidakis and Artavanis-Tsakonas 1992; Knust et al. 1992; Paroush et al. 1994; Lai et al. 2000). Importantly, many of the E(spl) genes in Drosophila melanogaster share the same transcription factor binding sites in their promoter regions yet show different patterns of expression during development, suggesting that additional differential coregulators are involved (De Celis et al. 1996; Nellesen et al. 1999; Wech et al. 1999). A subset of the E(spl) genes is regulated by a complex of regulatory proteins bound to a specific module containing binding sites for Suppressor of hairless [Su(H)] and proneural proteins (Nellesen et al. 1999; Cave et al. 2005).
Expression of the majority of the E(spl) genes is activated by Notch signaling (Jennings et al. 1994; Bailey and Posakony 1995; Eastman et al. 1997; Lai et al. 2000). The Notch pathway is conserved from worms to humans and regulates cell fate decisions in a wide variety of tissues (reviewed in Artavanis-Tsakonas et al. 1999; Baron 2003). The significance of the Notch pathway has been underscored by the discovery that altered forms of pathway members, including the E(spl) genes, cause a variety of human diseases including Alzheimer's, CADASIL, Alagille's syndrome, neoplasia, and neuroblastoma (reviewed in Joutel and Tournier-Lasserve 1998; Axelson 2004a,b). Notch codes for a transmembrane protein, which acts as the receptor in the signaling pathway. Notch and its ligands undergo post-translational events that are essential for proper signaling (reviewed in Chan and Jan 1998; Haines and Irvine 2003). The outcome of these events is the release of the intracellular domain of Notch (known variously as Nact, ICN, or NICD), which acts as a transcriptional activator (Struhl and Adachi 1998) and activates gene expression in combination with Su(H). In the absence of the Notch intracellular domain, Su(H) interacts with corepressors such as Hairless, dCtBP, Groucho, and SMRTR, to inhibit transcription (Morel et al. 2001; Barolo et al. 2002; Tsuda et al. 2002; Nagel et al. 2005). When Notch is activated, it binds to Su(H) and transforms it from a negative regulator to a positive regulator (Furriols and Bray 2001; Morel et al. 2001). Although NICD coupled with Su(H) induces the expression of many of the members of the E(spl)-C (Jennings et al. 1994; Bailey and Posakony 1995; Eastman et al. 1997; Lai et al. 2000), they are most likely not the only regulators of these genes. The distinct expression patterns of the different E(spl) genes, combined with the finding that activated Notch and Suppressor of Hairless are capable of eliciting only limited transcription of the genes (Cooper et al. 2000), indicate a requirement for additional transcriptional regulators that direct expression of the E(spl) genes (De Celis et al. 1996; Wech et al. 1999).
Cell-specific activation of some of the E(spl) genes requires interaction between NICD and proneural proteins (Cooper et al. 2000). Synergistic transcriptional activation by these proteins requires a specific organization of the upstream regulatory regions of the target genes (Cave et al. 2005). Since a specifically oriented pair of Su(H) binding sites [Su(H) paired site, SPS] and a bHLH activator binding site (A) are necessary for Notch–proneural synergy it has been proposed that the orientation and/or conformation of Su(H) bound to these sites allows direct interaction with Daughterless, a bHLH activator protein that interacts with Achaete–Scute proneural proteins (Cave et al. 2005). Only a subset of E(spl) promoters contains the SPS + A architecture, suggesting that other combinatorial factors may bind to different modules in the promoters of the E(spl) genes and direct their distinctive expression patterns during development (Nellesen et al. 1999; Cave et al. 2005).
To better understand the complex regulation of the E(spl) genes we analyzed the upstream regulatory regions of these genes using bioinformatic and phylogenetic footprinting approaches. The principle of phylogenetic footprinting is based on the fact that functional sequences tend to evolve at a slower rate than nonfunctional sequences. Thus sequences that are conserved in multiple and distantly related species are more likely to be functional than nonfunctional (Bergman et al. 2002). Recently, the fully sequenced, assembled, and annotated genomes of more than nine different Drosophila species have been released (Crosby et al. 2007). We have used multiple DNA alignment and scanning tools to compare and analyze the E(spl) regulatory regions in these different species. We have identified multispecies conserved sequences (MCSs) that relate to known functional binding sites including Su(H), proneural, and bHLH repressor sites. We have also discovered additional transcription factor binding sites and novel shared sequences that are conserved in nine different Drosophila species. These results provide insights into the evolution of the E(spl) locus and information about additional factors that regulate these genes.
MATERIALS AND METHODS
Reference DNA sequences:
D. melanogaster reference DNA for the E(spl) genes was obtained from NCBI entrez, http://www.ncbi.nlm.nih.gov/gquery/gquery.fcgi, or from the University of California Santa Cruz (UCSC) Genome Browser (Kent 2002), http://genome.ucsc.edu/cgi-bin/hgGateway (Assembly April 2004). The following promoter fragments were used as reference D. melanogaster sequences: m1, 1500 bp (Wurmbach et al. 1999; UCSC Genome Browser); m2, 1500 bp (Wurmbach et al. 1999; UCSC Genome Browser); HLHm3, 1700 bp (Nellesen et al. 1999); m4, 1384 bp (Singson et al. 1994); HLHm5, 1500 bp (Nellesen et al. 1999; UCSC Genome Browser), HLHm6 (Wurmbach et al. 1999; UCSC Genome Browser); HLHm7, 1700 bp (Singson et al. 1994; UCSC Genome Browser); HLHm8, 1500 bp (Klambt et al. 1989); mα, 1341 bp (Nellesen et al. 1999); HLHmβ, 1025 bp (Nellesen et al. 1999); HLHmγ, 1200 bp (Delidakis and Artavanis-Tsakonas 1992); and HLHmδ, 2000 bp (Nellesen et al. 1999; UCSC Genome Browser). Reference sequences from all other Drosophila species were accessed from FlyBase (Grumbling and Strelets 2006) using FlyBase BLAST (http://flybase.bio.indiana.edu/blas) and GBrowse (http://flybase.bio.indiana.edu/cgi-bin/gbrowse/dmel/).
Reference sequences for Apis mellifora and Anopheles gambiae E(spl) promoters were obtained from the Ensembl browser (Hubbard et al. 2005) (http://www.ensembl.org) and the UCSC Genome Browser (A. mellifera, Assembly January 2005 and A. gambiae, Assembly February 2003).
BLAT and EvoPrinter analysis:
D. melanogaster reference sequences for each of the E(spl) promoters were pasted into the BLAT search engine window (http://genome.ucsc.edu/cgi-bin/hgBlat) and individually compared to eight different test species: D. simulans (Assembly April 2005), D. yakuba (Assembly November 2005), D. erecta (Assembly August 2005), D. ananassae (Assembly August 2005), D. pseudoobscura (Assembly November 2004), D. virilis (Assembly August 2005), D. mojavensis (Assembly August 2005), and D. grimshawi (Assembly August 2005). Percentages of identities between species were determined by calculating the number of conserved sites from the highest-scoring BLAT alignment outputs. For multispecies alignments, the highest-scoring BLAT readout alignment for each test species was selected and pasted into an EvoPrinter (Odenwald et al. 2005) input window. (http://evoprinter.ninds.nih.gov). EvoPrinter outputs (EvoPrints) were generated using subsets of the BLAT inputs as well as BLAT readouts from all of the test species. On the basis of the Drosophila phylogenetic tree, species were sequentially added into EvoPrinter in the following order: D. simulans, D. yakuba, D. erecta, D. ananassae, D. pseudoobscura, D. virilis, D. mojavensis, D. grimshawi. We also used EvoDifference to identify sequences that are conserved in all but one of the above species.
Transcription factor binding site identification:
Su(H) binding sites, bHLH repressor sites, bHLH activator/proneural sites, and TATA boxes were identified in the Drosophila EvoPrint readouts for each additive species. The presence or absence of these sites was then confirmed by analyzing individual BLATs or by searching for the sites in the individual species sequences obtained from FlyBase (http://flybase.bio.indiana.edu/blast/). Identification of Su(H) and proneural binding sites in A. mellifera and A. gambia was done by scanning the promoters for consensus sequences. Additional transcription factor sites in Drosophila were first identified in the D. melanogaster reference sequences using MatInspector (http://www.genomatix.de/cgi-bin/matinspector_prof/mat_fam.pl). Identified sites were analyzed for conservation in eight other Drosophila species by analyzing BLAT readouts.
cis-Decoder analysis:
Shared conserved sequences between the E(spl) promoters were identified using the cis-Decoder programs, EvoPrint-Parser, CSB-aligner, and cDT-scanner programs (Brody et al. 2007; http://evoprinter.ninds.nih.gov/cisdecoder). EvoPrints for each promoter were parsed using EvoPrint parser to identify conserved sequence blocks (CSBs) that were ≥6 bases. To identify shared elements between E(spl) promoters, the parsed outputs were aligned using CSB-aligner. The outputs, cis-Decoder tags (cDTs), were scanned by hand to identify which cDTs were present in multiple E(spl) promoters. cis-Decoder:cDT-scanner was used to identify cDTs in the E(spl) sequences that are also found in promoters of genes known to be expressed in neuronal or mesodermal tissues during development.
RESULTS
Conservation in the upstream regulatory regions of the E(spl) genes:
To determine the general level of conservation within the upstream regulatory regions of the E(spl) genes we used the BLAST-like alignment tool (BLAT) (Kent 2002) to index D. melanogaster E(spl) promoter region sequences and scan orthologous sequences from eight other Drosophila species for matches. BLAT sequence comparisons of these nine different species of Drosophila show a range of conservation in the upstream regulatory regions of the different E(spl) genes (Table 1). We used the percentage of sequence identities between species to obtain a general indication of how many bases have been conserved between species during evolution. As expected, more closely related species generally have a higher percentage of identities for the E(spl) gene promoters. The m2, HLHmβ, m4, and HLHmγ genes have the most highly conserved promoters between D. melanogaster and the distantly related species D. mojavensis, with 41.4, 40.6, 36.4, and 35.0% identities, respectively. In fact, these genes were the most conserved in almost all the species we analyzed. Furthermore, they show the greatest conservation when all nine species are compared together using a recently available multigenomic comparative tool, EvoPrinter (Odenwald et al. 2005). The EvoPrinter algorithm overlays multiple BLAT readouts from species that are all aligned with a common reference sequence. The EvoPrinter output reveals sequences that are invariant in multiple species. We did observe several exceptions: HLHmγ, m4, and HLHm3 show higher identity between D. melanogaster and D. pseudoobscura, 62.3, 58.6, and 37.7%, respectively, than the more closely related D. melanogaster and D. anannasse, 58.1, 53.8, and 32.5%, respectively.
TABLE 1.
Comparison of E(spl) promoters between D. melanogaster and other Drosophila species
| Speciesa | m1 | m2 | HLHm3 | m4 | HLHm5 | m6 | HLHm7 | HLHm8 | mα | HLHmβ | HLHmγ | HLH mδ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D. simulans | 87.5 | —c | 87.3 | 75.3 | 94.0 | 93.9 | 82.1 | 76.8 | 99.0 | 94.0 | 96.0 | 89.6 |
| D. yakuba | 81.1 | 80.3 | 78.2 | 96.8 | 90.9 | 83.3 | 70.5 | 77.9 | 95.0 | 92.9 | 91.7 | 78.7 |
| D. erecta | 73.4 | 81.5 | 71.4 | 96.2 | 90.1 | 85.1 | 70.6 | 84.9 | 96.5 | 96.2 | 95.1 | 76.5 |
| D. ananassae | 32.3 | 56.2 | 32.5 | 53.8 | 51.7 | 51 | 31.1 | 40.5 | 56.3 | 73.1 | 58.1 | 49.9 |
| D. pseudoobscura | 22.1 | 55.1 | 37.6 | 58.6 | 49.0 | 36.5 | 27.3 | 30.4 | 57.0 | 55.1 | 62.8 | 44.2 |
| D. mojavensis | 8.3 | 41.4 | 22.0 | 36.4 | 22.5 | 18.4 | 23.7 | 21.5 | 22.5 | 40.6 | 35.0 | 23.0 |
| D. virilis | 10.7 | 40.6 | 23.1 | 37.4 | 24.9 | 17.8 | 19.5 | 20.1 | 23.6 | 31.4 | 38.4 | 19.4 |
| D. grimshawi | 3.2 | 43.0 | 29.4 | 40.3 | 21.8 | 20.1 | 22.3 | 21.3 | 24.7 | 41.4 | 30.3 | 22.3 |
| All 9 speciesb | 3.1 | 32.2c | 15.8 | 26.3 | 14.6 | 13.5 | 11.6 | 17.2 | 18.0 | 36.1 | 31.8 | 14.7 |
Numbers shown are percentage of identity.
Pairwise percentages of sequence identities were determined from BLAT alignments. The number of bases found to be identical in both species was divided by the total number of bases for the promoter sequence.
The percentages of sequence identities for all nine species were determined from EvoPrint alignments. The number of bases found to be identical in all nine species was divided by the total number of bases in the promoter sequence.
Publicly available sequence data for the D. simulans m2 promoter are incomplete and thus D. simulans comparisons are not included.
Further analysis of the sequence comparisons using EvoPrinter reveals interspersed regions referred to as MCSs (Odenwald et al. 2005), which have been conserved in the promoters of the E(spl) genes. Twelve conserved regions with sizes between 10 and 48 bp are found in both of the HLHmγ and HLHmβ promoters (Figure 1, A and B). HLHm8 has 9 conserved regions with sizes between 6 and 97 bp (Figure 1C). Our analysis of the other E(spl) genes showed a similar range of numbers and sizes of MCSs (data not shown).
Figure 1.—
Identification of multispecies conserved sequences in the promoters of E(spl) HLHmγ, HLHmβ, and HLHm8. EvoPrinter outputs for HLHmγ (A), HLHmβ (B), and HLHm8 (C) are shown. Uppercase letters in pink represent nucleotides in the 1200-bp D. melanogaster reference multispecies conserved sequences (MCSs) that are conserved in D. simulans, D. yakuba, D. erecta, D. annanasse, D. pseudoobscura, D. virilis, D. mojavensis, and D. grimshawi. TATAA boxes are underlined.
These results show that there are differences in the levels of conservation in the promoters of the different E(spl) genes, which may shed light on the evolution of these regulatory regions. In addition, specific sequences within these promoter regions have been conserved in at least nine different species of Drosophila, suggesting that they may play important functional roles in regulating the expression of these genes.
Su(H) site conservation in the E(spl) promoter regions:
Since three classes of Su(H) binding sites, high-affinity (YGTGRGAA) single, high-affinity paired, and low-affinity (RTGRGAR) single, have already been identified in the D. melanogaster E(spl) genes (Bailey and Posakony 1995; Eastman et al. 1997; Nellesen et al. 1999), we were interested in determining whether these sites are preferentially conserved in other Drosophila species. Although paired high-affinity sites have been shown to play a role during sensory organ precursor (SOP) formation (Nellesen et al. 1999; Cave et al. 2005), single high-affinity site and low-affinity site functionality during Drosophila development has not been as well characterized. We analyzed the E(spl) promoters to determine the level of conservation of all three types of Su(H) binding sites throughout nine species of Drosophila (Table 2). Approximately 70% (23 of 33) of the high-affinity Su(H) binding sites have been completely conserved in the E(spl) bHLH promoters whereas only 13% (3 of 23) of the low-affinity sites are conserved. Twelve of the 23 conserved high-affinity sites are part of a paired site.
TABLE 2.
Conservation of putative Su(H) binding sites
| Gene | Site | D. melanogaster | D. simulans | D. yakuba | D. erecta | D. ananassae | D. pseudoobsc. | D. virilis | D. mojavensis | D. grimshawi | aAll 8 species |
|---|---|---|---|---|---|---|---|---|---|---|---|
| m1 | Hpc | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | — | — |
| Lc | 6 | 5 | 5 | 5 | — | — | — | — | — | — | |
| Lpc | — | 1 | 1 | 1 | — | — | — | — | — | — | |
| m2 | Hc | 2 | —b | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 1b |
| Hpc | 2c | —b | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| Lc | 5 | —b | 4 | 4 | 2 | 3 | 1 | 2 | 2 | 1b | |
| Lpc | — | —b | — | — | 1 | — | 1 | — | — | —b | |
| HLHm3 | Hc | 3 | 3 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Hpc | — | — | — | 1 | — | — | — | — | — | — | |
| Lc | 3 | 3 | 3 | 2 | 1 | — | — | — | — | — | |
| Lpc | — | — | — | 1 | — | — | — | — | — | — | |
| m4 | Hc | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Lc | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | |
| HLHm5 | Hc | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 2 | 3 | 2 |
| Hpc | — | — | — | — | — | — | — | 1 | — | — | |
| Lc | 1 | 1 | 1 | 1 | — | 1 | 1 | 1 | 1 | — | |
| Lpc | 1 | ||||||||||
| m6 | Lc | 3 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 |
| Lpc | — | 1 | — | — | — | — | — | — | — | — | |
| HLHm7 | Hc | 4 | 4 | 4 | 4 | 2 | 4 | 4 | 4 | 4 | 2 |
| Lc | 2 | — | — | — | 1 | 1 | — | — | — | — | |
| Lpc | — | 1 | 1 | 1 | — | — | — | — | — | — | |
| HLHm8 | Hc | 3 | 3 | 3 | 3 | 3 | 3 | 2 | 2 | 2 | 2 |
| Lc | 3 | 3 | 3 | 3 | 1 | — | — | — | — | — | |
| mα | Hc | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 4 | 4 | 4 |
| Hpc | — | — | — | — | — | — | — | 1 | 1 | — | |
| Lc | 1 | 1 | 1 | 1 | — | — | — | — | — | — | |
| HLHmβ | Hc | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| HLHmγ | Hc | 4 | 4 | 4 | 4 | 4 | 4 | 3 | 3 | 3 | 3 |
| Lc | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| HLHmδ | Hc | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Lc | 2 | 1 | 1 | 1 | 2 | 1 | — | 1 | — | — |
Su(H) binding sites identified in D. melanogaster (Bailey and Posakony 1995; Eastman et al. 1997; Nellesen et al. 1999) are displayed in italics. Conservation of these sites was analyzed for each of eight other species from BLAT outputs and was checked with individual sequences for each species. Non-italic numbers correspond to the quantity of D. melanogaster sites conserved in each species and dashes represent the lack of sites. H, high-affinity sites, YGTGRGAA; L, low-affinity site, RTGRGAR. Sites that conform perfectly to the consensus are designated c and sites that vary from the consensus at 1 or 2 bases are designated pc for partial consensus. Consensus sequences that vary between species, but still maintain conformation to the consensus site are included in the rows designated “c.”
The number of D. melanogaster sites conserved in all eight other species analyzed.
Publicly available D. simulans m2 sequence data are incomplete and thus are not included in this analysis.
Partial consensus sites with the sequence CGTGTGAA, where the underlined T alters from the consensus, have been determined to be bound by Su(H) with high affinity (Nellesen et al. 1999).
Paired Su(H) binding sites are defined as two high-affinity Su(H) binding sites of a specific type, YGTGRGAAM, where the “Y” is a T in the upstream site and a C in the downstream site and M is an A or C, spaced 30 bp apart in an inverted repeat arrangement (Nellesen et al. 1999). All paired Su(H) sites found in D. melanogaster E(spl) promoter regions (m4, HLHm7, HLHm8, HLHmγ, HLHmδ, and HLHm3) are completely conserved in eight other Drosophila species (Figure 2). A hexamer sequence, GAAAGT/ACTTTC, is found between the paired Su(H) sites and has been previously identified in five of the D. melanogaster E(spl) promoters (Nellesen et al. 1999; Cave et al. 2005). This hexamer is fully conserved in m4, HLHm7, HLHm8, and HLHmδ, but is only partially conserved in HLHmγ (Figure 2). HLHm3 contains a paired site that is completely conserved, but no hexamer sequence is present between the Su(H) sites in any species. HLHm5 contains a modified pair of Su(H) sites that is assumed to be nonfunctional as a paired site (Nellesen et al. 1999; Cave et al. 2005). These sites were conserved in all nine Drosophila species. HLHm3 and HLHm5 share the sequence ACACACGAC (previously identified by Nellesen et al. 1999; Cave et al. 2005) that is located between two Su(H) binding sites and we also found that this sequence was conserved in all nine Drosophila species.
Figure 2.—
Conservation of Su(H) paired sites in Drosophila E(spl) promoters. Uppercase letters denote sequences conserved in D. melanogaster, D. simulans, D. yakuba, D. erecta, D. annanasse, D. pseudoobscura, D. virilis, D. mojavensis, and D. grimshawi. Lowercase letters denote sequences found in D. melanogaster, but not conserved in all nine species. Su(H) sites are in orange. Conserved hexamer sequences are underlined. The HLHm3/HLHm5 conserved internal sequence is underlined with a dashed line. A single-base-pair difference (Y is C rather than T) in HLHm5 determines the site as a nonpaired site (Nellesen et al. 1999) and is underlined with dots.
Although many D. melanogaster E(spl) bHLH promoter regions contain single high-affinity and low-affinity binding sites (Nellesen et al. 1999; Cave et al. 2005), we found that very few of them are conserved (Table 2). HLHmγ and HLHmβ are the only E(spl) bHLH genes with conserved single high-affinity Su(H) sites and HLHmγ is the only gene to have a conserved low-affinity site. mα, m2, m4, and m6, which are Su(H) dependent, and Notch responsive non-bHLH genes at the E(spl) locus (Nellesen et al. 1999; Wurmbach et al. 1999; Lai et al. 2000), have conserved high-affinity and low-affinity Su(H) binding sites (Nellesen et al. 1999).
In addition to analyzing the conservation of D. melanogaster Su(H) sites we determined whether nonconserved sites were present upstream of one bHLH gene, HLHmβ, and one Bearded-like gene, m4, in the same set of Drosophila species. The total number of Su(H) sites, not just those first identified in D. melanogaster, was identified for m4 and HLHmβ in nine species of Drosophila (supplemental Table S12 at http://www.genetics.org/supplemental/). In m4 only the conserved D. melanogaster sites are present in all the other species. However, in HLHmβ, which contains only one high-affinity site and no low-affinity sites in D. melanogster, one low-affinity site is present in D. anannasse and D. virilis and two low-affinity sites are present in D. mojavensis. Although no expression data are yet available for the E(spl) genes in these species, it is tempting to speculate that the extra low-affinity sites allow either for enhanced expression or for different patterns of expression in these species.
Overall, high-affinity Su(H) binding sites, particularly in the paired configuration, are highly conserved throughout the Drosophila genus, further supporting their critical roles in regulating E(spl) expression. There are fewer conserved single high-affinity and low-affinity sites; however, those that are conserved may very well be functional and regulate several of the E(spl) genes via a different mechanism than the paired sites.
bHLH activator and repressor site conservation in the E(spl) promoters:
Proneural bHLH activator proteins play a role in the regulation of E(spl) transcription and almost all of the E(spl) promoters in D. melanogaster contain binding sites (class A E boxes) for these proteins (Heitzler et al. 1996; Cooper et al. 2000; Li and Baker 2001). Several of these sites have already been shown to be functional and necessary for expression of the E(spl) genes in proneural clusters (Cooper et al. 2000). Two different types of proneural binding sites (class A E boxes) with consensus sequences of GCAGSTG and AWCAKGTG are preferentially bound by Achate–Scute (Ac/Sc) (Singson et al. 1994) and Atonal (Powell et al. 2004) proteins, respectively. We examined the presence of both types of bHLH activator sites first in D. melanogaster and then in nine species of Drosophila to determine whether they were conserved. In D. melanogaster, all of the proneural binding sites upstream of the E(spl) bHLH genes are of the Ac/Sc type. Atonal sites are only found upstream of m2 and m4 (Table 3). In all of the E(spl) promoters that contain proneural binding sites in D. melanogaster, except for HLHmβ, at least one site is conserved in all species (Table 3). In addition to the core consensus sequences, the flanking sequences are also highly conserved (supplemental Figure S1 at http://www.genetics.org/supplemental/). Although proneural sites were analyzed independently from the SPS module, many of the conserved sites were found to be located relatively close to the SPS sites (Figure 3; supplemental Figure S1) and thus part of the SPS + A module. Proneural sites that are part of the SPS + A module are all of the Ac/Sc type. Conserved proneural sites independent from SPS sites were identified in m2, m6, mα, and HLHmγ. The single, SPS-independent proneural site in D. melanogaster HLHmβ is partially conserved in D. simulans, D. yakuba, and D. erecta, but is completely absent in the other five species. Interestingly, in m6 a modified Ac/Sc site present in D. melanogaster and conserved in more closely related species becomes a consensus site in the more distantly related species D. virilis, D. mojavensis, and D. grimshawi.
TABLE 3.
Conservation of putative proneural binding sites
| Gene | Site | D. melanogaster | D. simulans | D. yakuba | D. erecta | D. ananassae | D. pseudoobscura | D. persimilis | D. virilis | D. mojavensis | D. grimshawi | All 9 Speciesa |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| m2 | Ato-c | 1 | —b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1b |
| Ac/Sc-pcc | 1 | —b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1b | |
| m4 | Ac/Sc-c | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 1 |
| Ato-c | 1 | 1 | 1 | 1 | — | — | — | — | — | — | — | |
| HLHm5 | Ac/Sc-c | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| m6 | Ac/Sc-c | — | — | — | — | — | — | — | 1 | 1 | 1 | — |
| Ac/Sc-pc | 1 | 1 | 1 | 1 | 1 | 1 | 1 | — | — | — | — | |
| HLHm7 | Ac/Sc-c | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Ac/Sc-pc | — | — | 1 | 1 | — | — | — | 1 | 1 | — | — | |
| HLHm8 | Ac/Sc | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| mα | Ac/Sc-c | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| HLHmβ | Ac/Sc-c | 1 | — | — | — | — | — | — | — | — | — | — |
| Ac/Sc-pc | — | 1 | 1 | 1 | — | — | — | — | — | — | — | |
| HLHmγ | Ac/Sc-c | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| HLHmδ | Ac/Sc-c | 1 | 11 | 1 | 1 | — | — | — | 1 | 1 | — | — |
| Ac/Sc-pcc | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 |
The number of proneural sites in D. melanogaster that conform to the consensus sequence for Ac/Sc binding, GCAGSTG where S is C or G (Singson et al. 1994), and to the consensus sequence for Atonal binding, AWCAKGTG where W is A or T and K is G or T (Powell et al. 2004), are displayed in italics. The numbers of the D. melanogaster sites that are conserved for each of eight other Drosophila species were determined from BLAT readouts and were checked with individual sequences for each species. Non-italic numbers correspond to the quantity of D. melanogaster sites conserved in each species and dashes represent the lack of sites. Sites that conform perfectly to the consensus are designated c and sites that vary from the consensus at one base are designated pc for partial consensus. Consensus sequences that vary between species, but still maintain conformation to the consensus site are included in the rows designated “c.” m1 and HLHm3 do not have proneural sites and thus are omitted.
D. melanogaster sites conserved in all nine other species analyzed.
Publicly available D. simulans m2 sequence data are incomplete and thus are not included in this analysis.
Sites in m2 and mδ identified as proneural by (Nellesen et al. 1999) according to the consensus RCAGSTG do not fit the Atonal or Ac/Sc consensus and are represented as partial consensus sites.
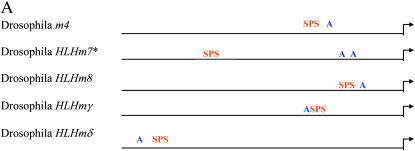
Figure 3.—
Conserved organization of the SPS + A module in E(spl) promoters. (A) General organization of the SPS and consensus proneural sites (RCAGSTG) upstream of m4, HLHm7, HLHm8, HLHmγ, and HLHmδ in D. melanogaster. Su(H) sites are in orange and proneural sites are in blue. (B) Conserved bases in the SPS + A module in the m4, HLHm7, HLHm8, HLHmγ, and HLHmδ promoters. (C) General organization of the SPS and proneural sites upstream of HLHm7 in nine different Drosophila species. Arrows depict transcription start sites. Su(H) sites are in orange and proneural sites are in blue. Modified A sites (YCAGSTG) are underlined.
The upstream regulatory regions of all of the E(spl) genes also contain bHLH repressor binding sites, and it has been postulated that they act to allow cross-regulation of these genes by one another (Nellesen et al. 1999; Cave et al. 2005). Three classes of bHLH repressor sites (N box, class C E box or Hairy site, and class B E box) are present, although none of these sites have yet been shown to be required for any functions in vivo. We examined these sites across 10 Drosophila species and identified at least one conserved bHLH repressor site in the promoters of all of the E(spl) genes, except for HLHm3 (Table 4). Conserved N boxes, which have low-affinity binding by E(spl) bHLH repressors (Jennings et al. 1999), are present upstream of m2, HLHm7, HLHm8, HLHmβ, HLHmγ, and HLHmδ. Class B E boxes, which are bound with higher affinity by E(spl) bHLH proteins (Jennings et al. 1999), are conserved only in m4 and HLHmδ. The most frequently occurring site in D. melanogaster is the N box; however, only 27% (7/26) of these sites are completely conserved in 8 other species whereas 83% (5/6) of the more rare class B E box sites are conserved. By comparison 33% (3/9) of the class C E box/Hairy sites are conserved.
TABLE 4.
Conservation of putative bHLH repressor binding sites in E(spl) promoter regions
| Gene | Site | D. melanogaster | D. simulans | D. yakuba | D. erecta | D. ananassae | D. pseudoobscura | D. persimilis | D. virilis | D. mojavensis | D. grimshawi | All 9 speciesa |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| m1 | EC | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1b | 1b | — | — |
| EB | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1c | 1c | 1 | |
| m2 | N | 2 | b | 2 | 2 | 2 | 1c | 1 | 1 | 1 | 1 | 1d |
| HLHm3 | N | 2 | 2 | 2 | 2 | — | — | — | — | — | — | — |
| EC | 2 | 2 | 2 | 2 | — | 1 | — | — | — | — | — | |
| EB | 1 | 1 | — | — | — | — | — | — | — | — | — | |
| m4 | EC | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| EB | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| HLHm5 | EC | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| m6 | N | 3 | 3 | 3 | 3 | 3 | 2 (1c, 1) | 1 | 1 | 1 | 1 | 1 |
| EC | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| EB | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| HLHm7 | N | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| EB | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| HLHm8 | N | 6 | 6 | 5 | 5 | 3 | 2 | 2 | 2 | 2 | 2 | 2 |
| EC | 1 | 1 | — | — | — | — | — | — | — | — | — | |
| mα | N | 2 | 1 | 1 | 1 | 1 | 1 | 1 | — | — | — | — |
| HLHmβ | N | 4 | 4 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| HLHmγ | N | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| EC | 1 | — | — | — | — | — | — | — | — | — | — | |
| HLHmδ | N | 5 | 5 | 5 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| EB | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
The numbers of bHLH repressor sites identified in D. melanogaster are displayed in italics. N, N box, CACNAG (Oellers et al. 1994); EB, class B E box, CACGTG (Dang et al. 1992); EC, Hairy site/class C E box, CACGCG (Rebeiz et al. 2005). The numbers of D. melanogaster sites that are conserved in each of eight other Drosophila species were identified in BLAT readouts and individual species sequence data. Non-italic numbers correspond to the quantity of D. melanogaster sites conserved in each species and dashes represent the lack of sites.
The number of D. melanogaster sites conserved in all nine other species analyzed.
Contains two modified bases.
Contains one modified base.
Publicly available D. simulans m2 sequence data are incomplete and thus are not included in this analysis.
Although nearly all of the E(spl) gene promoters contain at least one conserved bHLH repressor type site, the sites are not as well conserved as the proneural sites. Further analysis of the total number of bHLH repressors upstream of m4 and HLHmβ suggests that additional sites, some in different locations and some of different classes (supplemental Table S12 at http://www.genetics.org/supplemental/), may either supplant the sites that are not conserved or allow for variable expression patterns in different species.
SPS + A architecture is conserved across nine Drosophila species:
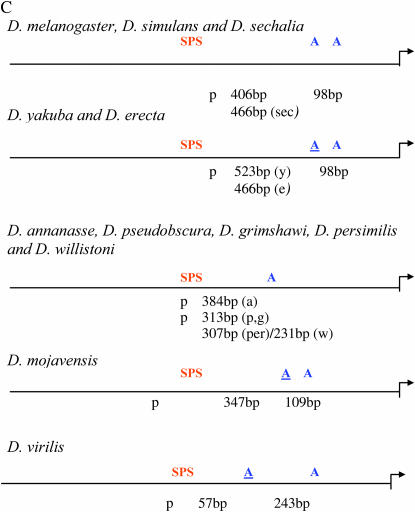
The SPS + A [paired Su(H) sites coupled with a proneural site] architecture has been shown to be critical for Notch–proneural synergistic induction of transcription (Nellesen et al. 1999; Cave et al. 2005). The spatial organization of these sites is in concordance with the direct interaction between Su(H) and proneural proteins (Cave et al. 2005). We found that this overall organization is conserved in all nine species of Drosophila that we analyzed (Figure 3A). A proneural site is located within 60 bp of the paired sites of m4, HLHm8, HLHmγ, and HLHmδ in all nine species (Figure 3B). m4 has an additional conserved proneural site further downstream. In the HLHm7 gene a more complicated evolutionary history is present (Figure 3C). In the closely related species D. melanogaster, D. simulans, and D. secchalia, there are two consensus proneural sites downstream of the paired sites. The site most proximal to the paired site in these species varies from 406 to 466 bp downstream and the more distal proneural site is 98 bp further downstream in all three species. In D. yakuba, D. erecta, D. mojavensis, and D. virilis there is a modified proneural site (C/TCAGGTG) most proximal to the paired sites. A more distal consensus site is found a further 98 bp downstream in D. yakuba and D. erecta, 109 bp downstream in D. mojavensis, and 243 bp downstream in D. virilis. In D. ananasse, D. pseudoobscura, D. persimilis, D. willistoni, and D. grimshawi there is a single proneural site that varies between 307 and 384 bp downstream.
In summary, the overall conservation and organization of the SPS + A architecture in nine different Drosophila species further supports its role as an important transcriptional module. It is tempting to speculate that the differences in proneural site location and number upstream of HLHm7 may reflect some constraint in the Su(H) proneural synergistic mechanism since HLHm7 is the only E(spl) gene with a paired site that does not have a proneural site very close to the paired Su(H) sites.
SPS + A architecture is conserved in honeybee and mosquito E(spl) gene promoter regions:
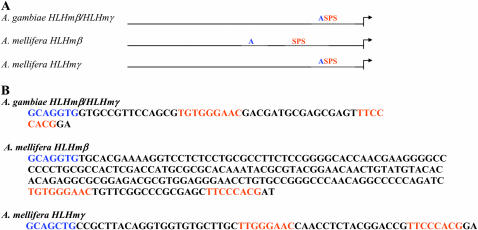
To further understand the evolution of the SPS + A architecture in Drosophila dipterans, we analyzed the E(spl) upstream regulatory regions in a more ancient Dipteran, the mosquito A. gambiae and a Hymenopteran, the honey bee A. mellifera. A. gambiae has one E(spl) bHLH gene, HLHmβ/γ and one Bearded gene while A. mellifera has three E(spl) bHLH genes, HLHmγ, HLHmβ, and HLHmβ′, and a single Bearded gene (Schlatter and Maier 2005). The A. gambiae and A. mellifera E(spl) bHLH gene products appear to be most closely related to the HLHmβ and HLHmγ pair of D. melanogaster (Schlatter and Maier 2005). Unlike the Drosophila HLHmβ promoter, we identified paired Su(H) sites and proneural sites in the E(spl) HLHmβ promoters from both honeybee and mosquito (Figure 4A). The SPS + A module was also present in the A. mellifera HLHmγ promoter, but not in the HLHmβ′ promoter. In A. mellifera the proneural site is 167 bp upstream of the paired site in the HLHmβ promoter and only 10 bp in the HLHmγ promoter, whereas in A. gambiae HLHmβ/mγ it is only 12 bp upstream of the paired site. Close analysis of the paired Su(H) sites reveals that they are very similar to those in Drosophila. The orientation is exactly the same, Su(H) forward–Su(H) reverse (Figure 4B). The spacing between the first G of each Su(H) site is 30 bp in the A. mellifera promoters and 32 bp in the A. gambiae promoter. In Drosophila, the paired sites have a 30-bp spacing in m4, HLHm7, HLHm8, and HLHmγ, while HLHmδ has a 28-bp spacing (Nellesen et al. 1999). Although Drosophila E(spl) promoters with the SPS motif do share a somewhat conserved hexamer sequence between the paired Su(H) sites (Nellesen et al. 1999) (Figure 3B), this sequence is not present in A. mellifera or A. gambiae E(spl) promoters.
Figure 4.—
SPS + A module in the E(spl) HLHmβ genes from honeybee and mosquito. (A) General organization of the SPS and proneural sites upstream of E(spl) bHLH genes in A. mellifora and A. gambiae. (B) Sequences of the SPS + A modules in A. mellifora and A. gambiae E(spl) bHLH genes. Arrows depict transcription start sites. Su(H) sites are in orange and proneural sites are in blue.
In addition to paired sites, we also analyzed the promoters for single Su(H) sites, proneural sites, and bHLH repressor sites. In A. gambiae HLHmβ/γ one low-affinity Su(H) site, one class C E box/Hairy site, and two N boxes were identified (supplemental Figure S2, A and B, at http://www.genetics.org/supplemental/). The A. mellifera HLHγ promoter contains six N boxes and three class C E box/Hairy sites (supplemental Figure S2, C and D); the HLHmβ promoter contains one low-affinity Su(H) site, one class C E box/Hairy site, and one N box (supplemental Figure S2, E and F); and the HLHmβ′ promoter contains three low-affinity Su(H) sites, one proneural Atonal site, and two class C E box/Hairy sites (supplemental Figure S2, G and H). The presence of these sites in many species of Drosophila as well as in the more ancient A. mellifera and A. gambiae suggests that they play an important role in the regulation of the E(spl) bHLH genes.
Identification of putative transcription factor binding sites:
Although Su(H) and proneural binding sites have been shown to be critical for the regulation of E(spl) expression, many lines of evidence suggest that there may be additional factors involved (Ligoxygakis et al. 1999; Nellesen et al. 1999; Wech et al. 1999; Cooper et al. 2000; our unpublished observations). The program MatInspector (Cartharius et al. 2005; Quandt et al. 1995) was used to identify transcription factor binding sites upstream of the E(spl) genes in D. melanogaster. MatInspector identifies putative binding sites and infers potential by scanning input sequences for matches to a library based on position weight matrices (PWM). The matrix library also contains information about the specificity and sensitivity of each nucleotide within all of the included consensus transcription factor binding sites. MatInspector calculates matrix similarity scores that range from 1 (sequence corresponds to most important nucleotides within a matrix site) to 0 (no correspondence to matrix site). Matrix similarity scores that are less than 0.8 are rejected as potential sites.
Using MatInspector, we identified potential transcription factor bindings sites in the D. melanogaster E(spl) promoters (Table 5). Paired homeodomain sites are present in all of the promoters, whereas Fushi tarazu, Snail, Dorsal, Zeste, and CF2-II sites are found in the majority of the promoters. We further analyzed the possible functionality of these sites by determining whether they were conserved in other Drosophila species. BLAT analysis revealed conservation of specific sites between D. melanogaster and individual Drosophila species (supplemental Tables S1–S11 and supplemental Figure 1 at http://www.genetics.org/supplemental/). We specifically searched for conserved sites in the promoter regions of the E(spl) bHLH genes to identify putative regulators that may be responsible for the distinctive expression patterns of these genes (Figure 5; supplemental Figure S1). Not surprisingly, many of the sites identified in D. melanogaster are not conserved in all nine other Drosophila species. However, a significant number of them are, suggesting that they be important for the regulation of these genes. HLHm7 and HLHmδ have the fewest conserved known transcription factor binding sites with six each. HLHm3, HLHm5, and HLHmβ all have multiple paired homeodomain sites. HLHm3, m4, and HLHm5 promoter regions contain multiple Fushi tarazu (Ftz) sites, whereas HLHm8, HLHmβ,, and HLHmγ have single Ftz sites. HLHm7 and HLHm8 each have two Dorsal sites in their promoters. HLHmβ, HLHmδ, and HLHm3 all have single Adf-1 sites. We also analyzed the promoters of the Bearded-like E(spl) genes for conservation of known transcription factor binding sites and identified many of the same conserved sites as well as several different sites not common to the bHLH gene promoters (supplemental Figure S1). Like many of the bHLH genes, m4 also has Paired and Ftz sites (Figure 5 and supplemental Figure S1). The conservation of these binding sites in the promoters of the E(spl) bHLH and Bearded-like genes for known developmental transcription factors suggests that they may play a functional role in regulating the expression of these genes during development.
TABLE 5.
Putative transcription factor binding sites in the E(spl) upstream regulatory region in D. melanogaster
| Binding site | m1 | m2 | HLHm3 | m4 | HLHm5 | HLHm6 | HLHm7 | HLHm8 | mα | HLHmβ | HLHmγ | HLHmδ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adf-1 | 1 | 5 | 1 | 8 | 4 | 2 | 1 | 2 | ||||
| Broad-complex Z1 | 3 | 1 | 1 | 2 | 1 | 1 | 1 | |||||
| Broad-complex Z2 | 1 | 1 | 1 | 1 | ||||||||
| Broad-complex Z3 | 1 | 2 | 1 | |||||||||
| Broad-complex Z4 | 3 | |||||||||||
| Caudal | 2 | 1 | 4 | 1 | 1 | 4 | ||||||
| CF2-II | 8 | 19 | 7 | 4 | 1 | 3 | 1 | 1 | 5 | |||
| Chorion factor 1 | 1 | 1 | 1 | |||||||||
| Crocodile | 2 | 1 | 1 | 1 | 3 | 2 | ||||||
| Dead ringer | 1 | 2 | ||||||||||
| Deformed | 2 | 2 | 1 | 2 | 1 | 4 | 2 | 2 | 2 | |||
| Dorsal | 3 | 1 | 1 | 2 | 3 | 7 | 3 | 2 | 1 | |||
| Doublesex | 1 | 2 | 2 | 1 | 1 | |||||||
| E74A | 1 | 1 | 1 | 3 | ||||||||
| Elf-1 (NTF-1, Grainyhead) | 1 | 1 | 2 | 2 | ||||||||
| Fushi tarazu | 1 | 2 | 4 | 8 | 3 | 5 | 8 | 6 | 2 | 1 | 1 | |
| GAGA factor | 1 | 1 | 2 | 1 | 2 | |||||||
| Glial cells missing | 1 | 3 | 2 | 1 | 1 | 1 | ||||||
| Heat shock factor | 3 | 3 | 9 | 3 | 1 | 5 | ||||||
| Hunchback | 1 | 1 | 2 | 1 | 2 | 1 | ||||||
| K50 type homeodomain | 1 | 1 | ||||||||||
| Knirp | 1 | 5 | 2 | 1 | ||||||||
| Krueppel | 2 | 5 | 1 | 1 | 2 | 1 | ||||||
| Paired homeodomain | 1 | 3 | 7 | 4 | 3 | 4 | 1 | 2 | 5 | 1 | 1 | |
| PAX6 P3 homeodomain | 2 | 2 | 2 | 1 | 3 | 9 | 1 | 1 | ||||
| PHO and PHO-like | 1 | 1 | 1 | |||||||||
| Snail | 3 | 1 | 2 | 2 | 3 | 2 | ||||||
| Tailless (gap gene) | 1 | 1 | 3 | 6 | 1 | 1 | ||||||
| T-cell factor | 4 | 1 | 4 | 1 | 1 | |||||||
| Ttk69 | 2 | 1 | 1 | 2 | 1 | 3 | ||||||
| Zeste | 2 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 |
Figure 5.—
Conserved transcription factor binding sites upstream of the E(spl) genes. Putative transcription factor binding sites identified upstream of the E(spl) bHLH, m4, and mα genes that are found to be completely conserved or partially conserved (two or fewer base substitutions) in at least nine Drosophila species are shown. Sites are color and number coded according to the list at the bottom. The spacing of the sites in the diagram reflects the general spacing in the actual sequences. Arrows depict transcription start sites. Actual sequences for all identified sites are presented in supplemental Figure S1 at http://www.genetics.org/supplemental/).
Shared novel sequences between Drosophila E(spl) promoter regions:
In addition to known transcription factor binding sites, previously uncharacterized sequences common to multiple E(spl) genes may play a role in the overlapping expression patterns observed for these genes. To identify such sequences programs called EvoPrint-parser and cis-Decoder:CSB-Alignment (Brody et al. 2007) were used to scan the conserved regions in the promoters of the E(spl) bHLH and m4 genes for sequences present in more than one of the genes. EvoPrint-parser identifies CSBs, which are defined as continuous strands of conserved bases that average ∼13 bp in length (Brody et al. 2007). cis-Decoder:CSB-Alignment was then used to compare the CSBs from all of the E(spl) genes and to identify sequences shared between two or more genes. The shared sequences identified by cis-Decoder:CSB-Alignment are called cDTs. More than 100 cDTs shared between all E(spl) promoter regions were identified (data not shown). When the analysis was focused to the E(spl) bHLH genes and the m4 gene, as a representative of Bearded-like E(spl) genes 49 cDTs between 6 and 10 bp in length were found to be present in at least two of the E(spl) genes (Table 6). The sequence CAATAA is present upstream of HLHm3, HLHm5, HLHm8, HLHmβ, and HLHmγ and the sequence TAATTG is present upstream of HLHm3, m4, HLHm5, HLHm8, and HLHmβ. GGATCG is upstream of three E(spl) genes: HLHm3, HLHmβ, and HLHmγ. Other possible coordinating sequences may be CACACG, which is found in front of HLHm3, HLHmγ, and HLHmδ, and AAATCT, which is upstream of HLHm3 and HLHm7. Some cDTs are present multiple times in individual promoters (data not shown).
TABLE 6.
Conserved upstream regulatory region sequences shared between E(spl) genes
| Sequence | HLHm3 | m4 | HLHm5 | HLHm7 | HLHm8 | HLHmβ | HLHmγ | HLHmδ |
|---|---|---|---|---|---|---|---|---|
| E(spl) specific | ||||||||
| ACGACAA | + | + | ||||||
| GGATCG | + | + | + | |||||
| TTGTCGT | + | + | ||||||
| GTGAGAAA | + | + | ||||||
| ACGACAA | + | + | ||||||
| Shared with mesodermal enhancers (Espl specific) | ||||||||
| GAAAGT | + | + | + | |||||
| AAGTGT | + | + | + | |||||
| GTGTGT (AGTGTGT) | + | + | + | + | ||||
| ATTATC (ATTATCCT) | + | + | + | + | ||||
| (CA)AATC(AT) | + | + | + | |||||
| CAATAA | + | + | + | + | + | |||
| GATGTT | + | + | ||||||
| AATCGA | + | + | ||||||
| TCATCA | + | + | ||||||
| AGCAAT | + | + | ||||||
| CACAAT | + | + | + | |||||
| ATTGCG | + | + | + | |||||
| TAATTG | + | + | + | + | + | |||
| YCAATY | ||||||||
| TGATAA | + | + | + | |||||
| CACACG | + | + | + | |||||
| AAATCT | + | + | ||||||
| AAAGTA | + | + | + | |||||
| AGGCAC | + | + | ||||||
| ACACAC (ACACACT) | + | + | + | + | ||||
| TTGCAG | + | + | + | |||||
| AACAAT | + | + | + | + | + | |||
| Shared with neural enhancers | ||||||||
| TTCTTGT | + | + | + | p | p | |||
| CACAATC | + | + | ||||||
| AATAACT | + | + | ||||||
| ACAAGA | + | + | + | |||||
| AAGTAAA | + | + | ||||||
| TTTAATTG | p | + | p | p | p | |||
| TTTCAG | + | + | p | p | ||||
| GATCGC | + | + | + | |||||
| CACAATC | + | + | + | |||||
| ATAAGGG | + | + | ||||||
| TCTTGT | + | + | + | |||||
| TGCAGCT | + | + | ||||||
| TGATGAA | p | p | p | + | ||||
| GAGGCA | + | + | ||||||
| GTGAGAA | + | + | ||||||
| TTTCAG | + | + | ||||||
| CACACGT | + | + | + | |||||
| AAGGATC | + | + | + | |||||
| CAGGATA | + | + | ||||||
| TTCTCAC | + | + | + | + | ||||
| GCACGA | + | + | ||||||
| CACACGT | + | + | p | p | p | p | ||
Underlined sequences refer to sites that are common to genes with overlapping expression patterns (as observed by Wech et al. 1999) during embryogenesis. + indicates conservation of sequence in nine species of Drosophila. p indicates partial conservation of consensus sequence in at least five species of Drosophila. Sequences in parentheses become E(spl) specific with additional flanking bases.
Drosophila tissue-specific cDT libraries that contain cDTs common to genes known to be expressed in particular tissues have been generated and are publicly accessible (Brody et al. 2007). The alignment program cDT-scanner was used to identify E(spl) bHLH cDTs that are also present in known neural or mesodermal expressed Drosophila genes. Many of the cDTs identified upstream of the E(spl) genes are also present in neural or mesodermal enhancers (Table 6). Interestingly, some of the cDTs appear to be specific to E(spl) promoter regions and are not found in mesodermal or neuronal regulatory regions. In fact, the sequence GGATCG described above appears to be E(spl) specific. Given the conservation in nine Drosophila species and presence in multiple E(spl) genes the conserved sequences identified using cis-Decoder are good candidates for regulatory transcription factor binding sites.
In addition to identifying shared novel sequences between the E(spl) genes, cis-Decoder analysis also revealed the presence of shared conserved sequences flanking Su(H) sites. For example, the high-affinity Su(H) site GTGG/AGAA is flanked at the 3′ end by a conserved AC in HLHmγ, HLHm5, HLHm3, HLHm7, and HLHm8. The reverse-oriented Su(H) site TTCTCAC is flanked at the 3′ end by conserved AT bases in m4, HLHm5, and HLHmδ while in HLHmγ and HLHmβ a conserved G flanks the site. Brody has also identified conserved flanking sequences that are shared among Su(H) sites in subsets of E(spl) genes (T. Brody, personal communication). These flanking sequences may indeed play critical roles in stabilizing different complexes bound to the Su(H) sites, thus playing a role in the differential expression patterns of the different E(spl) genes.
DISCUSSION
This comparative study of the upstream regulatory regions of the E(spl) genes in multiple species of Drosophila shows that sequences known to play critical functional roles in D. melanogaster during development have been completely conserved during Drosophila evolution. These results not only highlight the functional importance of these sites, but also shed light on less well understood sites and on the evolution of the E(spl) genes. Importantly, this analysis identified uncharacterized conserved sequences from all of the E(spl) bHLH promoters. These sequences may very likely play important functional roles in the regulation and diverse patterning of E(spl) gene expression.
Conservation of Su(H) binding sites:
One of the most striking findings from this study is that all the paired Su(H) sites identified in D. melanogaster E(spl) promoters are completely conserved, both in sequence and in orientation, in nine Drosophila species as well as in A. gambiae and A. mellifera. In addition, at least one proneural site is completely conserved, maintaining the SPS + A architecture. These results support previous findings showing that the synergistic effect of activated Notch and proneural proteins on E(spl) expression depends not just on the presence of Su(H) and proneural binding sites (Nellesen et al. 1999), but also on the correct orientation of paired Su(H) binding sites (Cave et al. 2005).
Cave et al. (2005) have proposed a model for Notch proneural synergistic activation that is dependent upon protein–protein interactions between Su(H) and proneural proteins. This model predicts that the proper orientation of Su(H) sites allows bound Su(H) to interact cooperatively with proneural proteins and that together they recruit NICD and/or enable activation by NICD. This model also predicts that proneural binding sites must be nearby the paired Su(H) sites. Results shown here confirm this since in all the E(spl) genes with an SPS + A architecture, except HLHm7, the conserved A site is <60 bp away from the SPS site in nine Drosophila species. The spacing of the paired Su(H) and proneural sites may allow ideal interaction conditions for the bound proteins and variations of this organization, such as that seen for HLHm7, may allow for differences in expression patterns between species. Recently, Zinzen et al. (2006) have suggested that shifted binding site spacing and organization in the upstream regulatory region of single-minded (sim) may result in varied expression patterns in the embryonic ventral midline in different species of Drosophila. The sim regulatory regions contain the same quantity and quality of single Su(H) and Twist binding sites in three different Drosophila species. However sim expression in D. melanogaster appears to be regulated by Notch and Twist together, whereas in D. pseuodoobscura and D. virilis the expression pattern suggests regulation by Notch alone. Unfortunately E(spl) expression data are currently limited for Drosophila species other than D. melanogaster and consequently we cannot yet make direct correlations between binding sites and expression patterns for these genes. However, the binding site analysis reported here has the potential to reveal important insights into the variations of E(spl) expression that may be seen in different species.
Interestingly, the required Notch-responsive Su(H) binding sites found upstream of sim are single high-affinity sites rather than paired sites (Cowden and Levine 2002; Zinzen et al. 2006). The few conserved single high-affinity sites in the E(spl) bHLH genes are present upstream of HLHmβ and HLHmγ. Further experiments in D. melanogaster and other Drosophila species are necessary to determine whether these sites are functional and whether expression of the HLHmβ and HLHmγ genes involves Su(H)/NICD bound to these sites alone or in coordination with other transcription factors.
Low level of bHLH repressor site conservation:
Although bHLH repressor binding sites have previously been identified in the E(spl) promoter regions, we are not aware of any critical analysis of the possible functionality of these sites before this study. It was originally proposed that bHLH repressor proteins block proneural bHLH protein activity by binding to E boxes (proneural sites) thereby inhibiting proneural DNA binding or by binding to N boxes [E(spl) bHLH sites] and recruiting corepressors to block the transcriptional activator function of the proneural proteins (Knust et al. 1992). Mutagenesis studies of N boxes in the scute promoter (Culi and Modolell 1998) and experiments showing that direct binding of E(spl) bHLH repressors to the C terminus of a proneural protein blocks its ability to activate expression (Giagtzoglou et al. 2005) suggest that direct DNA binding by the E(spl) bHLH repressors is not necessary for function. Our results support this as well, since we show that few bHLH repressor sites are conserved in the promoters of the E(spl) genes. Some of the sites that are conserved are those that overlap or are very near to the Su(H) paired sites in the SPS + A modules. As previously suggested by Culi and Modolell (1998), DNA binding by bHLH repressors to these sites may help to stabilize their interactions with proneural proteins and thus more effectively inhibit proneural activity.
Evolution of the E(spl) regulatory regions:
Homologs of the E(spl) sequences have been identified in many species, from mosquitos to humans; however, only in Drosophila is such an expansion of genes at the locus present (Maier et al. 1993; Schlatter and Maier 2005). It has been postulated that the presence of multiple genes at the Drosophila E(spl) locus was a result of duplications that not only enlarged the locus, but also allowed for diversification of function (Nellesen et al. 1999; Schlatter and Maier 2005). It is possible that the regulatory regions were not part of this duplication, but rather evolved independently. Our data argue against this, demonstrating the presence of the conserved SPS + A architecture in multiple E(spl) promoters in Drosophila species, A. mellifera, and A. gambiae as well as in the mouse Hairy/E(spl) (HES-1) promoters (Figures 3 and 4; Nellesen et al. 1999). We support an alternative postulate that duplications of the genes included the regulatory regions, but that there was a more rapid rate of evolution for the regulatory sequences than for the coding sequences (Schlatter and Maier 2005). This explains the low level of sequence identity in the regulatory regions of distantly related species compared to that in the coding regions (Nellesen et al. 1999; Schlatter and Maier 2005; our unpublished observations) as well as the conservation of the SPS + A module.
Studies in vertebrates suggest that there are several possible outcomes when gene duplication occurs (Walsh 2003). These include degenerative mutations with loss of one of the copies, an acquired novel function for one copy, or division of specializations from the original gene into each copy resulting in complementary functions or subfunctionalization (Force et al. 1999). The presence of paired sites in the more ancient E(spl) bHLH promoters of A. mellifera and A. gambiae and subsequent loss in some of the Drosophila promoters suggest that at least some subfunctionalization occurred.
Schlatter and Maier (2005) have proposed HLHmβ as the ancestral E(spl) bHLH gene since the single E(spl) bHLH gene product in A. gambiae shares the highest amino acid identity with Drosophila HLHmβ (75.7% compared to 67.4% with Drosophila HLHmγ). Our results support this since HLHmβ also showed higher promoter region sequence identity between multiple Drosophila species than HLHmγ. The presence of single high-affinity Su(H) sites upstream of only HLHmβ and HLHmγ provides further evidence for their ancient status and more general roles during development. Our identification of SPS + A modules upstream of the single A. gambiae E(spl) bHLH gene and the putative HLHmβ and HLHmγ genes in A. mellifera could suggest that HLHmβ may not be the progenitor gene since the Drosophila HLHmβ promoter region does not contain the SPS + A module. However, if subspecialization after duplication did occur it could be expected that the progenitor gene would lose specific sites retained by the new gene copy. Thus, we postulate that HLHmβ was indeed the progenitor gene and that HLHmγ, which does contain an SPS +A module as well as single high-affinity Su(H) and low-affinity Su(H) sites in its promoter region in Drosophila, originated from the first duplication and subspecialization.
Conserved E(spl) promoter sequences may be sites for regulatory transcription factors:
One of the critical questions in the understanding of the E(spl) genes is how their diverse expression patterns are achieved during development. Identification of conserved transcription factor binding sites and shared novel sequences in the promoters of the E(spl) genes presented in this study could elucidate the underlying molecules and mechanisms responsible. The high level of conservation of known functional sites for Su(H) and the proneural proteins suggests that other conserved sites may also play a role. Characterization of these sites and the proteins that bind to them may help uncover previously unidentified regulators of the E(spl) genes. Further confidence in the role of these conserved sites comes from the identification of Dorsal binding sites upstream of HLHm7, which has been shown to be expressed in the presumptive mesectoderm and to have altered expression patterns or levels in the mesectoderm in dorsal loss-of-function mutants in D. melanogaster (Gonzalez-Crespo and Levine 1993; Wech et al. 1999). The upstream regulatory regions of HLHm7 and HLHm8 each have one Dorsal site that is conserved in nine species of Drosophila and a second site that is partially conserved. Interestingly, conserved Dorsal sites are located immediately adjacent to the Su(H) paired sites upstream of HLHm7 and HLHm8. Studies on the role of Dorsal and Notch signaling on the regulation of sim and HLHm8 expression suggest that sim requires Dorsal for Notch activation, whereas HLHm8 does not (Cowden and Levine 2002). The presence of paired Su(H) sites upstream of HLHm8 and single sites upstream of sim may allow HLHm8 to be more sensitive to activation by Notch. It is possible that if low levels of activated Notch are present Dorsal could assist in HLHm8 expression.
Two other E(spl) bHLH genes, HLHmγ and HLHmδ are identically expressed in a subset of proneural clusters within wing imaginal discs (De Celis et al. 1996). Although both genes have conserved SPS + A modules in their promoters and their expression levels are affected by a loss of Su(H), their expression pattern in wing discs appears to be Su(H) independent (Nellesen et al. 1999). Thus, other regulatory transcription factors must be involved in their expression. It has previously been shown that a 234-bp promoter fragment of HLHmγ is sufficient to induce the wild-type expression pattern (Nellesen et al. 1999). Two of the shared conserved sites we identified in the HLHmγ and HLHmδ promoter regions, RCAATR and GAAAGT, are found within the 234-bp functional HLHmγ fragment. This analysis highlights the possible importance of these sites and directs future experiments. Accordingly, we plan to use site-directed mutagenesis to alter these sites to determine their involvement in regulating HLHmγ and HLHmδ expression in the wing disc.
HLHm3 is the most broadly expressed E(spl) bHLH gene during embryogenesis (Wech et al. 1999) and is thus most likely regulated by a variety of factors in addition to NICD and Su(H). Conserved sites upstream of HLHm3 may predict some of the factors that are involved in this regulation. HLHm3 is maternally provided and expressed in the developing embryonic gut, the embryonic midline, the brain, and the terminal sensory organ derivation sites (Wech et al. 1999). Accordingly, there are conserved binding sites for Caudal and Hunchback, two homeodomain proteins that are maternally expressed and play a role in early A–P axis formation (reviewed in Rivera-Pomar and Jackle 1996). Caudal also functions during gut development (Wu and Lengyel 1998), making it a possible regulator of HLHm3 during multiple stages of development. Conserved binding sites for two additional homeodomain-containing proteins, Paired and Fushi tarazu (Ftz), which function during later embryonic development, were also identified upstream of m3. Although best known for its role during segmetation, ftz is expressed in a subset of neuronal precursors in the developing CNS (Doe et al. 1988) and during hindgut formation (Krause et al. 1988). Thus, Ftz is another candidate regulator of HLHm3 during neurogenesis and gut development.
Although this study focused on identification of conserved sequences, further studies of E(spl) expression and divergent sequences in multiple Drosophila species will help to determine whether there are in fact functional differences of these genes between species. Since minimal expression and functional data are available for species other than D. melanogster, it is unknown whether changes of expression and function of the E(spl) genes occurred in Drosophila during evolution. If the expression patterns are indeed similar, then the conserved sites may reveal more factors that are responsible for regulating these genes. Alternatively, if expression varies in different species then nonconserved sequences such as those sites identified here may hold important insights into which factors are involved. In either case the analysis we have done in multiple and distantly related species has the potential to provide important answers to the regulation of the E(spl) genes. It will be interesting to determine the effects of altered binding sites and overexpression of relative transcription factors on E(spl) expression to confirm their proposed functions. In addition to these promoter sites, there are both transcription factor binding sites and micro-RNA sites in the 3′-UTR of these genes (Stark et al. 2003; Lai et al. 2005). Further analysis of the conservation of sequences in these regions may reveal additional sites that allow for the overlapping, but distinct, expression patterns of the E(spl) genes.
Acknowledgments
We thank Thomas Brody for sharing unpublished data and Efrain (Beto) Zuniga for technical assistance. We are grateful to Thomas Brody, Martha Grossel, and Michael Weir for comments on the manuscript. This work was supported by National Institutes of Health grant no. R15 GM067742-01 to D.E.
References
- Artavanis-Tsakonas, S., M. D. Rand and R. J. Lake, 1999. Notch signaling: cell fate control and signal integration in development. Science 284: 770–776. [DOI] [PubMed] [Google Scholar]
- Axelson, H., 2004. a Notch signaling and cancer: emerging complexity. Semin. Cancer Biol. 14: 317–319. [DOI] [PubMed] [Google Scholar]
- Axelson, H., 2004. b The Notch signaling cascade in neuroblastoma: role of the basic helix-loop-helix proteins HASH-1 and HES-1. Cancer Lett. 204: 171–178. [DOI] [PubMed] [Google Scholar]
- Bailey, A. M., and J. W. Posakony, 1995. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 9: 2609–2622. [DOI] [PubMed] [Google Scholar]
- Barolo, S., T. Stone, A. G. Bang and J. W. Posakony, 2002. Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes Dev. 16: 1964–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron, M., 2003. An overview of the Notch signalling pathway. Semin. Cell Dev. Biol. 14: 113–119. [DOI] [PubMed] [Google Scholar]
- Bergman, C. M., B. D. Pfeiffer, D. E. Rincon-Limas, R. A. Hoskins, A. Gnirke et al., 2002. Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biol. 3: RESEARCH0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody, T., W. Rasband, K. Baler, A. Kuzin, M. Kundu et al., 2007. cis-DECODER discovers constellations of conserved DNA sequences shared among tissue-specific enhancers. Genome Biol. 8: R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius, K., K. Frech, K. Grote, B. Klocke, M. Haltmeier et al., 2005. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942. [DOI] [PubMed] [Google Scholar]
- Cave, J. W., F. Loh, J. W. Surpris, L. Xia and M. A. Caudy, 2005. A DNA transcription code for cell-specific gene activation by notch signaling. Curr. Biol. 15: 94–104. [DOI] [PubMed] [Google Scholar]
- Chan, Y. M., and Y. N. Jan, 1998. Roles for proteolysis and trafficking in notch maturation and signal transduction. Cell 94: 423–426. [DOI] [PubMed] [Google Scholar]
- Cooper, M. T., D. M. Tyler, M. Furriols, A. Chalkiadaki, C. Delidakis et al., 2000. Spatially restricted factors cooperate with notch in the regulation of Enhancer of split genes. Dev. Biol. 221: 390–403. [DOI] [PubMed] [Google Scholar]
- Cowden, J., and M. Levine, 2002. The Snail repressor positions Notch signaling in the Drosophila embryo. Development 129: 1785–1793. [DOI] [PubMed] [Google Scholar]
- Crosby, M. A., J. L. Goodman, V. B. Strelets, P. Zhang and W. M. Gelbart, 2007. FlyBase: genomes by the dozen. Nucleic Acids Res. 35: D486–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culi, J., and J. Modolell, 1998. Proneural gene self-stimulation in neural precursors: an essential mechanism for sense organ development that is regulated by Notch signaling. Genes Dev. 12: 2036–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang, C. V., C. Dolde, M. L. Gillison and G. J. Kato, 1992. Discrimination between related DNA sites by a single amino acid residue of Myc-related basic-helix-loop-helix proteins. Proc. Natl. Acad. Sci. USA 89: 599–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis, J. F., J. de Celis, P. Ligoxygakis, A. Preiss, C. Delidakis et al., 1996. Functional relationships between Notch, Su(H) and the bHLH genes of the E(spl) complex: the E(spl) genes mediate only a subset of Notch activities during imaginal development. Development 122: 2719–2728. [DOI] [PubMed] [Google Scholar]
- Delidakis, C., and S. Artavanis-Tsakonas, 1992. The Enhancer of split [E(spl)] locus of Drosophila encodes seven independent helix-loop-helix proteins. Proc. Natl. Acad. Sci. USA 89: 8731–8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe, C. Q., Y. Hiromi, W. J. Gehring and C. S. Goodman, 1988. Expression and function of the segmentation gene fushi tarazu during Drosophila neurogenesis. Science 239: 170–175. [DOI] [PubMed] [Google Scholar]
- Eastman, D. S., R. Slee, E. Skoufos, L. Bangalore, S. Bray et al., 1997. Synergy between suppressor of Hairless and Notch in regulation of Enhancer of split m gamma and m delta expression. Mol. Cell. Biol. 17: 5620–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force, A., M. Lynch, F. B. Pickett, A. Amores, Y. L. Yan et al., 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furriols, M., and S. Bray, 2001. A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr. Biol. 11: 60–64. [DOI] [PubMed] [Google Scholar]
- Giagtzoglou, N., K. A. Koumbanakis, J. Fullard, I. Zarifi and C. Delidakis, 2005. Role of the Sc C terminus in transcriptional activation and E(spl) repressor recruitment. J. Biol. Chem. 280: 1299–1305. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Crespo, S., and M. Levine, 1993. Interactions between dorsal and helix-loop-helix proteins initiate the differentiation of the embryonic mesoderm and neuroectoderm in Drosophila. Genes Dev. 7: 1703–1713. [DOI] [PubMed] [Google Scholar]
- Grumbling, G., and V. Strelets, 2006. FlyBase: anatomical data, images and queries. Nucleic Acids Res. 34: D484–D488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines, N., and K. D. Irvine, 2003. Glycosylation regulates Notch signalling. Nat. Rev. Mol. Cell. Biol. 4: 786–797. [DOI] [PubMed] [Google Scholar]
- Halfon, M. S., Y. Grad, G. M. Church and A. M. Michelson, 2002. Computation-based discovery of related transcriptional regulatory modules and motifs using an experimentally validated combinatorial model. Genome Res. 12: 1019–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzler, P., M. Bourouis, L. Ruel, C. Carteret and P. Simpson, 1996. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development 122: 161–171. [DOI] [PubMed] [Google Scholar]
- Hubbard, T., D. Andrews, M. Caccamo, G. Cameron, Y. Chen et al., 2005. Ensembl 2005. Nucleic Acids Res. 33: D447–D453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings, B., A. Preiss, C. Delidakis and S. Bray, 1994. The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development 120: 3537–3548. [DOI] [PubMed] [Google Scholar]
- Jennings, B. H., D. M. Tyler and S. J. Bray, 1999. Target specificities of Drosophila enhancer of split basic helix-loop-helix proteins. Mol. Cell. Biol. 19: 4600–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joutel, A., and E. Tournier-Lasserve, 1998. Notch signalling pathway and human diseases. Semin. Cell Dev. Biol. 9: 619–625. [DOI] [PubMed] [Google Scholar]
- Kent, W. J., 2002. BLAT–the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klambt, C., E. Knust, K. Tietze and J. A. Campos-Ortega, 1989. Closely related transcripts encoded by the neurogenic gene complex enhancer of split of Drosophila melanogaster. EMBO J. 8: 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knust, E., H. Schrons, F. Grawe and J. A. Campos-Ortega, 1992. Seven genes of the Enhancer of split complex of Drosophila melanogaster encode helix-loop-helix proteins. Genetics 132: 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause, H. M., R. Klemenz and W. J. Gehring, 1988. Expression, modification, and localization of the fushi tarazu protein in Drosophila embryos. Genes Dev. 2: 1021–1036. [DOI] [PubMed] [Google Scholar]
- Lai, E. C., R. Bodner and J. W. Posakony, 2000. The enhancer of split complex of Drosophila includes four Notch-regulated members of the bearded gene family. Development 127: 3441–3455. [DOI] [PubMed] [Google Scholar]
- Lai, E. C., B. Tam and G. M. Rubin, 2005. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 19: 1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, M., and R. Tjian, 2003. Transcription regulation and animal diversity. Nature 424: 147–151. [DOI] [PubMed] [Google Scholar]
- Li, Y., and N. E. Baker, 2001. Proneural enhancement by Notch overcomes Suppressor-of-Hairless repressor function in the developing Drosophila eye. Curr. Biol. 11: 330–338. [DOI] [PubMed] [Google Scholar]
- Ligoxygakis, P., S. J. Bray, Y. Apidianakis and C. Delidakis, 1999. Ectopic expression of individual E(spl) genes has differential effects on different cell fate decisions and underscores the biphasic requirement for notch activity in wing margin establishment in Drosophila. Development 126: 2205–2214. [DOI] [PubMed] [Google Scholar]
- Maier, D., B. M. Marte, W. Schafer, Y. Yu and A. Preiss, 1993. Drosophila evolution challenges postulated redundancy in the E(spl) gene complex. Proc. Natl. Acad. Sci. USA 90: 5464–5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, V., M. Lecourtois, O. Massiani, D. Maier, A. Preiss et al., 2001. Transcriptional repression by suppressor of hairless involves the binding of a hairless-dCtBP complex in Drosophila. Curr. Biol. 11: 789–792. [DOI] [PubMed] [Google Scholar]
- Nagel, A. C., A. Krejci, G. Tenin, A. Bravo-Patino, S. Bray et al., 2005. Hairless-mediated repression of notch target genes requires the combined activity of Groucho and CtBP corepressors. Mol. Cell. Biol. 25: 10433–10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellesen, D. T., E. C. Lai and J. W. Posakony, 1999. Discrete enhancer elements mediate selective responsiveness of enhancer of split complex genes to common transcriptional activators. Dev. Biol. 213: 33–53. [DOI] [PubMed] [Google Scholar]
- Odenwald, W. F., W. Rasband, A. Kuzin and T. Brody, 2005. EVOPRINTER, a multigenomic comparative tool for rapid identification of functionally important DNA. Proc. Natl. Acad. Sci. USA 102: 14700–14705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oellers, N., M. Dehio and E. Knust, 1994. bHLH proteins encoded by the Enhancer of split complex of Drosophila negatively interfere with transcriptional activation mediated by proneural genes. Mol. Gen. Genet. 244: 465–473. [DOI] [PubMed] [Google Scholar]
- Paroush, Z., R. L. Finley, Jr., T. Kidd, S. M. Wainwright, P. W. Ingham et al., 1994. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79: 805–815. [DOI] [PubMed] [Google Scholar]
- Powell, L. M., P. I. Zur Lage, D. R. Prentice, B. Senthinathan and A. P. Jarman, 2004. The proneural proteins Atonal and Scute regulate neural target genes through different E-box binding sites. Mol. Cell. Biol. 24: 9517–9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt, K., K. Frech, H. Karas, E. Wingender and T. Werner, 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23: 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz, M., T. Stone and J. W. Posakony, 2005. An ancient transcriptional regulatory linkage. Dev. Biol. 281: 299–308. [DOI] [PubMed] [Google Scholar]
- Rivera-Pomar, R., and H. Jackle, 1996. From gradients to stripes in Drosophila embryogenesis: filling in the gaps. Trends Genet. 12: 478–483. [DOI] [PubMed] [Google Scholar]
- Schlatter, R., and D. Maier, 2005. The Enhancer of split and Achaete-Scute complexes of Drosophilids derived from simple ur-complexes preserved in mosquito and honeybee. BMC Evol. Biol. 5: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singson, A., M. W. Leviten, A. G. Bang, X. H. Hua and J. W. Posakony, 1994. Direct downstream targets of proneural activators in the imaginal disc include genes involved in lateral inhibitory signaling. Genes Dev. 8: 2058–2071. [DOI] [PubMed] [Google Scholar]
- Stark, A., J. Brennecke, R. B. Russell and S. M. Cohen, 2003. Identification of Drosophila microRNA targets. PLoS Biol. 1: E60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl, G., and A. Adachi, 1998. Nuclear access and action of notch in vivo. Cell 93: 649–660. [DOI] [PubMed] [Google Scholar]
- Tsuda, L., R. Nagaraj, S. L. Zipursky and U. Banerjee, 2002. An EGFR/Ebi/Sno pathway promotes delta expression by inactivating Su(H)/SMRTER repression during inductive notch signaling. Cell 110: 625–637. [DOI] [PubMed] [Google Scholar]
- Walsh, B., 2003. Population-genetic models of the fates of duplicate genes. Genetica 118: 279–294. [PubMed] [Google Scholar]
- Wech, I., S. Bray, C. Delidakis and A. Preiss, 1999. Distinct expression patterns of different enhancer of split bHLH genes during embryogenesis of Drosophila melanogaster. Dev. Genes Evol. 209: 370–375. [DOI] [PubMed] [Google Scholar]
- Wu, L. H., and J. A. Lengyel, 1998. Role of caudal in hindgut specification and gastrulation suggests homology between Drosophila amnioproctodeal invagination and vertebrate blastopore. Development 125: 2433–2442. [DOI] [PubMed] [Google Scholar]
- Wurmbach, E., I. Wech and A. Preiss, 1999. The Enhancer of split complex of Drosophila melanogaster harbors three classes of Notch responsive genes. Mech. Dev. 80: 171–180. [DOI] [PubMed] [Google Scholar]
- Zinzen, R. P., J. Cande, M. Ronshaugen, D. Papatsenko and M. Levine, 2006. Evolution of the ventral midline in insect embryos. Dev. Cell 11: 895–902. [DOI] [PubMed] [Google Scholar]