Abstract
A large portion of the annotated genes in Drosophila melanogaster show sex-biased expression, indicating that sex and reproduction-related genes (SRR genes) represent an appreciable component of the genome. Previous studies, in which subsets of genes were compared among few Drosophila species, have found that SRR genes exhibit unusual evolutionary patterns. Here, we have used the newly released genome sequences from 12 Drosophila species, coupled to a larger set of SRR genes, to comprehensively test the generality of these patterns. Among 2505 SRR genes examined, including ESTs with biased expression in reproductive tissues and genes characterized as involved in gametogenesis, we find that a relatively high proportion of SRR genes have experienced accelerated divergence throughout the genus Drosophila. Several testis-specific genes, male seminal fluid proteins (SFPs), and spermatogenesis genes show lineage-specific bursts of accelerated evolution and positive selection. SFP genes also show evidence of lineage-specific gene loss and/or gain. These results bring us closer to understanding the details of the evolutionary dynamics of SRR genes with respect to species divergence.
THE spectacular sexual dimorphisms observed in many species of insects, birds, and mammals were originally explained by Charles Darwin to be the result of competition for mates (i.e., sexual selection), which drives the evolution of sex-specific traits (Darwin 1871). Evolutionary biologists have since studied sexual dimorphisms, primarily at the phenotypic level, across a wide variety of species (Eberhard 1985; Andersson 1994; Möller 1994; Houde 1997; Markow 2002). With the advent of molecular techniques in the 1980s and their application in examining gene evolution among species, a pattern of rapid divergence of genes with sex-specific expression began to emerge (Coulthart and Singh 1988; Thomas and Singh 1992; Civetta and Singh 1995). We have since learned that sexual selection can influence not only the evolution of morphological sexual dimorphism but also the patterns and rates of molecular evolution and speciation (Civetta and Singh 1998a; Singh and Kulathinal 2000; Swanson et al. 2001; Swanson and Vacquier 2002; Coyne and Orr 2004). In Drosophila, sexual dimorphism occurs at morphological (e.g., body size, genitalia, abdomen pigmentation, sex combs in males but not females), behavioral (e.g., courtship and postmating behaviors), and molecular levels (e.g., yolk proteins and seminal fluid proteins) (see Markow 2002 for review). Interest in the evolution of sexual dimorphism at the molecular level and its consequences has received renewed attention with the recent reports that over half of the genes in Drosophila melanogaster and D. simulans exhibit sexually dimorphic expression (Ranz et al. 2003; Parisi et al. 2004). Within these sex-biased genes, those with male-biased expression tend to show greater difference in expression levels than do female-biased genes or non-sex-biased genes in both within- and between-species comparisons (Meiklejohn et al. 2003; Parisi et al. 2003, 2004; Ranz et al. 2003). In addition, rates of sequence evolution of sex and reproduction-related (SRR) and non-SRR genes differ quite dramatically: male and female SRR genes evolve faster than non-SRR genes (Begun et al. 2000; Swanson et al. 2001; Zhang et al. 2004; Jagadeeshan and Singh 2005; Mueller et al. 2005; Proschel et al. 2006; Lawniczak and Begun 2007; see Swanson and Vacquier 2002 for review).
Despite the importance of these conclusions, their generality remains questionable, as previous studies examined sequence divergence within a limited set of genes in a small number of species. Such studies reported that genes expressed in the testis and genes encoding seminal fluid proteins include many that evolve rapidly (Begun et al. 2000, 2006; Swanson et al. 2001; Holloway and Begun 2004; Kern et al. 2004; Zhang et al. 2004; Begun and Lindfors 2005; Mueller et al. 2005; Wagstaff and Begun 2005a; Schully and Hellberg 2006). Many female reproductive proteins have also been shown to evolve faster than non-sex-specific proteins (Civetta and Singh 1995; Jansa et al. 2003; Swanson et al. 2004; Jagadeeshan and Singh 2005). Certain genes encoding seminal fluid and testis proteins in D. melanogaster appear to have evolved so rapidly that they lack detectable orthologs in other Drosophila species (Swanson et al. 2001; Mueller et al. 2005; Begun et al. 2006; Levine et al. 2006).
The sequencing, assembly, and subsequent annotation of the 12 Drosophila genomes (Adams et al. 2000; Richards et al. 2005; Drosophila 12 Genomes Consortium 2007) have provided an important new tool for answering evolutionary questions. For example, we stand to learn how sexual systems have changed over time and what genes characterize male and female specialization. Moreover, profiling genetic divergence within the context of gene evolutionary history and function will help elucidate which, among the vast number of rapidly evolving reproductive genes, show adaptive evolution. Several rapidly evolving genes are expressed in more than one tissue (Jagadeeshan and Singh 2005) and it is known that there are differences in the evolutionary rates of tissue-specific and shared genes (Khaitovich et al. 2005). Therefore, it would be interesting to know how specificity, with respect to tissue expression and/or function, influences the rates of evolution of such genes.
In this study, we take advantage of the fully sequenced genomes of 12 Drosophila species (Drosophila 12 Genomes Consortium 2007) and of a larger, more comprehensive functional set of gene annotations (including genes that were recently identified through microarray and proteomic approaches) than previously analyzed. Using aligned orthologs from these species, we investigate the nature of divergence of the following categories of SRR genes among 12 species from the genus Drosophila, including 6 species of the melanogaster group (Table 1):
Genes encoding proteins secreted by male accessory glands (Acps), ejaculatory duct, and ejaculatory bulb and all major components of D. melanogaster seminal fluid (hereafter referred to collectively as seminal fluid proteins or SFPs). These proteins are particularly interesting because they are transferred from the male to the female along with sperm during mating and mediate a number of postmating events (for reviews, see Kubli 2003; Chapman and Davies 2004; Wolfner et al. 2005; Wong and Wolfner 2006; Ravi Ram and Wolfner 2007). From an evolutionary perspective, evidence for adaptive evolution has been found at several loci encoding D. melanogaster SFPs (Aguadé et al. 1992; Tsaur and Wu 1997; Aguadé 1999; Begun et al. 2000; Swanson et al. 2001; Mueller et al. 2005). While it has been suggested that a greater proportion of SFP genes may experience positive selection in comparison to nonreproductive genes, the 12 genomes provide the first opportunity for testing this hypothesis on a genomewide scale.
Genes encoding candidate female interactors of male SFPs and sperm proteins, collectively referred to here as female reproductive tract proteins (FRTPs). Due to the obligate interactions between female and male proteins for the success of fertilization, co-evolution of female proteins that interact with rapidly diverging male proteins is expected. Such a phenomenon was proposed to explain the observed evolutionary patterns of some genes in abalone and in mammals (Swanson et al. 2001; 2004; Galindo et al. 2003; Aagaard et al. 2006). In Drosophila, several candidate genes identified as expressed in the female reproductive tract and potentially secreted (Panhuis and Swanson 2006) or induced in the female reproductive tract by mating (Lawniczak and Begun 2004, 2007; McGraw et al. 2004) were observed to evolve rapidly. Adaptive evolution due to interaction with rapidly diverging seminal fluid proteins was invoked in both studies to explain the observed patterns of evolution.
Genes specifically inferred to be involved in gametogenesis according to Gene Ontology (GO) controlled vocabulary. This set of characterized genes, particularly genes regulating sperm development, is of interest because the prevalence of hybrid male sterility among closely related species of Drosophila might be affected by the rapid evolution of male reproductive genes, and spermatogenesis appears to be a target of F1 male hybrid fertility breakdown (Civetta and Singh 1998b; Kulathinal and Singh 1998; Michalak and Noor 2003; Presgraves et al. 2003; Haerty and Singh 2006; Moehring et al. 2007). Evidence of positive selection has been found for genes controlling key transitions during both spermatogenesis and oogenesis (Civetta et al. 2006; Bauer Dumont et al. 2007).
Genes with tissue-specific patterns of gene expression. These genes, annotated according to their presence in various EST libraries (see materials and methods), can be used to test the extent and details of molecular sexual dimorphism in expression (Andrews et al. 2000; Parisi et al. 2004) as well as the evolution of reproductive (testis and ovary) genes relative to presumably nonreproductive (head) genes (Coulthart and Singh 1988; Civetta and Singh 1995; Jagadeeshan and Singh 2005). Previous studies, which have used a limited number of genes, have suggested that the testis transcriptome may have a greater proportion of rapidly evolving genes relative to ovary or head (Civetta and Singh 1995; Jagadeeshan and Singh 2005) and that rates of evolution of testis-expressed genes are far higher than rapidly evolving genes expressed in ovary and head.
TABLE 1.
Sex- and reproduction-related gene classification
| Gene classification | Definition |
|---|---|
| Sex- and reproduction-related (SRR) genes | A broad classification that indicates genes linked, in a broad sense, to sex and/or reproduction. Genes that would fall under the SRR category are listed below. |
| Sex tissue expressed (STE) | Genes that are expressed in male or female reproductive tissues; as listed below. |
| Sex tissue shared | Genes that are expressed in both testis and ovary but not in head. |
| Testis specific | Genes that are expressed only in testis. |
| Ovary specific | Genes that are expressed only in ovary. |
| Testis related | Genes that are expressed in testis as well as in head. |
| Ovary related | Genes that are expressed in ovary as well as in head. |
| Seminal fluid proteins | Genes that are expressed in accessory glands and encoding seminal fluid proteins (SFPs) (includes proteins from ejaculatory bulb and ejaculatory duct also). |
| Female reproductive tract | Genes that are expressed in the female reproductive tract (FRTP). |
| Gametogenesis | Genes that are expressed in and involved in the generation/development of male and/or female gemates. |
| Male reproductive genes | A general classification of all genes with likely male-related function, including testis-specific, testis-related, sex-tissue-shared, SFP, and spermatogenesis genes. |
| Female reproductive genes | A general classification of all genes with likely female-related functions including ovary-specific, ovary-related, sex-tissue-shared, FRTP, and oogenesis genes. |
Not all the genes under these different categories are necessarily and directly involved in sex and reproduction but, due to their tissue of expression, may invariably influence the development and physiology of sex tissues and reproductive biology of the organism. Categorization is based on the expression patterns or functions of genes known in D. melanogaster. Very little is known about gene expression patterns in other Drosophila species and therefore at present we cannot rule out the possibility of change in expression pattern or function of some of these genes in other Drosophila species.
The comprehensive picture that we gain of SRR gene evolution through the analysis of gene orthology across the 12 Drosophila genomes and the study of evolutionary rates in six species of the D. melanogaster group validates previous observations from smaller-scale comparisons and extends them in several ways. We find significantly higher rates of evolution in genes with male-biased expression or male-specific function (SFP, testis specific, and spermatogenesis) relative to female-specific or non-SRR genes. Furthermore, several SFPs and spermatogenesis genes show lineage-specific bursts of accelerated evolution and positive selection. These results provide support for the broad role of sexual selection as an important evolutionary force driving the rapid evolution of the male SRR genome.
MATERIALS AND METHODS
Estimates of divergence and tests of positive selection:
A brief description of alignment methods, calculation of divergence rates, and inference of positive selection for the six sequenced species of the melanogaster group (D. melanogaster, D. simulans, D. sechellia, D. yakuba, D. erecta, and D. ananassae) is available on the AAAwiki website (http://rana.lbl.gov/∼venky/AAA/freeze_20061030/protein_coding_gene/GLEANR/alignment/).
Genes encoding seminal fluid proteins or expressed in the female reproductive tract:
For genes encoding proteins present in the seminal fluid, we focused on a stringently selected set of 68 SFP genes (supplemental Table 1 at http://www.genetics.org/supplemental/), the majority of which encode proteins with a predicted secretion signal sequence, and all of which show male-biased expression with enrichment either in accessory glands (Monsma and Wolfner 1988; Wolfner et al. 1997; Swanson et al. 2001; Holloway and Begun 2004; Mueller et al. 2005; Walker et al. 2006; Chintapalli et al. 2007) or in the ejaculatory duct or bulb (Est-6: Ludwig et al. 1993; Andropin: Samakovlis et al. 1991; Gld: Cavener et al. 1986; PEB-ME: Lung and Wolfner 2001; Dup99B: Saudan et al. 2002). Evolutionary rates of 25 of these 68 seminal protein genes were estimated using the six fully sequenced genomes of the melanogaster species group; orthologs of the other 43 genes either were not identified in one or more species or have undergone gene duplication and were therefore not included. In addition to analyses on this stringently selected set of SFP genes, we examined evolutionary rates for an independent set of 43 potential SFP genes found to have accessory-gland-enriched expression in microarray experiments (supplemental Table 1; Chintapalli et al. 2007; Ravi Ram and Wolfner 2007). These are considered “potential Acps” because their accessory-gland enrichment has not been verified by other means. Of these 43 loci, 18 were detectable in all six species and hence were included among the genes analyzed with PAML.
For female reproductive tract proteins, we analyzed two data sets of genes expressed in somatic tissues of the female reproductive tract: female reproductive tract ESTs (Swanson et al. 2004) and genes identified in microarray and proteomic screens of the female lower reproductive tract (Mack et al. 2006). The total list of FRTP genes (supplemental Table 1 at http://www.genetics.org/supplemental/) consisted of 958 genes; of these, 679 were analyzed using PAML (Drosophila 12 Genomes Consortium 2007; A. M. Larracuente, T. B. Sackton, A. J. Greenberg, A. Wong, N. D. Singh, D. Sturgill, Y. Zhang, B. Oliver and A. G. Clark, unpublished results). This set of genes includes many with non-tissue-specific expression, as well as a number of genes with nonreproductive functions (e.g., housekeeping genes) and as such is not strictly comparable to the SFP data set. Since proteins with extracellular or cell-surface localization are more likely to interact directly with male SFPs and sperm proteins, we divided female reproductive tract genes into those with (246/679) or without (433/679) a predicted secretion signal sequence and/or predicted transmembrane helices for some analyses. A total of 8 genes were found to be characterized in both SFP and FRTP data sets.
Genes involved in spermatogenesis and oogenesis:
A total of 417 genes involved in gametogenesis were identified on the basis of gene ontology searches via TermLink (FlyBase FB_2006_01). Of these, 102 are predicted to play a role during the development and/or maturation of sperm and are collectively referred to as spermatogenesis genes (of which 29 are included in our testis EST data set). A total of 292 are genes expected to function during the development of eggs and are collectively referred to as oogenesis genes (of which 30 are included in our ovary EST data set). Twenty-three genes are predicted to be part of both processes. Divergence data in the six D. melanogaster group species were available for a total of 73 spermatogenesis and 226 oogenesis genes among which 21 are found in both processes.
Genes expressed in testis, ovary, and/or head:
A total of 532,583 ESTs from various D. melanogaster cDNA libraries were collected and collated according to their tissue of expression (UniGene, http://www.ncbi.nlm.nih.gov/). Selecting only those sequences that map onto the 13,733 annotated genes, a total of 25,884 ESTs were from a testis library, 6442 ESTs were from an ovary library, and 21,372 ESTs were from a head library, respectively, representing 4439, 2386, and 5036 genes from each tissue (supplemental Table 1 at http://www.genetics.org/supplemental/). Genes were assigned to different categories according to their expression specificity: testis specific, ovary specific, head specific, testis related (testis and head), ovary related (ovary and head), sex tissue shared (ovary and testis), or expressed in all three organs. Using the D. melanogaster genome, a total of 2631 genes were classified as expressed in testis and/or in ovary, including 1741 testis-specific genes, 556 ovary-specific genes, and 334 genes expressed in both organs. Using only genes with orthologs in all six species of the melanogaster group, 5156 genes with tissue EST information (Table 2) were analyzed using PAML (Drosophila 12 Genomes Consortium 2007; A. M. Larracuente, T. B. Sackton, A. J. Greenberg, A. Wong, N. D. Singh, D. Sturgill, Y. Zhang, B. Oliver and A. G. Clark, unpublished results), including 1400 head-specific genes, 297 ovary-specific genes, 1102 testis-specific genes, and 246 genes shared by testis and ovary. Note that testis- or ovary-specific genes are not all necessarily or directly involved in reproduction and sex but do specifically relate to the male or female reproductive organs. Similarly, the head-specific gene set may include genes involved in courtship behavior and other reproductive functions, but will also include other genes with no role in reproduction. Evolutionary rates between the different organ categories were compared using a Tukey HSD test.
TABLE 2.
Genes used in the analysis of six species of the melanogaster group
| Gene classification | No. of genes used |
|---|---|
| Sex tissue expressed | A total of 5156 genes classified on the basis of their site of expression using the EST database (UniGene, supplemental Table 1 at http://www.genetics.org/supplemental/; 1400 head-specific, 397 ovary-specific, 1102 testis-specific, 372 ovary-related, 922 testis-related, 246 sex-tissue-shared, and 717 genes expressed in all three tissues). A total of 889 genes did not match any tissue category. |
| Seminal fluid proteins | A stringently selected set of 25 SFP genes (supplemental Table 1 at http://www.genetics.org/supplemental/) were analyzed (see materialsandmethods). |
| Female reproductive tract proteins | Divided into 246 genes with extracellular localization signals, which are more likely to interact directly with male SFPs and sperm proteins, and 433 genes without a predicted secretion signal sequence and/or predicted transmembrane helices (see materialsandmethods). A total of 8 genes were characterized in SFPs and FRTPs. |
| Gametogenesis | Identified on the basis of Gene Ontology searches via TermLink (FlyBase). |
| Spermatogenesis | Seventy-three genes involved in the development and/or maturation of sperm. |
| Oogenesis | A total of 226 genes involved in the development of eggs. Among the 73 and 226 genes, 21 genes are found to be involved in both spermatogenesis and oogenesis. |
| SRR genes | A total of 2505 genes are classified as SRR genes (testis-specific and ovary-specific genes, genes expressed in both testis and ovary, SFPs, FRTPs, and genes characterized in gametogenesis). A total of 2298 genes were found in only one category (testis/ovary/testis&ovary, SFP/FRTP, gametogenesis), 198 genes in at least two categories, and 8 in the three different categories (testis/ovary/ testis&ovary, SFP/FRTP and gametogenesis, supplemental Table 2 at http://www.genetics.org/supplemental/). |
Moreover, because of the different origins of the data sets used in this study, some overlap between the different categories (SFPs/FRTPs, gametogenesis, and genes expressed in testis, ovary, head) is expected. Among the 2505 genes classified as SRR, 198 are present in at least two classes, and 8 in the three different categories (see Table 2; supplemental Table 2 at http://www.genetics.org/supplemental/).
Orthology of sex and reproduction-related genes:
Due to lack of annotation or gene expression information for most of the genes in sequenced species other than D. melanogaster, we assigned function/expression for each gene on the basis of data available in D. melanogaster.
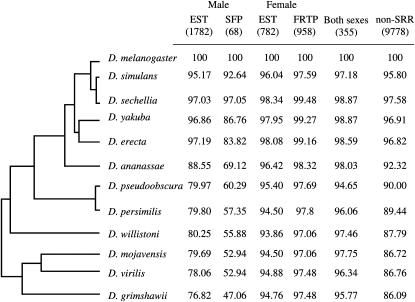
We used a reciprocal best-hit BLAST approach (using an e-value cutoff of 1e-03) to identify putative orthologs for male-biased genes (n = 1782, without SFPs), female-biased genes (n = 782, without FRTPs), and sex-biased genes (genes expressed in both testis and ovary and/or involved in both spermatogenesis and oogenesis; n = 355) (without SFP- or FRTP-encoding genes). Genes were classified according to either of two criteria: (1) the gene was associated with one of the GO terms, spermatogenesis, spermiogenesis (for male reproductive), or oogenesis (for female reproduction) or (2) when blasted against the tissue EST databases, the gene sequence had hits of at least one described EST in either testis or ovary but not in the head. A total of 9921 genes that did not fit these criteria and were not coding for SFPs or FRTPs were classified as non-SRR genes. In addition, we analyzed 68 SFP and 958 FRTP genes. Proportions of genes in the different categories (male, female, SFPs; FRTPs, and non-SRR) in D. grimshawii were compared using a χ2 test as this species is the sequenced species most distant from D. melanogaster.
RESULTS
Gene orthology: high turnover of male SRR genes:
Examining the gain and loss of orthologs across the genus Drosophila is a fitting starting point for understanding the extent of changes that have occurred at the whole-genome level in drosophilids. In a broad-spectrum scan across all 12 sequenced genomes using orthologs identified through a reciprocal best-hit BLAST approach, we compared sets of SRR and non-SRR genes, as defined in materials and methods. Using sequence similarity to male, female, and non-sex-specific EST libraries, a total of 2919 male and/or female-biased genes (see materials and methods) were collated, comprising of 1782 male-biased genes (testis-specific genes and/or genes involved in spermatogenesis), 782 female-biased genes (ovary-specific genes and/or genes involved in oogenesis), and 355 genes expressed in both sexes, in addition to 68 SFPs, 958 FRTPs, and 9921 genes not in these categories (see Table 1 for SRR gene classification; supplemental Table 3 at http://www.genetics.org/supplemental/).
A larger proportion of D. melanogaster testis, spermatogenesis genes, and seminal fluid protein-encoding genes lack detectable orthologs in other species relative to ovary and oogenesis or non-SRR genes (χ2 tests in D. grimshawii: P < 0.05 in all comparisons, a Bonferroni correction was applied; Figure 1), while differences in the proportion of genes lacking detectable orthologs in D. grimshawii were not significant between ovary and oogenesis genes and non-SRR genes (χ2 = 3.23, d.f. = 1, P = 0.362, a Bonferroni correction was applied; Figure 1). The percentage of SFP genes with orthologs decreases with increasing phylogenetic distance from D. melanogaster and falls below 50% in D. grimshawii (Figure 1). Since many SFP-encoding genes are small, and thus may not be detected using gene-prediction algorithms, we conducted additional tblastn searches against the genome assemblies of all 11 non-melanogaster species. At a fairly liberal BLAST cutoff of 0.1, additional best reciprocal hits were recovered for a number of SFPs in several species. In the most extreme case, an additional 11 hits (16%) were found for D. willistoni (supplemental Table 3 at http://www.genetics.org/supplemental/). Nonetheless, even if all of the additional hits indicate true orthologs, it is still the case that fewer melanogaster SFPs have detectable orthologs in each species in comparison to the other gene classes. In contrast, genes encoding FRTPs do not show such signs of decreasing orthology across divergence time as there is no difference in the rate of gain/loss of FRTP orthologs in comparison to ovary and oogenesis genes or non-SRR genes (χ2 = 4.73 and 0.02, d.f. = 1, P = 0.178 and 1, respectively; a Bonferroni correction was applied; Figure 1). Although we have not performed synteny tests here, our ortholog detection rate for a test case of a subset of the SFP genes (51 SFPs between D. melanogaster and D. pseudoobscura) matches the ortholog detection rate on the basis of synteny comparisons in a whole-genome analysis (Mueller et al. 2005). We therefore believe that different rates of gain or loss between species account for the majority of our observations, such that different species of Drosophila use different complements of SFPs and, to a lesser extent, testes-expressed genes. These data are consistent with previous studies finding novel SFPs in other Drosophila species (Begun and Lindfors 2005; Wagstaff and Begun 2005a; Begun et al. 2006).
Figure 1.—
Male SRR genes have fewer detectable orthologs in comparison to female SRR or non-SRR genes. Percentages of D. melanogaster male, SFP, female, FRTP, unbiased, and nonsex and non-SRR genes with detectable orthologs over phylogenetic distance. The total number of D. melanogaster genes in each class is given in parentheses. The representation of the phylogenetic relationship among Drosophila species is according to FlyBase.
Traffic in the fast lane: rates of evolution of SRR genes:
To further elucidate the nature of SRR gene evolution, we analyzed rates of sequence evolution for 8509 aligned orthologs (including 2505 SRRs genes; see Table 2 and supplemental Table 2 at http://www.genetics.org/supplemental/) from the D. melanogaster group using two major approaches: (1) analyses of sex-tissue-expressed (STE) vs. non-STE genes and (2) analyses of candidate sets of genes known or suggested to be involved in specific reproductive processes, such as genes encoding SFPs, as well as genes involved in or expressed during spermatogenesis and oogenesis (see Table 1 for classification of gene categories and Table 2 for details on the number of genes used in each analysis).
Sex-tissue-expressed genes:
Using 8509 genes with identified orthologs in the six D. melanogaster subgroup species, including 5156 with EST annotations (see materials and methods), we analyzed the proportions of genes under each tissue classification (supplemental Figure 1 at http://www.genetics.org/supplemental/). There is a greater proportion of head- and testis-specific genes as compared to ovary-specific genes (χ2 = 84.04 and 52.76, d.f. = 1, P = 0 and 0 for the comparison with head- and testis-specific genes, respectively; a Bonferroni correction was applied).
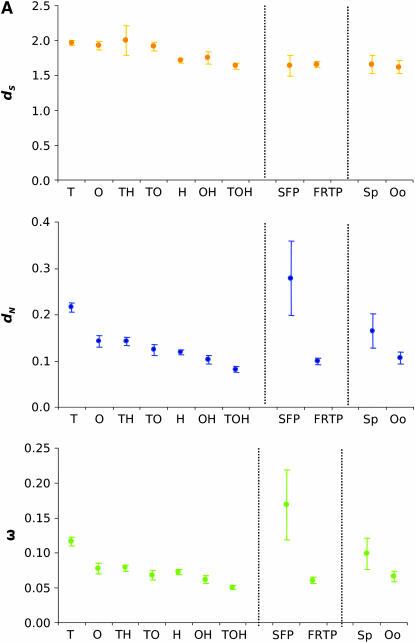
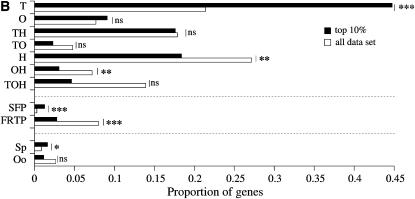
First, we looked at whether there was a relationship between tissue(s) of expression (testis, ovary, and head) and sequence divergence using the assumption of a single nonsynonymous substitution per nonsynomous site over synonymous substitution per synonymous site ratio (ω) per gene (PAML, model M0; Yang and Nielsen 2002). As previously reported in other organisms (Torgerson et al. 2002; Khaitovich et al. 2005), we found a significant relationship between tissue(s) of expression and patterns of sequence evolution. Tissue-specific genes (testis, ovary, and head specific) showed a higher average number of nonsynonymous substitutions per nonsynonymous site (dN) and ω across lineages relative to genes expressed in two or three tissues (Tukey HSD test, P < 0.001). Genes shared between two tissues are evolving faster than genes expressed in all three tissues (Figure 2A; Tukey HSD test, P < 0.001). In addition, among genes shared by two tissues, testis-related genes (i.e., testis and head) are evolving faster (higher average dN) than ovary-related genes (i.e., ovary and head) (Figure 2A; Tukey-HSD test, P < 0.001). Testis-specific genes are evolving far more rapidly than ovary- and head-specific genes as well as shared genes by both dN and ω (Figure 2A; Tukey HSD test, P < 0.01 in all comparisons). Testis-specific genes are overrepresented among the top 10% most rapidly evolving genes (by virtue of ω rank order) relative to the observed proportions among the 5156 genes with information on tissue expression (Figure 2B; χ2 = 76.78, d.f.= 1, P = 0; a Bonferroni correction was applied). In contrast, head-specific genes expressed in both ovary and head, as well as genes common to all three tissues, are significantly underrepresented in the top 10% (Figure 2B; χ2 = 11.41, 11.22, and 29.17; d.f. = 1; P = 5.11 × 10−3, 5.66 × 10−3, and 0 for head-specific genes, genes expressed in both ovary and head, and genes expressed in the three tissues, respectively; a Bonferroni correction was applied). These results reflect earlier reports of the molecular dimorphism between male and female transcriptomes at the divergence level (see Andrews et al. 2000). These results also indicate that breadth of tissue expression and rates of molecular evolution are tightly linked to each other and that testis genes, regardless of their breadth of expression, are evolving faster than genes expressed in head and/or ovary.
Figure 2.—
Higher evolutionary rates of male-specific genes in the melanogaster group. (A) dS, dN, and ω estimates of genes expressed in head, ovary, testis, expressed in seminal fluid and female reproductive tract, and involved in spermatogenesis and oogenesis. Error bars represent 95% confidence intervals. T, testis specific; O, ovary specific; TH, testis and head; TO, testis and ovary; H, head specific; OH, ovary and head; TOH, testis, ovary, and head; Sp, spermatogenesis; Oo, oogenesis. (B) Proportion of gene categories among the top 10% of the most rapidly evolving genes. Proportions of genes in each category among the top 10% genes were compared to the initial proportions in the whole data set using a χ2 test and a Bonferroni correction. ns, nonsignificant. *P < 0.05, ***P < 0.001. (C) Average dS and dN for testis-, ovary-, and head-specific genes across lineages. Testis-specific genes show significantly higher dN in all lineages (Tukey HSD test, P < 0.001).
Reproductive function:
Genes encoding SFPs exhibit higher rates of protein evolution when compared to non-SFP genes. The median ω for 25 SFP encoding loci examined in the melanogaster species group is significantly higher (0.169; 95% C.I.: 0.116–0.221) than the median for non-SFP genes (0.087; 95% C.I.: 0.086–0.089; P = 0.0004, Wilcoxon rank-sum test). Among the 10% most rapidly evolving genes, SFP genes are significantly overrepresented compared to the total data set (Figure 2B; χ2 = 19.83; d.f. = 1; P = 8.5 × 10−6).
In contrast to the above results, genes expressed in the female reproductive tract, excluding the ovary, show a lower rate of protein evolution than do genes expressed in male reproductive tissues or in nonreproductive tissues. The median ω for female reproductive tract genes across the melanogaster species group is 0.061 (95% C.I: 0.056–0.065) vs. 0.090 for the rest of the genome (0.088–0.092; P < 0.0001, Wilcoxon rank-sum test). Limiting the analysis to FRTP genes predicted to encode secreted or transmembrane proteins (which are more likely to interact with proteins transferred from the male) results in a slightly higher median ω (0.069, 0.061–0.076); however, this is still significantly lower than the genomewide average (P < 0.001). Similarly, FRTP genes are underrepresented among the 10% most rapidly evolving genes (Figure 2B; χ2 = 26.55; d.f. = 1; P = 3.0 × 10−7).
Our data also show that genes involved in spermatogenesis have experienced faster rates of evolution relative to genes involved in oogenesis (Figure 2A) due to a significantly greater relative proportion of nonsynonymous substitutions (Wilcoxon rank-sum test, P = 0.037). Spermatogenesis genes are overrepresented among the 10% most rapidly evolving genes (Figure 2B; χ2 = 5.08; d.f. = 1; P = 0.024), while oogenesis genes are not (Figure 2B; χ2 = 3.03; d.f. = 1; P = 0.081).
Lineage-specific effects:
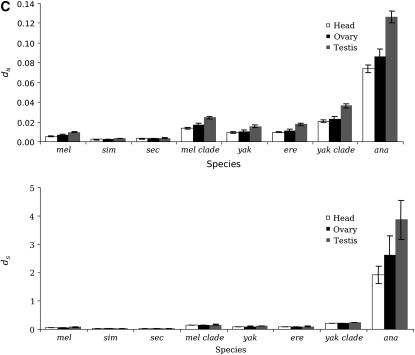
Testis-specific genes show significantly higher dN relative to ovary- or head-specific genes (Figure 2C; Tukey HSD test, P < 0.05) in all the lineages analyzed (D. melanogaster, D. simulans, D. sechellia, D. yakuba, D. erecta, and D. ananassae). It is also noteworthy that in older lineages (yakuba clade and D. ananassae), testis-specific genes also show a greater average rate of synonymous substitutions (dS, Figure 2C; Tukey HSD test, P < 0.05).
Using a free-ratio model allowing ω to vary in the different branches of the phylogeny (Yang and Nielsen 2002) and a 5% false discovery rate (FDR), we tested if the estimated ω for each branch was significantly higher than those estimated for the other branches. We observed a greater proportion of testis-specific genes with greater ω in the internal branch leading the melanogaster clade relative to head-specific genes in the same branch (Figure 3A; χ2 = 5.78, d.f. = 1, P = 0.016) as well as ovary-specific genes when compared to head-specific genes (Figure 3A, χ2 = 14.61, d.f. = 1, P = 1.32 × 10−4). Furthermore, the same proportions of testis- and ovary-specific genes are observed on this branch (χ2 = 3.57, d.f.= 1, P = 0.059). However, in the branch leading to D. yakuba and D. erecta, we observed no significant differences in the proportions of testis- or ovary-specific genes with greater ω when compared to head-specific genes (χ2 = 0 and 0.56; d.f. = 1; P = 1 and 0.45, respectively).
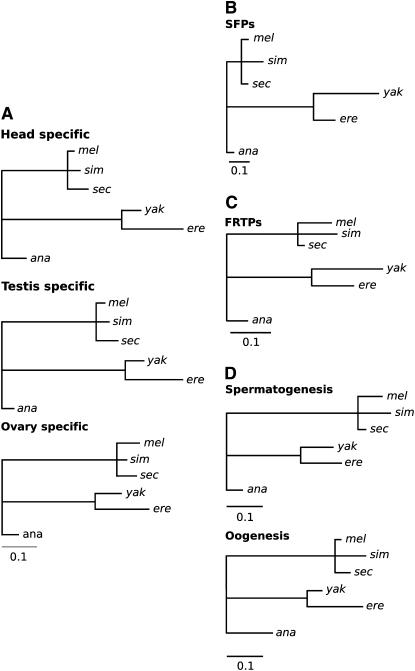
Figure 3.—
Lineage-specific evolutionary acceleration of testis-specific, SFP, and spermatogenesis genes. Branch lengths proportional to the number of genes with ω-values significantly higher on the foreground than background branch for genes (A) expressed specifically in head, testis, and ovary, (B) encoding SFPs, (C) encoding FRTPs, and (D) involved in spermatogenesis and oogenesis. Internal branch lengths indicate results for branch tests using the internal branch, as well as all daughter branches. Data are not available for the sim-sec branch.
The rate of molecular evolution of SFPs is heterogeneous, with accelerated protein evolution on branches leading to D. erecta and D. yakuba (Figure 3B). This observation is consistent with a genomewide trend toward elevated ω in these lineages, although this elevation of ω appears to be slightly more prevalent among SFP genes. The genomewide elevation in ω that we observed may have arisen if the common ancestor of D. yakuba and D. erecta had a smaller effective size (Ne), such that nearly neutral amino acid polymorphisms were fixed by drift at a higher rate than in other lineages with larger Ne. We emphasize, however, that this hypothesis is purely speculative for the time being. In a branch model, whereby ω is allowed to vary in one or more prespecified lineage(s), 7/25 SFP genes (28%) show evidence for an elevated ω in the D. erecta/D. yakuba clade at a 5% FDR, although this is not significantly different from a genomewide trend toward an acceleration in this clade [non-SFP or FRTP genes: 1432/7795 (18%); P = 0.20, Fisher's exact test]. By comparison, at most two SFP genes show similar evidence for elevated ω on any other branch.
In addition to analyses of this stringently selected set of SFP genes, we examined evolutionary rates of an independent set of 43 potential SFP genes (Chintapalli et al. 2007; Ravi Ram and Wolfner 2007). Using data available for the 18 that are found in all six species, we found results consistent with those for the 68 stringently selected genes above: a high fraction show evidence of accelerated divergence along the yakuba/erecta (4/18 at a 5% FDR) lineage. Female reproductive tract genes show a lower proportion of genes with accelerated evolution in the yakuba/erecta clade than do non-SFP and non-FRTP genes (Fisher's exact test: P = 0.03; Figure 3C); 101 of 671 FRTP genes (15%) show evidence for such an acceleration.
We found significant heterogeneity in the proportions of genes with elevated ω along branches for genes of oogenesis (χ2 = 63.59, d.f. = 7, P = 0) as well as for genes involved in spermatogenesis (χ2 = 30.18, d.f. = 7, P = 8.8 × 10−5) (Figure 3D). However, when applying a Bonferroni correction, only spermatogenesis genes show heterogeneity in the proportion of genes with elevated ω along branches (spermatogenesis: χ2 = 33.3, d.f.= 7, P = 2.33 × 10−5; oogenesis: χ2 = 11.9, d.f. = 7, P = 0.1039). We also found a greater proportion of spermatogenesis genes with higher ω in the branch ancestral to the melanogaster clade (D. melanogaster, D. simulans, and D. sechellia) (Figure 3D). This indicates a significant acceleration of ω for spermatogenesis genes along the branch ancestral to the melanogaster clade relative to the rest of the genome. We also observed a greater proportion of nonsynonymous changes in spermatogenesis genes due to accelerated evolution in deeper branches of the phylogeny but not along the D. simulans and D. sechellia branches (Figure 4). A comparison of P-values obtained from tests of selection along the branch ancestral to the melanogaster clade shows that genes involved in spermatogenesis have a significantly lower average P than all other nonspermatogenesis genes in the genome (randomization test: P = 0.0053; P = 0.73 when using only genes with ω higher in the foreground branch than in the background branches). This result indicates a significant acceleration of ω for spermatogenesis genes along the branch ancestral to the melanogaster clade relative to the rest of the genome.
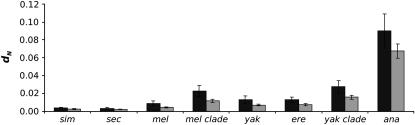
Figure 4.—
Faster evolution of genes involved in spermatogenesis over genes involved in oogenesis. Average dN for spermatogenesis (solid) and oogenesis genes (shaded) in different lineages. Genes involved in spermatogenesis show greater average dN and ω than oogenesis genes in the melanogaster group. This is particularly striking in the more distant members of the melanogaster subgroup.
Positive selection on SRR genes:
We found no significant differences in the proportion of testis-, ovary-, or head-specific genes that show evidence of positive selection when we implemented models M8 and M7 (PAML; Yang and Nielsen 2002; χ2 = 1.68, d.f. = 2, P = 0.432). When branch-site models were included, relatively similar proportions of genes in testis (16.61% at a 5% FDR), ovary (13.35%), and head (16.14%) showed evidence of positive selection (χ2 = 1.77, d.f. = 2, P = 0.413; supplemental Tables 4–6 at http://www.genetics.org/supplemental/). Among testes-specific genes demonstrating positive selection, there are 3 genes involved in meiosis (comr, mei-P26, sunz) and 16 genes involved in transcription. Likewise, among genes expressed in the ovary, genes involved in meiosis are also found to be evolving under positive selection (pan, rec, shtd) as well as ovo, which demonstrates evidence of positive selection in the yakuba–erecta lineage. Among genes expressed specifically in the head showing evidence for positive selection, we found some genes involved in behavior (learning, courtship, olfactory, or locomotor) such as Galpha49B, PKa-R1, Sh, slo, sol, or to. Furthermore, 21 genes are also described to be involved in the formation of the nervous system or in its activity (neurotransmitters, neuropeptides). According to gene ontologies (FB_2006_01), several genes expressed in the ovary, testis, or head are also involved in the defense response (ced-6, CG10359, CG31146, CG9631, Dcr-2, Dredd, Egfr, ics, r2d2).
Of the 25 SFP genes examined in the melanogaster species group, 4 showed evidence of positive selection at a 5% FDR (Table 3, “M8vsM7” in the “Model” column) under the assumption that evolutionary rates are the same in all branches (the M8 vs. M7 model comparison). The proportion of SFPs showing evidence for positive selection in this analysis (16%) is higher than the genomewide (non-SFP or FRTP) average of 5.98%, although not statistically significant (P = 0.06, one-tailed Fisher's exact test). At a less stringent 10% FDR, 6 of 25 SFP genes show evidence of positive selection, which is significantly more than expected, given the genomewide average (P = 0.04, one-tailed Fisher's exact test). We note that the non-SFP group also includes other classes of genes likely to have experienced high levels of positive selection, e.g., immunity genes. The four SFP genes inferred to be under positive selection in this analysis include two predicted regulators of proteolysis (CG4847, a predicted cysteine protease, and CG32203, a predicted protease inhibitor), as well as a predicted protein disulfide isomerase (Pdi). The fourth gene, Acp32CD, has no known function. Physiological or behavioral functions have not been assigned to any of these genes.
TABLE 3.
SFP genes inferred to have experienced positive selection under the site (M8 vs. M7) and/or branch-site models
| Gene | Symbol | Chromosome | Modela | P-valueb | ωc |
|---|---|---|---|---|---|
| FBgn0034229 | CG4847 | 2R | M8 vs. M7, BS (yak/ere) | 8.42E-05; 0.024 | 999; 7.44 |
| FBgn0023415 | Acp32CD | 2L | M8 vs. M7, BS (yak/ere) | 8.42E-05; 0.008 | 5.23; 4.96 |
| FBgn0014002 | Pdi | 3L | M8 vs. M7 | 0.0008 | 35.45 |
| FBgn0052203 | CG32203 | 3L | M8 vs. M7, BS (yak/ere) | 0; 0.001 | 2.65; 84.23 |
BS, branch site.
Model under which ω > 1 was inferred. Under M8, a single distribution of ω (following a β-distribution) is assumed for the entire tree. “yak/ere” refers to a model under which a class of sites with ω > 1 is assigned to the yakuba and erecta lineages but not to any other lineage. Where both M8 and yak/ere are indicated, a gene has been inferred to be under positive selection under both models.
P-value for the test(s) indicated under “Model,” with M7 used as the null for M8 and a model disallowing ω > 1 on any lineage used as the null for the yakuba–erecta lineage positive-selection model. Where two values are indicated, the first refers to the M8/M7 comparison, and the second refers to the branch-site yak/ere test.
ω estimated under the appropriate model.
Branch-site models, which allow variation in ω both across lineages and at different codons (Yang and Nielsen 2002), demonstrate evidence for a burst of positive selection in the yakuba–erecta lineage among SFPs. In tests where the internal branch or branches leading to D. erecta and D. yakuba, as well as the terminal branches ending in these species, are allowed to evolve independently of other lineages, 9 of 25 (36%) SFP genes show evidence for positive selection on a subset of codons in this lineage, while ∼7% of non-SFP or FRTP genes show similar patterns at a cutoff of P < 0.05 (P = 0.027, two-tailed Fisher's exact test) (Table 3, “BS (yak/ere)” in the “Model” column). Using a randomization test, we observed that SFP genes tend to have significantly smaller P-values from the yakuba/erecta branch-sites analysis than do FRTP genes (P < 2.2 × 10−16) or genes not expressed in reproductive tracts or seminal fluids (P = 0.001). Thus, positive selection appears to drive accelerated amino acid evolution along these branches. Given the hypothesis that sexual selection underlies positive selection on some SFP genes, it is tempting to postulate that differences in postcopulatory events along the erecta and/or yakuba lineages may account for the lineage-specific patterns that we observed. Interestingly, the yakuba lineage (D. yakuba and D. erecta) differs from the melanogaster lineage (D. melanogaster, D. simulans, D. sechellia) also with respect to the male reproductive morphology involved in genital coupling (Jagadeeshan and Singh 2006).
We find no differences in the proportion of genes that have undergone positive selection between genes expressed in the female reproductive tract and non-SRR genes. Female reproductive tract genes (6.26%) show evidence for positive selection in a subset of codons across the entire melanogaster species group (M8 vs. M7, at a 5% FDR), which does not differ from the nonfemale reproductive tract average of 5.98% (P = 0.74, two-tailed Fisher's exact test). The 42 FRTP genes inferred to have experienced positive selection encompass a broad range of predicted functions; of particular note are several predicted proteases (CG10824, CG11861, CG3074, CG7415).
Comparisons of the M8 vs. M7 site and the branch-site models in PAML provide evidence for positive selection for a variety of genes involved in gametogenesis (Table 4). While genes involved in oogenesis show lower rates of evolution than spermatogenesis genes (Figure 4), many show evidence of positive selection (supplemental Table 7 at http://www.genetics.org/supplemental/). Similarly, tests of selection using DNA sequence polymorphism in D. melanogaster and divergence to D. simulans have found that female-biased genes do not show the same consistent signal of positive selection that is observed for male-biased genes (Proschel et al. 2006). Of 16 genes involved in gametogenesis showing evidence of positive selection, 5 of them are involved in spermatid individualization or differentiation (poe, Fmr1, didum, Dredd, Nc) and 2 are involved in the early steps of meiosis (can, comr). Furthermore, according to gene ontology annotation, 16 of 47 oogenesis genes showing evidence of positive selection (supplemental Table 7) are involved in egg-shell formation among which we observed three chorion proteins (Cp15, Cp16, Cp18) and one vitelline membrane protein (Vm32E) that were previously reported to be rapidly evolving and for which interaction with sperm or environment have been suggested to be driving the observed rapid evolution (Jagadeeshan and Singh 2007).
TABLE 4.
Genes of gametogenesis showing evidence of positive selection under the site (M8 vs. M7) and/or branch-site models
| Gene | Symbol | Chromosome | Modela | P-valueb | ωc |
|---|---|---|---|---|---|
| FBgn0000320 | eya | 2L | BS (yakere) | 0.0029 | 9.56 |
| FBgn0002780 | mod | 3R | BS (yakere) | 0.0049 | 3.02 |
| FBgn0003731 | Egfr | 2R | BS (yakere) | 0.0049 | 22.35 |
| FBgn0003950 | unc | X | M8 vs. M7, BS (yakere) | 0.004 4E-4 | 2.139 999 |
| FBgn0011207 | pelo | 2L | M8 vs. M7 | 0.0036 | 1.55 |
| FBgn0011219 | Bsg | 2L | M8 vs. M7, BS (sec, simsecmel) | 0.0045 1.96E-13 1.41E-8 | 2.01 999 62.94 |
| FBgn0011230 | poe | 2L | BS (sim) | 6.63E-10 | 999 |
| FBgn0011569 | can | 3L | M8 vs. M7 | 0 | 2.05 |
| FBgn0011823 | Pen | 2L | BS (sim, yakere) | 1.30E-5 0.006 | 176.18 3.28 |
| FBgn0015933 | didum | 2R | BS (sim) | 2.19E-6 | 999 |
| FBgn0020381 | Dredd | X | BS (sim, simsecmel) | 1.40E-8 0.0073 | 999 3.21 |
| FBgn0026404 | Nc | 3L | M8 vs. M7, BS (sec) | 0.0012 1.25E-4 | 1.28 578 |
| FBgn0028734 | Fmr1 | 3R | M8 vs. M7 | 0.0034 | 1.79 |
| FBgn0028974 | xmas-2 | X | BS (sim) | 9.13E-4 | 999 |
| FBgn0034667 | comr | 2R | M8 vs. M7 | 0.0044 | 13.79 |
| FBgn0051711 | CG31711 | 2L | BS (sim, simsecmel) | 8.98E-10 1.2E-4 | 161.16 22.60 |
BS, branch site. P threshold value is Bonferroni corrected. Genes involved in both spermatogenesis and oogenesis are underlined.
Model under which ω > 1 was inferred. Under M8, a single distribution of ω (following a β-distribution) is assumed for the entire tree. “sim,” “sec,” “simsecmel,” and “yakere” refer to a model under which a class of sites with ω > 1 is assigned to the simulans, sechellia, simulans–sechellia–melanogaster, and yakuba–erecta lineages, respectively, but not to any other lineage.
P-value for the test(s) indicated under “Model,” with M7 used as the null for M8 and a model disallowing ω > 1 on any lineage used as the null for the lineage-specific positive-selection model. Where two or more values are indicated, the order is the M8 vs. M7 comparison and/or the branch-site test(s) with the order terminal branch (i.e., sim, sec, or mel) first and the internal branch (i.e., simsecmel or yakere) second.
ω estimated under the appropriate model.
DISCUSSION
The frequent observation of rapid and often adaptive evolution in a select set of genes, such as SRR and immune system genes across different taxonomic groups (Hughes and Yeager 1997; Singh and Kulathinal 2000; Panhuis et al. 2006), suggests vast heterogeneity in rates of molecular evolution across any genome. In view of the biological species concept (Dobzhansky 1951; Mayr 1963), rapidly evolving SRR genes have received much attention from evolutionary biologists due to their potential role in reproductive isolation. The extent of rapid SRR evolution from a genomewide perspective has only recently emerged (Swanson et al. 2001, 2004; Begun and Lindfors 2005; Mueller et al. 2005; Wagstaff and Begun 2005a,b), yet prior studies, of necessity, were restricted to only two or three species and to small numbers of SRR genes. Here, through the analysis of 8509 genes, including 2505 genes with likely and specific sex and reproductive functions (ESTs, SFP, FRTP, gametogenesis) across six species of the melanogaster species group whose genomes have been sequenced, we are able to gain a comprehensive view of the striking disparity in rates and patterns of evolution observed among male- and female-specific genes.
Our analyses show that there are distinct differences in the evolutionary dynamics of SRR genes. Among the various categories of SRR genes (see Table 1), genes expressed in testis and male reproductive tract secretory tissues (that contribute to seminal fluid) show higher rates of gain in D. melanogaster or loss in other species compared to genes not expressed in sex tissues. Male tissue-specific genes evolve faster on average than female tissue-specific genes with the former also showing apparent gain/loss of orthologs. The greater proportion of male D. melanogaster reproductive genes lacking detectable orthologs in distantly related Drosophila species suggests a higher rate of gain/loss. This may be due to lineage-specific duplications or faster rates of new genes being co-opted into reproductive functions, leading to loss of orthology due to various selective pressures (True and Carroll 2002; Ranz et al. 2003; Metta et al. 2006). Alternatively, lost orthology may be a result of faster sequence divergence of male reproductive genes as previously observed between D. melanogaster and D. pseudoobscura (Mueller et al. 2005; Musters et al. 2006). Nonetheless, as argued above (see results), we believe that different rates of gene gain and loss are the most likely explanation.
The recently reported D. melanogaster sperm proteome has revealed that there has been a significant proportion of duplications among sperm protein genes (Dorus et al. 2006), which could fuel rapid evolution across species. However, in comparisons between D. melanogaster and D. simulans, Dorus et al. (2006) found that more than half of the sperm proteome genes had dN values <0.01, suggesting that selection is constraining the evolution of structurally and functionally important genes. A total of 215 of 342 genes from Dorus et al.'s (2006) study are found in our data set. Our testis-specific genes have a significantly higher ω (0.116; 95% C.I.: 0.1100–0.1225) than the 215 genes coding for sperm proteins (0.0735; 95% C.I.: 0.0656–0.0816) (Wilcoxon rank-sum test, P < 0.001). We found results similar to Dorus et al. (2006) in comparisons across the six species of the melanogaster group, with SFP genes having significantly higher dN than other genes in our testis data set (Tukey HSD test, P < 0.05 in all comparisons) with the exception of genes involved in proteolysis (Tukey HSD test, P > 0.05) and genes of unknown functions (Tukey HSD test, P > 0.05). It is therefore clear that, although male SRR genes have evolved faster than other genes on average, genes with roles in specific sperm structures and functions remain constrained in their rates of evolution.
In view of previous studies showing rapid evolution of some genes expressed in ovary (Civetta and Singh 1995; Jagadeeshan and Singh 2005) or in the female reproductive tract (Swanson et al. 2004; Panhuis and Swanson 2006; Lawniczak and Begun 2007), it is surprising that the female reproductive transcriptome appears more constrained than the male reproductive transcriptome in terms of both evolutionary rates and preservation of detectable orthology across species. However, the majority of such comparative studies were performed between closely related species, and our genomewide comparison across six species shows that rates of nonsynonymous substitutions are more similar between testis–ovary and spermatogenesis–oogenesis genes along younger phylogenetic branches (D. simulans and D. sechellia). Differences between previous protein-based studies (Civetta and Singh 1995) and our protein sequence comparisons of the evolutionary rate of female SRR genes (more particularly, FRTP encoding genes) also raise the interesting possibility that perhaps the previous high proportion of protein changes found in the ovary might relate to an overrepresentation of species-specific post-translational changes. This is a particularly interesting possibility in light of recent studies showing that female physiological changes occurring shortly after mating are unlikely to be the result of changes in gene expression (McGraw et al. 2004; Mack et al. 2006).
Sexual selection has been shown to drive the rapid and divergent evolution of genital morphology (Eberhard 1985; Arnqvist 1998; Sirot 2003; Hosken and Stockley 2004; Jagadeeshan and Singh 2006), sperm morphology (Sivinski 1980; Miller and Pitnick 2002; Joly et al. 2004), seminal proteins transferred in the ejaculate (Swanson et al. 2001; Mueller et al. 2005), and genes coding for proteins involved directly in the process of fertilization (Swanson and Vacquier 2002). Two main hypotheses have been put forward to explain how interactions between the sexes could have shaped the rapid evolution of male reproductive traits and genes: cryptic female choice (Eberhard 1996) and the sexual arms race, in which a fitness advantage for one sex appears to be harmful to the other (Rice 2000; Chippindale et al. 2001). A recently proposed theory, male sex drive (Singh and Kulathinal 2005), attributes the faster evolution of male traits to the reasoning that any trait enhancing male fitness will be under strong positive selection. Newly evolved genital traits (Jagadeeshan and Singh 2006), retroposition of male-specific genes from the X to the autosomes (Betran et al. 2002; Long et al. 2003), and the evolution of seminal fluids to modify female reproductive behaviors (Chapman et al. 2003) may result from selection for male reproductive fitness. However, it must be noted that the evolution of such “male-fitness enhancing” traits is exposed to the scrutiny of female choice as well as conflict. Is it possible that one of these three mechanisms might have prevailed across the genome to drive the observed faster evolution of male SRR genes relative to female SRR genes? At present, it is not possible to use our data to distinguish between the cryptic female choice, sexual arms race, and male sex drive hypotheses, as we lack functional characterization of all the genes analyzed and cannot assert that a given testis- or ovary-expressed gene does, in fact, have a reproductive function. It is very likely that female choice, sexual conflict, and male sex drive are not mutually exclusive and may operate at various stages (independently or overlapping) of reproduction (see Hosken and Stockley 2004). Moreover, it might be misleading to evaluate fitness of males and females separately, when in reality the overall population's success depends on the interaction and co-adaptation of both sexes as separate sexes cannot maximize their fitness indefinitely (Civetta and Singh 2005). One example of the inadequacy of splitting fitness by sex comes from studies of the effect of some seminal proteins on the basis of a particular physiological response (e.g., increased female fecundity), which fail to consider the cost of mating [e.g., decreased female viability and therefore conflict (Chapman and Davies 2004)]. Therefore, although numerous authors have proposed that male/female co-evolution, driven by sexual selection, may underlie positive selection for some male reproductive genes (Cordero 1995; Eberhard 1996; Swanson and Vacquier 2002), formal tests of sexual selection will require the analyses of male and female molecular co-evolution of confirmed interacting proteins. Unfortunately, female interactors of seminal fluid and sperm proteins have not yet been identified in D. melanogaster. Several initial attempts to provide candidate interactors have focused on genes expressed in somatic (nonovarian) portions of the female reproductive tract. It is almost certain that this approach will fail to identify some interactors (e.g., those with expression in the head), but a number of interactors should be present in this set of organs (Swanson et al. 2004; Mack et al. 2006).
Selective forces unrelated to sexual selection could also explain the rapid evolution of SRR genes and the evidence of positive selection for a subset of them. Evidence of positive selection for some genes could be the consequence of non-sexual adaptive evolution, as several genes expressed in the seminal fluid or in the female reproductive tract are thought to be involved in immune functions (Samakovlis et al. 1991; Lung et al. 2001; Peng et al. 2005; Lawniczak and Begun 2007; Mueller et al. 2007). Therefore, host–pathogen interactions could be the primary cause of the observed rapid evolution. Ecological adaptation can also be proposed to explain the positive selection observed in genes involved in eggshell formation as previously suggested (Jagadeeshan and Singh 2007). Experimental evolution studies will provide insights into which selective forces can result in the patterns of rapid evolution and positive selection that we observe in these genes.
The results that we have obtained by using a systematic genome- and genus-wide approach over a comprehensive list of annotated genes in 12 Drosophila species suggest that sexual selection may impose distinct and different evolutionary pressures on the genome relative to natural selection, thereby resulting in a dichotomy in rates and patterns of evolution of SRR and non-SRR genes. Faster evolution in a wide variety of male SRR genes (SFPs, testis specific, and spermatogenesis) relative to female- and non-SRR genes (ovary-specific, oogenesis, unbiased, and head-specific genes) indicates that the male reproductive repertoire is under distinct evolutionary pressures. Although the lack of functional information for many genes precludes us from fully explaining the vast and higher rate of male divergence, our study provides relevant information regarding the nature of divergence among reproductive-related genes. This information will be valuable in formulating and designing further detailed investigations directed at testing specific hypotheses related to sexual selection.
Acknowledgments
We thank the Assembly/Alignment/Annotation group (especially Thom Kaufman, Bill Gelbart, Mike Eisen, and Andrew Clark) for making the data available to the Drosophila community and for encouraging us to undertake these genomewide comparative studies. We are also grateful to Tim Sackton, Amanda Larracuente,Venky Iyer, and Daniel Pollard for generating the consensus coding sequences, alignments, and PAML results. We also thank the following agencies for their support through research grants and fellowships: the National Institutes of Health (NIH) grant HD38921 to M.F.W.; the National Sciences and Engineering Research Council (NSERC) Individual Discovery grants to R.S.S. and A.C.; Smithsonian Institute postdoctoral Molecular Evolution Fellowship grant to S.J.; Howard Hughes Medical Institute Predoctoral Fellowship and National Science Foundation Doctoral Dissertation Improvement Grant to A.W.; NSERC Doctoral Post-Graduate Scholarship to C.G.A.; and the NIH National Research Service Award fellowship (F32GM074361) to L.K.S. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
References
- Aagaard, J. E., X. Yi, M. J. MacCoss and W. J. Swanson, 2006. Rapidly evolving zona pellucida domain proteins are a major component of the vitelline envelope of abalone eggs. Proc. Natl. Acad. Sci. USA 103: 17302–17307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Aguadé, M., 1999. Positive selection drives the evolution of the Acp29AB accessory gland protein in Drosophila. Genetics 152: 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé, M., N. Miyashita and C. H. Langley, 1992. Polymorphism and divergence in the Mst26A male accessory gland gene region in Drosophila. Genetics 132: 755–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, M., 1994. Sexual Selection. Princeton University Press; Princeton, NJ.
- Andrews, J., G. G. Bouffard, C. Cheadle, J. Lu, K. G. Becker et al., 2000. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 10: 2030–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnqvist, G., 1998. Comparative evidence for the evolution of genitalia by sexual selection. Nature 393: 784–786. [Google Scholar]
- Bauer Dumont, V. L., H. A. Flores, M. H. Wright and C. F. Aquadro, 2007. Recurrent positive selection at Bgcn, a key determinant of germ line differentiation, does not appear to be driven by simple coevolution with its partner protein Bam. Mol. Biol. Evol. 24: 182–191. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., and H. A. Lindfors, 2005. Rapid evolution of genomic Acp complement in the melanogaster subgroup of Drosophila. Mol. Biol. Evol. 22: 2010–2021. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., P. Whitley, B. L. Todd, H. M. Waldrip-Dail and A. G. Clark, 2000. Molecular population genetics of male accessory gland proteins in Drosophila. Genetics 156: 1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun, D. J., H. A. Lindfors, M. E. Thompson and A. K. Holloway, 2006. Recently evolved genes identified from Drosophila yakuba and D. erecta accessory gland expressed sequence tags. Genetics 172: 1675–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran, E., K. Thornton and M. Long, 2002. Retroposed new genes out of the X in Drosophila. Genome Res. 12: 1854–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavener, D., G. Corbett, D. Cox and R. Whetten, 1986. Isolation of the eclosion gene cluster and the developmental expression of the Gld gene in Drosophila melanogaster. EMBO J. 5: 2939–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, T., and S. J. Davies, 2004. Functions and analysis of the seminal fluid proteins of male Drosophila melanogaster fruit flies. Peptides 25: 1477–1490. [DOI] [PubMed] [Google Scholar]
- Chapman, T., G. Arnqvist, J. Bangham and L. Rowe, 2003. Sexual conflict. Trends Ecol. Evol. 18: 41–47. [Google Scholar]
- Chintapalli, V. R., J. Wang and J. A. T. Dow, 2007. Using FlyAtlas to identify better Drosophila models of human disease. Nat. Genet. 39: 715–720. [DOI] [PubMed] [Google Scholar]
- Chippindale, A. K., J. R. Gibson and W. R. Rice, 2001. Negative genetic correlation for adult fitness between sexes reveals ontogenetic conflict in Drosophila. Proc. Natl. Acad. Sci. USA 98: 1671–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civetta, A., and R. S. Singh, 1995. High divergence of reproductive tract proteins and their association with postzygotic reproductive isolation in Drosophila melanogaster and Drosophila virilis group species. J. Mol. Evol. 41: 1085–1095. [DOI] [PubMed] [Google Scholar]
- Civetta, A., and R. S. Singh, 1998. a Sex-related genes, directional sexual selection, and speciation. Mol. Biol. Evol. 15: 901–909. [DOI] [PubMed] [Google Scholar]
- Civetta, A., and R. S. Singh, 1998. b Sex and speciation: genetic architecture and evolutionary potential of sexual versus nonsexual traits in the sibling species of the Drosophila melanogaster complex. Evolution 52: 1080–1092. [DOI] [PubMed] [Google Scholar]
- Civetta, A., and R. S. Singh, 2005. Rapid evolution of sex-related genes. Sexual conflict or sex-specific adaptations?, pp. 13–21 in Selective Sweep, edited by D. Nurminsky. Kluwer Academic, Dordrecht, The Netherlands.
- Civetta, A., S. A. Rajakumar, B. Brouwers and J. P. Bacik, 2006. Rapid evolution and gene-specific patterns of selection for three genes of spermatogenesis in Drosophila. Mol. Biol. Evol. 23: 655–662. [DOI] [PubMed] [Google Scholar]
- Cordero, C., 1995. Ejaculate substances that affect female insect reproductive physiology and behaviour: Honest or arbitrary traits? J. Theor. Biol. 174: 453–461. [Google Scholar]
- Coulthart, M. B., and R. S. Singh, 1988. High level of divergence of male-reproductive-tract proteins between Drosophila melanogaster and its sibling species, D. simulans. Mol. Biol. Evol. 5: 182–191. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- Darwin, C., 1871. The Descent of Man and Selection in Relation to Sex. John Murray, London.
- Dobzhansky, T., 1951. Genetics and the Origin of Species, Ed. 3. Columbia University Press, New York.
- Dorus, S., S. A. Busby, U. Gerike, J. Shabanowitz, D. F. Hunt et al., 2006. Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat. Genet. 38: 1440–1445. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium, 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218. [DOI] [PubMed] [Google Scholar]
- Eberhard, W. G., 1985. Sexual Selection and Animal Genitalia. Harvard University Press, Cambridge, MA.
- Eberhard, W. G., 1996. Female Control: Sexual Selection by Cryptic Female Choice. Princeton University Press, Princeton, NJ.
- Galindo, B. E., V. D. Vacquier and W. J. Swanson, 2003. Positive selection in the egg receptor for abalone sperm lysin. Proc. Natl. Acad. Sci. USA 100: 4639–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty, W., and R. S. Singh, 2006. Gene regulation divergence is a major contributor to the evolution of Dobzhansky-Muller incompatibilities between species of Drosophila. Mol. Biol. Evol. 23: 1707–1714. [DOI] [PubMed] [Google Scholar]
- Holloway, A. K., and D. J. Begun, 2004. Molecular evolution and population genetics of duplicated accessory gland protein genes in Drosophila. Mol. Biol. Evol. 21: 1625–1628. [DOI] [PubMed] [Google Scholar]
- Hosken, D. J., and P. Stockley, 2004. Sexual selection and genital evolution. Trends Ecol. Evol. 19: 87–93. [DOI] [PubMed] [Google Scholar]
- Houde, A. E., 1997. Sex, Color and Mate Choice in Guppies. Princeton University Press, Princeton, NJ.
- Hughes, A. L., and M. Yeager, 1997. Molecular evolution of the vertebrate immune system. BioEssays 19: 777–786. [DOI] [PubMed] [Google Scholar]
- Jagadeeshan, S., and R. S. Singh, 2005. Rapidly evolving genes of Drosophila: differing levels of selective pressure in testis, ovary, and head tissues between sibling species. Mol. Biol. Evol. 22: 1793–1801. [DOI] [PubMed] [Google Scholar]
- Jagadeeshan, S., and R. S. Singh, 2006. A time-sequence functional analysis of mating behaviour and genital coupling in Drosophila: role of cryptic female choice and male sex-drive in the evolution of male genitalia. J. Evol. Biol. 19: 1058–1070. [DOI] [PubMed] [Google Scholar]
- Jagadeeshan, S., and R. S. Singh, 2007. Rapid evolution of outer egg membrane proteins in the Drosophila melanogaster subgroup: a case of ecologically driven evolution of female reproductive traits. Mol. Biol. Evol. 24: 929–938. [DOI] [PubMed] [Google Scholar]
- Jansa, S. A., B. L. Lundrigan and P. K. Tucker, 2003. Tests for positive selection on immune and reproductive genes in closely related species of the murine genus Mus. J. Mol. Evol. 56: 294–307. [DOI] [PubMed] [Google Scholar]
- Joly, D., A. Korol and E. Nevo, 2004. Sperm size evolution in Drosophila: inter- and intraspecific analysis. Genetica 120: 233–244. [DOI] [PubMed] [Google Scholar]
- Kern, A. D., C. D. Jones and D. J. Begun, 2004. Molecular population genetics of male accessory gland proteins in the Drosophila simulans complex. Genetics 167: 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich, P., I. Hellmann, W. Enard, K. Nowick, M. Leinweber et al., 2005. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309: 1850–1854. [DOI] [PubMed] [Google Scholar]
- Kubli, E., 2003. Sex-peptides: seminal peptides of the Drosophila male. Cell. Mol. Life Sci. 60: 1689–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathinal, R. J., and R. S. Singh, 1998. Cytological characterization of premeiotic versus postmeiotic defects producing hybrid male sterility among sibling species of the Drosophila melanogaster complex. Evolution 52: 1067–1079. [DOI] [PubMed] [Google Scholar]
- Lawniczak, M. K., and D. J. Begun, 2004. A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome 47: 900–910. [DOI] [PubMed] [Google Scholar]
- Lawniczak, M. K., and D. J. Begun, 2007. Molecular population genetics of female expressed mating-induced serine proteases in Drosophila melanogaster. Mol. Biol. Evol. 24: 1944–1951. [DOI] [PubMed] [Google Scholar]
- Levine, M. T., C. D. Jones, A. D. Kern, H. A. Lindfors and D. J. Begun, 2006. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis biased expression. Proc. Natl. Acad. Sci. USA 103: 9935–9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, M., E. Betran, K. Thornton and W. Wang, 2003. The origin of new genes: glimpses from the young and old. Nat. Rev. Genet. 4: 865–875. [DOI] [PubMed] [Google Scholar]
- Ludwig, M. Z., N. A. Tamarina and R. C. Richmond, 1993. Localization of sequences controlling the spatial, temporal, and sex-specific expression of the esterase 6 locus in Drosophila melanogaster adults. Proc. Natl. Acad. Sci. USA 90: 6233–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung, O., and M. F. Wolfner, 2001. Identification and characterization of the major Drosophila melanogaster mating plug protein. Insect Biochem. Mol. Biol. 31: 543–551. [DOI] [PubMed] [Google Scholar]
- Lung, O., L. Kuo and M. F. Wolfner, 2001. Drosophila males transfer antibacterial proteins from their accessory gland and ejaculatory duct to their mates. J. Insect Physiol. 47: 617–622. [DOI] [PubMed] [Google Scholar]
- Mack, P. D., A. Kapelnikov, Y. Heifetz and M. Bender, 2006. Mating-responsive genes in reproductive tissues of female Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 103: 10358–10363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow, T. A., 2002. Perspective: female remating, operational sex ratio, and the arena of sexual selection in Drosophila species. Evolution 56: 1725–1734. [DOI] [PubMed] [Google Scholar]
- Mayr, E., 1963. Animal Species and Evolution. Belknap Press/Harvard University Press, Cambridge, MA.
- McGraw, L. A., G. Gibson, A. G. Clark and M. F. Wolfner, 2004. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr. Biol. 14: 1509–1514. [DOI] [PubMed] [Google Scholar]
- Meiklejohn, C. D., J. Parsch, J. M. Ranz and D. L. Hartl, 2003. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. USA 100: 9894–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metta, M., R. Gudavalli, J. M. Gibert and C. Schlotterer, 2006. No accelerated rate of protein evolution in male-biased Drosophila pseudoobscura genes. Genetics 174: 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak, P., and M. A. Noor, 2003. Genome-wide patterns of expression in Drosophila pure species and hybrid males. Mol. Biol. Evol. 20: 1070–1076. [DOI] [PubMed] [Google Scholar]
- Miller, G. T., and S. Pitnick, 2002. Sperm-female coevolution in Drosophila. Science 298: 1230–1233. [DOI] [PubMed] [Google Scholar]
- Moehring, A. J., K. C. Teeter and M. A. Noor, 2007. Genome-wide patterns of expression in Drosophila pure species and hybrid males. II. Examination of multiple-species hybridizations, platforms, and life cycle stages. Mol. Biol. Evol. 24: 137–145. [DOI] [PubMed] [Google Scholar]
- Möller, A. P., 1994. Sexual Selection and the Barn Swallow. Oxford University Press, Oxford.
- Monsma, S. A., and M. F. Wolfner, 1988. Structure and expression of a Drosophila male accessory gland gene whose product resembles a peptide pheromone precursor. Genes Dev. 2: 1063–1073. [DOI] [PubMed] [Google Scholar]
- Mueller, J. L., K. Ravi Ram, L. A. McGraw, M. C. Bloch Qazi, E. D. Siggia et al., 2005. Cross-species comparison of Drosophila male accessory gland protein genes. Genetics 171: 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, J. L., J. L. Page and M. F. Wolfner, 2007. An ectopic expression screen reveals the protective and toxic effects of Drosophila seminal fluid proteins. Genetics 175: 777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters, H., M. A. Huntley and R. S. Singh, 2006. A genomic comparison of faster-sex, faster-X, and faster-male evolution between Drosophila melanogaster and Drosophila pseudoobscura. J. Mol. Evol. 62: 693–700. [DOI] [PubMed] [Google Scholar]
- Panhuis, T. M., and W. J. Swanson, 2006. Molecular evolution and population genetic analysis of candidate female reproductive genes in Drosophila. Genetics 173: 2039–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panhuis, T. M., N. L. Clark and W. J. Swanson, 2006. Rapid evolution of reproductive proteins in abalone and Drosophila. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361: 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M., R. Nuttall, P. Edwards, J. Minor, D. Naiman et al., 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299: 697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M., R. Nuttall, D. Naiman, G. Bouffard, J. Malley et al., 2004. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 5: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., P. Zipperlen and E. Kubli, 2005. Drosophila sex-peptide stimulates female innate immune system after mating via the Tool and Imd pathways. Curr. Biol. 15: 1690–1694. [DOI] [PubMed] [Google Scholar]
- Presgraves, D. C., L. Balagopalan, S. M. Abmayr and H. A. Orr, 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423: 715–719. [DOI] [PubMed] [Google Scholar]
- Proschel, M., Z. Zhang and J. Parsch, 2006. Widespread adaptive evolution of Drosophila genes with sex-biased expression. Genetics 174: 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz, J. M., C. I. Castillo-Davis, C. D. Meiklejohn and D. L. Hartl, 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300: 1742–1745. [DOI] [PubMed] [Google Scholar]
- Ravi Ram, K., and M. F. Wolfner, 2007. Seminal influences: Drosophila Acps and the molecular interplay between male and female during reproduction. Intgr. Comp. Biol. 47: 427–445. [DOI] [PubMed] [Google Scholar]
- Rice, W. R., 2000. Dangerous liaisons. Proc. Natl. Acad. Sci. USA 97: 12953–12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, S., Y. Liu, B. R. Bettencourt, P. Hradecky, S. Letovsky et al., 2005. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis element evolution. Genome Res. 15: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samakovlis, C., P. Kylsten, D. A. Kimbrell, A. Engstrom and D. Hultmark, 1991. The andropin gene and its product, a male-specific antibacterial peptide in Drosophila melanogaster. EMBO J. 10: 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudan, P., K. Hauck, M. Soller, Y. Choffat, M. Ottiger et al., 2002. Ductus ejaculatorius peptide 99B (DUP99B), a novel Drosophila melanogaster sex-peptide pheromone. Eur. J. Biochem. 269: 989–997. [DOI] [PubMed] [Google Scholar]
- Schully, S. D., and M. E. Hellberg, 2006. Positive selection on nucleotide substitutions and indels in accessory gland proteins of the Drosophila pseudoobscura subgroup. J. Mol. Evol. 62: 793–802. [DOI] [PubMed] [Google Scholar]
- Singh, R. S., and R. J. Kulathinal, 2000. Sex gene pool evolution and speciation: a new paradigm. Genes Genet. Syst. 75: 119–130. [DOI] [PubMed] [Google Scholar]
- Singh, R. S., and R. J. Kulathinal, 2005. Male sex drive and the masculinization of the genome. BioEssays 27: 518–525. [DOI] [PubMed] [Google Scholar]
- Sirot, L. K., 2003. The evolution of insect mating structures through sexual selection. Fla. Entomol. 86: 124–133. [Google Scholar]
- Sivinski, J., 1980. Sexual selection and insect sperm. Fla. Entomol. 63: 99–111. [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3: 137–144. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., A. G. Clark, H. M. Waldrip-Dail, M. F. Wolfner and C. F. Aquadro, 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl. Acad. Sci. USA 98: 7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W. J., A. Wong, M. F. Wolfner and C. F. Aquadro, 2004. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics 168: 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, S., and R. S. Singh, 1992. A comprehensive study of genic variation in natural populations of Drosophila melanogaster. VII. Varying rates of genic divergence as revealed by two-dimensional electrophoresis. Mol. Biol. Evol. 9: 507–525. [DOI] [PubMed] [Google Scholar]
- Torgerson, D. G., R. J. Kulathinal and R. S. Singh, 2002. Mammalian sperm proteins are rapidly evolving: evidence of positive selection in functionally diverse genes. Mol. Biol. Evol. 19: 1973–1980. [DOI] [PubMed] [Google Scholar]
- True, J. R., and S. B. Carroll, 2002. Gene co-option in physiological and morphological evolution. Annu. Rev. Cell Dev. Biol. 18: 53–80. [DOI] [PubMed] [Google Scholar]
- Tsaur, S. C., and C. I. Wu, 1997. Positive selection and the molecular evolution of a gene of male reproduction, Acp26Aa of Drosophila. Mol. Biol. Evol. 14: 544–549. [DOI] [PubMed] [Google Scholar]
- Wagstaff, B. J., and D. J. Begun, 2005. a Comparative genomics of accessory gland protein genes in Drosophila melanogaster and D. pseudoobscura. Mol. Biol. Evol. 22: 818–832. [DOI] [PubMed] [Google Scholar]
- Wagstaff, B. J., and D. J. Begun, 2005. b Molecular population genetics of accessory gland protein genes and testis-expressed genes in Drosophila mojavensis and D. arizonae. Genetics 171: 1083–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, M. J., C. M. Rylett, J. N. Keen, N. Audsley, M. Sajid et al., 2006. Proteomic identification of Drosophila melanogaster male accessory gland proteins, including a pro-cathepsin and a soluble gamma-glutamyl transpeptidase. Proteome Sci. 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfner, M. F., H. A. Harada, M. J. Bertram, T. J. Stelick, K. W. Kraus et al., 1997. New genes for male accessory gland proteins in Drosophila melanogaster. Insect Biochem. Mol. Biol. 27: 825–834. [DOI] [PubMed] [Google Scholar]
- Wolfner, M. F., S. Applebaum and Y. Heifetz, 2005. Insect gonadal glands and their gene products, pp. 179–212 in Comprehensive Insect Physiology, Biochemistry, Pharmacology and Molecular Biology, edited by L. Gilbert, K. Iatrou and S. Gill. Elsevier, Amsterdam.
- Wong, A., and M. F. Wolfner, 2006. Sexual behavior: a seminal peptide stimulates appetites. Curr. Biol. 16: R256–R257. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and R. Nielsen, 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19: 908–917. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., T. M. Hambuch and J. Parsch, 2004. Molecular evolution of sex-biased genes in Drosophila. Mol. Biol. Evol. 21: 2130–2139. [DOI] [PubMed] [Google Scholar]