Abstract
An ultimate objective of QTL mapping is cloning genes responsible for quantitative traits. However, projects seldom go beyond segments <5 cM without subsequent breeding and genotyping lines to identify additional crossovers in a genomic region of interest. We report on a QTL analysis performed as a preliminary step in the development of a resource for map-based cloning of domestication and improvement genes in corn. A large backcross (BC)1 population derived from a cross between maize (Zea mays ssp. mays) and teosinte (ssp. parviglumis) was grown for the analysis. A total of 1749 progenies were genotyped for 304 markers and measured for 22 morphological traits. The results are in agreement with earlier studies showing a small number of genomic regions having greater impact on the morphological traits distinguishing maize and teosinte. Despite considerable power to detect epistasis, few QTL interactions were identified. To create a permanent resource, seed of BC1 plants was archived and 1000 BC2S6 BC1-derived lines are in development for fine mapping and cloning. The identification of four BC1 progeny with crossovers in a single gene, tb1, indicated that enough derived lines already exist to clone many QTL without the need to generate and identify additional crossovers.
CORN and its wild progenitor, teosinte, differ dramatically in their overall plant architecture and the morphology of their female inflorescences. QTL mapping studies in maize–teosinte F2 populations have been utilized to determine the number, effect, and genomic distribution of loci responsible for differences in key traits related to domestication (Doebley and Stec 1991, 1993; Doebley et al. 1994). These earlier studies utilized low-density genetic maps and relatively few progeny, reducing the power to detect QTL and accurately estimate their location and effect (Beavis 1998). Subsequent advancements in the physical mapping of maize ESTs and SSRs have enabled the construction of genetic maps with more uniform genomic distribution and coverage. Furthermore, the development of inexpensive, high-throughput SNP and SSR assays has permitted the genotyping of greater numbers of progeny, improving mapping precision, estimation, and ability to detect smaller-effect QTL.
An additional drawback of experiments with smaller population sizes and sparse maps is that fewer crossovers can be identified in the vicinity of a locus. Fine mapping of QTL and the discovery of tightly linked markers for positional cloning require larger numbers of recombination events not found in typical F2, backcross, or recombinant inbred line mapping populations. Having a larger sample of genotyped lines and a reasonably dense map can shorten the time needed to hone in on a region and find a tightly linked marker, simply because the chance of identifying crossovers close to the underlying gene is greater. Map-based cloning could be accelerated by creating a large number of advanced inbred backcross lines containing overlapping introgressions. A repository of crossovers would then exist for identifying lines containing recombination events near a QTL in any region of the genome. Such a collection of lines would also provide a permanent resource for future mapping studies and allow the researcher to more quickly breed near-isogenic lines containing introgressions of agronomic or biological interest (Butruille et al. 1999; Doganlar et al. 2002).
In this study, we performed QTL analyses on a maize–teosinte backcross population with teosinte as the donor parent and maize as the recurrent parent. This population has a much larger size and a denser molecular marker map than previous maize–teosinte mapping studies. The population has been backcrossed a second time and is now being inbred by single-seed descent to isolate crossovers throughout the genome in a set of ∼1000 advanced backcross (BC) recombinant inbred lines (BC2S6 RILs). We performed a QTL analysis at the BC1 stage of this process for early identification of crossovers near critical loci involved in domestication, improvement, and environmental adaptation. The results presented here will expedite the identification of subsets of the RILs that are useful for map-based cloning of QTL.
MATERIALS AND METHODS
Plant material:
Pollen collected from a single F1 plant from a cross between U.S. maize inbred W22 and an accession of Balsas teosinte (Zea mays ssp. parviglumis) collected by Beadle and Kato from Valle de Bravo, Guerrero, Mexico, was used to pollinate several plants of W22. A total of 1749 BC1 progeny were grown and evaluated in two environments, 1123 plants in Madison, Wisconsin, during the summer of 2004 and 626 plants in Homestead, Florida, during the winter of 2005.
Molecular markers and genotyping:
The BC1 plants were genotyped for 294 polymorphic markers, including 270 SNPs, three indels, and 21 microsatellites, for construction of a genetic linkage map. We also genotyped the entire population for 3 additional SNPs and one polymorphic indel in the ORF and 3′-UTR of the domestication gene tb1, as well as 4 SNPs and one indel between the tb1 ORF and its 5′ neighbor gene. These markers were genotyped to evaluate our chances of detecting crossovers in close proximity to a single gene.
The SNP sites were selected primarily from alignments of low-copy EST sequences taken at random from ∼10,000 maize ESTs in the MMP-DuPont set (Gardiner et al. 2004). These ESTs were screened by overgo-hybridization against the maize B73 BAC library. Only ESTs that hybridized to a single BAC contig were used for SNP discovery (Wright et al. 2005). Sixteen SNP markers were selected from sequence alignments of domestication candidate genes. SNP genotyping was performed at Genaissance Pharmaceuticals using the Sequenome MassARRAY system (Jurinke et al. 2002). Indel and microsatellite marker genotypes were determined by separating PCR products on agarose gels. A complete list of the markers used in this study, including EST and candidate gene information, is included in supplemental Table 1 at http://www.genetics.org/supplemental/. Sequence alignments and SNP context sequences are available at http://www.panzea.org.
Marker map:
A linkage map of the molecular markers was constructed from genotypes of the full cohort of 1749 BC1 plants using MapMaker v. 2.0 (Lander et al. 1987). Marker order was confirmed using RECORD v. 1.0 (Van Os et al. 2005). Potential genotyping errors were identified using R/qtl (Broman et al. 2003), a QTL analysis module of the statistical software R (CRAN; cran.r-project.org). Genotypes identified as having a high error probability (LOD ≥ 3.1) using the methods of Lincoln and Lander (1992) were converted to missing data. Marker distances were recalculated with the revised data.
Phenotypic evaluation:
All of the BC1 plants were evaluated for 21 traits. These included 9 plant architecture traits (Table 1; BARE, BRLG, BRNO, CULM, LBIL, LBNN, PLHT, PROL, and TILL), 4 primary tassel traits (LCS, SPKLT, TBN, and TBS), 7 primary lateral inflorescence morphology traits (COBD, CUPR, GLUM, INFL, LIBN, RANK, and STAM), and days to pollen (POLL). The Florida plants were also evaluated for kernel weight (KERN). All of the primary lateral branch and primary lateral inflorescence traits were measured on the topmost branch.
TABLE 1.
List of the traits analyzed
| Trait | Description | Units |
|---|---|---|
| BARE (barren nodes) | Number of barren nodes on the main stalk above the uppermost primary lateral branch | Count |
| BRLG (branch length) | Length of the primary lateral branch | Centimeters |
| BRNO (branch number) | Number of primary lateral branches | Count |
| COBD | Ear diameter | Millimeters |
| CULM (culm diameter) | Diameter of the primary stalk | Millimeters |
| CUPRa (cupules per rank) | Number of cupules in a single rank | Count |
| GLUMa (glume score) | Hardness and protrusion of the outer glume | Score (1–7, 1 = maize) |
| INFL (inflorescence length) | Length of the primary lateral inflorescence | Millimeters |
| KERNa (kernel weight) | Average kernel weight | Milligrams |
| LBILa | Average length of the primary lateral branch internodes | Centimeters |
| LBNN | Number of nodes on the primary lateral branch | Count |
| LCS (length of central spike) | Length of the primary tassel central spike | Centimeters |
| LIBNa | Number of branches in the primary lateral inflorescence | Count |
| PLHT (plant height) | Length of the primary stalk from the ground to the tip of the primary tassel | Centimeters |
| POLL (days to pollen) | Days to first pollen shed | Days |
| PROLa (prolificacy) | Number of inflorescences on the primary lateral branch | Count |
| RANKa | Number of internode columns (ranks) on the primary lateral inflorescence | Count |
| SPKLT (spikelet length) | Average spikelet length of the primary tassel | Millimeters |
| TBN (tassel branch number) | Branch number of the primary tassel | Count |
| TBS (branching space) | Branching space of the primary tassel | Centimeters |
| TILL (tillering) | Ratio of the sum of all tiller lengths to PLHT | Ratio |
| STAMa (staminate score) | Fraction of the primary lateral inflorescence internodes that are male (staminate) | Number |
A trait evaluated in two maize × teosinte F2 populations (Doebley and Stec 1993; Doebley et al. 1994).
QTL analysis:
Data from the Wisconsin and Florida environments were analyzed separately. Each trait was analyzed independently. The search for QTL was initiated in R/qtl. One thousand permutations of the data were performed for each trait within each environment to identify a P < 0.05 LOD significance threshold level for QTL. Significant main-effect QTL were identified using maximum likelihood. A two-dimensional scan for epistasis was then run. Interactions having an interactive component of the joint two-locus LOD (LODint) > 2.8 were retained in the model. This LOD cutoff for epistatic interactions is the mean permutation threshold across traits for main-effect QTL. The chromosomal positions of the main-effect QTL were then used as an initial input for multiple-interval mapping (MIM) in Windows QTL Cartographer v. 2.5 (Wang et al. 2006). The positions of QTL were refined with MIM. Additional searches for QTL and significant QTL interactions were performed with MIM, using the Akaike's information criterion option for model selection. The final model fitting QTL and QTL interactions was analyzed in R/qtl, using a drop-one-term analysis of multiple loci to estimate the average effect of an allelic substitution at each locus (Sen and Churchill 2001). The estimation of additive effects and proportion of variation explained using this combined model enables summation of the estimates across loci.
RESULTS
Linkage map:
The molecular markers were assembled into a linkage map of 1474.9 cM (supplemental Figure 1 at http://www.genetics.org/supplemental/). As our marker density in some regions is high for a BC1, we took the precaution of identifying and removing potential genotyping errors using the method of Lincoln and Lander (1992), since even a few incorrect genotypes per marker can adversely expand the map. A small fraction of genotypes (0.039%) were identified as errors using this method and these were changed to missing data in the final mapping. The average distance between adjacent markers was 5.4 cM. The largest gap between adjacent markers was 18.9 cM. The physical map position of the most distal marker on each chromosome arm was queried (http://www.genome.arizona.edu/fpc/maize/) to determine the extent of coverage across the genome. The distance from the telomere to the most distal marker ranged from <0.1 to 14.2 Mb with an average of 2.75 Mb.
Phenotypic data:
The BC1 plants segregated into a range of phenotypes intermediate between teosinte and maize, with means and distributions tending more toward maize (supplemental Figure 2 at http://www.genetics.org/supplemental/). The majority of traits were normally distributed, with the exceptions of BRLG, LBIL, LIBN, PROL, RANK, STAM, and TILL, which were all skewed toward maize-like values. The plants grown in Florida differed greatly from those evaluated in Wisconsin for some traits. In general they were shorter, with thinner culms, very few tillers, and fewer male spikelets on the primary lateral inflorescence (STAM). They also had different tassel morphologies, having fewer branches, less branching space, longer central spikes, and longer spikelets. The Florida plants shedded pollen 25 days earlier on average.
QTL analysis:
We performed QTL mapping with the genotypic and trait data from the BC1 plants to identify loci responsible for the phenotypic differences between maize and teosinte. The genetic map position and effect of each QTL are reported in Table 2. The positions of QTL are also depicted on a linkage map in supplemental Figure 1 at http://www.genetics.org/supplemental/. The total numbers of QTL detected were 175 and 139 for the Wisconsin and Florida environments, respectively. Of these, 59 pairs of common QTL were identified where the 1-LOD interval of a Wisconsin QTL overlapped with the 1-LOD interval of a Florida QTL for the same trait. A larger fraction of the QTL were detectable in only one environment. The proportion of the phenotypic variance explained by a QTL (R2) was significantly larger (P < 0.0001) on average for QTL that were detected in both locations. The QTL detected in both environments had similar effects. There were only 2 QTL that had opposite effects on a trait depending on the location. In both of these cases, the Florida QTL (PLHT10.78f and POLL9.46f) were significantly epistatic with another locus, while the Wisconsin QTL showed no epistasis. When the epistatic contribution is added to the additive effect, the magnitude and direction of the Florida and Wisconsin QTL are similar.
TABLE 2.
Significant QTL and effects
| Wisconsin
|
Florida
|
||||
|---|---|---|---|---|---|
| QTLa | Effectb | % variancec | QTL | Effect | % variance |
| BARE1.119f | 0.21 | 2.1 | |||
| BARE3.74w | −0.60 | 2.1 | BARE3.71f | −0.23 | 2.5 |
| BARE4.74w | 0.64 | 2.5 | |||
| BARE4.100f | 0.38 | 7.3 | |||
| BARE5.70w | 0.55 | 1.8 | |||
| BARE7.127w | −0.42 | 1.0 | |||
| BARE8.50w | −0.57 | 1.8 | |||
| BARE10.46w | −0.81 | 3.1 | |||
| BARE10.67f | 0.21 | 2.1 | |||
| BRLG1.142f | −3.2 cm | 2.9 | |||
| BRLG3.70w | −5.7 cm | 5.0 | BRLG3.83f | −3.7 cm | 3.7 |
| BRLG4.66w | −4.0 cm | 2.5 | BRLG4.75f | −2.3 cm | 1.7 |
| BRLG7.0f | 3.5 cm | 3.6 | |||
| BRLG8.51f | 3.2 cm | 2.8 | |||
| BRLG9.59f | −2.1 cm | 1.3 | |||
| BRLG10.46w | −8.3 cm | 9.1 | |||
| BRNO1.13w | −0.40 | 2.0 | |||
| BRNO1.40f | −0.60 | 5.2 | |||
| BRNO1.78w | −0.32 | 1.0 | |||
| BRNO1.116w | −0.61 | 4.0 | |||
| BRNO1.196w | −0.33 | 1.4 | |||
| BRNO2.81w | −0.50 | 3.1 | BRNO2.82f | −0.84 | 8.8 |
| BRNO3.133f | −0.47 | 2.4 | |||
| BRNO4.29f | −0.71 | 4.9 | |||
| BRNO5.22w | −0.44 | 2.2 | |||
| BRNO5.75w | −0.61 | 4.3 | BRNO5.66f | −0.76 | 6.4 |
| BRNO7.2w | −0.12 | 0.4 | |||
| BRNO8.41w | −0.36 | 1.6 | |||
| BRNO9.24w | −0.14 | 0.6 | |||
| BRNO9.77f | −0.45 | 2.5 | |||
| BRNO10.48w | 0.34 | 1.2 | |||
| COBD1.64w | 1.4 mm | 6.5 | COBD1.38f | 0.8 mm | 3.9 |
| COBD1.153f | 0.7 mm | 2.8 | |||
| COBD2.136f | 0.6 mm | 2.5 | |||
| COBD3.72f | 0.3 mm | 0.6 | |||
| COBD3.93f | 0.5 mm | 1.1 | |||
| COBD4.31w | 0.5 mm | 0.5 | COBD4.30f | 0.8 mm | 4.2 |
| COBD4.48w | 0.7 mm | 2.0 | |||
| COBD4.102f | 1.0 mm | 6.5 | |||
| COBD5.73w | 1.1 mm | 4.9 | COBD5.68f | 0.9 mm | 5.8 |
| COBD6.36w | −0.1 mm | 0.9 | |||
| COBD7.100w | −0.1 mm | 1.4 | |||
| COBD8.40w | −0.8 mm | 2.3 | |||
| COBD9.52f | 0.6 mm | 2.8 | |||
| COBD10.45w | 0.9 mm | 2.4 | COBD10.47f | 0.8 mm | 5.0 |
| CULM1.0w | −1.2 mm | 2.4 | CULM1.0f | −1.5 mm | 7.1 |
| CULM1.92w | −1.7 mm | 4.1 | |||
| CULM1.186w | 1.2 mm | 1.9 | |||
| CULM2.87w | 1.8 mm | 5.0 | |||
| CULM3.43w | −1.3 mm | 2.5 | |||
| CULM3.83f | −1.0 mm | 2.8 | |||
| CULM4.70w | 1.0 mm | 1.5 | |||
| CULM5.75w | 2.3 mm | 8.2 | |||
| CULM7.89w | −1.0 mm | 1.6 | |||
| CULM7.123f | 1.0 mm | 3.3 | |||
| CULM9.59w | −1.2 mm | 2.2 | |||
| CULM10.46w | −2.5 mm | 7.1 | |||
| CUPR1.152w | 1.7 | 3.8 | CUPR1.146f | 1.7 | 6.2 |
| CUPR3.120f | 1.5 | 4.6 | |||
| CUPR3.149w | 1.5 | 3.7 | |||
| CUPR7.86f | 1.6 | 5.0 | |||
| CUPR9.28f | 1.5 | 5.0 | |||
| CUPR10.56w | 1.6 | 3.6 | CUPR10.42f | 1.7 | 6.2 |
| GLUM1.145w | −0.6 | 2.2 | GLUM1.146f | −0.8 | 3.8 |
| GLUM3.70w | −0.9 | 4.5 | GLUM3.51f | −0.5 | 1.9 |
| GLUM4.28f | −0.7 | 4.3 | |||
| GLUM4.61w | −1.7 | 24.4 | GLUM4.61f | −2.4 | 27.9 |
| GLUM4.108w | −0.7 | 2.6 | GLUM4.103f | −0.8 | 3.7 |
| GLUM5.104w | −0.4 | 1.1 | |||
| GLUM6.10w | 0.1 | 0.4 | |||
| GLUM9.56w | −0.7 | 2.6 | GLUM9.60f | −0.5 | 1.7 |
| GLUM10.46w | −1.3 | 10.2 | |||
| INFL1.59w | −10.6 mm | 2.0 | |||
| INFL1.100f | −7.4 mm | 3.2 | |||
| INFL1.148w | −15.4 mm | 4.0 | |||
| INFL3.47f | −5.3 mm | 1.5 | |||
| INFL3.70w | −18.3 mm | 5.7 | |||
| INFL4.52w | −9.5 mm | 1.4 | INFL4.63f | −7.0 mm | 2.7 |
| INFL4.98w | −10.5 mm | 1.8 | |||
| INFL5.161w | −9.8 mm | 1.7 | INFL5.165f | −6.2 mm | 2.1 |
| INFL7.116f | 7.0 mm | 2.8 | |||
| INFL9.48w | −16.7 mm | 4.7 | |||
| INFL10.42f | 10.9 mm | 7.0 | |||
| NA | KERN2.90f | 2.7 mg | 2.8 | ||
| NA | KERN3.122f | 9.1 mg | 23.9 | ||
| NA | KERN4.50f | 29.4 mg | 2.7 | ||
| NA | KERN4.115f | 11.5 mg | 1.8 | ||
| NA | KERN8.85f | 9.5 mg | 8.8 | ||
| NA | KERN9.66f | 1.6 mg | 7.2 | ||
| LBIL1.142f | −0.52 cm | 3.4 | |||
| LBIL2.82w | 0.31 cm | 2.6 | |||
| LBIL3.70w | −0.80 cm | 5.9 | LBIL3.83f | −0.69 cm | 4.9 |
| LBIL4.66w | −0.61 cm | 3.5 | LBIL4.77f | −0.62 cm | 5.6 |
| LBIL5.53f | −0.48 cm | 2.6 | |||
| LBIL5.82w | −0.02 cm | 1.7 | |||
| LBIL6.80f | −0.50 cm | 3.5 | |||
| LBIL9.76f | −0.53 cm | 3.2 | |||
| LBIL10.45f | −0.61 cm | 4.2 | |||
| LBIL10.63w | −0.74 cm | 5.2 | |||
| LBNN1.106w | 0.40 | 2.3 | LBNN1.102f | 0.28 | 1.8 |
| LBNN2.58w | 0.34 | 1.5 | LBNN2.57f | 0.45 | 4.7 |
| LBNN3.138f | 0.11 | 1.4 | |||
| LBNN4.80w | 0.38 | 2.4 | LBNN4.91f | 0.38 | 3.4 |
| LBNN7.26f | 0.05 | 1.1 | |||
| LBNN7.125w | −0.50 | 3.9 | |||
| LBNN9.55w | −0.31 | 1.6 | |||
| LBNN10.47w | −0.90 | 10.3 | |||
| LCS1.42w | 1.4 cm | 7.5 | LCS1.41f | 1.2 cm | 4.6 |
| LCS1.131f | −1.1 cm | 7.0 | |||
| LCS2.87w | −0.9 cm | 3.1 | |||
| LCS3.73f | 1.2 cm | 6.1 | |||
| LCS4.124w | −0.3 cm | 1.2 | |||
| LCS5.24f | 0.6 cm | 2.6 | |||
| LCS5.102w | 0.7 cm | 1.9 | |||
| LCS5.158f | 0.4 cm | 1.7 | |||
| LCS6.46f | 0.5 cm | 1.6 | |||
| LCS6.96w | 0.7 cm | 1.9 | LCS6.94f | 0.9 cm | 2.5 |
| LCS7.24w | 0.8 cm | 2.0 | |||
| LCS7.127 | 1.0 cm | 2.4 | |||
| LCS8.0w | 0.4 cm | 1.7 | |||
| LCS9.16 | −0.1 cm | 1.4 | |||
| LCS9.108w | 0.0 cm | 1.2 | |||
| LCS10.38 | 0.1 cm | 1.2 | |||
| LCS10.56w | −0.1 cm | 1.2 | |||
| LIBN2.48w | −0.48 | 2.0 | LIBN2.57f | −0.19 | 3.2 |
| LIBN3.78w | −0.79 | 5.1 | |||
| LIBN6.98w | −0.55 | 2.5 | |||
| LIBN7.6w | −0.37 | 2.0 | |||
| LIBN7.121f | 0.16 | 2.1 | |||
| LIBN9.51w | −0.54 | 2.2 | |||
| LIBN10.59w | −0.77 | 6.5 | |||
| PLHT1.47f | −5.5 cm | 2.2 | |||
| PLHT1.84w | −6.6 cm | 2.0 | |||
| PLHT1.129w | −10.9 cm | 5.2 | PLHT1.134f | −9.4 cm | 5.9 |
| PLHT2.70f | −3.6 cm | 2.0 | |||
| PLHT2.87w | −4.3 cm | 0.9 | |||
| PLHT3.77w | −12.4 cm | 7.7 | PLHT3.71f | −12.3 cm | 10.7 |
| PLHT3.165w | −3.4 cm | 0.8 | |||
| PLHT4.75w | −7.8 cm | 2.9 | PLHT4.71f | −5.9 cm | 2.2 |
| PLHT5.23f | −9.5 cm | 6.2 | |||
| PLHT5.34w | −9.2 cm | 3.7 | |||
| PLHT6.91w | −5.2 cm | 1.9 | |||
| PLHT7.14w | −8.1 cm | 3.8 | PLHT7.13f | −6.6 cm | 2.9 |
| PLHT7.113w | −7.6 cm | 2.8 | |||
| PLHT8.28w | −8.0 cm | 3.7 | |||
| PLHT9.67w | −10.5 cm | 6.8 | PLHT9.59f | −6.1 cm | 2.3 |
| PLHT10.42w | −9.0 cm | 3.2 | |||
| PLHT10.79w | −4.1 cm | 0.8 | PLHT10.78f | 0.6 cm | 1.1 |
| POLL1.5f | −1.1 days | 1.9 | |||
| POLL1.84w | −1.5 days | 2.8 | |||
| POLL1.155w | −1.5 days | 3.0 | |||
| POLL2.37w | −1.2 days | 1.8 | POLL2.42f | −0.5 days | 2.4 |
| POLL3.77w | −2.2 days | 5.3 | POLL3.70f | −1.8 days | 6.2 |
| POLL3.165f | −0.9 days | 1.4 | |||
| POLL4.118w | −0.4 days | 1.5 | |||
| POLL5.22w | −0.3 days | 1.3 | |||
| POLL5.168f | 0.2 days | 2.1 | |||
| POLL6.56f | −0.5 days | 1.9 | |||
| POLL6.88w | −0.9 days | 1.7 | |||
| POLL7.44w | 1.3 days | 2.0 | POLL7.42f | 1.3 days | 3.1 |
| POLL7.106f | 1.0 days | 2.1 | |||
| POLL8.52w | −0.1 days | 0.6 | |||
| POLL9.59w | −2.1 days | 9.4 | POLL9.46f | 0.2 days | 1.4 |
| POLL10.47w | −5.4 days | 22.9 | |||
| PROL1.80w | −0.16 | 3.8 | PROL1.67f | −0.14 | 5.2 |
| PROL2.123f | −0.12 | 4.6 | |||
| PROL3.80w | −0.08 | 1.9 | |||
| PROL4.116w | −0.09 | 2.8 | |||
| PROL5.53w | −0.12 | 2.7 | PROL5.64f | −0.15 | 3.4 |
| PROL7.120w | 0.04 | 1.1 | |||
| PROL10.52w | −0.14 | 2.0 | |||
| RANK1.53w | 0.3 | 4.3 | RANK1.50f | 0.3 | 3.6 |
| RANK3.77w | 0.3 | 3.9 | |||
| RANK3.166w | 0.3 | 2.8 | |||
| RANK4.67w | 0.4 | 3.6 | |||
| RANK4.107w | 0.3 | 3.1 | RANK4.102f | 0.5 | 11.6 |
| RANK5.94w | 0.1 | 1.8 | |||
| RANK8.29w | −0.3 | 2.1 | |||
| RANK10.37w | 0.3 | 2.8 | RANK10.47f | 0.4 | 8.9 |
| SPKLT1.9w | 0.15 mm | 1.0 | SPKLT1.9f | 0.34 mm | 6.2 |
| SPKLT1.52w | 0.24 mm | 2.6 | |||
| SPKLT1.153w | 0.12 mm | 0.7 | |||
| SPKLT1.194w | 0.14 mm | 1.3 | |||
| SPKLT2.23w | 0.33 mm | 7.0 | SPKLT2.10f | 0.23 mm | 2.2 |
| SPKLT2.45f | 0.27 mm | 3.4 | |||
| SPKLT2.122w | 0.22 mm | 3.3 | |||
| SPKLT3.145w | −0.15 mm | 1.4 | |||
| SPKLT4.77w | 0.21 mm | 2.6 | |||
| SPKLT6.81w | 0.29 mm | 4.5 | SPKLT6.73f | 0.25 mm | 3.0 |
| SPKLT8.85w | −0.13 mm | 1.0 | |||
| SPKLT9.50w | 0.25 mm | 2.4 | |||
| SPKLT9.78w | 0.10 mm | 0.5 | |||
| SPKLT10.7w | 0.18 mm | 1.8 | |||
| SPKLT10.46f | −0.24 mm | 3.1 | |||
| SPKLT10.77w | −0.13 mm | 1.0 | |||
| STAM1.148w | −0.20 | 10.0 | STAM1.154f | −0.03 | 5.0 |
| STAM3.66w | −0.26 | 16.3 | STAM3.68f | −0.03 | 6.4 |
| STAM4.50w | −0.09 | 2.4 | |||
| STAM4.110w | −0.08 | 1.7 | |||
| STAM5.70w | −0.13 | 4.1 | |||
| STAM5.158f | −0.02 | 3.2 | |||
| STAM9.49w | −0.12 | 4.0 | STAM9.51f | −0.03 | 6.8 |
| STAM10.104f | −0.02 | 1.7 | |||
| TBN1.0f | −4.0 | 3.2 | |||
| TBN1.38w | −3.7 | 1.2 | |||
| TBN1.112w | −3.9 | 1.7 | |||
| TBN1.150f | −2.6 | 1.6 | |||
| TBN1.178w | 3.5 | 1.3 | |||
| TBN1.196f | 3.1 | 1.8 | |||
| TBN2.41w | −7.4 | 5.3 | TBN2.39f | −3.8 | 4.6 |
| TBN2.117w | −2.7 | 0.9 | |||
| TBN3.6f | −2.3 | 2.1 | |||
| TBN3.40w | −3.4 | 1.3 | |||
| TBN3.72f | −9.0 | 18.3 | |||
| TBN3.89w | −6.8 | 4.4 | |||
| TBN4.63f | −2.4 | 1.9 | |||
| TBN4.100w | 1.8 | 0.5 | |||
| TBN5.19f | −3.1 | 1.7 | |||
| TBN5.105w | −11.8 | 13.5 | TBN5.110f | −4.2 | 4.0 |
| TBN6.35f | −2.8 | 1.6 | |||
| TBN6.100w | −8.0 | 6.7 | TBN6.94f | −2.2 | 1.7 |
| TBN6.116f | −2.9 | 3.1 | |||
| TBN7.43w | −4.0 | 1.7 | TBN7.34f | −1.4 | 2.3 |
| TBN7.111w | 3.1 | 0.9 | |||
| TBN8.25w | −5.8 | 3.3 | TBN8.25f | −3.6 | 4.4 |
| TBN8.114f | −2.5 | 2.6 | |||
| TBN9.23f | 2.3 | 1.4 | |||
| TBN9.72f | −4.0 | 3.2 | |||
| TBN10.48w | −5.5 | 2.7 | |||
| TBS1.0f | −0.7 cm | 2.1 | |||
| TBS1.38w | −0.8 cm | 1.6 | TBS1.33f | −0.6 cm | 1.5 |
| TBS1.125w | −0.9 cm | 2.4 | |||
| TBS1.146f | −1.1 cm | 4.3 | |||
| TBS1.198f | 0.9 cm | 2.8 | |||
| TBS3.72f | −1.6 cm | 10.5 | |||
| TBS3.91w | −1.2 cm | 3.7 | |||
| TBS4.53w | −0.8 cm | 1.7 | TBS4.55f | −1.1 cm | 3.3 |
| TBS5.16f | −1.1 cm | 5.2 | |||
| TBS5.113w | −1.0 cm | 2.7 | TBS5.116f | −0.9 cm | 3.3 |
| TBS5.154w | −0.7 cm | 1.3 | |||
| TBS6.11w | 0.7 cm | 1.2 | |||
| TBS7.126w | 1.0 cm | 2.6 | TBS7.117f | 1.0 cm | 4.6 |
| TBS9.51w | −0.7 cm | 2.7 | |||
| TBS9.78w | −1.0 cm | 1.7 | TBS9.78f | −1.7 cm | 11.4 |
| TBS10.47w | −1.4 cm | 5.7 | |||
| TILL1.32w | 0.25 | 1.4 | |||
| TILL1.146w | −0.61 | 7.7 | TILL1.158f | −0.05 | 2.2 |
| TILL2.77w | −0.39 | 2.3 | |||
| TILL2.100w | −0.42 | 1.8 | |||
| TILL3.141w | −0.31 | 1.9 | |||
| TILL5.80w | −0.41 | 3.6 | |||
| TILL6.35f | −0.04 | 1.5 | |||
| TILL7.7w | −0.37 | 3.0 | |||
| TILL8.58w | −0.31 | 2.1 | |||
| TILL10.49w | −0.49 | 4.2 | |||
QTL from Wisconsin and Florida with overlapping 1-LOD drop-off intervals are listed on the same line.
The QTL names are a concatenation of the trait, chromosome number, and chromosome position of each locus.
Average effect of substituting a teosinte allele with a second maize allele.
Proportion of the phenotypic variance explained by the locus (R2).
The numbers of QTL that were significant by permutation (P < 0.05) for each trait are reported in Table 3. The mean numbers of QTL per trait were 8.3 for Wisconsin and 6.3 for Florida. The number of loci per trait ranged from 2 for TILL in Florida to 17 for TBN in Florida. Heritability for the overall genetic model was estimated from the drop-one-term analysis of multiple-loci model as the sum of the proportion of the variance explained by the significant main-effect and epistatic QTL detected for a trait. Heritabilities ranged from 0.05 for LIBN in Florida to 0.64 for GLUM in Florida.
TABLE 3.
Number of QTL and heritability of the genetic model for each trait
| Wisconsin
|
Florida
|
|||
|---|---|---|---|---|
| Trait | No. additive QTL + epistatic interactions | h2 | No. additive QTL + epistatic interactions | h2 |
| BARE | 6 | 0.12 | 4 | 0.14 |
| BRLG | 3 | 0.16 | 6 | 0.16 |
| BRNO | 11 + 1 | 0.29 | 6 | 0.28 |
| COBD | 8 + 2 | 0.22 | 10 + 1 | 0.41 |
| CULM | 10 | 0.38 | 3 | 0.13 |
| CUPR | 3 | 0.12 | 5 | 0.26 |
| GLUM | 8 + 2 | 0.55 | 6 + 1 | 0.64 |
| INFL | 7 | 0.22 | 6 | 0.21 |
| KERN | NA | NA | 6 + 1 | 0.37 |
| LBIL | 5 + 2 | 0.17 | 7 + 1 | 0.25 |
| LBNN | 6 | 0.22 | 5 + 1 | 0.15 |
| LCS | 9 + 2 | 0.20 | 10 + 5 | 0.31 |
| LIBN | 6 + 2 | 0.17 | 2 | 0.05 |
| PLHT | 14 + 1 | 0.47 | 9 + 1 | 0.38 |
| POLL | 11 + 3 | 0.42 | 9 + 2 | 0.23 |
| PROL | 6 + 3 | 0.12 | 3 + 1 | 0.10 |
| RANK | 8 + 1 | 0.27 | 3 | 0.23 |
| SPKLT | 14 | 0.38 | 5 | 0.20 |
| STAM | 6 + 2 | 0.36 | 5 + 4 | 0.19 |
| TBN | 14 + 1 | 0.47 | 17 + 1 | 0.56 |
| TBS | 11 + 1 | 0.32 | 10 + 1 | 0.50 |
| TILL | 9 | 0.34 | 2 | 0.04 |
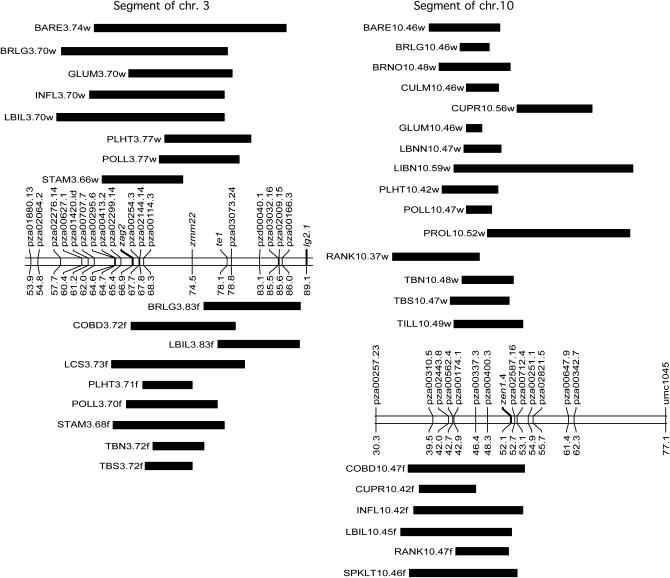
Several QTL with relatively large effects (R2 > 0.1) were detected. The QTL explaining the largest proportion of the variance (R2) for any trait in a single environment was KERN4.50f, which was responsible for 29.4% of the variance for kernel weight (Table 2). The QTL with the largest R2 for both environments was at the domestication gene tga1 (GLUM4.61w and GLUM4.61f). Individuals carrying a teosinte allele for this gene had harder and longer glumes. The trait with the largest number of QTL was TBN. A single large-effect QTL was detected for this trait in each environment. TBN5.105w and TBN3.72f were each responsible for >13% of the variance in their respective locations. Interestingly, these QTL were detected in one environment but not in the other. Another large-effect QTL detected in just one location was POLL10.47w. The presence of an additional maize allele at this locus shortened the time to pollen shed by 5.4 days in Wisconsin. This region showed no effect on days-to-pollen in Florida, suggesting that one or more loci in this genomic region are responsive to day length. Two genomic regions contained a relatively large number of QTL in both locations. Within the vicinity of the POLL10.47w locus, QTL for 14 other traits in Wisconsin and for 6 traits in Florida were detected, all within 5 cM (Figure 1). A segment of chromosome 3 also affected many traits in both locations. STAM3.66w is the strongest locus for primary lateral inflorescence sex determination. In close proximity are QTL for 7 other traits in Wisconsin and 8 traits in Florida, including several of strong effect (Table 2; Figure 1).
Figure 1.—
Regions on chromosomes 3 (left) and 10 (right) affecting several traits. A segment of the genetic map is depicted, showing molecular marker names and positions on the chromosome in centimorgans. QTL detected in Wisconsin are shown as solid bars above the chromosome. The placement and length of the bar indicate the position and extent of the 1-LOD drop-off interval. The names of the loci, which include the trait, chromosome, and position, are listed to the left of the bars. QTL detected in Florida are depicted in the same fashion below the chromosome.
The large size of the BC1 population provides ample power for detecting additive × additive epistasis. The expected number of individuals for each of the four two-locus genotypic classes in Wisconsin is 280. Totals of 20 and 19 significant epistatic interactions (LODint > 2.8) were detected in Wisconsin and Florida, respectively (Table 4). Only one pair of epistatic loci was detected in both locations. This interaction was between STAM1.148w or STAM1.154f, a QTL on chromosome 1 in very close proximity to the tb1 domestication gene, and STAM3.66w or STAM3.68f. Addition of a second maize allele at either position reduces the fraction of male spikelets in the primary lateral inflorescence. The interactive effect of these QTL is less than additive. The largest epistatic interaction detected was between GLUM4.61w, the tga1 locus, and GLUM10.46w. Neither an additive nor an epistatic QTL was detected for GLUM on chromosome 10 in Florida. In Wisconsin, the interaction effect of a maize allele substitution at both loci was synergistic.
TABLE 4.
Epistatic loci and effects
| Locus 1 | Locus 2 | Effectb | Directionc |
|---|---|---|---|
| BRNO7.2w | BRNO9.24w | 0.32 | <A |
| COBD4.48w | COBD7.100w | −1.2 mm | <A |
| COBD5.73w | COBD6.36w | −1.1 mm | <A |
| COBD3.72f | COBD10.47f | 0.8 mm | >A |
| GLUM4.61w | GLUM10.46w | −1.3 | >A |
| GLUM6.10w | GLUM6.95wa | 0.5 | <A |
| GLUM4.28f | GLUM4.61f | −0.5 | >A |
| KERN2.90f | KERN9.66f | 11.1 mg | >A |
| LBIL2.82w | LBIL5.82w | −0.86 | >A |
| LBIL3.70w | LBIL10.63w | 0.80 | <A |
| LBIL4.77f | LBIL6.80f | 0.68 | <A |
| LBNN3.138f | LBNN7.26f | 0.43 | >A |
| LCS4.124w | LCS10.56w | 1.2 cm | <A |
| LCS8.0w | LCS9.108w | 1.1 cm | >A |
| LCS1.131f | LCS2.86fa | −1.2 cm | >A |
| LCS1.131f | LCS3.73f | 1.6 cm | >A |
| LCS1.131f | LCS5.24f | −1.5 cm | >A |
| LCS5.158f | LCS10.38f | 1.3 cm | >A |
| LCS6.46f | LCS9.16f | −1.0 cm | <A |
| PLHT9.67w | PLHT10.42w | −5.4 cm | >A |
| PLHT2.70f | PLHT10.78f | −9.0 cm | >A |
| POLL4.118w | POLL5.22w | −1.9 days | >A |
| POLL6.88w | POLL8.52w | −1.2 days | >A |
| POLL2.42f | POLL5.168f | 2.1 days | <A |
| POLL6.56f | POLL9.46f | −1.7 days | >A |
| PROL1.86w | PROL5.53w | 0.15 | <A |
| PROL3.80w | PROL4.116w | 0.19 | <A |
| PROL4.116w | PROL7.120w | −0.16 | >A |
| PROL1.67f | PROL2.123f | 0.26 | <A |
| RANK3.77w | RANK5.94w | 0.4 | >A |
| STAM1.148w | STAM3.66w | 0.21 | <A |
| STAM3.66w | STAM4.50w | 0.14 | <A |
| STAM1.154f | STAM3.68f | 0.05 | <A |
| STAM1.154f | STAM9.51f | 0.04 | <A |
| STAM5.158f | STAM9.51f | 0.04 | <A |
| STAM9.51f | STAM10.104f | 0.05 | <A |
| TBN3.89w | TBN4.100w | −3.0 | >A |
| TBN6.116f | TBN7.34f | 4.7 | <A |
| TBS9.51w | TBS10.47w | −1.3 cm | >A |
A QTL that does not have a significant additive effect.
The interactive effect of the two QTL.
Direction of the epistasis: >A or <A indicates that the interactive effect is greater than or less than the magnitude of the sum of the additive effects of the two loci.
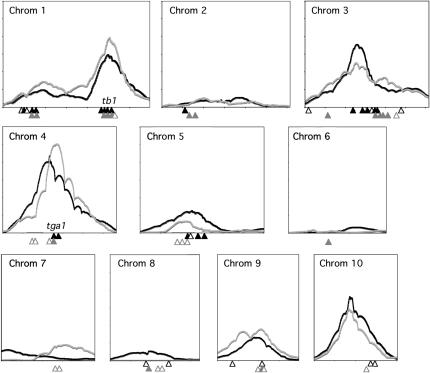
QTL for the seven domestication traits analyzed in this study and in previous research by Doebley and Stec (1993) are restricted to just a few regions of the genome. In the present study, we constructed a visual overview of the sum of the standardized effects of these traits across each chromosome (Figure 2). Seven regions of large effect are apparent using both the Wisconsin and the Florida data. These include segments of the short and long arms of chromosome 1, the middle of chromosome 3, the short arms of chromosomes 4 and 5, and the middle of chromosomes 9 and 10. There was similar localization of effects on chromosome arms 1S and 1L, on chromosome 3, and on the short arms of chromosomes 4 and 5 in earlier F2 studies (Doebley and Stec 1993). However, those studies both had a large-effect QTL for RANK on 2 S that was not detected here. Furthermore, there was little effect attributed to chromosomes 9 and 10 in previous populations.
Figure 2.—
Genomic distribution of the summed significance levels of seven domestication traits (LIBN, PROL, GLUM, RANK, CUPR, STAM, and LIBN). Each chromosome is depicted as a plot with the x-axis representing genetic position. The likelihood ratio (LR) of each position on the chromosome was standardized for each trait by dividing it by the sum of all LR values across the entire genome. The height of the curve (y-axis) is the sum of the standardized values across the seven traits. The solid and shaded curves are from the Wisconsin and Florida data, respectively. QTL for these seven traits detected in two previous studies of maize–teosinte F2 populations (Doebley and Stec 1993) are depicted with solid and shaded triangles below each plot. Loci with R2-values greater than or <0.1 are shown as solid or open triangles, respectively.
Crossovers at tb1:
One of the objectives for genotyping a large BC1 population was the generation and detection of greater numbers of crossovers near QTL. The seed archived from these crossovers will facilitate the introgression of key genomic regions into isogenic backgrounds for the eventual positional cloning of domestication genes. To evaluate our chances of detecting crossovers in the vicinity of a single gene, we genotyped the 1749 BC1 plants for 10 polymorphic markers within the tb1 gene and its nearest 5′ neighbor. Crossovers were detected in 4 individuals. Two of these were in the tb1 ORF and two were between 65 and 163 kb upstream from the ORF. The total physical distance surveyed in the region of tb1 was 164.4 kbp. The genetic distance was 0.23 cM. Given a maize genome size of 2500 Mbp and the BC1 genetic map length of 1474.9 cM, the average expected genetic to physical map ratio is 5.9 × 10−4 cM/kbp. The recombination rate across tb1 was 1.4 × 10−3 cM/kbp, over twice the genomewide average.
DISCUSSION
Segregation distortion and flowering time:
The Wisconsin sample had many more loci with deviations from the expected BC1 segregation ratio of 1:1, including a 61.7-cM stretch on chromosome 10 with much higher homozygous maize frequencies. Maize–teosinte first-generation backcross individuals tend to flower 2–3 weeks later on average than U.S. Corn Belt varieties during the long days of summer in Wisconsin (W. H. Briggs, personal observations). Many plants flower too late to be used successfully for crosses in the field. The plants that were genotyped and phenotyped were biased in favor of those that were used in crosses to initiate the BC2S6 RILs. It is expected that the subset of individuals we selected would have a higher frequency of maize alleles at regions of the genome affecting flowering time. This is particularly likely for the region on chromosome 10, where we detected a large-effect QTL for days to pollen. The maize allele at POLL10.47w shortens the flowering time in Wisconsin by >5 days. This region of chromosome 10 has been shown to condition days to pollen shed (Koester et al. 1993), possibly through the action of putative homologs of known flowering-time genes in rice and Arabidopsis (Chardon et al. 2004).
There were fewer markers with significant deviations from the expected segregation ratio in Florida. These BC1 plants flowered a month earlier than their sibs in Wisconsin. There was no threat of cold weather to affect the viability of crosses made later in Florida, so the individuals genotyped and phenotyped were not selected on the basis of flowering time. The comparatively smaller regions of the genome showing distorted segregation did not overlap with those detected in Wisconsin with the exception of a segment on the long arm of chromosome 6. These results suggest that most of the segregation distortion we observed is due to postzygotic mechanisms such as a bias for early flowering.
Genomic regions with large effects on domestication traits:
The parents used to construct our study population differ from those used in previous reports on the genetic control of morphological traits in maize–teosinte intercross populations (Doebley and Stec 1991, 1993). The earlier F2 studies utilized primitive tropical landraces instead of an improved U.S. inbred for the maize parent and different accessions of Z. mays ssp. parviglumis or ssp. mexicana for the teosinte parents. We used W22 as the maize parent for three reasons. First, a Midwest inbred will facilitate the advancement of RILs at the latitude of Wisconsin. Second, it is necessary to have an inbred recurrent parent to produce a series of advanced backcross lines that have an isogenic background. Third, teosinte phenotypes might express more strongly in a W22 background than they do in some other widely utilized public inbreds such as B73 (J. Doebley, personal observations).
Despite differences in the pedigrees of the mapping populations, the large-effect loci that are essential for the transformation of teosinte into maize should be detectable by analysis of virtually any segregating maize–teosinte population, although the strengths of the effects might vary. Five genomic regions, segments on 1L, 3, 4S, and to a lesser extent 1S and 5S had strong effects in all of the studies to date (Figure 2; Doebley and Stec 1993). Consistencies in the traits affected by each of these regions are described. The QTL interval on 1L straddles the cloned domestication gene tb1. This region consistently affects STAM, TILL, and CUPR. Plants having teosinte alleles of tb1 introgressed into a maize background express the teosinte phenotype for these traits (Doebley et al. 1995; Clark et al. 2006). Depending on the study, the 1L segment may also affect LBIL and GLUM. The chromosome 3 segment contains QTL for STAM and LBIL in both Wisconsin and Florida as well as GLUM in Wisconsin only. QTL for these traits were also detected in the earlier F2 studies (Doebley and Stec 1993). This region had QTL for several other traits that were not reported previously (Figure 1). Both STAM3.66w and STAM3.68f were epistatic with the chromosome 1L region containing tb1. Studies of the 1L and 3 segments from maize and teosinte introgressed into teosinte and maize backgrounds, respectively, also showed evidence for epistasis between these two loci for STAM (Doebley et al. 1995). The large-effect QTL on 4S overlaps the domestication gene tga1 that conditions GLUM, the formation of the hard teosinte cupulate fruitcase (Wang et al. 2005). The impetus to positionally clone this gene came from the detection of a large-effect QTL in previous studies (Doebley and Stec 1993). We also detected QTL for LBIL and RANK within this interval, in agreement with earlier analyses. Although the QTL on 1S has a smaller effect on the key domestication phenotypes, it has been significant in this study and previous studies. We detected QTL for RANK in both Wisconsin and Florida and TILL in Wisconsin, consistent with Doebley and Stec (1991, 1993). The QTL on 5S contained loci for LBIL and PROL in both environments as well as STAM in Wisconsin. QTL for these traits were also detected in the F2 populations.
Multiple studies have shown a large-effect QTL for RANK on 2S (Maguire 1961; Galinat 1973; Doebley and Stec 1991, 1993) for which zfl2, the maize homolog of the LEAFY gene in Arabidopsis, is a candidate gene (Bomblies and Doebley 2006). We did not detect a QTL significant for RANK near zfl2 in the BC1 population. One possible explanation for this is that the W22 allele for this QTL is dominant to the teosinte allele in our backcross population. The ability to detect a QTL in a backcross would be reduced or eliminated if the recurrent parent allele was dominant. An interesting observation in this regard is that no BC1 plants were two ranked, while this phenotype is common in maize–teosinte F2 populations. This suggests that the maize phyllotaxy of four or more ranks (polystichous) is dominant over two-ranked (distichous) teosinte. zfl2 has been shown to control the conversion of ear phyllotaxy from distichous, found in teosinte, to polystichous (consisting of four or more ranks) found in maize.
Alternatively, the maize and teosinte alleles at this QTL captured in our BC1 may confer the same phenotypic effects on phyllotaxy in a predominantly maize background. The absence of a rank QTL on 2S and the presence of a rank QTL on 10 in the BC1 population is an interesting coincidence. There are comparatively fewer QTL on chromosome 10 in the earlier studies of F2 populations (Doebley and Stec 1993). Portions of 2S and 10 are orthologous (Helentjaris et al. 1988) and carry duplicate genes, such as zfl1 and zfl2 (Bomblies et al. 2003). Loci having a significant effect on rank were detected on 10 in Wisconsin and Florida. Perhaps the lineages from teosinte to the maize lines used in prior studies all involved an allele change at a paralogous gene on 2 S.
Epistasis:
Although the large BC1 population provides ample power for detecting additive-by-additive epistasis, few significant interactions were identified. We used a genomewide scan that did not require that a QTL have a significant main additive effect for consideration. Of the 29 cases of epistasis detected, only one was consistent for the two locations, the interaction between the tb1 and chromosome 3 QTL for STAM. This interaction was detected in previous studies (Doebley et al. 1995). The overall small amount of epistasis detected is consistent with previous larger-sized QTL and biometric studies of quantitative traits in maize (Stuber et al. 1973; Edwards et al. 1987). The extent of epistasis may also be limited in a backcross population compared to an F2-derived population because of the absence of the homozygous teosinte class and the inability to evaluate non-additive-by-additive interactions. As we restricted the detection threshold for epistatic interactions to those in the range of main-effect QTL, it is likely that many effects were undetected. These unreported interactions may account for a sizable portion of the variation for some traits.
Candidate genes:
Two known domestication genes have been cloned in maize, tb1 and tga1. These genes underlie genomic regions involved in maize domestication on 1L and 4S. A 30-cM segment of chromosome 3 is similarly critical in the magnitude of its effects on multiple domestication traits (Figures 1 and 2). The region encompasses several candidate genes that may be responsible for variation in one or more traits.
Two MADS-box genes, zag2 and zmm22, that show evidence of selection during domestication or improvement (Zhao 2006) map in the major-effect region on 3. Genes of the MADS-box class are particularly interesting candidates for inflorescence traits such as STAM because of their involvement in floral organ identity and patterning. zag2 is homologous to the Arabidopsis stamen and carpel identity gene AGAMOUS (Schmidt et al. 1993). Sequence diversity for zag2 is significantly lower in maize landraces and maize inbreds compared to teosinte, suggesting that it was under strong selection during domestication. Little is known about the function of zmm22. In close proximity to zag2 is ts4, another gene involved in inflorescence sex determination. ts4 was recently cloned and is involved in meristem fate and sexual identity (Chuck et al. 2006). Three other genes that were previously considered candidates for the chromosome 3 QTL are ba1, lg2, and te1. lg2 and ba1 were located beyond the major QTL intervals in our analysis, which provides more accurate positioning than prior studies. Furthermore, sequence diversity analyses of ba1 and lg2 show somewhat ambiguous evidence for selection during domestication (Gallavotti et al. 2004; Zhao 2006). te1 is within the 1-LOD interval of our QTL. Initial analysis of the coding sequence of te1 did not show evidence for selection (White and Doebley 1999), but further study revealed that maize diversity is significantly reduced relative to teosinte in the 5′-UTR (Zhao 2006). This gene is also involved in floral development and sexual identity (Veit et al. 1993).
A number of candidate genes are located within the segment of chromosome 10 that contains several QTL. As the locus with the largest effect for this segment, POLL10.47w, affects flowering time and because many more trait associations were significant in Wisconsin than in Florida, we believe there might be a day-length responsive gene in this region. This possibility is supported by the detection of flowering-time QTL in other maize studies and the projection of rice candidate genes onto this position of the maize map (Chardon et al. 2004). These candidates encode a putative circadian clock protein (OsCCA_LHY) and cryptochrome (OsCRY12). zen1, a maize homolog of the Antirrhinum floral development gene centroradialis, is also located in this interval. Comparison of the maize and teosinte sequence diversities in zen1 shows evidence of selection during domestication (Zhao 2006).
Population utility:
The impetus for this preliminary analysis of a large BC1 population from which we are deriving RILs is the identification of potential QTL candidates for positional cloning. Our intention is to identify enough recombination events surronding a QTL to fine map it without the need to breed and genotype for additional crossovers. We identified four crossovers within a single gene, tb1, by genotyping the BC1 population for 10 markers. The recombination rate at tb1 is approximately twice the genomewide average. Rates are even higher in other intragenic recombination hotspots, extending up to two orders of magnitude higher than the genome average at the bronze and a1 genes (Brown and Sundaresan 1991; Dooner and Martinez-Ferez 1997). If we assume that our survey of tb1 is representative of genic regions, then enough crossovers have already been captured in the resource to make it possible to dissect a QTL and positionally clone an underlying gene, particularly if the QTL contributes a large effect to the variance of a trait. The majority of the BC1 plants used in the study have been backcrossed a second time to W22 and are being self-pollinated for six generations to produce ∼1000 advanced inbred backcross lines. During this process, 50% of the crossovers will be lost during the second backcross and another ∼50% will be lost during the selfing process. If there are insufficient numbers of crossovers in a QTL region of interest in the BC2S6 lines, remnant seed of the BC2 will enable recovery of additional crossovers. The chromosome 3 QTL interval of STAM3.66w and the segment of chromosome 10 encompassing POLL10.47w are of particular interest for fine mapping and positional cloning of genes involved in the evolution of maize during domestication, adaptation, and improvement.
In this study, we identified genomic regions that contain genes important in the evolution of maize from teosinte through linkage with molecular markers. Another approach being used to find genes that were critical during maize domestication and improvement is the comparison of the levels of polymorphism in inbreds, landraces, and teosinte. This approach has been applied to large numbers of EST sequences in maize (Vigouroux et al. 2002; Wright et al. 2005; Yamasaki et al. 2005). A subset of genes with reduced polymorphism in maize relative to teosinte displays a signature of selection during domestication or improvement. Some candidates may be overlooked in these studies using EST sequences. Evidence for selection may not be detectable in cases where the extent of the selective sweep was short and the target of selection was distant from the transcribed region, such as an upstream cis regulatory element (Clark et al. 2006). Domestication and improvement genes that have such a pattern of polymorphism may be more readily identified in larger mapping populations such as our maize–teosinte backcross resource.
Candidates identified by the “signature of selection” method may have no polymorphism within maize, making them difficult to map in populations with maize parents. The maize–teosinte BC1 population is useful for mapping such genes. Once mapped, it is possible to visualize whether they are located near QTL identified in this study. The signature and QTL mapping strategies are complementary. Wright et al. (2005) showed that candidate genes identified by a signature of selection during domestication clustered near previously identified QTL for morphological traits differentiating maize and teosinte (Doebley and Stec 1993). Conversely, researchers have looked for evidence of selection by sequence analysis of genes previously identified through QTL mapping to track down causative sites within them (Wang et al. 2005; Clark et al. 2006). Together, the large maize–teosinte RIL resource and the expanding volume of sequence data available across diverse Z. mays (www.panzea.org) will enhance our ability to understand the genetic basis of the morphological changes accompanying maize domestication.
Acknowledgments
We are grateful for statistical guidance from Brian Yandell and technical assistance from Traci Dusso, Jesse Rucker, and Janet Steffen. This work was supported by the National Science Foundation.
References
- Beavis, W. D., 1998. QTL analyses: power, precision, and accuracy, pp. 145–162 in Molecular Dissection of Complex Traits, edited by A. H. Patterson. CRC Press, Boca Raton, FL.
- Bomblies, K., and J. F. Doebley, 2006. Pleiotropic effects of the duplicate maize FLORICAULA/LEAFY genes zfl1 and zfl2 on traits under selection during maize domestication. Genetics 172: 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies, K., R. L. Wang, B. A. Ambrose, R. J. Schmidt, R. B. Meeley et al., 2003. Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development 130: 2385–2395. [DOI] [PubMed] [Google Scholar]
- Broman, K. W., H. Wu, S. Sen and G. A. Churchill, 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Brown, J., and V. Sundaresan, 1991. A recombination hotspot in the maize A1 intragenic region. Theor. Appl. Genet. 81: 185–188. [DOI] [PubMed] [Google Scholar]
- Butruille, D. V., R. P. Guries and T. C. Osborn, 1999. Linkage analysis of molecular markers and quantitative trait loci in populations of inbred backcross lines of Brassica napus L. Genetics 153: 949–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardon, F., B. Virlon, L. Moreau, M. Falque, J. Joets et al., 2004. Genetic architecture of flowering time in maize as inferred from quantitative trait loci meta-analysis and synteny conservation with the rice genome. Genetics 168: 2169–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G., R. Meeley, E. Irish, S. Hajime and S. Hake, 2006. The tasselseed4 gene encodes a negative regulator of floral homeotic gene expression. 48th Annual Maize Genetics Conference, Programs and Abstracts, Asilomar, Pacific Grove, CA p. 94.
- Clark, R., T. Nussbaum Wagler, P. Quijada and J. Doebley, 2006. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescence architecture. Nat. Genet. 38: 594–597. [DOI] [PubMed] [Google Scholar]
- Doebley, J., and A. Stec, 1991. Genetic analysis of the morphological differences between maize and teosinte. Genetics 129: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., and A. Stec, 1993. Inheritance of the morphological differences between maize and teosinte: comparison of results for two F2 populations. Genetics 134: 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., A. Bacigalupo and A. Stec, 1994. Inheritance of kernel weight in two maize-teosinte hybrid populations: implications for crop evolution. J. Hered. 85: 191–195. [Google Scholar]
- Doebley, J., A. Stec and C. Gustus, 1995. teosinte branched 1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141: 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doganlar, S., A. Frary, H. M. Ku and S. D. Tanksley, 2002. Mapping quantitative trait loci in inbred backcross lines of Lycopersicon pimpenellifolium (LA1589). Genome 45: 1189–1202. [DOI] [PubMed] [Google Scholar]
- Dooner, H. K., and I. M. Martinez-Ferez, 1997. Recombination occurs uniformly within the bronze gene, a meiotic recombination hotspot in the maize genome. Plant Cell 9: 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, M. D., C. W. Stuber and J. F. Wendel, 1987. Molecular-marker facilitated investigations of quantitative-trait loci in maize. I. Numbers, genomic distribution and types of gene action. Genetics 116: 113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinat, W. C., 1973. Intergenomic mapping of maize, teosinte and Tripsacum. Evolution 27: 644–655. [DOI] [PubMed] [Google Scholar]
- Gallavotti, A., Q. Zhao, J. Kyozuka, R. B. Meeley, M. K. Ritter et al., 2004. The role of barren stalk1 in the architecture of maize. Nature 432: 630–635. [DOI] [PubMed] [Google Scholar]
- Gardiner, J., S. Schroeder, M. L. Polacco, H. Sanchez-Villeda, Z. Fang et al., 2004. Anchoring 9,371 maize expressed sequence tagged unigenes to the bacterial artificial chromosome contig map by two-dimensional overgo hybridization. Plant Physiol. 134: 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helentjaris, T., D. Weber and S. Wright, 1988. Identification of genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics 118: 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurinke, C., D. van den Boom, C. R. Cantor and H. Koster, 2002. The use of MassARRAY technology for high throughput genotyping. Adv. Biochem. Eng./Biotech. 77: 58–74. [DOI] [PubMed] [Google Scholar]
- Koester, R. P., P. H. Sisco and C. W. Stuber, 1993. Identification of quantitative trait loci controlling days to flowering and plant height in two near isogenic lines of maize. Crop Sci. 33: 1209–1216. [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Lincoln, S. E., and E. S. Lander, 1992. Systematic detection of errors in genetic linkage data. Genomics 14: 604–610. [DOI] [PubMed] [Google Scholar]
- Maguire, M. P., 1961. Divergence in Tripsacum and Zea chromosomes. Evolution 15: 394–400. [Google Scholar]
- Schmidt, R. L., B. Veit, M. A. Mandel, M. Mena, S. Hake et al., 1993. Identification and molecular characterization of ZAG1, the maize homolog of the Arabidopsis floral homeotic gene AGAMOUS. Plant Cell 5: 729–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, S., and G. A. Churchill, 2001. A statistical framework for quantitative trait mapping. Genetics 159: 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuber, C. W., W. P. Williams and R. H. Moll, 1973. Epistasis in maize (Zea mays L.). III. Significance in predictions of hybrid performances. Crop Sci. 13: 195–200. [Google Scholar]
- Veit, V., R. J. Schmidt, S. Hake and M. F. Yanofsky, 1993. Maize floral development: new genes and old mutants. Plant Cell 5: 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., T. Nussbaum-Wagler, B. Li, Q. Zhao, Y. Vigouroux et al., 2005. The origin of the naked grains of maize. Nature 436: 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., C. J. Basten and Z. B. Zeng, 2006. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm.
- Wright, S. I., I. Vroh Bi, S. G. Schroeder, M. Yamasaki, J. F. Doebley et al., 2005. The effects of artificial selection on the maize genome. Science 308: 1310–1314. [DOI] [PubMed] [Google Scholar]
- Van Os, H., P. Stam, R. G. F. Visser and H. J. Van Eck, 2005. RECORD: a novel method for ordering loci on a genetic map. Theor. Appl. Genet. 112: 30–40. [DOI] [PubMed] [Google Scholar]
- Vigouroux, Y., M. McMullen, C. T. Hittinger, K. Houchins, L. Schulz et al., 2002. Identifying genes of agronomic importance in maize by screening microsatellites for evidence of selection during domestication. Proc. Natl. Acad. Sci. USA 99: 9650–9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, S., and J. F. Doebley, 1999. The molecular evolution of terminal ear 1, a regulatory gene in the genus Zea. Genetics 153: 1455–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki, M., M. I. Tenaillon, I. Vroh Bi, S. G. Schroeder, H. Sanchez-Villeda et al., 2005. A large-scale screen for artificial selection in maize identifies candidate agronomic loci for domestication and crop improvement. Plant Cell 17: 2859–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Q., 2006. Molecular population genetics of maize regulatory genes during domestication. Ph.D. Dissertation, University of Wisconsin, Madison, WI.