Abstract
Human UNG2 is a multifunctional glycosylase that removes uracil near replication forks and in non-replicating DNA, and is important for affinity maturation of antibodies in B cells. How these diverse functions are regulated remains obscure. Here, we report three new phosphoforms of the non-catalytic domain that confer distinct functional properties to UNG2. These are apparently generated by cyclin-dependent kinases through stepwise phosphorylation of S23, T60 and S64 in the cell cycle. Phosphorylation of S23 in late G1/early S confers increased association with replication protein A (RPA) and replicating chromatin and markedly increases the catalytic turnover of UNG2. Conversely, progressive phosphorylation of T60 and S64 throughout S phase mediates reduced binding to RPA and flag UNG2 for breakdown in G2 by forming a cyclin E/c-myc-like phosphodegron. The enhanced catalytic turnover of UNG2 p-S23 likely optimises the protein to excise uracil along with rapidly moving replication forks. Our findings may aid further studies of how UNG2 initiates mutagenic rather than repair processing of activation-induced deaminase-generated uracil at Ig loci in B cells.
Keywords: cell cycle, phosphodegron phosphorylation, replication forks, uracil-DNA glycosylase
Introduction
Uracil is one of the most common base lesions in DNA and may result from misincorporation of dUMP instead of dTMP during replication or from deamination of cytosine (Kavli et al, 2007). While U:A pairs generated from misincorporation of dUMP are not directly mutagenic, deamination of cytosine results in mutagenic U:G mispairs. In most cells, deamination occurs spontaneously. However in B cells, DNA cytosine at Ig loci may also be enzymatically deaminated to uracil by activation-induced deaminase (AID) (Rada et al, 2002; Dickerson et al, 2003). The importance of sanitation of genomic uracil is indicated by the ubiquitous presence of uracil DNA glycosylases (UDGs) (Krokan et al, 2000). In humans, UNG2 constitutes the quantitatively dominating UDG activity in the nucleus, and is the major enzyme for removal of both misincorporated uracil and deaminated cytosines (Kavli et al, 2002). Moreover, UNG2 is the only known glycosylase involved in antibody diversification, by processing of AID-induced U:G pairs in B cells (Kavli et al, 2005; Di Noia et al, 2006). Finally, UNG2 appears to be involved in the assembly of CENP-A at centromeres (Zeitlin et al, 2005), although its precise role in this context remains to be determined. Given the diverse roles of UNG2 in mammalian nuclei and its ability to initiate either repair or mutagenic processing of uracil, it is reasonable that the protein is subject to precise regulation. At the transcriptional level, two mRNAs are expressed from the UNG gene by the use of different promoters and alternative splicing (Nilsen et al, 1997; Haug et al, 1998). The UNG1 mRNA encodes a 304 aa protein translocated to mitochondria, while the UNG2 mRNA encodes a 313 aa protein that is translocated intact to nuclei (Otterlei et al, 1998). Both mRNAs are cell cycle regulated, reaching maximum levels just prior to the S phase (Slupphaug et al, 1991; Haug et al, 1998). The C-terminal 220 residues of UNG2 form a compact, globular catalytic domain (Mol et al, 1995; Slupphaug et al, 1995, 1996), while the N-terminal region constitutes a distinct regulatory domain that is not necessary for enzymatic activity (Kavli et al, 2002). The 86 aa N-terminal non-catalytic domain furthermore confers biochemical properties to UNG2 that are not displayed by the catalytic domain alone, including a strong single-strand DNA preference of UNG2 in the presence of physiological concentrations of Mg2+ (Kavli et al, 2002), supporting a role for UNG2 in uracil removal from ssDNA, for example, in front of the replicative polymerase complex and in R-loops at Ig loci. A role for UNG2 in processing of uracil in ssDNA is substantiated by the presence of at least one replication protein A (RPA)-binding motif in the UNG2 N-terminal domain. Furthermore, a PCNA-binding motif located at the N terminus is likely important for the role of UNG2 in rapid postreplicative removal of misincorporated dUMP at the replication fork (Otterlei et al, 1999).
UNG2 is the initiating factor in uracil-BER in the nucleus, as well as a central mediator in affinity maturation of antibodies. It is thus highly relevant to investigate to what extent the non-catalytic domain modulates UNG2 function, and the present work addresses the significance of phosphorylation in this context. It has previously been demonstrated that UNG2 exists as a phosphoprotein in vivo (Muller-Weeks et al, 1998) and that it apparently may be phosphorylated at T6 and T126 subsequent to UV irradiation (Lu et al, 2004). Here, we report three novel, major phosphorylation forms of UNG2 present in freely cycling HeLa cells, and demonstrate that these are regulated throughout the cell cycle. Combined with functional analysis of phosphomimicking and phosphoinhibiting UNG2 mutants and activity analysis of true UNG2 phosphoforms, our results support a model in which the total cellular level, the activity and the association of UNG2 with proteins at the replication fork are regulated by three consecutive phosphorylations in the non-catalytic N-terminal domain.
Results
UNG2 in freely cycling HeLa cells is stepwise phosphorylated at three Ser/Thr residues in the N-terminal non-catalytic domain
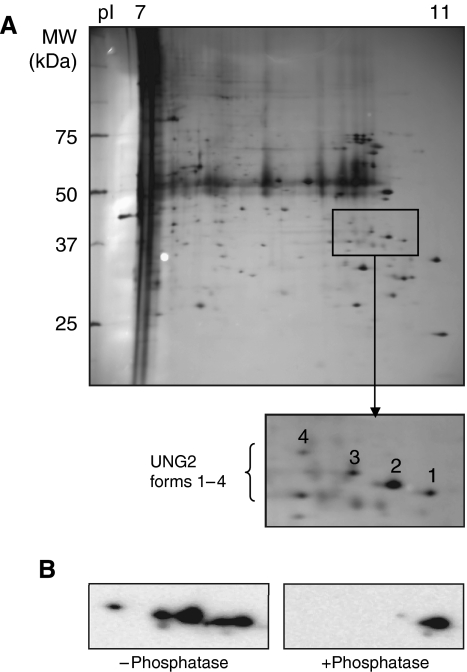
To identify potential UNG2 phosphoforms, UNG2 was precipitated from nuclear extracts of proliferating HeLa cells using UNG antibody PU059. Captured proteins were separated by isoelectric focusing in the pI range 7–11 and subjected to second dimension SDS–PAGE. Silver-stained spots in the expected pI and MW range of UNG2 (Figure 1A) were excised and subjected to trypsination and MALDI-TOF MS peptide mass fingerprinting. Four of the spots were identified as UNG2 (forms 1–4; Figure 1A). The peptide fingerprints revealed mass shifts corresponding to one phosphate in residues 20–49 (in forms 2–4), one phosphate in residues 51–73 (in form 3) and two phosphates in the latter peptide in form 4. The presence of phosphates in forms 2–4 was further verified by pretreatment of the immunoprecipitated UNG2 with calf intestine phosphatase prior to 2D PAGE and western analysis. This resulted in loss of all UNG2 forms except the most positively charged form 1 representing unphosphorylated UNG2 (Figure 1B).
Figure 1.
Isolation of UNG2 phosphoforms. (A) UNG2 was immunoprecipitated from HeLa nuclear extract using PU059 antibodies and separated by 2D PAGE (18 cm IPG strip pH 7–11). Spots representing UNG2 were identified by MALDI-TOF MS fingerprinting. Spot 1: unphosphorylated; spot 2: phosphorylated in peptide 20–49; spot 3: phosphorylated in peptides 20–49 and 50–73/51–73 (expected missed cleavage site) and spot 4: phosphorylated in peptides 20–49 and doubly phosphorylated in peptides 50–73/51–73 (B) 2D PAGE (7 cm IPG strip pH 7–11) and western analysis of immunoprecipitated UNG2 without phosphatase pretreatment (left panel) and after phosphatase pretreatment (right panel).
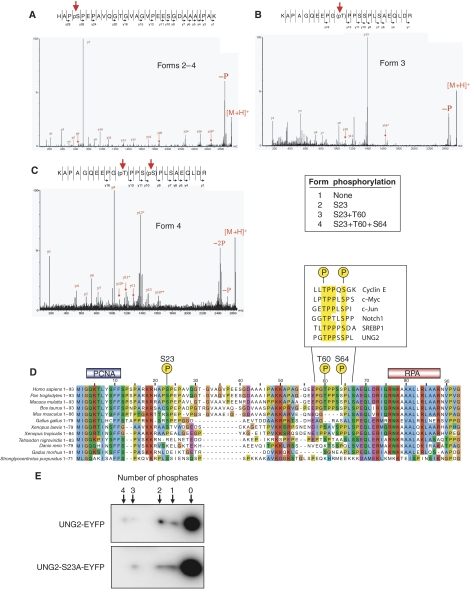
To identify the phosphorylated residues more precisely, peptides from the four spots were analysed by MALDI Q-TOF MS/MS (Stensballe and Jensen, 2004) (Figure 2A–C). The analyses revealed the following UNG2 isoforms: form 1: unphosphorylated; form 2: monophosphorylated at S23; form 3: double phosphorylated at S23 and T60 and form 4 triple phosphorylated at S23, T60 and S64. Moreover, the lack of observed single phosphorylations at T60 and S64 suggests that the phosphorylations occur in a stepwise fashion from the N-terminus to the C-terminus of the regulatory domain. The localisation of the phosphorylated residues within the human UNG2 N-terminus is shown in Figure 2D together with known N-terminal sequences from other eukaryotic UNG2 proteins. The known PCNA- and RPA-binding regions in hUNG2 are also illustrated. The MS/MS sequencing results were entirely in agreement with the MALDI-TOF results and were also confirmed using the program DISPHOS 1.3 (http://core.ist.temple.edu/pred/information.html) using the entries from the Phospho.ELM database (Diella et al, 2004). This stringent predictor takes into consideration that intrinsic structural disorder in and around the potential target is a prerequisite for phosphorylation, and notably identifies S23, T60 and S64 in addition to S63 as potential phosphorylation sites. These Ser/Thr residues are the most conserved in the eukaryotic sequences outside the highly conserved PCNA- and RPA-binding regions (Figure 2D).
Figure 2.
Characterisation of phosphorylation sites in UNG2 by MALDI Q-TOF MS/MS. (A) Spots 2–4 contain phosphate on Ser23. (B) Spot 3 contains phosphate on Thr60. (C) Spot 4 contains phosphates at both Thr60 and Ser64. (D) Alignment of N-terminal amino-acid sequences in UNG2 from higher eukaryotes using ClustalX (Thompson et al, 1997). Individual residues are coloured according to ClustalX colour coding. Highest degree of conservation is observed in the regions corresponding to the PCNA- and RPA-binding motifs of hUNG2. Among the serines and threonines outside these motifs, S23, T60 and S64 are the most conserved. The boxed sequences in (D) illustrate phosphodegrons in other human proteins, which share sequence homology to UNG2. (E) Phosphorylation of T60/S64 may occur in the absence of S23 phosphorylation. WT UNG2 and UNG2 S23A were expressed as EYFP fusion proteins in HeLa cells, and different phosphoforms visualised subsequent to 2D PAGE and western analysis using antibodies against EYFP.
Interestingly, the second and third phosphorylations (at T60 and S64, respectively) create a motif that is very similar to the phosphodegrons observed in cyclin E, c-Myc, c-Jun, Notch1 and SREBP1 (Welcker and Clurman, 2007 and references therein) that serves as recognition motifs for their ubiquitinylation and proteasomal degradation subsequent to phosphorylation (Figure 2D). To investigate whether this apparent phosphodegron could be formed in vivo in the absence of a phosphorylated S23 residue, UNG2-EYFP and the phospho-negative mutant UNG2 S23A-EYFP were expressed in HeLa. 2D western analysis of total cell extracts using GFP antibodies revealed increased accumulation of the unphosphorylated proteins relative to that observed for endogeneous UNG2 (Figure 2E). Moreover, WT UNG2-EYFP appeared to harbour a fourth and not yet identified phosphorylation. Nevertheless, UNG2 S23A-EYFP lacked only one of the phosphorylations observed in the unmutated protein, indicating that phosphorylation of S23 is not essential for phosphorylation at the T60 and S64 positions.
Phosphorylation of UNG2 at S23, T60 and S64 is cell cycle regulated and directs ubiquitinylation and proteolytic breakdown in late S/G2
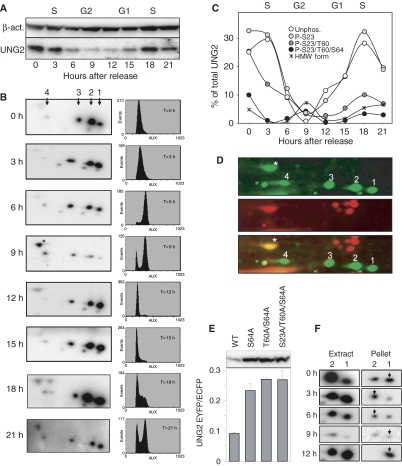
UNG2 is several fold upregulated during late G1 and early S phases (Slupphaug et al, 1991; Haug et al, 1998; Fischer et al, 2004). To investigate whether the UNG2 phosphorylations were also cell cycle regulated, HeLa cells were synchronised at the G1/S border by double thymidine block, and analysed at defined time points after release. Analysis of total cellular UNG2 by 1D PAGE and western analysis demonstrated maximum UNG2 protein in early S phase, while a pronounced reduction at the protein level was observed in late S and G2 (Figure 3A). Moreover, separation of the proteins by 2D PAGE revealed a marked shift in the relative amounts of each of the UNG2 phosphoforms through the cell cycle (Figure 3B). In early S (3 h), the un- and monophosphorylated forms of UNG2 dominate. Towards the end of S (6 h), the relative amount of the di- and triphosphorylated forms increases, while in late G2 (9 h) the latter forms dominate over the un- and monophosphorylated forms. Entry into the next G1 (12 h) is accompanied by reappearance of the un- and monophosphorylated forms, and the cycle is repeated (15–21 h). The cell cycle data are summarised in Figure 3C. Notably, the triphosphorylated form 4 increases in late S, and peaks in G2 concomitantly with the appearance of a fifth form of UNG2 (marked with an asterisk in Figure 3B at 9 h) of higher molecular weight (HMW). Although the total UNG2 level in the cells is very low in G2, this HMW form dominates in late G2. The order of appearance of the HMW form suggests that it is generated by further modification of the triple-phosphorylated form 4. This is also supported by the migration in 2D PAGE of the HMW form relative to the triple-phosphorylated form 4, corresponding to an MW and pI shift that would be expected from a mono-ubiquitinylated form 4 UNG2. Ubiquitinylation of UNG2 has previously been reported to induce proteasome-mediated degradation of UNG2 in S phase (Fischer et al, 2004). To further analyse the nature of the HMW form, western blots from 2D gels were doubly labelled with polyclonal anti-UNG PU059 and monoclonal anti-ubiquitin antibodies prior to incubation with both Cy3- and Cy5-labelled anti-mouse and anti-rabbit secondary antibodies, respectively (Figure 3D). A strong signal was obtained with both Cy3 and Cy5 over the HMW form of UNG2, strongly supporting that this form is mono-ubiquitinylated and triply phosphorylated UNG2. Notably, in contrast to UNG2 forms 1–4, ubiquitinylated UNG2 could not be immunoprecipitated from cell extracts using anti-UNG PU059 antibodies. UNG2 was also not accessible when ubiquitin was targeted for immunoprecipitation, indicating that this form is unstable or transient in nature, or that affinity capture is hampered by ubiquitin or by other proteins binding to UNG2.
Figure 3.
Cell cycle regulation of UNG2 protein level and phosphorylation status. (A) HeLa cells were synchronised by double thymidine block and whole-cell extracts prepared at the indicated time points after release and separated by 1D PAGE prior to western analysis using anti-UNG PU059 and β-actin primary antibodies. (B) The same extracts as in (A) were subjected to 2D PAGE and western analysis using anti-UNG PU059 primary antibodies (left panels). FACS analyses of the cell cycle distribution at each time point are illustrated in the right panels. (C) Quantitative distribution of the different UNG2 forms through the cell cycle. Since different 2D gels cannot be directly quantitatively compared, the sum of all UNG2 forms at each time point is given relative to the sum of all forms at the time of release (100%) based on western quantification after 1D PAGE. The individual distribution of the UNG2 forms at each time point (each 2D gel) is based on western quantification after 2D PAGE. (D) 2D western analysis of HeLa whole-cell G2 extract using a mixture of PU059 and anti-ubiquitin primary antibodies and a mixture of Cy3- and Cy5-labelled anti-mouse and anti-rabbit secondary antibodies. Green signal: UNG2; red signal: ubiquitin. The merged image demonstrates ubiquitinylation of the HMW UNG2 form (asterisk). (E) Phosphoinhibiting (S/T → A) mutations in the putative UNG2 phosphodegron result in increased cellular accumulation of UNG2. HeLa cells were transfected with pUNG2-EYFP and phosphoinhibiting mutants thereof in the putative phosphodegron. Co-transfection with equal amount of vector expressing ECFP was used as internal control to monitor transfection efficiency. Expression of UNG2-EYFP proteins was quantified relative to ECFP subsequent to western analysis using anti-EGFP antibodies (mean of two independent experiments). (F) UNG2 forms 1 and 2 extracted at 200 mM KCl (left) and remaining in chromatin (right). Arrows indicate that the association of UNG2 p-S23 (form 2) with chromatin is increased during S phase relative to unphosphorylated UNG2 (form 1). Note that western images in the right panels were obtained by several fold longer exposure times compared to those in the left panels.
The doubly phosphorylated pT380/pS384 degron in cyclin E was recently demonstrated to be an optimal, high-affinity degron (Hao et al, 2007). To investigate whether the corresponding sequence in UNG2 also constitutes a degron, UNG2 T60 and S64 were mutated to alanine, and expressed as EYFP fusions in HeLa. The concentrations of the transfection constructs were reduced to avoid too high expression of the fusion proteins and saturation of the UNG2 processing machinery. The cells were co-transfected with ECFP as an internal standard to correct for potential differences in transfection efficiency. Subsequent to western analysis of total cell lysates using GFP antibodies, the Ala mutants were all detectable at 2.5- to 3-fold higher levels compared to WT UNG2 (Figure 3E). These results strongly indicate that the sequence encompassing T60 and S64 in UNG2 constitutes a phosphodegron.
Finally, to investigate whether specific phosphoforms of UNG2 were poorly extracted by our procedure, pellets remaining after the first extraction step were sonicated in high salt buffer to release proteins strongly binding to chromatin. This consistently revealed small amounts of unphosphorylated (form 1) and S23-phosphorylated (form 2) UNG2 that were not extracted in buffer containing 200 mM KCl (Figure 3F, right panels). Moreover, which of these two forms were most tightly associated with chromatin appeared to be dependent on the cell cycle stage. Thus, outside the S phase, unphosphorylated UNG2 was retained more in the chromatin relative to UNG2 p-S23, while in S phase a more selective binding of UNG2 p-S23 was observed (Figure 3F, arrows). These results indicate that S23-phosphorylated UNG2 has an increased affinity for replicating chromatin relative to the other UNG2 forms.
Phosphomimicking mutations in the UNG2 regulatory domain modulate catalytic activity and association with RPA
To analyse functional implications of the observed UNG2 phosphorylations, each of the phosphoforms were mimicked by mutation of the corresponding Ser/Thr residues to Asp, and purified to apparent homogeneity (S23A and S23A/T60A/S64A were included as controls) (Figure 4A). The first attempts to purify the triple phosphomimicking UNG2 S23D/T60D/S64D as well as a UNG2 S64D mutant resulted in catalytically impaired proteins. However, by reducing the sonication energy during bacterial lysis, and performing all steps during purification on ice (including the Talon column step), we obtained fully active proteins. These findings indicate that a negative charge introduced at residue 64 in the UNG2 N-terminus introduces a local structural instability in the protein that may be of functional relevance, and further studies are now underway to determine the overall structural change in UNG2 following S64D mutation.
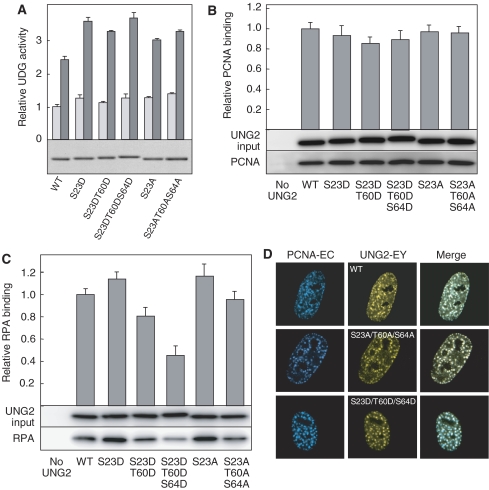
Figure 4.
Phosphomimicking mutations markedly affect UNG2 activity and binding to RPA. (A) Catalytic activity of UNG2 mutants relative to WT measured against ssDNA (dark grey bars) and dsDNA (light grey bars) substrates. Each bar represents the mean of three experiments with standard deviations as indicated. Lower panel illustrates the purified proteins after 1D PAGE and SimplyBlue staining. (B, C) WT UNG2 and mutants were incubated with extracts from UNG-deficient B cells and binding to PCNA (B) or RPA (C) quantified after UNG2 IP, 1D PAGE and western analysis (lower panels). Each bar represents PCNA (or RPA) signal divided by UNG2 input signal, and averaged from six independent experiments with standard deviations as indicated. (D) Phosphomimicking and phosphoinhibiting mutations do not affect localisation of UNG2 to replication foci. UNG2-EYFP WT and mutants were co-transfected in HeLa cells with PCNA-ECFP to verify localisation in replication foci.
Interestingly, an overall increase in enzymatic activity was observed with the phosphomimicking mutants relative to WT, and the increase was most pronounced with ssDNA substrate. The increase was less pronounced with the Ala mutants, demonstrating that introduction of negative charges at the distinct positions in the UNG2 regulatory domain specifically stimulates the activity of UNG2. To analyse the activity stimulation in more detail, the phosphomimicking mutants were subjected to kinetic analyses. These analyses revealed that the mutations had a mixed effect upon affinity (Km) and turnover (kcat) of UNG2 (Table I). In general, the introduced negative charges mediated a marked increase in the catalytic turnover against both types of substrate, although also accompanied by a somewhat increased Km. Notably, the single S23D mutation resembling the dominant S-phase phosphoform of UNG2 increased the catalytic turnover about two- and three-fold relative to WT against dsDNA and ssDNA substrates, respectively. Increased catalytic turnover could be of biological significance to optimise uracil excision at rapidly moving replication forks. Additional mutation of T60 and S64 mediated somewhat reduced catalytic turnover compared to the single S23D mutant. The primary function of these phosphorylations is thus likely to form a functional phosphodegron, although specific functions in uracil processing cannot be excluded.
Table 1. Kinetic constants of UNG2 and N-terminal UNG2 phosphomimicking mutants.
| Enzyme | Substrate | Km (μM) | kcat (min−1) |
|---|---|---|---|
| Without RPA | |||
| WT | U:A | 1.0±0.08 | 449±9 |
| S23D | U:A | 1.8±0.06 | 870±6 |
| S23D, T60D | U:A | 1.4±0.06 | 641±6 |
| S23D, T60D, S64D | U:A | 1.2±0.07 | 726±5 |
| WT | Uss | 0.05±0.00 | 663±10 |
| S23D | Uss | 0.14±0.01 | 1981±13 |
| S23D, T60D | Uss | 0.09±0.01 | 1141±15 |
| S23D, T60D, S64D | Uss | 0.11±0.01 | 1360±10 |
| With RPA | |||
| WT | U:A | 0.59±0.09 | 143±5 |
| S23D | U:A | 0.70±0.06 | 187±7 |
| S23D, T60D | U:A | 0.66±0.06 | 162±7 |
| S23D, T60D, S64D | U:A | 0.67±0.07 | 272±6 |
| WT | Uss | 0.04±0.005 | 773±18 |
| S23D | Uss | 0.05±0.004 | 1204±11 |
| S23D, T60D | Uss | 0.03±0.004 | 546±11 |
| S23D, T60D, S64D | Uss | 0.02±0.003 | 882±10 |
To test whether phosphomimicking mutations affected binding to PCNA and/or RPA, purified UNG2 proteins were incubated with extracts from UNG-deficient human B-cell lines derived from hyper-IgM patients (Imai et al, 2003; Kavli et al, 2005), immunoprecipitated using PU059 and analysed for coprecipitation of PCNA and RPA. Binding of UNG2 to PCNA was not markedly influenced by any of the mutations (Figure 4B). This was also supported by activity analyses of the UNG2 proteins in the presence of PCNA, in which a two-fold stimulation of all the UNG2 forms was observed (data not shown).
Conversely, the binding of UNG2 to RPA was markedly and differentially affected by the mutations (Figure 4C). Notably, the S23D mutation mediated a moderate but consistent increase in the binding of UNG2 to RPA in agreement with preferential binding of monophosphorylated UNG2 P-S23 to chromatin in S-phase cells (Figure 3F). A similar increase in RPA binding of the S23A mutant suggests that alterations of the local UNG2 secondary structure, rather than the specific charge mediate increased binding. The additional T60D and S64D mutations mediated progressive loss of RPA binding, and the triple S23D/T60D/S64D mutant retained only about 40% of the RPA-binding capacity relative to WT UNG2. Importantly, a corresponding loss was not observed with the triple Ala mutant, indicating that the negative charge in the putative phosphodegron mediates reduced RPA binding. In the solution NMR structure of the UNG2 peptide encompassing residues 73–88 bound to the RPA32 subunit (Mer et al, 2000), the UNG2 peptide adopts a helical structure upon binding to RPA32. Here, interaction of three arginines in UNG2 with four acidic residues in RPA32 contributes to the binding. Our results indicate that the region of UNG2 encompassing residues T60 and S64 may also contact RPA32, and that phosphorylation of these residues may contribute repulsive forces between the two proteins.
Given the high local concentration and crucial importance of RPA near the replication fork (reviewed in Fanning et al, 2006), the above findings prompted us to investigate if the presence of RPA modulated the kinetic properties of the UNG2 proteins. Whereas RPA stimulated catalytic turnover of WT UNG2 turnover against ssDNA substrate, the three phosphomimicking mutants displayed reduced turnover (Table I). Nevertheless, the S23D mutant still maintained the highest turnover among the four proteins. Conversely, RPA mediated a larger increase in the affinity (lowered Km) of the phosphomimicking mutants against ssDNA substrate, compared to WT.
Given the markedly reduced binding of UNG2 S23D/T60D/S64D to RPA, we wanted to investigate whether the phosphorylation status of UNG2 affected its localisation to replication foci. WT UNG2, phosphomimicking mutants as well as phosphoinhibiting (Ala) mutants were thus transiently expressed in HeLa cells as EYFP fusion proteins together with ECFP-PCNA (Figure 4D). The intracellular localisation of UNG2-EYFP, UNG2 S23D/T60D/S64D-EYFP and UNG2 S23A/T60A/S64A-EYFP together with ECFP-PCNA is illustrated in Figure 4D. Notably, the subcellular localisation of the mutants in replication foci was not altered compared to WT UNG2. Identical results were obtained subsequent to transfection with single (S23) and double (S23/T60) mutants to Asp/Ala (data not shown). Thus, the overall localisation of UNG2 to nuclei and replication foci is apparently not directly controlled by cell cycle-regulated N-terminal phosphorylations. This is in accordance with the fully competent PCNA binding of the phosphomimicking mutants (Figure 4B), which likely contributes to their localisation in replication foci. No overt reduction in the overall level of the triple phosphomimicking mutant could be observed. However, this likely reflects that under conditions of overexpression, the specific ubiquitinylation apparatus becomes saturated. Alternatively, phosphomimicking mutants cannot directly substitute phosphoserines or phosphothreonines in the contact of the docking site to the ubiquitin ligase.
Finally, it should be mentioned that we have not been able to produce stably transfected HeLa cell lines producing UNG2 under the strong CMV promoter, whereas we have successfully produced such a cell line producing UNG2 under it own cell cycle-regulated promoter (Kavli et al, 2002) and under an inducible promoter (Akbari et al, 2004). However, induction of high UNG2 expression results in an increased fraction of cells in the S/G2 stage of the cell cycle compared with the control cell line (data not shown). This underscores that tightly regulated expression of UNG2 is of vital importance for the cells.
In vitro phosphorylation of UNG2 by cyclin-dependent kinases stimulates UNG2 catalytic activity
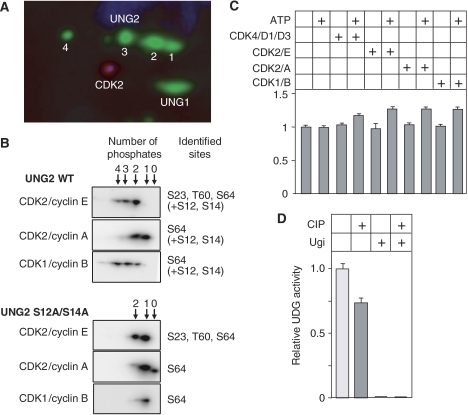
Both their regulation in the cell cycle and their position immediately N-terminus to prolines in the primary amino-acid sequence (Figure 2D) indicate that the N-terminal UNG2 phosphorylation sites may be targeted by cyclin-dependent kinases (CDKs). To investigate this in more detail, we first reprobed 2D western blots of UNG2 immunoprecipitates using CDK1 and CDK2 antibodies. Of these, CDK2 was clearly identified (Figure 5A). We then tested the effect of the CDK inhibitors olomoucine and roscovitine on the phosphorylation pattern of UNG2. However, since this treatment also affected the distribution of cells in the cell cycle, the results could not be interpreted in terms of direct effects of individual CDKs on UNG2 activity (data not shown). As an alternative approach, we investigated the ability of four active CDK/cyclin pairs to phosphorylate UNG2 in vitro. Of these, CDK4/cyclin D1/D3 is active in G1, CDK2/cyclin E in late G1/S, CDK2/cyclin A in late S/G2 and CDK1/cyclin B in M. When UNG2 was subjected to in vitro phosphorylation by either of these sets of CDK/cyclins, multiple phosphorylated forms of UNG2 were observed (Figure 5B). Mass spectrometry analyses revealed that the ‘early' CDK4/cyclin D and CDK2/cyclin E kinases were able to phosphorylate all three UNG2 Ser/Thr residues observed in vivo (only CDK2/cyclin E shown). Of these, only S64 was phosphorylated by the ‘late' CDK2/cyclin A and CDK1/cyclin B kinases (Figure 5B, upper panels) in agreement with the accumulation of this phosphorylation in G2 (Figure 3B). In addition, (unspecific) phosphorylation by all kinases was observed at S12 and S14. To avoid interference from phosphorylations at these residues, an S12A/S14A mutant was purified and subjected to in vitro phosphorylation by the above kinases. This yielded the same pattern of phosphorylations as in WT UNG2, except at the mutated sites. PCNA was also added in parallel reactions since PCNA has been shown to act as a connector to target CDK/cyclins to specific DNA replication proteins (Koundrioukoff et al, 2000). No change in the phosphorylation pattern was observed in the presence of PCNA (data not shown). Although differential quantification of the phosphorylations was not possible by our analyses, the results indicate an overlapping function of the various CDK/cyclins in the cell cycle regulation of UNG2.
Figure 5.
CDK2 coprecipitates with UNG2 from cell extracts and phosphorylation by CDK/cyclins enhances UNG2 catalytic activity. (A) CDK2 is detected after 2D PAGE and western analysis of UNG2 immunoprecipitates. Numbering refers to UNG2 forms given in Figure 2. (B) Phosphorylation pattern of WT UNG2 and UNG2 S12A/S14A subsequent to in vitro phosphorylation with various CDK/cyclins, visualised by 2D western. The number of phosphates within each form is indicated above the panels, and phosphoresidues identified by Q-TOF MS/MS are indicated to the right. (C) Catalytic activity of UNG2 S12A/S14A subsequent to in vitro phosphorylation. Each bar represents activity against [3H]dUMP-containing (U:A) substrate relative to WT UNG2 in the absence of kinase and ATP, and represents the mean of three experiments with standard deviations as indicated. (D) Dephosphorylation significantly reduces UNG2 activity (measured against [3H]dUMP-containing (U:A) substrate) in extracts from G1/S-phase cells in which the p-S23 phosphoform dominates. The UNG inhibitor Ugi was included in parallel controls to monitor potential contribution to the measured activity from other uracil-DNA glycosylases. No such activity was detected. Each bar represents the mean of three experiments with standard deviations as indicated.
As shown in Figure 5C, phosphorylation by each of the kinases mediated a significant (20–30%) increase in UNG2 activity. Although these analyses did not allow quantification of the precise activity of each of the true UNG2 phosphoforms as observed in vivo, the results clearly demonstrate that phosphorylation of the N-terminal regulatory domain stimulates the catalytic activity of UNG2, in support of the results from phosphomimicking mutants.
Finally, the activity increase observed in the phosphomimicking mutants was corroborated by phosphatase treatment of HeLa cell extracts. When total extracts from cells in G1/S phase were subjected to dephosphorylation using calf intestine phosphatase, the total UDG activity was reduced by about 25% (Figure 5D). In this extract, the p-S23 form of UNG2 is the quantitatively dominating form (Figure 3B; 0 h) and the major part of the activity reduction is likely attributed to dephosphorylation of this form. However, the actual reduction in UNG2 p-S23 activity is likely higher, since the cell extracts also contain appreciable amounts of non-phosphorylated UNG1, which would remain unaffected by the phosphatase treatment. In summary, the in vitro phosphorylation and the phosphatase experiments strongly support that UNG2 phosphomimicking mutants are valid models for studying the functional consequences of in vivo UNG phosphorylation.
Discussion
Accumulating evidence suggests that phosphorylation of DNA base excision repair proteins plays a key role in fine tuning of DNA repair as well as in coordination of DNA repair with other cellular processes such as DNA replication (reviewed in Fan and Wilson, 2005). However, the precise phosphoforms of the proteins existing in vivo, the kinases involved and the functional implications of the phosphorylations in BER, remain poorly understood for most BER proteins.
Here, we present the first precise identification of three cell cycle-regulated phosphoforms of UNG2, and provide evidence that these may be generated by distinct CDK/cyclins. Moreover, the phosphorylations regulate cellular turnover of the UNG2 protein, association with RPA and modulates catalytic activity. Ubiquitin-dependent breakdown of UNG2 observed by Fischer et al (2004) and their finding that breakdown is inhibited by the CDK inhibitor roscovitine is corroborated by our results. We furthermore find that the maximum level of apparently mono-ubiquitinylated and triple-phosphorylated form of UNG2 occurs in G2, concomitantly with the lowest level of total UNG2. This, the migration in 2D gels of ubiquitinylated UNG2 relative to triple-phosphorylated UNG2, and the increased cellular accumulation of the S64A-containing mutants in the putative phosphodegron, strongly indicate that the triple-phosphorylated protein serves as a precursor for ubiquitinylation and breakdown in late S/G2. Interestingly, very recent work from the laboratory of Primo Schär has demonstrated that TDG, another member of the UDG superfamily, is downregulated upon entry of the S phase and upregulated in G2 (Hardeland et al, 2007). This inverse regulation of UNG2 and TDG implies that the two uracil-DNA glycosylases have non-redundant functions that must be coordinated through the cell cycle. Apparently, the presence of TDG in the S phase is incompatible with cell cycle progression and proliferation (Hardeland et al, 2007). One potential explanation for this is the exceptional strong binding of TDG to product AP sites (Waters et al, 1999) that could obstruct replication. It is less clear however, why UNG2 is degraded in late S/G2, since deamination of cytosine to uracil also is likely to occur during G2. One clue might come from our recent findings that removal of uracil at Xenopus centromeres apparently mediates loading of CENP-A, a central component in kinetochore formation (Zeitlin et al, 2005). Although speculative, it is possible that a strictly regulated level of UNG2, or specific phosphoforms of UNG2, may be necessary to confine uracil excision to centromere regions during G2. Alternatively, the cellular level of UNG2 is reduced during G2 to allow temporary persistence of deaminated cytosines serving as initiator sites for CENP-A loading.
The increase in the UNG2 catalytic activity subsequent to phosphorylation by CDK/cyclins may be of special functional importance, since UNG2 attached to the replication machinery should be able to cope with the speed of the replication fork to efficiently recognise and remove uracils. Association of UNG2 to PCNA (Otterlei et al, 1999) and the doubling of the catalytic turnover of monophosphorylated relative to WT UNG2 against dsDNA likely support rapid removal of misincorporated uracil (U:A) from the newly synthesised strand. Conversely, virtually nothing is known about uracil processing in the single-stranded DNA region in front of the replicative polymerase. Deamination of cytosine occurs 200- to 300-fold faster in ssDNA than in dsDNA (Lindahl, 1993), and likely poses a significant mutagenic threat when occurring immediately in front of the replicative polymerases. Increased binding of monophosphorylated (p-S23) UNG2 to RPA during S phase, concomitant with markedly (three-fold) increased catalytic turnover against single-stranded uracils conform with a role of UNG2 in prereplicative removal of uracil. It is less clear if the di- and triphosphorylated forms of UNG2 possess additional functional characteristics that aid replicative uracil removal. Rather, we hypothesise that the main function of these phosphorylations is to create a phosphodegron that initiates ubiquitinylation. This is also supported by the reduced binding of the fully phosphorylated UNG2 to RPA, that likely release UNG2 and facilitates recognition by an E3 ubiquitin ligase. It can furthermore not be excluded that ubiquitinylation may be regulated by additional factors than mere phosphorylation status. One possibility is that fully phosphorylated UNG2 released from chromatin upon termination of replication forks is targeted for ubiquitinylation. Alternatively, PCNA may serve as an adaptor for UNG2 ubiquitinylation, since the PCNA-binding capacity of UNG2 is retained in the phosphomimicking mutants. PCNA-dependent ubiquitinylation and proteolysis were recently reported for the Xenopus replication licensing factor Cdt1 (Arias and Walter, 2006). Notably, Cdt1 binds to PCNA via the same conserved PCNA-binding motif as in UNG2. Work is now in progress to identify the E3 ligase responsible for UNG2 ubiquitinylation. This work may shed new light upon the mechanisms underlying UNG2 protein turnover during the cell cycle, and the potential consequences of dysregulated turnover.
In activated B cells, excision of AID-deaminated cytosines from the template strand of the replication fork was recently proposed to act as a specific trigger of translesion synthesis (Di Noia et al, 2006). This could explain the mutagenic outcome of uracil excision rather than faithful repair. However, it is not clear why such translesion synthesis across AP sites should be triggered in activated B cells and not in other replicating cells. Given the results presented here, the role of distinct functional modifications of UNG2 in activated B cells should provide an attractive area for future investigations.
Materials and methods
Reagents
Primary antibodies were PU059 recognising the catalytic domain of UNG2 (0.5 μg/ml), anti-RPA (Abcam ab2175), anti-PCNA (Abcam ab29), anti-actin (Abcam ab8226), anti-ubiquitin (Santa Cruz sc-8017), anti-CDK1 (Santa Cruz sc-747), anti-CDK2 (Santa Cruz sc-748) and anti-GFP (prepared in-house). Secondary antibodies were swine anti-rabbit, HRP (Dako Chemicals), rabbit anti-mouse, HRP (Dako Chemicals), Cy3 anti-mouse/Cy5 anti-rabbit (GE Healthcare). The expression vector pET11d-tRPA encoding the trimeric RPA complex was kindly provided by Professor Marc Wold at The University of Iowa. Expression and purification were essentially as described (Henricksen et al, 1994).
Cell culture, synchronisation and preparation of extracts
HeLa cells were grown in suspension in HEPES-buffered DMEM, 10% FCS, 0.03% glutamine, 0.1 mg/ml gentamicin and 2.3 μg/ml fungizone. Nuclear extracts were prepared from 2.5 × 108 cells. After pelleting and washing in PBS, cells were resuspended in five volumes of hypotonic buffer (10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 1 mM DTT, Complete® EDTA-free protease inhibitors (Roche) and phosphatase inhibitor cocktails 1 and 2 (Sigma) and incubated for 10 min on ice. Cells were lysed by 15 strokes in a Kontes glass homogeniser (B-type pestle) and nuclei pelleted by centrifugation at 680 g for 5 min. Nuclear pellets were resuspended in two volumes of extraction buffer (10 mM Tris–HCl pH 7.8, 200 mM KCl, 20% glycerol, 0.25% NP40, 1 mM DTT and protease and phosphatase inhibitors) and proteins were extracted at 4°C for 2 h. Finally, extracts were clarified at 16 000 g for 20 min. HeLa cells were synchronised by the double thymidine block essentially as described (Richardson et al, 2000). Verification of cell cycle stage was performed by standard flow cytometry. At defined time points post-release, cells were washed twice in ice-cold PBS, resuspended in two volumes of extraction buffer, and proteins were extracted and clarified as above. When stated, the pellets were further extracted by sonication in a buffer containing 400 mM KCl, and clarified as above. UNG−/− lymphoblastoid cells were grown, and total extracts thereof prepared as described (Akbari et al, 2004). All extracts were snap-frozen in liquid N2 and stored at −80°C prior to use. For analysis of UDG activity subsequent to dephosphorylation, HeLa G1/S-phase extract (5 μg protein) was treated with 40 U of calf intestine phosphatase for 45 min at 37°C prior to analysis.
Immunoprecipitation, electrophoresis and western analysis
UNG2 was isolated using polyclonal PU059 that recognises the UNG1/2 common catalytic domain. PU059 (20 μg) was covalently coupled to 100 μl magnetic Dynabeads® Protein-A (Dynal, Norway) according to the manufacturer's instructions. After incubation with protein extract for 4 h at 4°C, the beads were thoroughly washed and proteins were eluted overnight in Destreak rehydration solution (GE Healthcare), 1% IPG buffer for 2D electrophoresis. For dephosphorylation, the beads were treated with 40 U of calf intestinal phosphatase (New England Biolabs) prior to elution of bound proteins.
1D PAGE was performed in 10% NuPage Novex Bis-Tris gels using MOPS run buffer. For 2D PAGE, proteins were focused in 7–11 IPG NL strips at 20°C and 50 μA (18 cm for silver-stained gels and 7 cm for gels prior to western analysis) in the IPGphor II unit (GE Healthcare). Strips were equilibrated in DTT and iodoacetamide prior to second dimension SDS–PAGE. Gels were either electroblotted on PVDF (Immobilon™; Millipore) or Hybond LFP (GE Healthcare) or silver stained. Membranes were blocked for 1 h in PBS, 0.1% Tween (PBST), 5% fat-free dry milk and incubated in primary antibody for 1 h in blocking buffer. After 3 × 10 min washing in PBST, membranes were further incubated for 1 h in secondary HRP-conjugated antibodies in PBST, washed 3 × 10 min in PBST, developed using SuperSignal West Femto (Pierce) and visualised using Kodak Image station 2000R. Membranes incubated with Cy3/Cy5-labelled secondary antibodies were visualised using a Typhoon Trio imager (GE Healthcare).
In-gel digestion and mass spectrometric analysis
Silver-stained spots were excised from gels and in-gel digested with trypsin as described (Shevchenko et al, 1996). Dried peptides were resuspended in 10 μl of 0.5% acetic acid, 0.02% heptafluorobutyric acid, desalted as described (Rappsilber et al, 2003) and matrix (0.3 g/l α-cyano-4-hyroxycinnamic acid (CHCA) was added in EtOH:acetone (2:1) on an anchor chip plate. Peptide fingerprints were acquired with a Reflex IV MALDI-TOF mass spectrometer (Bruker Daltonics) in reflectron mode. The human portion of the NCBI database was interrogated using the Mascot software (Matrix Science).
For MS/MS analysis, tryptic peptides were prepared as above. To reduce the complexity of the collision-induced fragmentation pattern, the C-terminal lysine in the phosphopeptide encompassing aa 20–49 was chemically modified (Peters et al, 2001). Peptides were incubated in 0.4 M 2-methoxy-4,5-dihydro-1H-imidazole pH 10.5 for 3 h at 55°C, the reaction was quenched by 5% (v/v) FA and desalted using in-house-prepared PorosR2 (Applied Biosystems) micro-columns. Peptides were directly eluted on MALDI plates with 2.5-dihydroxybenzoic acid (DHB) in 50% acetonitrile, 1% phosphoric acid (Stensballe and Jensen, 2004). MS/MS spectra were acquired using a Q-TOF Ultima MALDI (Waters Micromass) mass spectrometer, equipped with an in-source camera and raw data were manually processed.
In vitro phosphorylations were identified by nano LC-MS/MS using gradient elution from an in-house-prepared ReproSil-Pur C18-AQ column (75 μm × 50 mm, 3 μm particle size) (Dr Maisch GmbH). Injection volume was 5 μl, and samples were desalted on a C18 PepMap100 Nano-Precolumn (300 μm × 1 mm, 5 μm particle size) (LC Packings). Eluted peptides were analysed online by a QStar XL (Applied Biosystems) mass spectrometer, using a PicoTip Emitter needle and applied ionising voltage of 2400 V. Only ions of charge states +2, +3 and +4 were analysed in MS/MS mode. MS/MS data were subsequently analysed by Mascot software (Matrix Science).
UNG2 mutants, co-immunoprecipitation, in vitro phosphorylation and UDG activity assays
Mutations in pET28a-UNG2 (Scaramozzino et al, 2003) were introduced by Quick-change site-directed mutagenesis (Stratagene). His-tagged WT UNG2 and mutants were expressed in E. coli BL21-Codon plus® (DE3)-RIPL (Stratagene). LB cultures (1 l) were grown to OD 0.6 at 37°C, induced by 1 mM IPTG and UNG2 expressed at 16°C overnight. Cells were sonicated on ice in the presence of 1 mg/ml lysozyme and Complete (Roche) protease inhibitors. Nucleic acids were precipitated with 0.4% protamine sulphate. His-tagged UNG2 mutants were purified by TALON Superflow Resin (Clontech) and dialysed against UNG buffer containing 20 mM Tris–HCl pH 7.5, 60 mM NaCl, 1 mM EDTA and 1 mM DTT. After addition of glycerol to 50%, proteins were snap-frozen in liquid N2 and stored as aliquots at −80°C. Initial protein concentration was measured using the Bio-Rad assay using BSA as a standard. To allow precise relative quantification, all purified UNG2 proteins were subjected to polyacrylamide gel electrophoresis (six gels), stained with SimplyBlue (Invitrogen), and subjected to densitometric analysis. For co-immunoprecipitation analysis, 100 ng UNG2 WT or phosphomimicking mutants were incubated with 1 mg UNG−/− lymphoblastoid cell extract for 2 h at 4°C and complexes were immunoprecipitated using PU059-coupled Dynabeads. For precipitation of PCNA, extracts were treated with Omnicleave endonuclease (Epicentre) to release PCNA from chromatin. After washing in 3 × 0.5 ml 10 mM Tris pH 7.8, bound proteins were eluted in 1 × LDS loading buffer supplemented with 50 mM DTT and separated in 10% NuPage Novex Bis-Tris gels using MOPS run buffer and subjected to western analysis as described above. UNG2 proteins were in vitro phosphorylated with CDK4/cyclin D1 and CDK4/cyclin D3 (Cell Signalling), CDK2/cyclin A and CDK2/cyclin B (Upstate) and CDK2/cyclin E (BIOMOL International) according to the manufacturer's instructions. UDG activity of the UNG2 proteins was assayed against [3H]dUMP-containing calf thymus DNA (U:A) as described (Kavli et al, 2002). For enzyme kinetic analyses, UNG2 proteins were analysed in the presence of [3H]dUMP-containing ds (U:A) at concentrations ranging from 0.02 to 1.8 μM, and single-stranded [3H]dUMP-containing DNA at concentrations ranging from 0.1 to 8.4 μM. When stated, 0.85 pmol of RPA trimer was added to the reactions. Enzyme kinetic parameters were calculated using the Enzpack for Windows version 1.4 software package (Biosoft) using the method of Wilkinson.
Mammalian expression constructs and confocal microscopy
pUNG2-EYFP and pNLS-ECFP-PCNA were published previously (Kavli et al, 2005). Site-specific mutations were introduced as above and verified by DNA sequencing. HeLa cells were transfected using FuGENE 6 (Roche) according to the manufacturer's protocol. Fluorescent images (1 μm thickness) of transfected, freely cycling cells were recorded 24 h post-transfection using a Zeiss LSM 510 laser scanning microscope equipped with a Plan-Apochromate 63 × /1.4 oil immersion objective.
Acknowledgments
We thank Petter B Aslaksen, Karin M Gilljam, Mona Fenstad and Joachim Frost for valuable technical assistance. This research was supported by the National Programme for Research in Functional Genomics in Norway (FUGE) in the Research Council of Norway, the Norwegian Cancer Association, the Cancer Fund at St Olav's Hospital, Trondheim and the Svanhild and Arne Must Fund for Medical Research and EU Integrated Project on DNA Repair.
References
- Akbari M, Otterlei M, Peña-Diaz J, Aas PA, Kavli B, Liabakk NB, Hagen L, Imai K, Durandy A, Slupphaug G, Krokan HE (2004) Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells. Nucleic Acids Res 32: 5486–5498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC (2006) PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol 8: 84–90 [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Rada C, Neuberger MS (2006) SMUG1 is able to excise uracil from immunoglobulin genes: insight into mutation versus repair. EMBO J 25: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson SK, Market E, Besmer E, Papavasiliou FN (2003) AID mediates hypermutation by deaminating single stranded DNA. J Exp Med 197: 1291–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diella F, Cameron S, Gemund C, Linding R, Via A, Kuster B, Sicheritz-Ponten T, Blom N, Gibson TJ (2004) Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics 5: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Wilson DM III (2005) Protein–protein interactions and posttranslational modifications in mammalian base excision repair. Free Radic Biol Med 38: 1121–1138 [DOI] [PubMed] [Google Scholar]
- Fanning E, Klimovich V, Nager AR (2006) A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res 34: 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JA, Muller-Weeks S, Caradonna S (2004) Proteolytic degradation of the nuclear isoform of uracil-DNA glycosylase occurs during the S phase of the cell cycle. DNA Repair 3: 505–513 [DOI] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP (2007) Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell 26: 131–143 [DOI] [PubMed] [Google Scholar]
- Hardeland U, Kunz C, Focke F, Szadkowski M, Schar P (2007) Cell cycle regulation as a mechanism for functional separation of the apparently redundant uracil DNA glycosylases TDG and UNG2. Nucleic Acids Res 35: 3859–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug T, Skorpen F, Aas PA, Malm V, Skjelbred C, Krokan HE (1998) Regulation of expression of nuclear and mitochondrial forms of human uracil-DNA glycosylase. Nucleic Acids Res 26: 1449–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henricksen LA, Umbricht CB, Wold MS (1994) Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem 269: 11121–11132 [PubMed] [Google Scholar]
- Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, Krokan HE, Ochs HD, Fischer A, Durandy A (2003) Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol 4: 1023–1028 [DOI] [PubMed] [Google Scholar]
- Kavli B, Andersen S, Otterlei M, Liabakk NB, Imai K, Fischer A, Durandy A, Krokan HE, Slupphaug G (2005) B cells from hyper-IgM patients carrying UNG mutations lack ability to remove uracil from ssDNA and have elevated genomic uracil. J Exp Med 201: 2011–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavli B, Otterlei M, Slupphaug G, Krokan HE (2007) Uracil in DNA-general mutagen, but normal intermediate in acquired immunity. DNA Repair 6: 505–516 [DOI] [PubMed] [Google Scholar]
- Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F, Aas PA, Hagen L, Krokan HE, Slupphaug G (2002) hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J Biol Chem 277: 39926–39936 [DOI] [PubMed] [Google Scholar]
- Koundrioukoff S, Jonsson ZO, Hasan S, de Jong RN, van der Vliet PC, Hottiger MO, Hubscher U (2000) A direct interaction between proliferating cell nuclear antigen (PCNA) and Cdk2 targets PCNA-interacting proteins for phosphorylation. J Biol Chem 275: 22882–22887 [DOI] [PubMed] [Google Scholar]
- Krokan HE, Nilsen H, Skorpen F, Otterlei M, Slupphaug G (2000) Base excision repair of DNA in mammalian cells. FEBS Lett 476: 73–77 [DOI] [PubMed] [Google Scholar]
- Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362: 709–715 [DOI] [PubMed] [Google Scholar]
- Lu X, Bocangel D, Nannenga B, Yamaguchi H, Appella E, Donehower LA (2004) The p53-induced oncogenic phosphatase PPM1D interacts with uracil DNA glycosylase and suppresses base excision repair. Mol Cell 15: 621–634 [DOI] [PubMed] [Google Scholar]
- Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM, Chazin WJ (2000) Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell 103: 449–456 [DOI] [PubMed] [Google Scholar]
- Mol CD, Arvai AS, Slupphaug G, Kavli B, Alseth I, Krokan HE, Tainer JA (1995) Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell 80: 869–878 [DOI] [PubMed] [Google Scholar]
- Muller-Weeks S, Mastran B, Caradonna S (1998) The nuclear isoform of the highly conserved human uracil-DNA glycosylase is an Mr 36 000 phosphoprotein. J Biol Chem 273: 21909–21917 [DOI] [PubMed] [Google Scholar]
- Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus TA, Skorpen F, Krokan HE (1997) Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res 25: 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterlei M, Haug T, Nagelhus TA, Slupphaug G, Lindmo T, Krokan HE (1998) Nuclear and mitochondrial splice forms of human uracil-DNA glycosylase contain a complex nuclear localisation signal and a strong classical mitochondrial localisation signal, respectively. Nucleic Acids Res 26: 4611–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, Aas PA, Steinsbekk K, Bakke O, Krokan HE (1999) Post-replicative base excision repair in replication foci. EMBO J 18: 3834–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters EC, Horn DM, Tully DC, Brock A (2001) A novel multifunctional labeling reagent for enhanced protein characterization with mass spectrometry. Rapid Commun Mass Spectrom 15: 2387–2392 [DOI] [PubMed] [Google Scholar]
- Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS (2002) Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol 12: 1748–1755 [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75: 663–670 [DOI] [PubMed] [Google Scholar]
- Richardson RT, Batova IN, Widgren EE, Zheng LX, Whitfield M, Marzluff WF, O'Rand MG (2000) Characterization of the histone H1-binding protein, NASP, as a cell cycle-regulated somatic protein. J Biol Chem 275: 30378–30386 [DOI] [PubMed] [Google Scholar]
- Scaramozzino N, Sanz G, Crance JM, Saparbaev M, Drillien R, Laval J, Kavli B, Garin D (2003) Characterisation of the substrate specificity of homogeneous vaccinia virus uracil-DNA glycosylase. Nucleic Acids Res 31: 4950–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vor O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Slupphaug G, Eftedal I, Kavli B, Bharati S, Helle NM, Haug T, Levine DW, Krokan HE (1995) Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry 34: 128–138 [DOI] [PubMed] [Google Scholar]
- Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA (1996) A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature 384: 87–92 [DOI] [PubMed] [Google Scholar]
- Slupphaug G, Olsen LC, Helland D, Aasland R, Krokan HE (1991) Cell cycle regulation and in vitro hybrid arrest analysis of the major human uracil-DNA glycosylase. Nucleic Acids Res 19: 5131–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensballe A, Jensen ON (2004) Phosphoric acid enhances the performance of Fe(III) affinity chromatography and matrix-assisted laser desorption/ionization tandem mass spectrometry for recovery, detection and sequencing of phosphopeptides. Rapid Commun Mass Spectrom 18: 1721–1730 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters TR, Gallinari P, Jiricny J, Swann PF (1999) Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J Biol Chem 274: 67–74 [DOI] [PubMed] [Google Scholar]
- Welcker M, Clurman BE (2007) Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div 2: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin SG, Patel S, Kavli B, Slupphaug G (2005) Xenopus CENP-A assembly into chromatin requires base excision repair proteins. DNA Repair 4: 760–772 [DOI] [PubMed] [Google Scholar]