Abstract
Recombination is a crucial component of evolution and breeding, producing new genetic combinations on which selection can act. Rates of recombination vary tremendously, not only between species but also within species and for specific chromosomal segments. In this study, by examining recombination events captured in recombinant inbred mapping populations previously created for maize, wheat, Arabidopsis, and mouse, we demonstrate that substantial variation exists for genomewide crossover rates in both outcrossed and inbred plant and animal species. We also identify quantitative trait loci (QTL) that control this variation. The method that we developed and employed here holds promise for elucidating factors that regulate meiotic recombination and for creation of hyperrecombinogenic lines, which can help overcome limited recombination that hampers breeding progress.
ALTHOUGH natural selection is a powerful evolutionary process, it utilizes only the existing variation present in a population. Recombination of alleles is required to efficiently evolve new genetic varieties. Not surprisingly, theoretical predictions (Otto and Michalakis 1998) and empirical studies (Saleem et al. 2001) indicate that populations experiencing directional or strong selection pressures evolve increased recombination rates. Similarly to the natural evolutionary processes, combining many positive alleles into a single germplasm is the main objective of plant and animal breeding. The stacking of the favorable alleles is limited by the time and the number of meioses required to recombine numerous alleles from multiple parents. Consequently, a better understanding of the factors controlling recombination holds numerous implications for both academic and applied realms.
To date, many of the genes involved in meiotic recombination have been identified and the mechanistic basis of recombination have begun to emerge (Krogh and Symington 2004; Cohen et al. 2006; Li and Ma 2006). However, the mechanisms that regulate recombination are poorly understood. Particularly, little is known about the control of genomewide recombination rates. Variation in recombination rates has been documented both within and between species, as well as between particular chromosomal regions (Rees 1961; Säll 1990; Beavis and Grant 1991; Tulsieram et al. 1992; Fatmi et al. 1993; Korol et al. 1994; Williams et al. 1995; Sanchez-Moran et al. 2002; Anderson et al. 2003; De Massy 2003; Myers et al. 2005; Yandeau-Nelson et al. 2006). A minimum of one obligatory crossover per chromosome, or chromosome arm, occurs during meiosis as a requirement for proper chromosome segregation (Pardo-Manuel De Villena and Sapienza 2001). However, factors that control whether just this one or multiple crossovers occur per chromosome are poorly understood. Even though the idea that recombination frequencies can be genetically dissected, as any other quantitative trait, was first proposed long ago (Rasmusson 1927), to our knowledge, no quantitative trait loci (QTL) affecting recombination rates have been reported in any species.
In this study, we applied a quantitative genetics approach to identify factors controlling meiotic recombination frequencies. We developed a simple and straightforward method of using genotyped recombinant inbred line (RIL) mapping populations, which are available now for a large number of species, as sources of data on recombination frequencies. Subsequently, we used the crossover numbers as a quantitative trait. Utilizing standard QTL mapping approaches, we were able to identify genomic regions that control genomewide recombination rates in three plant and one animal species.
CONCEPT
Our approach to identify quantitative trait loci influencing global meiotic recombination frequencies utilizes recombinant inbred (RI) mapping populations that have been developed for many plants and animals and where segregation of genes influencing the recombination frequency across the entire genome can be observed.
RI line populations are standard tools for gene mapping and are made by crossing two homozygous parents and then selfing or sib mating the progeny for several generations without selection. When two homozygous parents differing in alleles of genes influencing the global recombination frequency are crossed to generate an RI population they will give rise to an F2 progeny segregating for those genes. The F2 individuals themselves are not informative for the global recombination frequency, as the gametes that produce an F2 individual were produced in identical F1 individuals and therefore all experience the same global recombination factors. However, when F2 individuals are selfed or intercrossed to produce RI lines, genetic differences in recombination frequency are segregating and become fixed in individual lines. The resultant RI lines differ in their global recombination frequencies and, consequently, in the number of recombinations accumulated during their creation.
Molecular marker genotypes of individual RI lines can be used to measure the number of recombination events that accumulated during their creation. Subsequently, the number of recombinations can be treated as a quantitative trait using standard QTL mapping methods to identify the controllers of global recombination frequency. This approach is applicable to a wide range of important plant and animal species for which RI mapping populations are readily available.
There are some limitations to this approach:
Because the number of recombinations is analyzed after several generations of selfing, or intercrossing, less than perfect linkage disequilibrium (LD) is present between the functional loci and the genomic evidence of recombination. In every generation, meiosis takes place and the QTL could segregate away from the linked markers. Consequently, all estimates of effects are likely to be substantial underestimates. This also reduces the statistical power to detect the recombination QTL.
The numbers of recombination events accumulated in the RI lines are sums of meioses in males and females, which may differ in their recombination frequencies. However, because all RI lines have had to go through an equal number of male and female meioses, differences in male and female recombination rates do not affect the ability to detect recombination QTL or lead to artifacts.
Multiple crossover events occurring between the same two markers in different generations will be underscored.
Not all recombination events are scorable. Of the events generated during the formation of F2 gametes, 50% are scorable because the F2 individuals are 50% homozygous. With the progressing inbreeding, the fraction of scorable events declines rapidly. However, as long as all RI lines genotyped are the same generation, these “silent crossovers” do not constitute a problem because the process is systematic.
Finally, because the majority of scorable recombination events take place in early generations during the formation of RI lines, when they are relatively heterozygous, the eventual genotype may not always accurately reflect the recombination history of the line.
However, despite these issues, the approach we propose here requires considerably less effort than other methods of measuring crossover rates as it relies on mapping data that already exist in a large number of species.
Correct marker order on the genetic map is critical for counting crossovers. Misplaced markers can disrupt scoring by generating false additional recombination events and leading to a decrease in the detection power. Therefore, we directed special attention to excluding potentially misplaced markers from the analysis (see materials and methods).
The production of the RI populations over several generations bears the risk of contamination caused by unwanted outcrossing among the RIs, which would cause an increase in the number of recombination events counted for the affected lines. Such accidental outcrossing would increase the variance but would not, in general, interfere with the analysis, unless the frequency of outcrossing depends on the genetics of the lines and was the dominant cause of variation in recombination. In general, accidental outcrossing events would dramatically increase the number of recombinations, and these lines might appear as outliers. We have tested whether our QTL mapping results are robust by repeating the analysis, omitting potential outliers showing an increased number of recombination events.
Although not demonstrated here, global recombination QTL could also be mapped in multiple heterozygous families by association mapping (e.g., in humans).
MATERIALS AND METHODS
QTL mapping:
In Arabidopsis thaliana we used the following data sets: a Landsberg (Ler) × Cape Verde Islands (Cvi) core map data set (http://www.dpw.wau.nl/natural/resources/populations/CVI/) (Alonso-Blanco et al. 1998) comprising 162 recombinant inbred lines advanced to the F8 generation analyzed for 99 markers; Ler × Columbia (Col) (http://arabidopsis.info/new_ri_map.html) (Lister and Dean 1993), including 101 RI lines and 261 framework markers that were reduced to 95 evenly distributed markers; and Ler × Kas-2, Ler × An-1, and Ler × Kond recombinant inbred line populations (http://www.genetics.org/supplemental/) (El-Lithy et al. 2006) in the F9 generation consisting of 164, 120, and 121 lines, respectively, analyzed for 77, 64, and 75 markers, respectively.
For mouse, the LXS panel of recombinant inbred strains (Williams et al. 2004), which is the largest well-genotyped RI lines sample available in mice, consisting of 77 lines (sib mating for 22 generations) genotyped with 4826 SNPs, was used (SNPs from Build 34, Wellcome–CTC Mouse Strain SNP Genotype Set, http://www.well.ox.ac.uk/mouse/INBREDS). Markers rs3673049 and rs4223605 were excluded as they appeared to provide erroneous data.
For maize, we used the intermated B73 × Mo17 (IBM) population (Lee et al. 2002) of recombinant inbred lines obtained by four generations of intermating among F2 plants before selfing. A data set consisting of 2176 markers analyzed in 302 lines was downloaded from the MaizeGDB database (http://maizegdb.org/ibm302scores.html, accessed on May 23, 2005).
For wheat, we used data from the International Triticeae Mapping Initiative (ITMI) W7984 × Opata 85 recombinant inbred line population (Song et al. 2005) (wheat maps: Synthetic × Opata, BARC, http://wheat.pw.usda.gov/graingenes/) consisting of 1475 markers analyzed in 115 lines.
In all data sets markers and lines with >20% missing values were excluded from the analysis. Since marker order is crucial for counting crossovers, we extracted a framework map from the maize and wheat data sets comprising a huge number of markers. This was done by selecting markers with a least number of missing values at distances of ∼15 and 5 cM in maize and wheat, respectively. Using MapMaker's (Lander et al. 1987; Lincoln et al. 1993) “ripple” command (5 loci, LOD 3.0), the stability of the marker order was verified. Selected markers were replaced by new markers until the ripple command indicated a stable order, until marker intervals did not exceed 20 cM, where possible, and until no conflicts showing a high LOD compared to the overall marker order were present in the three-point linkage data. Finally, the average marker distances were 12.6 and 14.5 cM for maize and wheat, respectively. For chromosome 7A in wheat no stable marker order could be found. The numbers of markers and lines finally used after these clean-up steps are given in Table 1.
TABLE 1.
Arabidopsis, mouse, maize, and wheat RI populations analyzed for genomewide recombination events and the QTL detected for this trait
| Genomewide recombination events
|
QTL detected
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RI population | No. of RI lines | No. of markers | Mean | Standard deviation | Min–max | Chromosome | LOD | r2 (%) | Effecta | Position/flanking markers |
| Arabidopsis thaliana Ler × Cvi | 162 | 99 | 8.87 | 2.97 | 3–17 | 1 | 4.95 | 15.1 | −1.86 | CD.173L/175C-Col; GH.127L-Col/ADH |
| A. thaliana Ler × Col | 95 | 95 | 7.48 | 2.54 | 3–14 | None | ||||
| A. thaliana Ler × Kas-2 | 142 | 74 | 7.06 | 2.86 | 1–18 | None | ||||
| A. thaliana Ler × An-1 | 112 | 64 | 6.30 | 2.30 | 0–13 | 1 | 2.84 | 9.7 | −0.73 | nga128; SNP301 |
| A. thaliana | 115 | 73 | 6.23 | 2.24 | 2–14 | 1 | 2.48 | 6.2 | 0.88 | At SNP110 |
| Ler × Kond | 2 | 2.69 | 6.8 | −0.61 | At SNP233 | |||||
| 3 | 3.28 | 8.2 | 1.05 | At CHIB | ||||||
| 5 | 2.71 | 7.3 | −0.80 | SNP236; SNP193 | ||||||
| Mouse | 77 | 4826 | 45.35 | 6.59 | 33–58 | 1 | 3.12 | 11.3 | −4.7 | At rs13475815 |
| LXS | 13 | 3.96 | 15.5 | −5.4 | At rs13481798 | |||||
| Maize | 285 | 399 | 66.68 | 10.77 | 44–112 | 3 | 4.31 | 8.3 | −3.31 | lim66; mmp79 |
| B73 × Mo17 (IBM) | 3 | 5.03 | 7.1 | 2.88 | umc1973; umc1539 | |||||
| Wheat W7984 × Opata 85 | 113 | 248 | 39.17 | 6.78 | 25–56 | 3B | 3.98 | 12.5 | −2.51 | Xtam61; Xpsr689 |
Positive and negative effect means the allele increasing the number of recombinations coming from the first and the second parent, respectively.
To determine the total number of crossovers for each individual RI line, we compared the alleles present at adjacent markers, which were ordered according to their map position. A difference in parental origin for adjacent markers was counted as a recombination event that happened during the generation of the respective RI line.
We used the total number of recombinations per RI line to map QTL for global recombination frequency, applying the composite-interval mapping (CIM) method of QTL Cartographer (Wang et al. 2005) (model 6). For a genomewide significance level of 0.05 the LOD thresholds were determined via 1000 permutations: 2.6 for Ler × Cvi, 2.5 for Ler × Col, 2.4 for Ler × Kas-2, 2.5 for Ler × An-1, 2.45 for Ler × Kond, 3.2 for maize, and 3.3 for wheat. For the Arabidopsis populations, the parameters chosen within the standard model (model 6) other than the defaults were 10 control markers and a window size of 5 cM. In mice, given the density of markers, CIM was not used. Rather, stepwise regression was used to identify the most significant markers.
Sequencing of MEI1:
A 9-kb region surrounding the gene MEI1 was sequenced in three distinct Arabidopsis lines (Col, Ler, and Cvi), using >20 overlapping primer sets with amplicons of 600–1000 bp each. Each amplicon was amplified by PCR using Sigma (St. Louis) Jumpstart Red Taq. Using a Dyad theromcycler (MJ Research Watertown, MA), they were denatured at 94° for 3 min and then denatured for 30 cycles at 94° for 1 min, annealed at 59° for 1 min, and extended at 72° for 1.5 min, with a final extension of 72° for 10 min. The amplicons were checked on a 2% agarose gel and then cleaned up via SAP and exonuclease 1 digestion and an ethanol plus MgCl2 precipitation. Samples were then sequenced using the ABI BigDye Terminator3 system and read on an ABI 3730 sequencer. Contig alignments were created using PHRED and PHRAP software, as well as manual alignment and contig joining within Biolign (Tom Hall; http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
Immunolocalization of MLH1:
Arabidopsis plants were grown in a growth chamber at a 16 hr/8 hr day/night regime. Immature flowers were collected and fixed in a buffer containing 4% formaldehyde (Pawlowski et al. 2003). Anthers at appropriate stages of meiosis were dissected from fixed flowers. Sample preparation and immunolocalization procedures were performed as described previously (Pawlowski et al. 2003). Polyclonal antibodies produced in rabbits against the Arabidopsis MLH1 protein were used at a dilution of 1:1000. Three-dimensional stacks of images were collected using a DeltaVision RT restoration microscopy workstation (Applied Precision, Issaquah, WA) with optical sections 150 nm apart, subjected to deconvolution, and analyzed using the SoftWoRx software (Applied Precision). MLH1 protein foci were counted manually in the three-dimensional image stacks.
RESULTS
Distribution of crossover events:
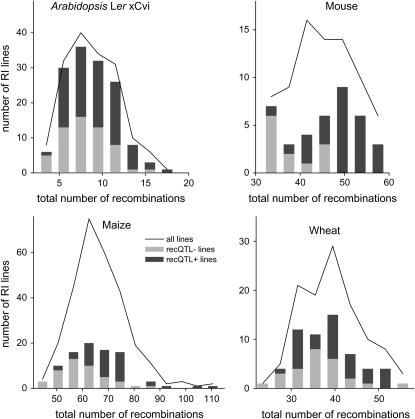
We measured the frequency of recombination by counting breakpoints between stretches of marker alleles from one parent and the other parent in the RI mapping data with markers ordered according to their map position. Using the total number of crossovers accumulated in individual lines in RI populations of maize, wheat, A. thaliana, and mouse, we revealed substantial variation existing for the genomewide recombination frequency in all four species. The total number of recombinations showed a typical distribution of a quantitative trait (Figure 1, Table 1). The mean numbers of crossovers (Table 1) are quite different for each of the species. However, it would be difficult to draw conclusions from these between-species differences given the different number and structure of chromosomes in each species as well as the different history of the RI populations, which these differences most likely reflect. Compared to Arabidopsis, mouse, and wheat, additional recombination events occurred during intermating to produce the maize RI lines.
Figure 1.—
Variation of the genomewide total number of recombinations of individual RI lines for Arabidopsis, mouse, maize, and wheat. In all graphs the solid line (all lines) indicates the distribution of the entire RI population. The RI lines were also classified according to their genotype at the QTL and the flanking markers. The “recQTL+” lines carry the alleles conferring an increase in the number of recombinations at all considered QTL (one single QTL for Arabidopsis and wheat, respectively, and two QTL for maize and mouse, respectively). The “recQTL−” lines contain the alleles conferring a decrease in number of recombinations.
QTL mapping:
By QTL mapping of the total number of crossovers, we detected significant QTL in all four species (Figure 2, Table 1). In A. thaliana, we found significant QTL in three of the five populations analyzed. A QTL on chromosome 1 was present in all three populations. The positions of the flanking markers in the Arabidopsis sequence (http://www.arabidopsis.org/) revealed that the QTL composed the same chromosomal region in Ler × Cvi and Ler × Kond, but a different region in Ler × An-1.
Figure 2.—
QTL for total number of recombinations in Arabidopsis, mouse, maize, and wheat. LOD curves for chromosomes carrying the significant QTL: a single QTL on chromosome 1 in Arabidopsis (dotted line: analysis omitting recombinations on chromosome 1), QTL on chromosomes 1 and 13 in mouse, two QTL on chromosome 3 in maize, and a single QTL on chromosome 3B in wheat. The horizontal lines show the experiment-specific threshold values (genomewide significance level of 0.05) estimated by permutation tests in QTL Cartographer for Arabidopsis, maize, and wheat. For these species the positions of the markers used in the QTL analysis are given as vertical marks above the x-axis. In mouse the QTL on the other chromosome was fixed in stepwise regression analysis.
Location of a recombination QTL may reflect a position of a gene, whose product regulates recombination frequency, or may indicate the presence of an unusually strong recombination hotspot. To separate between cis and trans effects of the detected QTL, we subtracted the recombination events accumulated on the chromosomes carrying the QTL from the total genomewide crossover number and repeated the QTL analysis. In the Arabidopsis Ler × Cvi population, chromosome 1 exhibited 2.4 crossovers, the highest mean number of recombinations per chromosome of all five chromosomes (overall mean 1.8). When we excluded the crossovers on this chromosome from the analysis, the chromosome 1 QTL was no longer significant (Figure 2). Moreover, no significant QTL could be detected on any of the chromosomes. A similar situation was found in Ler × An-1. The QTL on chromosome 1 was no longer significant when the recombinations on this chromosome were excluded from the QTL mapping (chromosome 1 again had the most events). However, in the Arabidopsis Ler × Kond population, omitting recombination events detected on the QTL-carrying chromosomes had different consequences. For chromosomes 1 and 5, the LOD of the respective QTL increased, for chromosome 2 the LOD only slightly changed, and for chromosome 3 the LOD decreased below the threshold. In addition, the LOD profile for the remaining chromosomes also changed, resulting in disappearance of some QTL and appearance of new QTL. Overall, this suggests that some of the QTL are trans-acting factors, but some may be cis-acting. However, with Arabidopsis's small genetic map size, insufficient statistical power could also result in the apparent cis-like results.
We also looked for possible cis effects in the mouse, maize, and wheat data. In the mouse, we excluded crossovers on chromosomes 1 and 13 where significant QTL were located. In each case, the QTL became slightly less significant when the effects of their chromosome were excluded, but the complete model became more significant. In maize and wheat, only minor differences were seen when excluding the recombinations on the QTL-carrying chromosomes.
Candidate genes:
In mouse and Arabidopsis, genome sequence was available to suggest candidate genes beneath the broad QTL peak. In Arabidopsis, the QTL on chromosome 1 in the Ler × Cvi population included the MEI1 gene (He and Mascarenhas 1998; Grelon et al. 2003). A 9-kb region surrounding the gene MEI1 was sequenced in three distinct Arabidopsis lines (Col, Ler, and Cvi). In all, there were 13 polymorphisms, indicating that this is a region of low diversity. Of these 13 polymorphisms, 11 either were in noncoding regions or were silent substitutions. From the two nonsilent polymorphisms, one, between (Col, Cvi) > (Ler), leads to a threonine-to-isoleucine substitution. This residue is in a region that is not conserved among sequenced plants. The other polymorphism, between (Col, Ler) > (Cvi) leads to a serine-to-phenylalanine change. The serine residue is in a SKK motif, which is fairly conserved across the sequenced plants, except for rice.
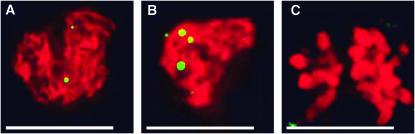
We then used a cytological approach to evaluate two RI lines with double crossovers that dissected the MEI1 region. To measure crossover frequencies in these lines, we counted chromosomal foci of the MLH1 recombination protein during meiosis. MLH1 is required for formation of the interference-dependent type I crossovers in fungi, mammals, and plants (Argueso et al. 2002; Higgins et al. 2004; Kolas et al. 2005) and localizes to the sites of the forming crossovers. In Arabidopsis, type I crossovers constitute at least 85% of all crossovers (Higgins et al. 2004). Numbers of MLH1 foci are routinely used as a proxy for global crossover rates in the mouse (Anderson et al. 1999; Koehler et al. 2002). We quantified MLH1 foci in meiocytes at late zygotene/early pachytene, when the number of foci is the highest (Figure 3; J. M. Szymaniak and W. P. Pawlowski, unpublished data). We found that the MLH1 foci numbers were significantly different between the two lines: in CVL44, we detected 9.4 ± 0.7 (mean ± SE) foci per nucleus (n = 17) and in CVL46, 12.6 ± 1.1 foci per nucleus (n = 14). These data corroborate the analysis of crossover breakpoints in revealing significant within-species variation in crossover frequencies. On the other hand, they did not agree with the MEI1 predictions. Consequently, it is likely that the QTL on Arabidopsis chromosome 1 does not include MEI1.
Figure 3.—
MLH1 immunostaining in Arabidopsis meiocytes in zygotene (A), pachytene (B), and diplotene (C). 4′,6-Diamidino-2-phenylindole (DAPI)-stained chromatin is shown in red, and MLH1 protein is in green. Images represent fragments of three-dimensional nuclei and are flat projections from several consecutive optical sections. Bar, 5 μm.
DISCUSSION
Our approach to score meiotic crossovers documents substantial within-species variation for genomewide recombination rates in both outcrossed and inbred plant and animal species and shows that QTL that underlie this variation can be identified. Such variation is likely to be ubiquitous, especially considering that the statistical power to map this trait is not high.
Our data corroborate previous studies of a number of species of plants and animals, which reported existence of strong genetic background effects on the frequency of meiotic recombination (Roberts and Roberts 1921; Rees 1961; Säll 1990; Williams et al. 1995; Koehler et al. 2002; Sanchez-Moran et al. 2002; Anderson et al. 2003). Although most of these studies considered crossover frequencies in specific chromosome intervals, two reports in plants, one in maize (Anderson et al. 2003) and one in Arabidopsis (Sanchez-Moran et al. 2002), indicated significant differences in global recombination rates among several different genotypes. The eventual cloning of QTL underlying the natural variation in global recombination rates will provide insight into the molecular mechanisms regulating meiotic recombination.
Understanding the recombination rate variation will have obvious practical applications by facilitating construction of highly recombinogenic lines. Such lines will be of major interest for plant and animal breeding, where particular traits are incorporated within a population by repeatedly crossing individuals with beneficial alleles—often a long and costly process. It will also accelerate the removal of linkage drag during the introgression of valuable genes from genetic resources. Identifying recombination QTL and, later, the recombination regulator genes that underlie them, will lead to these goals by providing targets for genetic engineering efforts. In addition, naturally occurring alleles that convey increased recombination frequencies could be used directly to produce hyperrecombinogenic lines. Although the gains in the latter case will most likely be moderate, increasing recombination frequencies severalfold may not always be desirable because high recombination in a long run could be detrimental to genome stability. Increasing meiotic recombination frequencies will also aid development of methods for improving genetic maps and positional cloning techniques.
Both cis- and trans-acting factors are known to affect crossover frequencies (Dooner and Martinez-Ferez 1997; Timmermans et al. 1997; Gerton et al. 2000; Yao et al. 2002; De Massy 2003; Myers et al. 2005; Przeworski 2005; Yandeau-Nelson et al. 2006). However, the majority of the QTL that we detected appear to be trans-acting, and the few that may be cis-acting may be the result of lack of statistical power. It is possible that this reflects specificity of the RIL populations that we selected for this study. Alternatively, cis-acting factors could be more important for localization of crossovers in specific chromosomal intervals, while the global number of crossover events may be mostly regulated by trans-acting proteins. There are also likely to be differences in male and female controllers of meioses, and while this approach is most likely to map consistent QTL in both sexes, other mapping designs could be used to differentiate the effects.
The number of QTL that we were able to detect in the individual populations was relatively low, presumably because of the low power of our approach. Despite this obvious lack of power, we still identified QTL with large effects explaining up to 15% of the observed variation in total recombination frequency, suggesting that the regions we revealed have huge impacts on the control of genomewide recombination rates. Moreover, we did no special selection of the RI populations used; i.e., we did not have any information if the parents were different for the genes we were interested in. However, we were able to detect significant QTL in six of the eight populations analyzed, suggesting that large natural variation exists for the genes controlling genomewide recombination rates. The lack of power is most likely the result of a small sample size: a population of <300 RI lines shows only the results of a few hundred meioses.
The lack of power problem is likely to be resolved in the next few years. “Mapping as you go” (Podlich et al. 2004) approaches are becoming common for many crops and animals, and they involve genotyping at numerous steps in the breeding process. Across an entire breeding program, where tens of thousands of individuals are being genotyped per year, it should be possible to map with much higher power and resolution. Additionally, in the next few years, the public maize nested association-mapping population with 7000 RI lines and the mouse complex trait consortium panels with 1000 RI lines should provide unprecedented resolution of these QTL.
Acknowledgments
The authors thank Christopher Franklin (University of Birmingham, United Kingdom) for the gift of the anti-MLH1 antibody and Teresa Pawlowska for comments on the manuscript. This work was supported by the National Science Foundation (DBI-0321467), the United States Department of Agriculture–Agricultural Research Services, Cornell University, Binational Agricultural Research and Development Fund (IS-3723-05), and a travel grant from Leibniz Universität Hannover.
References
- Alonso-Blanco, C., A. J. M. Peeters, M. Koornneef, C. Lister, C. Dean et al., 1998. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 14: 259–271. [DOI] [PubMed] [Google Scholar]
- Anderson, L. K., A. Reeves, L. M. Webb and T. Ashley, 1999. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151: 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, L. K., G. G. Doyle, B. Brigham, J. Carter, K. D. Hooker et al., 2003. High-resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics 165: 849–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso, J. L., D. Smith, J. Yi, M. Waase, S. Sarin et al., 2002. Analysis of conditional mutations in the Saccharomyces cerevisiae MLH1 gene in mismatch repair and in meiotic crossing over. Genetics 160: 909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis, W. D., and D. Grant, 1991. A linkage map based on information from four F2 populations of maize (Zea mays L.). Theor. Appl. Genet. 82: 636–644. [DOI] [PubMed] [Google Scholar]
- Cohen, P. E., S. E. Pollack and J. W. Pollard, 2006. Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endocr. Rev. 27: 398–426. [DOI] [PubMed] [Google Scholar]
- De Massy, B., 2003. Distribution of meiotic recombination sites. Trends Genet. 19: 514–522. [DOI] [PubMed] [Google Scholar]
- Dooner, H. K., and I. M. Martinez-Ferez, 1997. Recombination occurs uniformly within the bronze gene, a meiotic recombination hotspot in the maize genome. Plant Cell 9: 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Lithy, M. E., L. Bentsink, C. J. Hanhart, G. J. Ruys, D. Rovito et al., 2006. New Arabidopsis recombinant inbred line populations genotyped using SNPWave and their use for mapping flowering-time quantitative trait loci. Genetics 172: 1867–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatmi, A., C. G. Poneleit and T. W. Pfeiffer, 1993. Variability of recombination frequencies in the Iowa Stiff Stalk Synthetic (Zea mays L). Theor. Appl. Genet. 86: 859–866. [DOI] [PubMed] [Google Scholar]
- Gerton, J. L., J. DeRisi, R. Shroff, M. Lichten, P. O. Brown et al., 2000. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 11383–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon, M., G. Gendrot, D. Vezon and G. Pelletier, 2003. The Arabidopsis MEI1 gene encodes a protein with five BRCT domains that is involved in meiosis-specific DNA repair events independent of SPO11-induced DSBs. Plant J. 35: 465–475. [DOI] [PubMed] [Google Scholar]
- He, C., and J. P. Mascarenhas, 1998. MEI1, an Arabidopsis gene required for male meiosis: isolation and characterization. Sex. Plant Reprod. 11: 199–207. [Google Scholar]
- Higgins, J. D., S. J. Armstrong, F. C. H. Franklin and G. H. Jones, 2004. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 18: 2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler, K. E., J. P. Cherry, A. Lynn, P. A. Hunt and T. J. Hassold, 2002. Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. Genetics 162: 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas, N. K., A. Svetlanov, M. L. Lenzi, F. P. Macaluso, S. M. Lipkin et al., 2005. Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J. Cell Biol. 171: 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korol, A. B., I. A. Preygel and S. I. Preygel, 1994. Recombination Variability and Evolution. Chapman & Hall, London.
- Krogh, B. O., and L. S. Symington, 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38: 233–271. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Lee, M., N. Sharopova, W. D. Beavis, D. Grant, M. Katt et al., 2002. Expanding the genetic map of maize with the intermated B73 x Mo17 (IBM) population. Plant Mol. Biol. 48: 453–461. [DOI] [PubMed] [Google Scholar]
- Li, W. X., and H. Ma, 2006. Double-stranded DNA breaks and gene functions in recombination and meiosis. Cell Res. 16: 402–412. [DOI] [PubMed] [Google Scholar]
- Lincoln, S. E., M. J. Daly and E. S. Lander, 1993. Constructing genetic linkage maps with MAPMAKER/EXP version 3.0. Whitehead Institute for Biomedical Research Technical Report. Whitehead Institute, Cambridge, MA.
- Lister, C., and C. Dean, 1993. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 4: 745–750. [DOI] [PubMed] [Google Scholar]
- Myers, S., L. Bottolo, C. Freeman, G. McVean and P. Donnelly, 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310: 321–324. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., and Y. Michalakis, 1998. The evolution of recombination in changing environments. Trends Ecol. Evol. 13: 145–151. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel De Villena, F., and C. Sapienza, 2001. Recombination is proportional to the number of chromosome arms in mammals. Mamm. Genome 12: 318–322. [DOI] [PubMed] [Google Scholar]
- Pawlowski, W. P., I. N. Golubovskaya and W. Z. Cande, 2003. Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition. Plant Cell 15: 1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlich, D. W., C. R. Winkler and M. Cooper, 2004. Mapping as you go: an effective approach for marker-assisted selection of complex traits. Crop Sci. 44: 1560–1571. [Google Scholar]
- Przeworski, M., 2005. Motivating hotspots. Science 310: 247–248. [DOI] [PubMed] [Google Scholar]
- Rasmusson, J., 1927. Genetically changed linkage values in Pisum. Hereditas 10: 1–152. [Google Scholar]
- Rees, H., 1961. Genotypic control of chromosome form and behaviour. Bot. Rev. 27: 288–318. [Google Scholar]
- Roberts, F. A., and E. Roberts, 1921. Studies on crossing over. I. The effect of selection on crossover values. J. Exp. Zool. 32: 333–354. [Google Scholar]
- Saleem, M., B. C. Lamb and E. Nevo, 2001. Inherited differences in crossing over and gene conversion frequencies between wild strains of Sordaria fimicola from “Evolution Canyon.” Genetics 159: 1573–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Säll, T., 1990. Genetic control of recombination in barley: II. Variation in linkage between marker genes. Hereditas 112: 171–178. [Google Scholar]
- Sanchez-Moran, E., S. J. Armstrong, J. L. Santos, F. C. H. Franklin and G. H. Jones, 2002. Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics 162: 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Q. J., J. R. Shi, S. Singh, E. W. Fickus, J. M. Costa et al., 2005. Development and mapping of microsatellite (SSR) markers in wheat. Theor. Appl. Genet. 110: 550–560. [DOI] [PubMed] [Google Scholar]
- Timmermans, M. C. P., O. P. Das, J. M. Bradeen and J. Messing, 1997. Region-specific cis- and trans-acting factors contribute to genetic variability in meiotic recombination in maize. Genetics 146: 1101–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulsieram, L., W. A. Compton, R. Morris, M. Thomas-Compton and K. Eskridge, 1992. Analysis of genetic recombination in maize populations using molecular markers. Theor. Appl. Genet. 84: 65–72. [DOI] [PubMed] [Google Scholar]
- Wang, S., C. J. Basten and Z.-B. Zeng, 2005. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm.
- Williams, C. G., M. M. Goodman and C. W. Stuber, 1995. Comparative recombination distances among Zea mays L. inbreds, wide crosses and interspecific hybrids. Genetics 141: 1573–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R. W., B. Bennett, L. Lu, J. Gu, J. C. DeFries et al., 2004. Genetic structure of the LXS panel of recombinant inbred mouse strains: a powerful resource for complex trait analysis. Mamm. Genome 15: 637–647. [DOI] [PubMed] [Google Scholar]
- Yandeau-Nelson, M. D., B. J. Nikolau and P. S. Schnable, 2006. Effects of trans-acting genetic modifiers on meiotic recombination across the a1-sh2 interval of maize. Genetics 174: 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, H., Q. Zhou, J. Li, H. Smith, M. Yandeau et al., 2002. Molecular characterization of meiotic recombination across the 140-kb multigenic a1-sh2 interval of maize. Proc. Natl. Acad. Sci. USA 99: 6157–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]