Abstract
The size of eukaryotic genomes can vary by several orders of magnitude, yet genome size does not correlate with the number of genes nor with the size or complexity of the organism. Although “whole”-genome sequences, such as those now available for 12 Drosophila species, provide information about euchromatic DNA content, they cannot give an accurate estimate of genome sizes that include heterochromatin or repetitive DNA content. Moreover, genome sequences typically represent only one strain or isolate of a single species that does not reflect intraspecies variation. To more accurately estimate whole-genome DNA content and compare these estimates to newly assembled genomes, we used flow cytometry to measure the 2C genome values, relative to Drosophila melanogaster. We estimated genome sizes for the 12 sequenced Drosophila species as well as 91 different strains of 38 species of Drosophilidae. Significant differences in intra- and interspecific 2C genome values exist within the Drosophilidae. Furthermore, by measuring polyploid 16C ovarian follicle cell underreplication we estimated the amount of satellite DNA in each of these species. We found a strong correlation between genome size and amount of satellite underreplication. Addition and loss of heterochromatin satellite repeat elements appear to have made major contributions to the large differences in genome size observed in the Drosophilidae.
THE evolutionary processes associated with the wide spectrum of eukaryotic genome sizes have eluded biologists for decades. The so-called “C-value paradox” refers to our lack of understanding as to how and why there is so much variation in eukaryotic genome size (for reviews see Hartl 2000; Petrov 2001). For example, the mountain grasshopper Podisma has an estimated genome size 100-fold that of the fruit fly Drosophila melanogaster and ∼6-fold larger than the human genome (Hartl 2000; Bensasson et al. 2001; Petrov 2001). Genome size in these examples clearly does not correlate with the number of genes found in each genome or with the complexity of the organism. It appears, instead, that the vast differences in genome size are a result of repetitive DNA sequences that litter eukaryotic genomes in one form or another (Hartl 2000). These observations raise several interesting questions: First, how have genomes of closely related species changed and have repetitive sequences contributed to the evolution of closely related genomes and distantly related species alike? Second, what are the molecular mechanisms through which genomes change their DNA content? Finally, and most interestingly, are such changes in eukaryotic genome size under selection? The availability of genome sequences, especially of closely related species such as the 12 Drosophila genomes, now make it possible to compare whole genomes and address some of these questions.
How have genomes changed? Various models have attempted to describe how genomes have evolved to contain more or less DNA (for reviews see Britten and Davidson 1971; Hartl 2000; Petrov 2001, 2002). Using Drosophila, studies attempting to detect global trends in genome size have focused on measurements of transposable elements, pseudogenes, intron, exon, and intergenic lengths (Petrov et al. 1996; Moriyama et al. 1998; Petrov and Hartl 1998). Such studies have been illuminating and suggest that global forces determine the growth and contraction of disparate genomic elements. For example, large genomes tend to have larger intergenic distances, introns, and exons (Moreau et al. 1985). However, repetitive DNA sequences account for the bulk of the vast differences that have been reported (Hartl 2000). For example, the closely related D. nasutoides and D. simulans have been reported to have 56 and 5% satellite repeat DNA, respectively (Zacharias 1986; Lohe and Brutlag 1987).
By what mechanisms have these genomes changed size? Random deletions/insertions, polyploidization, and proliferation of transposable elements are thought to contribute to genome change (for review see Hartl 2000). Also, certain sequences, for example, repetitive elements typical of heterochromatin, may have repeat-specific shrinkage mechanisms, such as unequal meiotic exchange between sister chromatids or replication errors (Britten and Kohne 1968; Southern 1975; Smith 1976; Stephan and Cho 1994; Petrov 2001). Understanding the levels and distributions of heterochromatic repetitive elements across a range of related species will aid in discriminating among the potential responsible mechanisms.
Given that most eukaryotic genomes contain vast amounts of repetitive sequences (Hartl 2000), understanding how these sequences contribute to genome evolution is critical. Moreover, it is becoming increasingly clear that heterochromatic repeats and tandem array repeats are not “junk DNA,” but rather serve critical functions, such as meiotic chromosome pairing, epigenetic maintenance of centromere function, and other epigenetic processes (Hawley et al. 1993; Dernburg et al. 1996; Sun et al. 1997; Allshire 2002; Reinhart and Bartel 2002; Cam et al. 2005; Chandler 2007). However, the repetitive nature of heterochromatic and other DNAs makes them difficult to clone and sequence (Sun et al. 2003). Consequently, assembled genome sequences often do not accurately represent heterochromatic content and thus underestimate total genome size as well as repeat sequence content.
Genome size estimates are available for 70 species of the family Drosophilidae (Powell 1997; Ashburner et al. 2005; http://www.genomesize.com) and clearly exhibit large differences among and within species. Multiple estimates exist for several species and suggest intraspecific genome size differences of up to 50% for some. In strains of D. melanogaster, the intraspecific genome size variation was attributed to differences in heterochromatin content (Halfer 1981). Scant information is available, however, regarding the heterochromatin satellite DNA content of many other species, and thus available genome size estimates have limited usefulness in addressing evolutionary questions. The majority of existing estimates are from unpublished studies and thus details regarding the methodology, tissues, and strains used cannot be ascertained. Remaining estimates were performed with a range of different techniques, such as flow cytometry, Feulgen densitometry, molecular weight determinations, and sequencing, and employed different tissue types such as ovaries, sperm, testes, brains, whole bodies, and hemacytes. These methodological inconsistencies, coupled with an absence of information on the contribution of various repeat sequences to the observed genome size variability, necessitate a new approach that will provide accurate simultaneous measures of both genome size and satellite DNA content across the Drosophilidae. Of special interest are those 12 species for which whole-genome sequences are now available (http://rana.lbl.gov/drosophila/).
In this study, we address the following questions: (1) What is the range of genome sizes across the Drosophilidae?, (2) What is the range of variation within species for genome size?, and (3) What is the contribution of heterochromatic satellite DNA to intra- and interspecific variability in genome size? To address these questions, we ascertained the genome sizes of 91 strains from 38 species within the Drosophilidae, including the 12 sequenced species (http://rana.lbl.gov/drosophila/). Using flow cytometry, we determined the genome sizes and the fraction of each of these genomes that is underreplicated in ovarian follicle cells. Although follicle cells from all 38 species terminate with 16 complement (16C) ploidy, we observed dramatic differences in the fraction of the 2 complement (2C) genome that is actually replicated in each species. This indicates measurable differences in underreplicated satellite content. We also found a strong correlation between genome size and amount of satellite DNA, suggesting that variation in heterochromatic DNA contributes significantly to genome size evolution in the Drosophilidae.
MATERIALS AND METHODS
Species and strains used:
To identify potential strain differences, we examined more than one strain of each species—a total of 91 different strains from 38 species. All strains and species are available for future analysis and most are banked in the Tucson Drosophila Species Stock Center and the Bloomington (Bl) Drosophila Stock Center (supplemental Table 3 at http://www.genetics.org/supplemental/). One strain (H2AvD-GFP; Clarkson and Saint 1999) and one D. virilis strain (no. 2465, origin unknown but likely from M. Pardue, Massachusetts Institute of Technology) are available upon request from G. Bosco. Since Bloomington stock numbers can change over time, genotypes for each D. melanogaster strain are shown in supplemental Table 3 at http://www.genetics.org/supplemental/.
Preparation of nuclei and flow cytometry:
We dissected 10–20 ovary pairs in Grace's insect medium (GIBCO, Grand Island, NY) and placed them into 1.7-ml tubes with 0.8 ml of medium. Grace's medium was removed and 700 μl filtered ice-cold PARTEC buffer (200 mm Tris–HCI ph 7.4, 4 mm MgCl2, 0.1% Triton X-100) was added to the 1.7-ml tube with the ovaries and then placed into a 60-mm petri dish and homogenized with a single-edged razor blade. Chopped ovaries were filtered twice over cheesecloth (∼3 cm2) and once through a 30-μm mesh (Sefar) and collected in a flow cytometry tube (Sarstedt). Another 700 μl of PARTEC buffer was used to wash the petri dish, filtered, and pooled into flow cytometry tubes.
Two nucleic-acid-binding fluorescent dyes were used, propidium iodide (PI) and 4′,6-diamidino-2-phenylindole (DAPI). For DAPI staining, nuclei in tubes were placed on ice and 20 μl of DAPI (100 μg/ml) were added. Samples were analyzed on a PARTEC CCA-II flow cytometry machine (PARTEC). For PI staining, we used the same protocol as above with the addition of 50 μl RNase A (1 mg/ml) and 100 μl PI (1 mg/ml) to each sample. PI measurements were done on a FACScan flow cytometer (Becton Dickinson) at several thousand nuclei per second.
For both DAPI and PI measurements, each sample was compared to a D. melanogaster control (y1w1 Bloomington no. 1495, hereafter referred to as D.m. yw) that was prepared at the same time for each sample. Both PARTEC CCA-II and FACScan machines were calibrated to flow rates and gain settings for the D.m. yw control. In all cases, a minimum of three biological replicates was performed on each strain, and a minimum of 104 nuclei was measured for each replicate.
Determination of flow cytometry values and statistical analysis:
Histograms exhibiting four peaks (2C, 4C, 8C, and 16C) were obtained for polyploid follicle cells (Figure 1). The mean fluorescence intensity for each peak was obtained and this fluorescence value is proportional to DNA content as previously described for follicle cell nuclei (Lilly and Spradling 1996; Leach et al. 2000; Bosco et al. 2001). As ANOVA revealed no significant differences among replicates for a given strain, they were averaged (data not shown and Table 1). This average fluorescent intensity was divided by its D.m. yw control, yielding a normalized estimate of 2C DNA content, relative to D.m. yw. For each of the three biological replicates for each strain, 16C/2C ratios were determined and then averaged to obtain an average 16C/2C ratio for each strain.
Figure 1.—
Drosophila polyploid follicle cells underreplicate satellite DNA repeats. Proliferating follicle cells duplicate their entire genomes and cycle from 2C to 4C and after mitotic division back to 2C (A). 2C cells enter their polyploid state by replicating their euchromatic sequences and replicate little or no centric/pericentric satellite repeat sequences (B). Consequently, 4c-p cells have less 4C DNA content, and a second and third round of polyploid S-phases produce 16C cells with vastly underreplicated satellite DNA. Flow cytometry histograms of follicle cell nuclei from (C) D. melanogaster, (D) D. grimshawi, (E) D. immigrans, and (F) D. virilis are shown by illustrating the four major 2C, 4C, 8C, and 16C ploidy peaks where the x-axis represents arbitrary fluorescent units and the y-axis is the number of nuclei. Note that the 4C peak can be resolved into two peaks (see insets in C and F), where the 4C peak from mitotic proliferating cells has more DNA content than the 4C-p peak. This is because follicle cells undergoing polyploidization fail to replicate the centric and pericentric satellite repeats and thus have less DNA than mitotic 4C cells, as described in A. In larger genomes such as (D) D. grimshawi, (E) D. immigrans and (F) D. virilis, the extent of underreplication can be seen by a dramatic shift of all polyploid peaks to the left. The most extreme example is seen in (F) D. virilis where the 8C peak nearly overlaps the normal mitotic cell 4C peak (see inset), suggesting that about half of the genome fails to replicate. This is consistent with measurements of ∼48% heterochromatin content in D. virilis (see Table 5). We observed underreplication in all 91 strains from all 38 species that we examined.
TABLE 1.
Fold difference for multiple D. melanogaster and D. virilis strains
| PI
|
DAPI
|
|||
|---|---|---|---|---|
| Strain no. | 2C ± SE | 16C/2C ± SE | 2C ± SE (A:T corrected) | 16C/2C ± SE |
| D. melanogaster | ||||
| Bl 2057 | 1.01 ± 0.09 | 6.15 ± 0.29 | 1.67 ± 0.01 (1.25) | 6.06 ± 0.04 |
| Bl 1495 | 1.00 ± 0 | 6.30 ± 0.03 | 1.00 ± 0.07 (0.99) | 9.31 ± 0.13 |
| Bl 4455 | 0.99 ± 0.01 | 6.09 ± 0.01 | 1.19 ± 0.05 (1.06) | 8.83 ± 0.10 |
| Bl 6599 | 1.32 ± 0.03 | 5.49 ± 0.17 | 1.29 ± 0.05 (1.10) | 9.17 ± 0.17 |
| Bl 1785 | 0.97 ± 0.01 (0.98) | 10.65 ± 0.07 | ||
| Bl 576 | 1.13 ± 0.08 (1.04) | 9.06 ± 0.08 | ||
| Bl 1633 | 1.08 ± 0.06 (1.02) | 8.85 ± 0.03 | ||
| H2AvD-GFP | 1.11 ± 0.02 (1.03) | 9.09 ± 0.10 | ||
| Bl 4269 | 1.12 ± 0.04 (1.03) | 9.01 ± 0.05 | ||
| Bl 189 | 1.04 ± 0.11 (1.00) | 9.00 ± 0.14 | ||
| D. virilis | ||||
| 15010-1051.00 | 1.97 ± 0.03 | 4.44 ± 0.07 | 2.71 ± 0.01 (1.64) | 6.12 ± 0.03 |
| 15010-1051.45 | 2.09 ± 0.04 | 4.73 ± 0.04 | 2.54 ± 0.03 (1.58) | 6.08 ± 0.02 |
| 15010-1051.46 | 2.25 ± 0.08 (1.47) | 5.87 ± 0.01 | ||
| 15010-1051.87 | 1.78 ± 0.06 | 4.57 ± 0.02 | 2.38 ± 0.15 (1.52) | 5.86 ± 0.03 |
| 2465 | 1.70 ± 0.30 | 5.40 ± 1.16 | 2.34 ± 0.11 (1.50) | 5.21 ± 0.02 |
2C and 16C/2C values were obtained for multiple strains of D. melanogaster and D. virilis using either PI or DAPI dyes in flow cytometric measures of the genome size of ovarian follicle cell nuclei. All values represent averages of three biological replicates, except for D. melanogaster Bl 1495 and Bl 2057, which were measured in four and six biological replicates, respectively. Standard error (±SE) is shown for each value. DAPI values corrected for A:T bias fluorescence as described in Figure 2A and in the materials and methods are shown in parentheses (A:T corrected). Note that, before bias correction, the DAPI values for D. virilis are much higher than the PI 2C values whereas this dye effect is minimal in D. melanogaster 2C values. This reflects a greater total A:T content in D. virilis.
Conversion of 2C values to picograms and megabases:
To convert relative genome sizes to megabase values, we produced a best-fit regression line for experimentally measured 2C flow cytometry values and the corresponding published genome sizes for D. melanogaster and D. virilis. (Laird 1971, 1973; Rasch et al. 1971; Kavenoff and Zimm 1973; Mulligan and Rasch 1980; Celniker et al. 2002; Hoskins et al. 2002; Bennett et al. 2003). Two best-fit curves (one for PI and another for DAPI) were obtained, which then were used to convert 2C measurements into megabase values. The advantage of this method is that it takes into account complex relationships between 2C flow cytometry values and DNA content for different species. One disadvantage is the lack of information on the D. virilis strains used previously for genome size estimates. Consequently, we used an average from two different studies (Kavenoff and Zimm 1973; Laird 1973) and must assume that these D. virilis strains are sufficiently close to the five strains examined in this study.
Relative 2C values used for conversion to megabases are shown in supplemental Tables 1 and 2 at http://www.genetics.org/supplemental/. DAPI relative 2C values were first corrected for A:T bias as described below and in Figure 2A. Picograms were calculated from megabases based on the conversion 0.1 pg = 97.8 Mb.
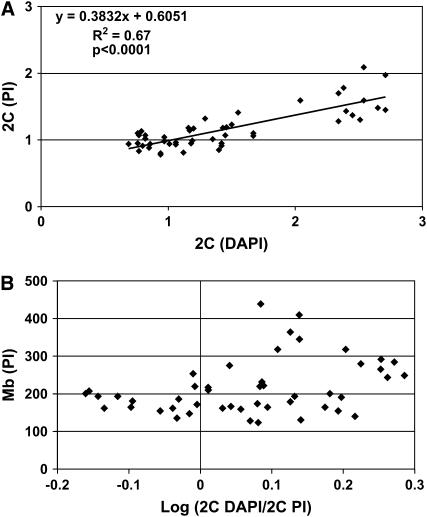
Figure 2.—
DAPI measurements overestimate DNA content. (A) 2C values relative to D.m. yw control for DAPI (x-axis) were plotted against their corresponding 2C values for PI (y-axis). A trend line was fit to ascertain how DAPI values change relative to PI values. A slope that is <1 shows that DAPI values increase at a greater rate than PI values. This indicates that as genomes become larger DAPI overestimates DNA content (see text for details) and thus must be corrected. A two-tailed P-value was calculated from the correlation coefficient (R) and 45 degrees of freedom (d.f.) using Graphpad software. (B) DAPI fluorescence has a A:T bias whereas PI does not. The 2C DAPI/2C PI ratio values for each strain reflect the overall A:T/G:C content of each genome. The log (2C DAPI/2C PI) values (x-axis) and the corresponding haploid genome size (y-axis) values, as determined by PI 2C, are shown. Note that these measurements are for total genomic A:T/G:C content and may differ substantially from estimates of euchromatic A:T/G:C sequence content.
Estimates of underreplicated satellite content:
The expected DNA content of 16C polyploid follicle cells is eight times the raw 2C value (8 × 2C). Observed raw 16C values obtained from PI flow cytometry are less than the expected values because heterochromatic sequences do not replicate completely if at all in follicle cells (Figure 1) (Gall et al. 1971; Hammond and Laird 1985a; Lilly and Spradling 1996; Leach et al. 2000). Thus, the difference between the expected and the observed 16C values reflects the fraction of each genome that is underreplicating satellite repeats [(8 × 2C) − 16C]. For values obtained by PI fluorescence, the following formula was used to calculate the percentage of underreplication in 16C follicle cells: [(8 × 2C) − 16C]/(8 × 2C) × 100. The percentage of underreplication is an estimate for the heterochromatic satellite DNA content in each genome.
Determination of the expected 16C ploidy DNA contents (i.e., 8 × 2C) with DAPI data is confounded by the fact that DAPI values are skewed by A:T content, and therefore 2C values and 16C values reflect DNA content plus A:T richness. Consequently, estimates of underreplication determined by DAPI will be less precise than those derived from PI measurements, and DAPI values must first be normalized for the A:T bias. To normalize DAPI 16C/2C values, we used the following formula: normalized DAPI 16C/2C = [(PI 16C/2C D.m. yw)/(DAPI 16C/2C D.m. yw)] × DAPI 16C/2C for each strain. Normalized DAPI percentages of underreplication values were determined by multiplying the normalized DAPI 16C/2C by 26%. Because we determined a mean 26% underreplication for four D. melanogaster strains by using PI (Table 4), the mean 26% value was used to convert 16C/2C values that were normalized to D. melanogaster.
TABLE 4.
Genome size and predicted percentage of satellite DNA
| Species strain no. | pg ± SE | Mb ± SE | Assembly sizea | % satellite DNA |
|---|---|---|---|---|
| D. sechellia 14021-0248.25 | 0.17 ± 0.004 | 171 ± 4 | 167 | 24 ± 0 |
| D. simulans 14021-0251.195 | 0.17 ± 0.002 | 162 ± 2 | 142 | 17 ± 1 |
| D. melanogaster 14021-0231.36 | 0.20 ± 0.017 | 200 ± 18 | 130 | 24 ± 3 |
| D. yakuba 14021-0261.01 | 0.19 ± 0.011 | 190 ± 11 | 169 | 23 ± 2 |
| D. erecta 14021-0224.01 | 0.14 ± 0.004 | 135 ± 4 | 153 | 9 ± 2 |
| D. ananassae 14024-0371.13 | 0.22 ± 0.009 | 217 ± 9 | 231 | 23 ± 2 |
| D. pseudoobscura 14011-0121.94 | 0.20 ± 0.004 | 193 ± 4 | 153 | 14 ± 4 |
| D. persimilis 14011-0111.49 | 0.20 ± 0.005 | 193 ± 5 | 188 | 14 ± 1 |
| D. willistoni 14030-0811.24 | 0.23 ± 0.008 | 222 ± 7 | 237 | 12 ± 1 |
| D. virilis 15010-1051.87 | 0.37 ± 0.013 | 364 ± 13 | 206 | 44 ± 1 |
| D. mojavensis 15081-1352.22 | 0.13 ± 0.0 | 130 ± 0 | 194 | 2 ± 1 |
| D. grimshawi 15287-2541.00 | 0.24 ± 0.005 | 231 ± 5 | 200 | 32 ± 0.4 |
Predicted genome sizes for the 12 sequenced Drosophila species. Values, in picograms and megabases, and standard error (±SE) for each strain from propidium iodide flow cytometry measurements are shown. The predicted percentage and standard error (±SE) of underreplicated heterochromatic satellite DNA is shown for each specific strain.
For comparison, the total assembled sequenced genomes in megabases are shown (http://insects.eugenes.org/species/data). All percentage of satellite DNA estimates are from this study.
Chromocenter measurements and immunofluorescence:
Ovaries were dissected and prepared for DAPI (0.05 μg/ml final) and immunofluorescence (Hartl et al. 2007). Rabbit antidimethyl lysine-9 on histone H3 (Upstate) was used at 1:100 dilution and visualized with Cy3-goat anti-rabbit (Jackson ImmunoResearch) at 1:250 dilution. Stage 13 follicle cell nuclei were imaged with a Nikon Eclipse E800 microscope and a ×40 objective using a RT Monochrome SPOT Model 2.1.1 camera. All settings were kept identical for all samples although background signal varied among samples. Nuclear and chromocenter areas were determined with the Adobe Photoshop 7.0 Polygonal Lasso tool, and the total areas for each nucleus and chromocenter were determined in pixels using the Image histogram function. The area of the chromocenter, as determined by DAPI and histone H3 dimethyl-lysine-9, was normalized to the total nuclear area. An average normalized chromocenter area for each species was calculated. For each of the three species examined, 35 different cells were measured. Standard errors and P-values using a two-tailed test were determined using MS Excel.
RESULTS
Fluorescent flow cytometry can accurately estimate genome size:
As genome size estimates were previously available for D. melanogaster (Laird 1971; Rasch et al. 1971; Kavenoff and Zimm 1973; Mulligan and Rasch 1980; Celniker et al. 2002; Bennett et al. 2003) and D. virilis (Kavenoff and Zimm 1973; Laird 1973), we assessed the ability of PI and DAPI flow cytometry to accurately reproduce the previously described genome size differences for these two species. For example, previous estimates described the D. virilis genome to be much larger than D. melanogaster and to have a higher heterochromatin content (Gall et al. 1971; Schweber 1974). We conducted a set of preliminary studies on multiple strains of D. melanogaster and of D. virilis and determined the fluorescence intensity for follicle cell nuclei with 2C and 16C ploidy, relative to D. melanogaster yw controls (Table 1). We performed flow cytometry using PI fluorescence for four D. melanogaster and four D. virilis strains. Using PI as the dye, ANOVA detected significant species differences, but not strain or replicate differences in 2C values or 16C/2C values (Table 2A). ANOVA performed on measurements of the same 4 plus 6 additional D. melanogaster strains (10 total) and on the same 4 plus 1 additional (5 total) D. virilis strains with DAPI revealed significant species and strain, but not replicate, differences (Table 2B). We conclude that both dyes detect interspecific genome size differences. Comparison of the DAPI 2C values for each of the D. virilis strains to D.m. yw revealed a 2.25- to 2.71-fold difference. For PI 2C values, there was a 1.7- to 2.09-fold difference between D. virilis and D.m. yw (Table 1). Our 2C values fit very well with values for D. virilis genome sizes previously estimated to be 1.75- to 2.26-fold larger than D. melanogaster (Kavenoff and Zimm 1973; Laird 1973; J. Spencer Johnston as referenced in Table 5.2 of Ashburner et al. 2005). This and previously published work demonstrate that flow cytometry provides a valid method for determining genome size when an appropriate control is used (Johnston et al. 1999; Bennett et al. 2003).
TABLE 2.
ANOVA analysis of species and strains
| Mean square | Sum-of-squares | F-ratio | P-value | |
|---|---|---|---|---|
| A. PI: D. melanogaster and D. virilis | ||||
| 2C/2C | ||||
| Species | 2.88491 | 2.8849099 | 53.9511 | <0.001 |
| Strain | 0.021983 | 0.0659505 | 0.0975 | 0.9604 |
| Replicate | 0.029462 | 0.0589232 | 0.1380 | 0.8720 |
| 16C/2C | ||||
| Species | 7.66103 | 7.661029 | 12.7873 | 0.0020 |
| Strain | 0.21726 | 0.651793 | 0.2008 | 0.8944 |
| Replicate | 0.582162 | 1.164325 | 0.5861 | 0.5668 |
| B. DAPI: D. melanogaster and D. virilis | ||||
| 2C/2C | ||||
| Species | 16.4818 | 16.481775 | 362.8012 | <0.001 |
| Strain | 0.709896 | 6.389066 | 2.0626 | 0.0610 |
| Replicate | 0.018108 | 0.036216 | 0.0413 | 0.9565 |
| 16C/2C | ||||
| Species | 94.6818 | 94.68176 | 111.4805 | <0.001 |
| Strain | 5.52161 | 49.69453 | 2.3710 | 0.0327 |
| Replicate | 0.01223 | 0.02446 | 0.0039 | 0.9961 |
Effects of dye on genome size measures:
In general, 2C DAPI values for most strains, relative to D. melanogaster, were elevated when compared to 2C values obtained by PI (Table 1 and supplemental Tables 1 and 2 at http://www.genetic.org/supplemental/). DAPI binding preference for A:T sequences has been physically documented (Wilson 1990; Colson et al. 1995, 1996), and its preferential fluorescence for A:T-rich DNA in flow cytometry also has been described (Johnston et al. 1999; Meister 2005). Moreover, cytological changes in DAPI fluorescent intensity accurately correlate with physical changes in A:T-rich repeat content in D. melanogaster polyploid cells (Lilly and Spradling 1996; Royzman et al. 2002). Discrepancies between DAPI and PI 2C values therefore suggest that most, but not all, species have A:T-rich genomes.
Given the A:T content bias, we plotted DAPI 2C values against PI 2C values to assess whether only some or most species exhibit a DAPI bias (Figure 2A). If DAPI and PI values are equivalent, we would expect a linear relationship with a slope approximately equal to 1. Interestingly, although DAPI values increased with PI values, DAPI values increased at a greater rate (Figure 2A). The trend was highly significant (P < 0.0001), consistent with larger genomes having more A:T-rich satellite DNA, thus leading to an exaggerated DAPI signal. A simple conversion from DAPI-derived values to picograms or megabases thus was not possible, especially for A:T-rich genomes, without first performing a correction. A linear regression predicts that PI (y) values change in relation to DAPI (x) values as described by the equation y = 0.3832x + 0.6051 (Figure 2A). We employed a DAPI correction factor that allowed us to account for A:T bias in DAPI fluorescence values where the corrected DAPI 2C value = 0.3832(observed DAPI 2C) + 0.6051. The linear regression shown in Figure 2A was then utilized to determine the A:T-content-corrected DAPI 2C values (Table 1, A:T corrected). These corrected values were then used to determine genome sizes (Table 3).
TABLE 3.
Mean genome size and range
| Species | Mean ± SE (n) PI (Mb) | Range of PI | Mean ± SE (n) DAPI (Mb) | Range of DAPI | Previous estimate (Mb) |
|---|---|---|---|---|---|
| C. pararufithorax | 284 ± 6 (1) | — | 429 ± 6 (1) | — | — |
| C. procnemis | 318 ± 6 (1) | — | 260 ± 7 (1) | — | — |
| C. rufithorax | 292 ± 6 (1) | — | 420 ± 13 (1) | — | — |
| D. acutilabella | 172 ± 4 (2) | 168–176 | — | ||
| D. americana | 275 ± 4 (1) | — | 240 ± 14 (2) | 226–254 | 328a (MW) |
| D. ananassae | 215 ± 5 (3) | 210–217 | 198 ± 2 (3) | 195–202 | 205b (CY) |
| D. buskii | 194 ± 5 (1) | — | 144b (CY) | ||
| D. equinoxialis | 304 ± 9 (2) | 295–313 | 248b (CY) | ||
| D. erecta | 145 ± 10 (2) | 135–154 | 139 ± 2 (2) | 137–141 | 159b (CY) |
| D. funebris | 330 ± 22 (1) | — | 269c (KI) | ||
| D. grimshawi | 231 ± 5 (1) | — | 247 ± 13 (1) | — | 247d (FD) |
| D. guttifera | 160 ± 21 (2) | 140–181 | 188 ± 44 (2) | 144–232 | — |
| D. hydei | 164 ± 16 (1) | — | 177 ± 22 (2) | 155–199 | 197–246e (CY, KI, MW) |
| D. immigrans | 299 ± 19 (2) | 279–318 | 347 ± 18 (3) | 328–382 | — |
| D. littoralis | 238 ± 5 (1) | — | — | ||
| D. melanogaster | 201 ± 16 (4) | 174–253 | 195 ± 10 (10) | 167–272 | 176-180f (CY) |
| D. mercatorum | 128 ± 5 (1) | — | 166 ± 4 (2) | 162–170 | — |
| D. mimica | 257 ± 6 (4) | 243–270 | 387 ± 8 (3) | 373–399 | — |
| D. mojavensis | 152 ± 11 (3) | 130–166 | 183 ± 3 (3) | 180–189 | 215g (BC) |
| D. nannoptera | 236 ± 35 (3) | 173–295 | — | ||
| D. novamexicana | 244 ± 20 (2) | 224–265 | — | ||
| D. persimilis | 183 ± 10 (3) | 164–193 | 170 ± 34 (3) | 135–239 | 197b (CY) |
| D. pseudoobscura | 185 ± 12 (3) | 162–200 | 135 ± 6 (3) | 125–144 | 168b (CY) |
| D. repleta | 167 ± 13 (3) | 153–192 | — | ||
| D. sechellia | 166 ± 5 (2) | 162–171 | 170 ± 3 (2) | 167–173 | 167b (CY) |
| D. simulans | 160 ± 11 (6) | 123–207 | 170 ± 18 (7) | 119–235 | 139–153bc (CY, KI) |
| D. virilis | 404 ± 21 (4) | 364–438 | 389 ± 12 (5) | 373–429 | 307–394bh (CY, MW) |
| D. willistoni | 206 ± 14 (3) | 178–222 | 234 ± 5 (3) | 224–241 | 235i (UN) |
| D. yakuba | 188 ± 2 (2) | 186–190 | 220 ± 53 (2) | 167–272 | 173b (CY) |
| Hirtodrosophila duncani | 333 ± 9 (1) | — | — | ||
| S. latifasciaeformis | 313 ± 27 (2) | 286–340 | 195b (CY) | ||
| S. lebanonensis | 259 ± 2 (2) | 257–260 | 210b (CY) | ||
| S. palmae | 168 ± 9 (1) | — | — | ||
| S. stonei | 300 ± 11 (2) | 289–311 | 207b (CY) | ||
| Zaprionus badyi | 253 ± 6 (1) | — | — | ||
| Z. ghesquerei | 153 ± 7 (1) | — | — | ||
| Z. sepsoides | 352 ± 71 (2) | 281–423 | — | ||
| Z. tuberculatus | 299 ± 74 (3) | 247–384 | — |
Mean values for PI and corrected DAPI measurements are for haploid genome size and are from this study. Standard error (SE), the range (lowest and highest values), and the number of strains for each species (n) are shown. See supplemental data at http://www.genetics.org/supplemental/ for specific strain values. DAPI values for larger genomes tend to be less accurate than PI values (see text). For comparison, previously reported genome size estimates are listed in the right-most column. Methods used for determining previous estimates are biochemical analysis (BC), cytometry (CY), kinetics (KI), Feulgen densitometry (FD), and molecular weight (MW), or method unknown (UN).
From Kavenoff and Zimm (1973).
From J. Spencer Johnston as quoted in Table 5.2 of Ashburner et al. (2005).
From Rasch (1985).
From Schulze and Lee (1986).
From Kavenoff and Zimm (1973) and Laird (1971).
From Powell (1997).
Total A:T content is positively correlated with genome size:
The relative A:T/G:C content of different species can be estimated from the 2C DAPI/2C PI ratio (Meister 2005). We took advantage of this DAPI bias to ask how A:T content varies among these Drosophila species and whether A:T content was correlated to genome size as suggested by the trend in Figure 2A. Of 48 strains tested (from 30 different species), 33 had log DAPI/PI values greater than zero, indicating that most genomes are A:T rich (Figure 2B, supplemental Table 4 at http://www.genetics.org/supplemental/). We observed that, although some smaller genomes are A:T rich, the largest genomes (>250 Mb) are the most A:T rich. Fourteen strains exhibited log DAPI/PI values less than zero, indicating a relatively high G:C content. Interestingly, these G:C-rich genomes were almost exclusively the smallest genomes (<200 Mb), consisting of multiple strains of D. persimilis, D. pseudoobscura, D. simulans, and D. erecta (Figure 2B, supplemental Table 4 at http://www.genetics.org/supplemental/). This was most pronounced in D. persimilis and D. pseudoobscura. Consequently, DAPI measurements may underestimate sizes of these genomes and are expected to be lower than PI-derived values, which is in fact what we observed (Table 3).
Genome size estimates:
After establishing the efficacy of flow cytometry measurements of 2C Drosophila follicle cells for predicting genome size, we then estimated genome sizes for 91 strains from 38 different species of Drosophilidae. For some species, only 1 strain was available, while for others as many as 10 were tested. Values obtained for individual strains are available in supplemental Tables 1 and 2 at http://www.genetics.org/supplemental/). All 38 species were measured with DAPI and 21 also were measured with PI (Table 3). Using PI, the smallest genomes were seen in D. mercatorum, D. mojavensis, and D. erecta while D. virilis had the largest. While this pattern was also seen with DAPI, Chymomyza pararufithorax's and C. rufithorax's genomes were slightly larger than that of D. virilis.
Follicle cell underreplication is inversely proportional to genome size in all species:
We took advantage of the fact that D. melanogaster follicle cells that normally become polyploid and have 16C do not completely replicate the centric- and peri-centric heterochromatic satellite DNA (Gall et al. 1971; Hammond and Laird 1985a,b; Lilly and Spradling 1996; Leach et al. 2000). Follicle cells undergo three rounds of endoreduplication and terminate with 16C ploidy, as indicated by four major peaks when nuclei are analyzed by fluorescence flow cytometry (Figure 1A). Using this method, we determined that in all 91 strains of 38 species follicle cells terminate DNA replication with 16C ploidy (Figure 1; supplemental Tables 1 and 2 at http://www.genetics.org/supplemental/; data not shown). Regulation of follicle cell ploidy thus is a well-conserved developmental process.
An additional 4C-polyploid (4C-p, Figure 1) peak is also evident in the flow cytometry histograms because the majority of the heterochromatic sequences are not completely replicated as these cells progress from 2C through their first polyploid S-phase, resulting in a 4C-p content that has less DNA than a 4C cell undergoing mitosis (Hammond and Laird 1985a; Lilly and Spradling 1996; Leach et al. 2000). The 4C-p peak, since it emits less fluorescence, is always shifted to the left, relative to the 4C peak (Figure 1). The extreme example is satellite repeats that have been estimated to remain at their 2C copy number as polyploidy ensues (Gall et al. 1971; Lilly and Spradling 1996; Leach et al. 2000). For some species, where extensive underreplication occurs, a distinct additional 4C-p peak is evident (Figure 1, C and F, insets). Moreover, the 16C peaks from different species such as D. melanogaster and D. virilis are only slightly shifted from one another (compare Figure 1, C and F), indicating that the actual 16C DNA content of these species is not that different. This similarity is observed despite the fact that D. virilis 2C DNA content is ∼1.8-fold greater than that of D. melanogaster (Tables 1 and 4).
We found that the mean PI fluorescence ratio of the 16C/2C values is always <8 (Table 1; Figure 3A; supplemental Table 1 at http://www.genetics.org/supplemental/). The 16C/2C ratio indicates that portion of a given genome that does not fully replicate in follicle cells. Species with larger genomes are expected to have more heterochromatic repeats and therefore replicate a smaller fraction of their 2C genomes in their follicle cells. Indeed, when we plot 2C and 16C/2C values, a clear trend is revealed where larger genomes have smaller 16C/2C ratios (Figure 3A). When we plot DAPI 16C/2C ratios against their corresponding 2C values, we also see a clear negative correlation (Figure 3B; supplemental Tables 1 and 2 at http://www.genetics.org/supplemental/). Taken together, these data indicate that Drosophila species other than D. melanogaster also underreplicate their satellite sequences in follicle cells.
Figure 3.—
The 16C/2C ratios are inversely proportional to 2C values. 16C/2C ratios were compared to their corresponding 2C values for PI (A) and DAPI (B) values. In each case, 16C/2C values decrease as genomes increase in size, indicating that a larger fraction of the genome is being underreplicated. A two-tailed P-value was calculated from the correlation coefficient (R) using Graphpad software. PI values (A) had 45 d.f. and DAPI values (B) had 90 d.f.
Estimates for underreplicating the percentage of satellite DNA:
Although 20% of the D. melanogaster genome is estimated to be satellite sequence (Lohe and Brutlag 1986), cytological methods and recent heterochromatin sequencing efforts place the heterochromatin content at ∼33% (Gatti et al. 1976; Hoskins et al. 2002). By using the 16C/2C ratio we were able to estimate the genomic fraction of each genome that is underreplicated. PI values for 16C/2C indicate underreplication of ∼20–31% of the D. melanogaster genome in 16C follicle cells while DAPI values show 23–40% (Table 5). These values are surprisingly close to those reported for D. melanogaster satellite DNA and heterochromatin content (Gatti et al. 1976; Hoskins et al. 2002).
TABLE 5.
% of underreplication of heterochromatin satellite DNA
| PI
|
DAPI
|
|||
|---|---|---|---|---|
| Species | Mean ± SE (n) | Range | Mean ± SE (n) | Range |
| C. pararufithorax | 18 (1) | — | 31 (1) | — |
| C. procnemis | 30 (1) | — | 30 (1) | — |
| C. rufithorax | 21 (1) | — | 31 (1) | — |
| D. acutilabella | 22 ± 0 (2) | 22 | ||
| D. americana | 28 (1) | — | 26 ± 1 (2) | 25–28 |
| D. ananassae | 21 ± 2 (3) | 17–23 | 25 ± 0 (3) | 25 |
| D. buskii | 22 (1) | — | ||
| D. equinoxialis | 28 ± 0 (2) | 28 | ||
| D. erecta | 11 ± 2 (2) | 9–13 | 22 ± 1 (2) | 20–21 |
| D. funebris | 30 (1) | — | ||
| D. grimshawi | 32 ± 0 (1) | — | 32 (1) | — |
| D. guttifera | 3 ± 2 (2) | 1–5 | 19 ± 2 (2) | 17–21 |
| D. hydei | 1 (1) | — | 22 ± 2 (2) | 21–24 |
| D. immigrans | 30 ± 3 (2) | 27–33 | 38 ± 1 (3) | 37–39 |
| D. littoralis | 26 (1) | — | ||
| D. melanogaster | 26 ± 3 (4) | 19–33 | 28 ± 1 (10) | 23–40 |
| D. mercatorum | 12 (1) | — | 19 ± 3 (2) | 16–22 |
| D. mimica | 27 ± 1 (4) | 25–30 | 35 ± 1 (3) | 34–37 |
| D. mojavensis | 8 ± 4 (3) | 2–16 | 20 ± 1 (3) | 17–22 |
| D. nannoptera | 37 ± 2 (3) | 34–41 | ||
| D. novamexicana | 26 ± 1 (2) | 25–27 | ||
| D. persimilis | 13 ± 1 (3) | 11–14 | 22 ± 1 (3) | 20–23 |
| D. pseudoobscura | 12 ± 4 (3) | 4–16 | 20 ± 1 (3) | 19–21 |
| D. repleta | 19 ± 1 (3) | 18–20 | ||
| D. sechellia | 24 ± 0 (2) | 23–25 | 27 ± 0 (2) | 26–27 |
| D. simulans | 20 ± 1 (6) | 14–23 | 28 ± 3 (7) | 23–38 |
| D. virilis | 44 ± 1 (4) | 40–48 | 42 ± 1 (5) | 40–46 |
| D. willistoni | 14 ± 1 (3) | 12–15 | 23 ± 0 (3) | 22–23 |
| D. yakuba | 21 ± 2 (2) | 19–23 | 32 ± 9 (2) | 23–41 |
| H. duncani | 30 ± 0 (1) | — | ||
| S. latifasciaeformis | 32 ± 0 (2) | 32 | ||
| S. lebanonensis | 25 ± 0 (2) | 25 | ||
| S. palmae | 25 ± 0 (1) | — | ||
| S. stonei | 26 ± 0 (2) | 25–26 | ||
| Z. badyi | 38 (1) | — | ||
| Z. ghesquerei | 24 (1) | — | ||
| Z. sepsoides | 46 ± 5 (2) | 39–53 | ||
| Z. tuberculatus | 35 ± 2 (3) | 31–43 | ||
Mean percentage of underreplication of satellite DNA in 16C follicle cells is shown as measured by PI and DAPI. Standard error (SE), the range (lowest and highest values), and the number of strains for each species (n) are shown.
Since D. virilis has one of the largest genomes (Table 3), we expected this species to have the largest underreplicated DNA content. PI values for 16C/2C D. virilis indicate 40–48% underreplication while DAPI values suggest 40–46% (Table 5). These values fit very well with those previously described for D. virilis heterochromatin content of 40–42% (Gall et al. 1971; Schweber 1974).
To further confirm that underreplication estimates correlate with heterochromatin satellite DNA content, we stained follicle cells with two heterochromatin markers. Centric and pericentric heterochromatin aggregate into a chromocenter in these cells. Chromocenter size and DAPI staining intensity have been found to reflect satellite DNA content (Lilly and Spradling 1996; Royzman et al. 2002). As shown in Figure 4A, DAPI-stained D. melanogaster follicle cell nuclei exhibit bright subnuclear chromocenters. C. pararufithorax chromocenters are smaller than in D. melanogaster, whereas those of D. virilis are larger (Figure 4, D and G). When we used antidimethyl-histone H3 lysine-9 (dmH3-K9) antibodies that recognize methylated H3-K9, a heterochromatin-specific histone modification (Allshire 2002), the same pattern was observed for these three species (Figure 4, B, E, and H). Species differences in chromocenter size are highly significant (Figure 4J), which is congruent with flow cytometry estimates of 18, 26, and 44% satellite DNA in C. pararufithorax, D. melanogaster, and D. virilis, respectively (Table 5). In summary, follicle cell underreplication and 16C/2C ratios are good predictors of satellite sequence and possibly heterochromatin content. Moreover, a clear trend exists in which larger genomes tend to have more underreplicated satellite DNA (Figure 5 and Table 5). For example, D. virilis, with the largest genome (364–438 Mb), has among the highest (40–48%) underreplicated content. Conversely, D. mojavensis, with one of the smallest genomes (130–166 Mb), also has the least amount of underreplicated DNA (2–16%). In addition, some species, such as D. pseudoobscura, D. melanogaster, and D. mojavensis, exhibited a large range of intraspecific differences in underreplication (Table 5).
Figure 4.—
Chromocenter size reflects satellite content. Stage 13 follicle cell nuclei were stained with DAPI (A, D, and G) and with antidimethyl histone H3 (B, E, and H). Chromocenters (arrows) stain as a bright DAPI dot within the nucleus and are enriched for lysine-9 dimethyl H3. The merged images (C, F, and I) show colocalization of DAPI and lysine-9 dimethyl H3. Bars, 10 μm. The area from each chromocenter was measured and normalized for nuclear area (J). Both DAPI (solid bars) and lysine-9 dimethyl H3 (shaded bars) area measurements show that, compared to D. melanogaster, D. virilis chromocenters are significantly larger (P < 0.0001) whereas C. pararufithorax has significantly smaller chromocenters (P < 0.0001). Standard error bars are shown and values represent averages from 35 cells (see materials and methods).
Figure 5.—
Larger genomes have greater underreplication. The percentage of underreplication (y-axis) was calculated on the basis of 16C/2C values (see materials and methods) and is shown plotted against haploid genome size in megabases, as determined by PI (A) and DAPI (B) flow cytometry. A trend line was added to show that, as genomes become larger, a greater fraction of the total DNA content is underreplicated. Note that the same trend is observed regardless of the dye used. Two-tailed P-values were calculated as in Figure 3.
DISCUSSION
We provide the first systematic and replicated estimates of genome size and satellite DNA content in multiple species of Drosophilidae, revealing both intra- and interspecific differences in genome size. Of particular interest are the sequenced genomes of the 12 Drosophila species and how whole-genome sequence and accurate size estimates now allow us to more completely understand how these genomes have evolved and function.
Ploidy regulation and underreplication during oogenesis is conserved:
Ploidy regulation in endoreduplicating ovarian follicle cells is evolutionarily conserved as all species we examined complete follicle cell DNA replication with 16C ploidy. Strict ploidy control appears critical for proper development of this tissue type. In D. melanogaster, hypomorphic mutations in the Rbf/E2F pathway allow ectopic DNA replication in follicle cells and disrupt ploidy control, but these mutations also lead to female sterility, indicating a more central function for Rbf/E2F than just control of ploidy(Royzman et al. 1999; Bosco et al. 2001; Cayirlioglu et al. 2001). The evolutionary conservation of 16C follicle cell ploidy in all 38 species argues for a critical role for ploidy level in proper follicle cell function.
We also determined that all 38 species, and not just D. melanogaster, underreplicate their genomes in polyploid follicle cells. Underreplication also constitutes a conserved feature of all species examined in this study. Underreplication is a pervasive but poorly understood process with important implications for DNA replication fork barriers and transcription in diploid cells (Leach et al. 2000; Belyakin et al. 2005). Structural features of heterochromatin satellite repeats, as opposed to specific sequences, have been proposed to act as replication barriers (Leach et al. 2000). The fact that underreplication is conserved, despite great differences in satellite DNA content and species-specific repeat sequence motifs, implies that structural and possibly epigenetic factors act as fork barriers (Demakova et al. 2007). The availability of additional Drosophila genome sequences will allow a more thorough analysis of underreplicated genomic regions and genetic elements such as fork barriers that may control this conserved process.
Furthermore, by exploiting underreplication of satellite repeats, we detected surprising variation in satellite DNA content and in its contribution to genome size differences. The amount of underreplication fits well with cytological assays of heterochromatic regions as well as with previously described heterochromatin content estimates. Thus we propose that follicle cell underreplication values may be good predictors of heterochromatin content.
The variation in satellite DNA content and its significance for changes in genome size is consistent with previous ideas that genomes have expanded/contracted mainly by addition/deletion of repeat sequences (for review see Hartl 2000). In light of the 12 Drosophila genome sequences and our satellite DNA estimates, we can speculate as to the mechanisms by which these species have modified their satellite repeats. Unequal sister-chromatid exchange and replication errors have been suggested as possible molecular mechanisms that can produce variation in satellite DNA content (Britten and Kohne 1968; Southern 1975; Smith 1976; Stephan and Cho 1994; Petrov 2001). However, unequal exchange of meiotic sister chromatids as well as replication errors are expected to give rise to both deletions and/or duplications. Unless meiotic drive or other species-specific selection acts upon these meiotic events, gametes bearing either deletions or duplications should be recovered in equal proportions, generating large intra- and interspecific variation. Our data suggest exactly the opposite: Intraspecific satellite DNA content differences are small whereas interspecific differences can be large (Table 5).
This raises an important question: Is genome size, and more specifically satellite DNA content, under selection? One obvious constraint on the contraction of heterochromatin repeats is centromeric function. In D. melanogaster, the minimum satellite DNA for a fully functional centromere has been measured to be ∼420 kb (Sun et al. 1997). Other species are likely to have similar lower-limit constraints to ensure proper chromosome segregation. Among the species with the smallest genomes, D. erecta, D. hydei, D. mercatorum, and D. mojavensis, ∼150 Mb or smaller (Table 3), none have <2% satellite DNA (Table 5 and Figure 5A). Of the 12 sequenced species, we estimate D. mojavensis (strain 15081-1352.22) to have the smallest genome at 130 Mb and the least satellite DNA (2%). Interestingly, if the 2% satellite DNA (2.6 Mb) were distributed evenly among the six D. mojavensis chromosomes, then each chromosome would have ∼430 kb of satellite heterochromatin. It will be informative to determine the chromosomal distributions of these repeats in different species and to ascertain whether these species adhere to the ∼420-kb limit seen in D. melanogaster.
Do Drosophila satellite and heterochromatin contents have upper limits? Several transcription factors have been shown to also bind satellite repeat sequences. Species-specific upper limits to heterochromatin content may be determined by threshold levels of euchromatic DNA-binding proteins that also bind satellite repeats (for review see Ashburner et al. 2005, p. 67). This model is attractive because it suggests that the species-specific genomic arrangements that place specific genes within the influence of heterochromatin dictates how much expansion/deletion is tolerated.
Estimates of total genome size and A:T content:
Genome sizes for a number (20) of the Drosophila species examined here were reported upon in the earlier studies mentioned above (Table 3). In many of those species, the genome sizes appear similar, although some deviate substantially. Unfortunately, the Drosophila species for which there were earlier genome size estimates came either from unpublished citations or from different investigations that used a wide range of methodologies or tissue types. Moreover, there is no strain origin information available for these estimates. Thus, for those species, the difference between previous estimates and ours cannot be evaluated (Table 3). For 12 of these species, our estimates differ by <50 Mb of previous values; we found two species (D. americana and D. mojavensis) to be ∼50 Mb lower and four species (D. buskii, D. equinoxialis, D. funebris, and Scaptodrosophila lebanonensis) to be ∼50 Mb greater than previous estimates; we found two species (S. latifasciaeformis and S. stonei) to be ∼90 Mb greater than previous estimates (Table 3). DAPI fluorescence alone was used in all six cases where our values are greater than previous reported estimates, and thus these higher values may not be as accurate as those previously determined by PI flow cytometry. We would predict therefore that these genomes are likely to have A:T-rich genomes because the DAPI values are higher than expected (Tables 3 and 5).
The smallest genome, 128 Mb for D. mercatorum, and the largest, 404 Mb for D. virilis, differ by as much as 3.2-fold (Table 3). Although our estimates suggest that up to 48% of D. virilis could be heterochromatic satellite DNA, this still does not account for the 3.2-fold difference in genome size with D. mercatorum. This difference is consistent, however, with a previous report that the D.virilis euchromatic genome has also expanded (Moriyama et al. 1998). By contrast, D. virilis is 1.6- to 1.9-fold larger than the 231-Mb genome of D. grimshawi, a difference that can be accounted for by an ∼1.6-fold difference in satellite DNA estimates (Tables 3 and 5). In the close relatives D. melanogaster, D. simulans, and D. erecta, satellite DNA content differences are sufficient to explain the small but significant differences in our genome size measurements (Tables 3 and 5).
The importance of dye type is underscored by the genome size estimates for the Chymomyza species with PI vs. DAPI. While DAPI values are intrinsically less accurate when estimating total DNA content, they are, nevertheless, informative. It is noteworthy that some species with relatively high 2C DAPI values, such as the three Chymomyza, did not have correspondingly high PI 2C values relative to D.m. yw (Table 3, supplemental Tables 1 and 2 at http://www.genetics.org/supplemental/). This suggests that the Chymomyza lineage is characterized by relatively high A:T-rich sequences. High Chymomyza A:T content could reflect high levels of AT-rich centric heterochromatin or indicate that Chymomyza euchromatin is more A:T rich than that of D. melanogaster. Chymomyza PI 16C/2C ratios (supplemental Table 1 at http://www.genetics.org/supplemental/) show no significant difference from the 16C/2C ratio observed for D.m. yw. Levels of underreplicating DNA in Chymomyza therefore appear similar to that of D. melanogaster (Table 5), and thus the relatively high A:T content in Chymomyza is likely a function of euchromatic, as opposed to heterochromatic, A:T sequences. Furthermore, cytological staining of at least one Chymomyza species chromocenter (Figure 4) indicates that high DAPI values are likely due to A:T-rich euchromatin. Digestion with restriction enzymes that recognize either A:T- or G:C-rich sequences have confirmed that Chymomyza genomic DNA is more A:T rich than D. melanogaster and D. virilis (data not shown; P. Campbell and G. Bosco, unpublished data).
Species with both small and large genomes had DAPI/PI ratios >1, although the general trend was that larger genomes were more A:T rich (Figure 2B). This pattern is most apparent when comparing the relative 2C values derived from DAPI and PI in one of the largest genomes, D. virilis (Table 1, Figure 2B, supplemental Tables 1 and 2 at http://www.genetics.org/supplemental/). Because heterochromatin repeats are generally more A:T rich than euchromatic sequences (Gall et al. 1971), we conclude that material contributing to relatively high DAPI values is largely A:T-rich heterochromatin, except in the case of the Chymomyza discussed above.
A:T content and dye effects on small genomes:
Genomes with DAPI/PI ratios <1, or relatively G:C-rich genomes, were all < ∼200 Mb/haploid genome (Figure 2B). D. persimilis, D. pseudoobscura, and D. simulans were the most notable examples (supplemental Table 4 at http://www.genetics.org/supplemental/). These genomes exhibited significant underreplication, indicating that, although small, they still contain considerable amounts of satellite repeats. The question then arises as to the nature of these repeats and why the DAPI/PI ratio is low. One possibility is that these genomes have low repeat content and repeats are not A:T rich. This is a likely explanation because DAPI 16C/2C ratios greater than the expected value of 8 were observed for smaller genomes (Figure 3B). DAPI thus appears to underestimate underreplication in smaller genomes because underreplicated satellite sequences are more G:C rich than satellite sequences of larger genomes. For example, in D. melanogaster >50% of satellite sequences (AAGAC, AAGAG, AAGAGAG, 1.688 satellite) are 28–40% G:C rich whereas the entire satellite sequences of D. virilis (ACAAACT, ATAAACT, and ACAAATT) are ∼28% G:C rich (Gall et al. 1971; Schweber 1974; Lohe and Brutlag 1986, 1987). For closely related sibling species, such as D. melanogaster, D. simulans, and D. erecta with nearly identical satellite repeat sequences, this trend is not apparent, although D. melanogaster contains more satellite repeats than D. simulans, which has more than D. erecta (Table 5), as previously described (Lohe and Brutlag 1987). Unfortunately, since the actual sequence identity of satellite repeats and their abundance in most species are unknown, a more thorough and inclusive analysis cannot be performed.
In one case, a D. melanogaster strain (Bl 2057), we observed a discrepancy between the PI and the DAPI 2C values (Table 1). This suggested that this strain, unlike the other D. melanogaster strains, had acquired some additional A:T-rich DNA. However, the PI 16C/2C ratio (6.15) for this strain does not differ significantly from the other strains (Table 1). If additional A:T sequences are present, they are unlikely to consist of underreplicating satellite repeats. Without further molecular analysis it is difficult to say what might underlie the cause of this discrepancy.
We also found statistically significant differences in genome size among strains of a given species, although these differences in many cases were small. Examination of more strains from these species, especially strains freshly derived from nature, may be necessary to reveal more substantial differences. Specific examples with significant intraspecific variation in heterochromatin content have been described previously (Halfer 1981). Any phylogenetic analyses of genome size (G. Bosco, T. Markow and B. McAllister, unpublished results) therefore will need to account for intraspecific variation as well as for the influence of dye.
The 12 Drosophila species genomes:
Genomes of 12 Drosophila species have been sequenced, allowing us to compare the sizes of the euchromatic assembled portions of the sequenced genomes to sizes estimated with our methods and the contributions of heterochromatin to those sizes (Table 4). In four species, D. ananassae, D. erecta, D. willistoni, and D. mojavensis assembled genome sizes (http://rana.lbl.gov/drosophila/ and Drosophila 12 Genomes Consortium 2007) are larger than those measured by flow cytometry. Size differences, when they exist, are expected to be in the opposite direction: heterochromatin and satellite sequences should not be represented in the sequenced genomes and thus sequenced genomes should be the same or smaller than the estimates reported here. The largest discrepancy is in D. mojavensis, which has the lowest amount of underreplicated satellite DNA (Table 4). For D. ananassae, previous genome size estimates (Ashburner et al. 2005) are identical to ours, and our estimates do not differ with dye type, making it unlikely that this discrepancy reflects errors intrinsic to cytometric measurements of DNA content. In the case of these four species, it is possible that assembly sizes do not accurately represent euchromatic genome sizes as assembly errors have been reported for previous genome releases, including Drosophila, mouse, and human genomes (Benos et al. 2001; Celniker et al. 2002; Cheung et al. 2003a,b).
The differences in genome size and heterochromatin content point to specific and testable evolutionary questions. For example, is loss and or gain of heterochromatic repeat elements the same for different repeat types and for different chromosomes, as has been shown for D. melanogaster and closely related species? Surprisingly little is known about the repeat sequences, abundance, and distribution of satellite sequences in all but a handful of Drosophila species. What are the costs, if any, of the possession of higher amounts of heterochromatin in one vs. another strain of the same species? In D. melanogaster, varying amounts of heterochromatin such as Y chromosome translocations have been shown to be a potent suppressor of position-effect variegation, thus raising the question as to how different strains and different species with vast differences in heterochromatin could use or cope with large differences (Becker 1977). Aside from the known structural roles that heterochromatin plays in centromere function (Sun et al. 1997, 2003) and meiotic chromosome pairing (Hawley et al. 1993; Dernburg et al. 1996), are there other important functions for heterochromatin, such as epigenetic modification, that are under selection and possibly driving genome expansion? Our genome size and heterochromatin estimates complement the Drosophila genome sequences and will allow a more in-depth exploration of the possible mechanisms and evolutionary forces by which genomes have expanded and contracted.
Acknowledgments
We are grateful to David Galbraith, Georgina Lambert, and Brian Larkins for access to their PARTEC flow cytometer and Barb Carolous for assistance in FacScan flow cytometry. We also thank Sergio Castrezana, Stacy Mazzalupo, and the entire staff of the Tucson Drosophila Stock Center for assistance with Drosophila species, food, and technical expertise. We thank the Bloomington Drosophila Stock Center for providing flies and Jodi Mosely, Vivian Lien, and Airlia Thompson for technical assistance in dissecting ovaries. We are grateful to Erin Kelleher, Luciano Matzkin, Carlos Machado, and Bryant McAllister for critical reading of the manuscript. This work was supported by a grant to G.B. from the National Institutes of Health (RO1 GM069462) and by grants from the National Science Foundation (DBI-9910562 and DBI-0450644) to T.A.M.
This article is dedicated to Joao Torres Leiva-Neto (1974–2005), who was one of our most enthusiastic and dedicated students and without whom this study would not have been possible.
References
- Allshire, R., 2002. Molecular biology. RNAi and heterochromatin—a hushed-up affair. Science 297: 1818–1819. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., and G. K. Golic and R. S. Hawley, 2005. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Becker, H. J., 1977. Heterochromatin of the Drosophila melanogaster Y chromosome as modifier of position effect variegation: the time of its action. Mol. Gen. Genet. 151: 111–114. [DOI] [PubMed] [Google Scholar]
- Belyakin, S. N., G. K. Christophides, A. A. Alekseyenko, E. V. Kriventseva, E. S. Belyaeva et al., 2005. Genomic analysis of Drosophila chromosome underreplication reveals a link between replication control and transcriptional territories. Proc. Natl. Acad. Sci. USA 102: 8269–8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M. D., I. J. Leitch, H. J. Price and J. S. Johnston, 2003. Comparisons with Caenorhabditis (approximately 100 Mb) and Drosophila (approximately 175 Mb) using flow cytometry show genome size in Arabidopsis to be approximately 157 Mb and thus approximately 25% larger than the Arabidopsis genome initiative estimate of approximately 125 Mb. Ann. Bot. 91: 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benos, P. V., M. K. Gatt, L. Murphy, D. Harris, B. Barrell et al., 2001. From first base: the sequence of the tip of the X chromosome of Drosophila melanogaster: a comparison of two sequencing strategies. Genome Res 11: 710–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensasson, D., D. A. Petrov, D. X. Zhang, D. L. Hartl and G. M. Hewitt, 2001. Genomic gigantism: DNA loss is slow in mountain grasshoppers. Mol. Biol. Evol. 18: 246–253. [DOI] [PubMed] [Google Scholar]
- Bosco, G., W. Du and T. L. Orr-Weaver, 2001. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol. 3: 289–295. [DOI] [PubMed] [Google Scholar]
- Britten, R. J., and E. H. Davidson, 1971. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q. Rev. Biol. 46: 111–138. [DOI] [PubMed] [Google Scholar]
- Britten, R. J., and D. E. Kohne, 1968. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science 161: 529–540. [DOI] [PubMed] [Google Scholar]
- Cam, H. P., T. Sugiyama, E. S. Chen, X. Chen, P. C. FitzGerald et al., 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37: 809–819. [DOI] [PubMed] [Google Scholar]
- Cayirlioglu, P., P. C. Bonnette, M. R. Dickson and R. J. Duronio, 2001. Drosophila E2f2 promotes the conversion from genomic DNA replication to gene amplification in ovarian follicle cells. Development 128: 5085–5098. [DOI] [PubMed] [Google Scholar]
- Celniker, S. E., D. A. Wheeler, B. Kronmiller, J. W. Carlson, A. Halpern et al., 2002. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, V. L., 2007. Paramutation: from maize to mice. Cell 128: 641–645. [DOI] [PubMed] [Google Scholar]
- Cheung, J., X. Estivill, R. Khaja, J. R. MacDonald, K. Lau et al., 2003. a Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biol. 4: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, J., M. D. Wilson, J. Zhang, R. Khaja, J. R. MacDonald et al., 2003. b Recent segmental and gene duplications in the mouse genome. Genome Biol. 4: R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson, M., and R. Saint, 1999. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 18: 457–462. [DOI] [PubMed] [Google Scholar]
- Colson, P., C. Houssier and C. Bailly, 1995. Use of electric linear dichroism and competition experiments with intercalating drugs to investigate the mode of binding of Hoechst 33258, berenil and DAPI to GC sequences. J. Biomol. Struct. Dyn. 13: 351–366. [DOI] [PubMed] [Google Scholar]
- Colson, P., C. Bailly and C. Houssier, 1996. Electric linear dichroism as a new tool to study sequence preference in drug binding to DNA. Biophys. Chem. 58: 125–140. [DOI] [PubMed] [Google Scholar]
- Demakova, O. V., G. V. Pokholkova, T. D. Kolesnikova, S. A. Demakov, E. N. Andreyeva et al., 2007. The SU(VAR)3–9/HP1 complex differentially regulates the compaction state and degree of underreplication of X chromosome pericentric heterochromatin in Drosophila melanogaster. Genetics 175: 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg, A. F., J. W. Sedat and R. S. Hawley, 1996. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 86: 135–146. [DOI] [PubMed] [Google Scholar]
- Dickson, E., J. B. Boyd and C. D. Laird, 1971. Sequence diversity of polytene chromosome DNA from Drosophila hydei. J. Mol. Biol. 61: 615–627. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium, 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218. [DOI] [PubMed] [Google Scholar]
- Gall, J. G., E. H. Cohen and M. L. Polan, 1971. Reptitive DNA sequences in Drosophila. Chromosoma 33: 319–344. [DOI] [PubMed] [Google Scholar]
- Gatti, M., S. Pimpinelli and G. Santini, 1976. Characterization of Drosophila heterochromatin. I. Staining and decondensation with Hoechst 33258 and quinacrine. Chromosoma 57: 351–375. [DOI] [PubMed] [Google Scholar]
- Halfer, C., 1981. Interstrain heterochromatin polymorphisms in Drosophila melanogaster. Chromosoma 84: 195–206. [DOI] [PubMed] [Google Scholar]
- Hammond, M. P., and C. D. Laird, 1985. a Chromosome structure and DNA replication in nurse and follicle cells of Drosophila melanogaster. Chromosoma 91: 267–278. [DOI] [PubMed] [Google Scholar]
- Hammond, M. P., and C. D. Laird, 1985. b Control of DNA replication and spatial distribution of defined DNA sequences in salivary gland cells of Drosophila melanogaster. Chromosoma 91: 279–286. [DOI] [PubMed] [Google Scholar]
- Hartl, D. L., 2000. Molecular melodies in high and low C. Nat. Rev. Genet. 1: 145–149. [DOI] [PubMed] [Google Scholar]
- Hartl, T., C. Boswell, T. L. Orr-Weaver and G. Bosco, 2007. Developmentally regulated histone modifications in Drosophila follicle cells: initiation of gene amplification is associated with histone H3 and H4 hyperacetylation and H1 phosphorylation. Chromosoma 116: 197–214. [DOI] [PubMed] [Google Scholar]
- Hawley, R. S., H. Irick, A. E. Zitron, D. A. Haddox, A. Lohe et al., 1993. There are two mechanisms of achiasmate segregation in Drosophila, one of which requires heterochromatic homology. Dev. Genet. 13: 440–467. [DOI] [PubMed] [Google Scholar]
- Hoskins, R. A., C. D. Smith, J. W. Carlson, A. B. Carvalho, A. Halpern et al., 2002. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 3: RESEARCH0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, J. S., M. D. Bennett, A. L. Rayburn, D. W. Galbraith and H. J. Price, 1999. Reference standards for determination of DNA content of plant nuclei. Am. J. Bot. 86: 609. [PubMed] [Google Scholar]
- Kavenoff, R., and B. H. Zimm, 1973. Chromosome-sized DNA molecules from Drosophila. Chromosoma 41: 1–27. [DOI] [PubMed] [Google Scholar]
- Laird, C. D., 1971. Chromatid structure: relationship between DNA content and nucleotide sequence diversity. Chromosoma 32: 378–406. [DOI] [PubMed] [Google Scholar]
- Laird, C. D., 1973. DNA of Drosophila chromosomes. Annu. Rev. Genet. 7: 177–204. [DOI] [PubMed] [Google Scholar]
- Laird, C. D., and B. J. McCarthy, 1969. Molecular characterization of the Drosophila genome. Genetics 63: 865–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, T. J., H. L. Chotkowski, M. G. Wotring, R. L. Dilwith and R. L. Glaser, 2000. Replication of heterochromatin and structure of polytene chromosomes. Mol. Cell. Biol. 20: 6308–6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly, M. A., and A. C. Spradling, 1996. The Drosophila endocycle is controlled by cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 10: 2514–2526. [DOI] [PubMed] [Google Scholar]
- Lohe, A. R., and D. L. Brutlag, 1986. Multiplicity of satellite DNA sequences in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 83: 696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe, A. R., and D. L. Brutlag, 1987. Identical satellite DNA sequences in sibling species of Drosophila. J. Mol. Biol. 194: 161–170. [DOI] [PubMed] [Google Scholar]
- Meister, A., 2005. Calculation of binding length of base-specific DNA dyes by comparison of sequence and flow cytometric data. Application to Oryza sativa and Arabidopsis thaliana. J. Theor. Biol. 232: 93–97. [DOI] [PubMed] [Google Scholar]
- Moreau, P. J. F., D. Zickler and G. Leblon, 1985. One class of mutants with disturbed centromere cleavage and chromosome pairing in Sordaria macrospora. Mol. Gen. Genet. 198: 189–197. [Google Scholar]
- Moriyama, E. N., D. A. Petrov and D. L. Hartl, 1998. Genome size and intron size in Drosophila. Mol. Biol. Evol. 15: 770–773. [DOI] [PubMed] [Google Scholar]
- Mulder, M. P., P. van Duijn and H. J. Gloor, 1968. The replicative organization of DNA in polytene chromosomes of Drosophila hydei. Genetica 39: 385–428. [DOI] [PubMed] [Google Scholar]
- Mulligan, P. K., and E. M. Rasch, 1980. The determination of genome size in male and female germ cells of Drosophila melanogaster by DNA-Feulgen cytophotometry. Histochemistry 66: 11–18. [DOI] [PubMed] [Google Scholar]
- Petrov, D. A., 2001. Evolution of genome size: new approaches to an old problem. Trends Genet. 17: 23–28. [DOI] [PubMed] [Google Scholar]
- Petrov, D. A., 2002. Mutational equilibrium model of genome size evolution. Theor. Popul. Biol. 61: 531–544. [DOI] [PubMed] [Google Scholar]
- Petrov, D. A., and D. L. Hartl, 1998. High rate of DNA loss in the Drosophila melanogaster and Drosophila virilis species groups. Mol. Biol. Evol. 15: 293–302. [DOI] [PubMed] [Google Scholar]
- Petrov, D. A., E. R. Lozovskaya and D. L. Hartl, 1996. High intrinsic rate of DNA loss in Drosophila. Nature 384: 346–349. [DOI] [PubMed] [Google Scholar]
- Powell, J. R., 1997. Progress and Prospects in Evolutionary Biology: The Drosophila Model. Oxford University Press, New York.
- Rasch, E. M., 1985. DNA “standards” and the range of accurate DNA estimates by Feulgen absorption microspectrophotometry, pp. 137–166 in Advances in Microscopy, edited by R. R. Cowden and S. H. Harrison. Alan R. Liss, New York. [PubMed]
- Rasch, E. M., H. J. Barr and R. W. Rasch, 1971. The DNA content of sperm of Drosophila melanogaster. Chromosoma 33: 1–18. [DOI] [PubMed] [Google Scholar]
- Reinhart, B. J., and D. P. Bartel, 2002. Small RNAs correspond to centromere heterochromatic repeats. Science 297: 1831. [DOI] [PubMed] [Google Scholar]
- Royzman, I., R. J. Austin, G. Bosco, S. P. Bell and T. L. Orr-Weaver, 1999. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev. 13: 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman, I., A. Hayashi-Hagihara, K. J. Dej, G. Bosco, J. Y. Lee et al., 2002. The E2F cell cycle regulator is required for Drosophila nurse cell DNA replication and apoptosis. Mech. Dev. 119: 225–237. [DOI] [PubMed] [Google Scholar]
- Schulze, D. H., and C. S. Lee, 1986. DNA sequence comparison among closely related Drosophila species in the mulleri complex. Genetics 113: 287–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweber, M. S., 1974. The satellite bands of the DNA of Drosophila virilis. Chromosoma 44: 371–382. [DOI] [PubMed] [Google Scholar]
- Smith, G. P., 1976. Evolution of repeated DNA sequences by unequal crossover. Science 191: 528–535. [DOI] [PubMed] [Google Scholar]
- Southern, E. M., 1975. Long range periodicities in mouse satellite DNA. J. Mol. Biol. 94: 51–69. [DOI] [PubMed] [Google Scholar]
- Stephan, W., and S. Cho, 1994. Possible role of natural selection in the formation of tandem-repetitive noncoding DNA. Genetics 136: 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X., J. Wahlstrom and G. H. Karpen, 1997. Molecular structure of a functional Drosophila centromere. Cell 91: 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X., H. D. Le, J. M. Wahlstrom and G. H. Karpen, 2003. Sequence analysis of a functional Drosophila centromere. Genome Res. 13: 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, W. D., 1990. Nucleic Acids in Chemistry and Biology, edited by G. M. Blackburn and M. J. Gait. Oxford University Press, Oxford.
- Zacharias, H., 1986. Tissue-specific schedule of selective replication in Drosophila nasutoides. Rouxs Arch. Dev. Biol. 195: 378–388. [DOI] [PubMed] [Google Scholar]