Abstract
Mef2 genes encode highly conserved transcription factors involved in somitic and cardiac mesoderm development in diverse bilaterians. Vertebrates have multiple mef2 genes. In mice, mef2c is required for heart and vascular development. We show that a zebrafish mef2c gene (mef2ca) is required in cranial neural crest (CNC) for proper head skeletal patterning. mef2ca mutants have head skeletal phenotypes resembling those seen upon partial loss-of-function of endothelin1 (edn1). Furthermore, mef2ca interacts genetically with edn1, arguing that mef2ca functions within the edn1 pathway. mef2ca is expressed in CNC and this expression does not require edn1 signaling. Mosaic analyses reveal that mef2ca is required in CNC for pharyngeal skeletal morphogenesis. Proper expression of many edn1-dependent target genes including hand2, bapx1, and gsc, depends upon mef2ca function. mef2ca plays a critical role in establishing the proper nested expression patterns of dlx genes. dlx5a and dlx6a, known Edn1 targets, are downregulated in mef2ca mutant pharyngeal arch CNC. Surprisingly, dlx4b and dlx3b are oppositely affected in mef2ca mutants. dlx4b expression is abolished while the edn1-dependent expression of dlx3b is ectopically expressed in more dorsal CNC. Together our results support a model in which CNC cells require mef2ca downstream of edn1 signaling for proper craniofacial development.
Keywords: mef2c, edn1, neural crest, pharyngeal skeleton, pharyngeal arch, dlx, hand2, zebrafish
Introduction
Mef2 (myocyte enhancing factor) genes encode MADS domain-containing transcription factors that play critical roles in mesoderm development in invertebrates and vertebrates (reviewed in Black and Olson, 1998). Mammals have four mef2 genes (mef2a, mef2b, mef2c, and mef2d). Mice lacking mef2c function die during early embryonic development due to severe heart and vascular defects (Bi et al., 1999; Lin et al., 1998; Lin et al., 1997). In addition to cardiac and vascular expression domains, mef2c is expressed in postmigratory cranial neural crest (CNC) within pharyngeal arch primordia (Dodou et al., 2004; Edmondson et al., 1994). The early embryonic lethality has, to date, precluded functional analysis in later mef2c expression domains such as CNC.
Most of the cartilages and bones of the vertebrate head are derived from CNC cells. In vertebrate embryos, three bilateral streams of CNC populate the first (mandibular), second (hyoid) and the set of more posterior branchial arches, respectively. Within each pharyngeal arch in gnathostome embryos, prominent dorsal and ventral cartilages form, separated by a dorsal-ventral joint. For example, in the first arch, CNC cells form an upper (dorsal) and lower (ventral) jaw, articulating at the jaw joint. One potential function of the mef2c CNC expression domain could be to regulate aspects of pharyngeal skeletal development. A critical signaling pathway that patterns CNC at the stages that mef2c has been reported to be expressed is Endothelin1 (Edn1),
Development of the lower jaw in mice, chicks, and fish requires Endothelin signaling (Clouthier et al., 1998; Kempf et al., 1998; Kurihara et al., 1994; Miller et al., 2000; Nair et al., 2007). Edn1, a secreted signaling molecule, is expressed in pharyngeal arch epithelia and mesoderm, and signals to CNC cells, which express the G-protein-coupled transmembrane Endothelin receptor EdnrA (reviewed in Clouthier and Schilling, 2004). Genetic work in mice and zebrafish have revealed other molecules involved in regulating or transducing the Edn1 signal during craniofacial development. In mice, mutation of Edn1, EdnrA, or an endothelin converting enzyme (ECE1) gives similar lethal craniofacial phenotypes where the lower jaw is hypoplastic and malformed (Clouthier et al., 1998; Kurihara et al., 1994; Yanagisawa et al., 1998). The proprotein convertase Furin has been shown to biochemically cleave Edn1, and in zebrafish, reducing furin function causes phenotypes resembling edn1 mutants (Denault et al., 1995; Walker et al., 2006a). These studies suggest that Edn1 is cleaved twice, first by Furin and second by ECE1 and that these cleavages are required for Edn1 bioactivity. Further upstream of Edn1, the transcription factor Tbx1 is required in zebrafish for edn1 pharyngeal arch expression (Piotrowski et al., 2003). In humans, mutation of Tbx1 can cause one of the most common craniofacial defects, DiGeorge syndrome, highlighting the clinical importance of Edn1 signaling in craniofacial development (Jerome and Papaioannou, 2001; Lindsay et al., 2001; Merscher et al., 2001).
Downstream of EdnrA, G-proteins transduce the Edn1 signal, as mice doubly mutant for GalphaQ and Galpha11 have Edn1-like craniofacial defects (Ivey et al., 2003). G-protein activation by EdnrA likely leads to activation of Phospholipase C (Plc), because in zebrafish, mutation of plcβ3 causes phenotypes similar to edn1 mutants and plcβ3 and edn1 interact genetically (Walker et al., 2006b). The Edn1 signal eventually activates a transcriptional hierarchy in CNC involving Dlx5, Dlx6, and Hand2(dHAND), and nearly all CNC expression of these three genes requires Edn1 signaling (Miller et al., 2003; Ruest et al., 2004; Walker et al., 2006a). Dlx5 and Dlx6 regulate the CNC expression of Hand2 (dHAND), an effector of edn1 signaling in mice and zebrafish (Charite et al., 2001; Depew et al., 2002; Miller et al., 2003; Ruest et al., 2004; Thomas et al., 1998). A Hand2 pharyngeal arch enhancer requires Edn1 signaling, as enhancer expression is absent in the arches of EdnrA mutant mice (Charite et al., 2001). Targeted deletion of this Edn1-responsive Hand2 enhancer reveals it to be required for craniofacial development (Yanagisawa et al., 2003). Biochemical analyses of this Hand2 pharyngeal arch enhancer found that this enhancer binds Dlx6 (but not Dlx2, Dlx3, or Dlx5) (Charite et al., 2001). Thus in mice, Dlx6 appears to participate in transducing the Edn1 signal to Hand2.
Dlx genes have nested expression patterns within each pharyngeal arch in mice, many aspects of which are conserved in zebrafish (Depew et al., 2002; Walker et al., 2006a). Dlx1 and Dlx2 are expressed broadly throughout seemingly all postmigratory pharyngeal arch CNC. Dlx5 and Dlx6 are more ventrally restricted within the pharyngeal arch primordia. Dlx3 and Dlx4 are even more ventrally restricted. Genetic experiments in the mouse have revealed this Dlx code to be of paramount importance in establishing regional identity within each pharyngeal arch (Depew et al., 1999; Depew et al., 2002; Depew et al., 2005; Qiu et al., 1995). Dlx6, together with Dlx5, specifies ventral identity within the pharyngeal arches. Mice doubly mutant for Dlx5 and Dlx6 have a fantastic homeotic phenotype in which the lower jaw is transformed into an upper jaw (Depew et al., 2002).
A similar ventral-to-dorsal transformation is seen in Edn1 and EdnrA mutants (Ozeki et al., 2004; Ruest et al., 2004). In zebrafish mutant for Edn1 pathway genes or with reduced levels of Edn1, a ventral hyoid bone can be homeotically transformed into a more dorsal bone (Kimmel et al., 2003). Gene expression studies show that the dorsally restricted eng2 expression ectopically expands ventrally in edn1 mutants (Miller et al., 2003). Furthermore, overexpression of Edn1 protein can cause the reciprocal transformation, where dorsal cartilages appear transformed into ventral cartilages (Kimmel et al., 2007). Thus, Edn1 is a master regulator of lower jaw and other ventral pharyngeal arch fates and in its absence the ventral arch assumes dorsal fates. Edn1 seems to exert this regulation primarily through Dlx genes. Despite this fundamental importance of the Dlx genes, little is known about what establishes their nested expression patterns.
Large scale genetic screens in the zebrafish have identified over 100 loci required for head skeletal patterning (Neuhauss et al., 1996; Nissen et al., 2003; Piotrowski et al., 1996; Schilling et al., 1996). One class of mutations was put into an “anterior arch” class for phenotypically similar defects in the anterior pharyngeal arch skeleton, particularly the first and second arches. This anterior arch class contains four loci, sucker, schmerle, sturgeon, and hoover (Piotrowski et al., 1996). We've previously shown these first three loci to encode Endothelin1, FurinA, and Plcβ3, respectively (Miller et al., 2000; Walker et al., 2006a; Walker et al., 2006b).
Within this “anterior arch” class, both strong and weak phenotypes are seen. Strong phenotypes, represented by typical edn1 and plcβ3 mutants, include highly penetrant severe reduction of lower jaw and other ventral pharyngeal cartilages, absence of the jaw joint and other dorsal-ventral pharyngeal joints, and loss of ventral pharyngeal bones (Kimmel et al., 1998; Kimmel et al., 2003; Miller et al., 2000; Walker et al., 2006b). Weak phenotypes, represented by typical mutants for furinA and hoover mutants, or partial reduction of Edn1 signaling by morpholino knockdown, include only mild, if any, reductions in ventral cartilage, and incompletely penetrant absence of joints and expansion of hyoid pharyngeal bones (Kimmel et al., 1998; Kimmel et al., 2003; Miller and Kimmel, 2001; Walker et al., 2006a).
Here we present fine mapping, sequencing, and morpholino phenocopy data showing that hoover is the zebrafish mef2ca gene. mef2ca mutants have skeletal phenotypes resembling those seen upon partial reduction of edn1 function. In addition to these striking phenotypic similarities, we find strong genetic interactions between mef2ca and edn1 including dominant enhancement and an unpredicted partial rescue of the edn1 phenotype in the double homozygote. mef2ca is expressed transiently in early postmigratory pharyngeal arch CNC. Mosaic analyses reveal mef2ca to function autonomously in CNC for pharyngeal skeletal patterning. mef2ca mutants exhibit similar, but less severe, gene expression defects as edn1 mutants, such as reduced pharyngeal arch expression of hand2, bapx1, gsc, dlx5a, and dlx6a. Unexpectedly, we find opposite effects of mef2ca on the adjacent and convergently transcribed dlx gene pair dlx3b and dlx4b. dlx4b expression, normally restricted to the ventral most CNC, is almost completely lost in mef2ca mutants. In stark contrast, the edn1-dependent ventrally-restricted dlx3b expression expands dorsally in mef2ca mutants. Together our results reveal a role for mef2c in effecting the response to the Edn1 signal within CNC cells.
Methods
Fish maintenance and gynogenetic screens
Fish were raised and staged as described (Westerfield, 1993; Kimmel et al., 1995). mef2ca(hoover)tn213 mutant fish were generously provided by Drs. Tatjana Piotrowski and Christiane Nusslein-Volhard. mef2cab631 and mef2cab1086 were identified by screening ENU mutagenized gynogenetic clutches stained for head cartilage in fixed larvae and live Alizarin red staining, respectively (Kimmel et al., 2003; Miller et al., 2004). For phenotypic analyses, the strongest allele (see Table 1), mef2cab1086 was used unless noted otherwise. fli1:GFP albino transgenic fish have been previously described (Lawson and Weinstein, 2002).
Table 1.
mef2ca mutant phenotypes resemble partial loss of edn1 function phenotypes
| % with Joint Loss in: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| arch 1 |
arch 2 |
||||||||
| unilateral |
unilateral |
||||||||
| allele | n | bilateral | Left | Right | bilateral | Left | Right | % with BHN | % with PQP |
| b1086 | 64 | 97 | 0 | 2 | 95 | 0 | 3 | 52 | 77 |
| tn213 | 63 | 87 | 6 | 6 | 70 | 9 | 8 | 78 | 30 |
| b631 |
365 | 38 | 7 | 28 | 5 | 1 | 3 | 26 | 73 |
| mef2ca-MO: |
|||||||||
| 5 ng | 135 | 81 | 6 | 6 | 47 | 5 | 7 | 55 | 15 |
| 1.5 ng |
29 | 0 | 14 | 17 | 0 | 0 | 0 | 7 | 0 |
| edn1-MO |
|||||||||
| 0.5 ng | 105* | 23 | 3 | 7 | 32 | 4 | 10 | 21 | 21 |
Pharyngeal cartilage phenotypes seen in 4.5 dpf larvae for all three alleles of mef2ca, mef2ca morphants, and low-level edn1 morphants. Joint loss refers to absence of a joint between dorsal and ventral pharyngeal cartilages. BHN, ectopic nubbin of cartilage around basihyal; PQP, ectopic medial process emanating from palatoquadrate (see Fig. 2G-I).
Of 242 fish injected with 0.5 ng edn1-MO, 119 had no detectable cartilage phenotype and 18 had very severe phenotypes resembling edn1 mutant larvae. The percentages listed here are of the 105 larvae with phenotypes weaker than the edn1 mutant phenotype.
Mapping
For mapping, the b631 mutation, which arose on an AB background, was crossed onto the wik mapping strain (Knapik et al., 1998). b631/wik heterozygotes were incrossed, generating clutches of mutant fish. At 4.5 days postfertilization (dpf), fish were anesthetized with MS222, then scored for facial phenotypes. A subset of each cross, including all fish with any facial aspects of the b631 phenotype, was then euthanized with MS222 and bisected. The heads were arrayed on one set of 96-well plates, and the tails were arrayed on another. DNA was isolated from the tails while the heads were stained with Alcian green to visualize cartilage. Each stained head was then inspected under a dissecting scope for the characteristic aspects of the mutant phenotype (loss of dorsal/ventral pharyngeal joints, ectopic nodules of cartilage around the basihyal and/or ectopic medial processes extending from the palatoquadrate). This process identified over 1000 fish with the characteristic b631 cartilage phenotypes, which we used as our mapping panel. Of these, 504 fish were polymorphic and succesfully genotyped for all of the markers shown in Fig. 1A. To map mef2ca, a polymorphism in the 3′UTR was revealed by PCR amplification with primers ATGTCCTGAACAATGCTAAG and CACATCCGCTCGTAATTGAGT and cutting with RsaI. Marker z9701 was amplified using primers GTGTGTGCTGCAAGAGTTCAG and ATGTAAAACGCTGGTGTTTGTG.
Fig. 1.
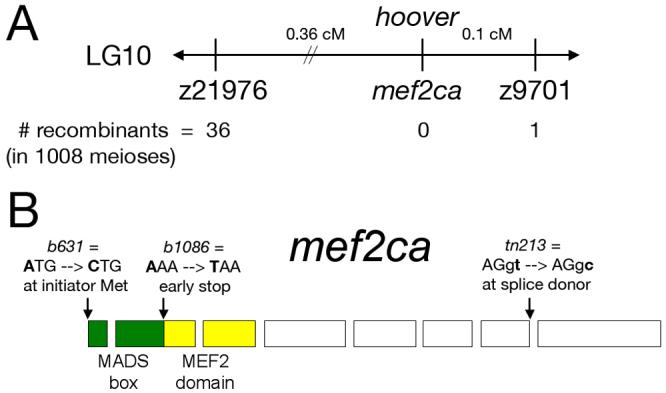
Molecular identification of the lesions in hoover(mef2ca) mutants. (A) Schematic showing the map position of hoover, which is non-recombinant with mef2ca in 1008 meioses. (B) mef2ca is mutated in all three hoover alleles. In b631 mutants, an A-to-C mutation is predicted to change the initiator methionine to leucine. In b1086 mutants, an A-to-T mutation creates an early stop in exon two, predicted to create a truncated protein lacking a MEF2 domain. In tn213 mutants, a T-to-C mutation alters the conserved second nucleotide of the intron 7 splice donor.
Sequencing
We sequenced mef2ca from wild-type and mutant genomic DNA by amplifying the previously reported exons by RT-PCR or with primers designed to flanking intronic sequences designed from the zebrafish genome assembly (http://www.ensembl.org/Danio_rerio/index.html). We note that the mef2ca sequence we obtained from both the AB and tlf backgrounds differs from the published sequence (Ticho et al., 1996, Genbank accession #U66569) at amino acid position 163, with their sequence having an ATG or methionine codon, and all of our sequences having a CTG or leucine codon.
Morpholino injections
For morpholino injections, a mef2ca morpholino oligonucleotide (MO) (TTCCTTCCTCTTCCAAAAGTACAGT), and an edn1 MO (GTAGTATGCAAGTCCCGTATTCCAG) were injected as described, (Miller and Kimmel, 2001). Roughly 5 nL was pressure-injected into the yolks of 1-4 cell embryos. Surviving larvae at day 4 were Alcian stained and scored for head skeletal phenotypes.
Tissue labeling procedures.
Alcian Green was used to stain the cartilage of fixed larvae as described (Miller et al., 2003). Facial cartilages were dissected out and prepared as flat mounts (Kimmel et al., 1998). Double Alcian Blue (cartilage) and Alizarin Red (bone) staining was done on fixed larvae using a modified “acid-free” protocol (Walker and Kimmel, in press). Wholemount RNA in situ hybridizations were performed as described (Miller et al., 2000) using previously described probes: mef2ca (Ticho et al., 1996), hand2 (Angelo et al., 2000), bapx1 (Miller et al., 2003), goosecoid (Schulte-Merker et al., 1994), dlx2a and dlx3b (Akimenko et al., 1994), dlx4b (Liu et al., 2003), and dlx5a and dlx6a (Walker et al., 2006a).
For in situ hybridization, embryos were raised in 0.0015% PTU (1-phenyl 2-thiourea) to inhibit melanogenesis (Westerfield, 1993). The lesion in mef2cab1086 mutants was genotyped by PCR amplification with primers TGCGGCTCGTTGTACTCGAAGTATT and TCATCAGGTTACGTTTACAAAGAGGAAGTTT, followed by digestion XmnI. See (Neff et al., 1998) for dCAPS genotyping method. edn1tf216b mutants were PCR genotyped as described (Miller et al., 2000).
Mosaic analyses
Neural crest cell transplants were performed between wild-type and mef2cab1086 embryos in a Tg(fli1:egfp)y1 background as previously described (Crump et al., 2004). Labeled cells were taken from donor embryos and placed 90 degrees from dorsal in host embryos at shield stage. Embryos were imaged at 30 hpf and re-imaged at 5dpf, followed by cartilage and bone double staining and flat mounting for imaging.
Results
Molecular identification of lesions in zebrafish mef2ca
By screening mutagenized gynogenetic diploid clutches for larval head skeletal phenotypes, we identified two new mutations, b631 and b1086, which both cause phenotypes similar to those seen upon mutation of the locus hoover (see below). A single allele of hoover, tn213, was previously identified in a large scale mutagenesis screen (Piotrowski et al., 1996). Complementation analyses revealed that b631, tn213 and b1086 are all allelic (data not shown).
Using bulked segregant analyses, we genetically mapped b631 to distal LG10 closely linked to microsatellite z9701. We refined the map position to 0.36 cM distal to z21976 and 0.1 cM proximal to z9701 (Fig. 1A). A previously published zebrafish mef2c gene had been mapped close to z9701 (Kelly et al., 2000; Ticho et al., 1996). We refer to this gene as mef2ca, as we subsequently identified an unlinked duplicate zebrafish mef2c gene, mef2cb (M.B. W., unpublished data). We mapped mef2ca to the same interval as hoover, non-recombinant in 1008 meioses (Fig. 1A). Thus, mef2ca is an outstanding positional candidate for hoover.
Sequencing mef2ca in all three hoover mutant alleles revealed lesions predicted to result in loss-of-function alleles (Fig. 1B). An A-to-T transversion at nucleotide 172 introduces a predicted early stop codon upstream of the MEF2 domain in b1086 mutants. In tn213 mutants, A T-to-C transition at the second nucleotide of the intron seven splice donor results in a predicted transcript that would be aberrantly spliced. In b631 mutants, an A-to-C transversion changes the start methionine codon to a leucine codon. Together, these mapping and sequencing data strongly indicate that hoover is zebrafish mef2ca, and we hereafter refer to this locus as mef2ca for clarity and simplicity.
Mutation of mef2ca causes phenotypes resembling partial reduction-of-function of Edn1 phenotypes
All three mef2ca mutant alleles are characterized by larval craniofacial phenotypes resembling partial reduction of function of edn1 (Fig. 2; Table 1) (Kimmel et al., 2003; Miller and Kimmel, 2001; Piotrowski et al., 1996; Walker et al., 2006a). The mouth is displaced ventrally, and joints between dorsal and ventral cartilages in the anterior pharyngeal arches are lost (Fig. 2, Table 1). Ectopic cartilage nodules are typically seen located around the second arch midline cartilage, the basihyal. Ectopic cartilage processes are typically seen emanating medially off of the first arch upper jaw cartilage, the palatoquadrate (Fig. 2, Table 1). The dorsal and ventral hyoid bones, the opercle and branchiostegal ray, are expanded and often fused to each other, often causing the ventral branchiostegal ray to appear homeotically transformed into an opercle (Fig. 2C,D and see Kimmel et al., 2003). Phenotypically, the three alleles form an allelic series, with b1086 being the most severe, and b631 being the least severe (Table 1). Reducing mef2ca function with morpholino oligonucleotides (MOs) robustly phenocopies multiple aspects of the mef2ca phenotype in a dose-dependent fashion (Fig. 2E-F, Table 1). These mef2ca mutant and mef2ca-MO-injected cartilage phenotypes strikingly resemble the phenotypes seen upon partial reduction of edn1 or strong reduction in furinA function (see Intro). Injecting low doses of edn1-MOs spares ventral cartilage formation but abolishes joint formation and results in cartilage nodules and processes similar to those seen in mef2ca mutants (Table 1, Fig. 2G-I).
Fig. 2.
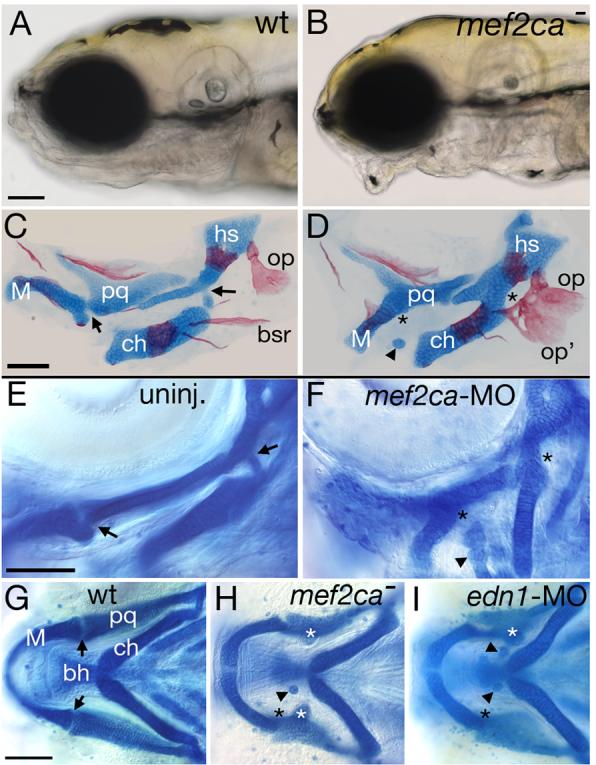
Reduction of mef2ca function causes facial deformities which phenocopy partial edn1 loss-of-function. (A-B) Live phenotypes of mef2ca mutants at 5 dpf. mef2ca mutants have malformed faces, open mouths, and ventrally displaced jaws. (C-D) Cartilage and bone phenotypes in the pharyngeal arches of mef2ca mutants. Dorsal/ventral joints (arrows) are missing in mef2ca mutants (asterisks). Mutants also have ectopic cartilage nodules (arrowhead), distinctive ectopic medial processes emanating from the upper jaw cartilage (see Table 2) and homeotic transformations of hyoid dermal bone identity. In mef2ca mutants, a ventral hyoid bone, the branchiostegal ray, is enlarged, assumes the shape of and fuses to the dorsal hyoid bone, the opercle. (E-F) mef2ca-MO injection phenocopies mef2ca mutations. Lateral views of wholemount 4.5 dpf wild-type larvae, uninjected (E) and injected with 5 ng of mef2ca morpholino (F). mef2ca-MO injected fish display characteristic mef2ca mutant cartilage phenotypes. Joints are lost (asterisks) and ectopic cartilage nodules (arrowhead) are seen near the basihyal. (G-I) mef2ca mutation phenocopies gradual reduction in edn1 function. Ventral views of 4.5 dpf Alcian stained wild-type (G), mef2cab631 mutant (H), and low level edn1-MO injected (I) larvae. mef2ca mutants and low level edn1-MO morphants display joint loss (black asterisks), ectopic medially-projecting processes on the upper jaw cartilage, the palatoquadrate (white asterisks), and ectopic cartilage nodules near the basihyal (arrowheads). bh, basihyal; bsr, branchiostegal ray; ch, ceratohyal; hs, hyosymplectic; M, Meckel's; op, opercle; pq, palatoquadrate.
mef2ca interacts genetically with edn1
Our analyses of the facial skeletal phenotype show that strong loss of function of mef2ca produces phenotypes resembling partial reduction of edn1 function or reductions of Edn1-pathway genes described in the Introduction. The resemblances include the losses of the DV joints, the enlarged opercular-branchiostegal dermal bones, and weak reductions of the ventral cartilages. To learn if these similarities are because the genes function in a common genetic pathway, we analyzed skeletal phenotypes of mef2ca; edn1 double mutants. We observe prominent epistasis that allow ordering the two genes in a hierarchical pathway.
Surprisingly, reduction or loss of mef2ca function results in partial rescue of the edn1−/− skeletal phenotypes. We illustrate with individual cases (Fig. 3), before coming to quantitative analyses (Fig. 4). We focus on the hyoid arch for these analyses: The edn1−/− homozygous single mutant shows invariable loss of the joint between dorsal (HS) and ventral (CH) cartilage, and invariable severe reduction in the size of CH. Sometimes, the CH can appear to be missing altogether (Fig. 3G, especially on one side of this mutant). The hyoid dermal bone is often severely reduced as well (Fig. 3G, especially on the other side in this instance). The double mutant shows the joint loss, and in comparison with edn1−/− single mutant shows a larger CH (Fig. 3I, arrowhead) and expanded dermal bone (the OP), i.e., more like the mef2ca−/− single mutant (Fig. 3C) and the wild-type condition (Fig. 3A).
Fig. 3.
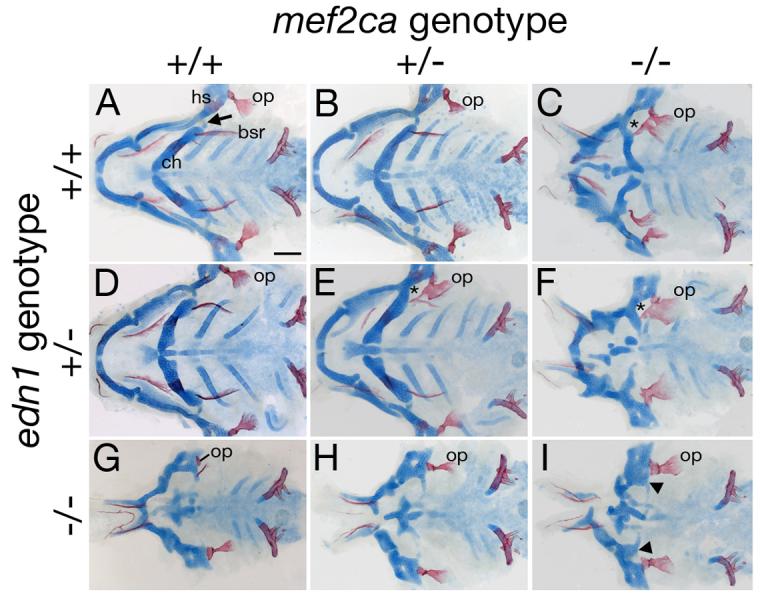
Complex genetic interactions between mef2ca and edn1. Flat-mounted pharyngeal skeletons of 5 dpf fish stained for cartilage in blue and bone in red for fish of different mef2ca and edn1 genotypes. (A) In the wild-type second arch, a prominent fan-shaped opercle bone (op) articulates with the hyosymplectic (hs). Its serial homolog, the branchiostegal ray (bsr) appears saber-shaped and articulates with the ceratohyal (ch). We typically detect no phenotypes in edn1 or mef2ca single heterozygous classes (B and D), although rarely very mild phenotypes are seen in edn1 mutant heterozygotes (Table 2). (C) In mef2ca homozygous mutants, the opercle is enlarged, and the ventral branchiostegal ray is enlarged and transformed towards opercle morphology. The dorsal/ventral joint is lost (asterisk showing joint loss in the hyoid arch). The hyoid ventral cartilage is reduced. (E) Fish heterozygous for both mef2ca and edn1 (mef2ca+/−; edn1+/−) display mef2ca mutant phenotypes such as joint loss (asterisk) and enlarged opercles fused to malformed branchiostegal rays. (F) Heterozygosity of edn1 enhances the mef2ca cartilage and bone phenotypes (compare to C, and see Table 2). (G-I) Heterozygosity for mef2ca mutation (H) partially and subtly rescues cartilage and bone phenotypes in edn1−/− homozygous mutants (G), while homozygosity for mef2ca mutation more significantly partially rescues hyoid cartilage and bone phenotypes of edn1 homozygous mutants (I).
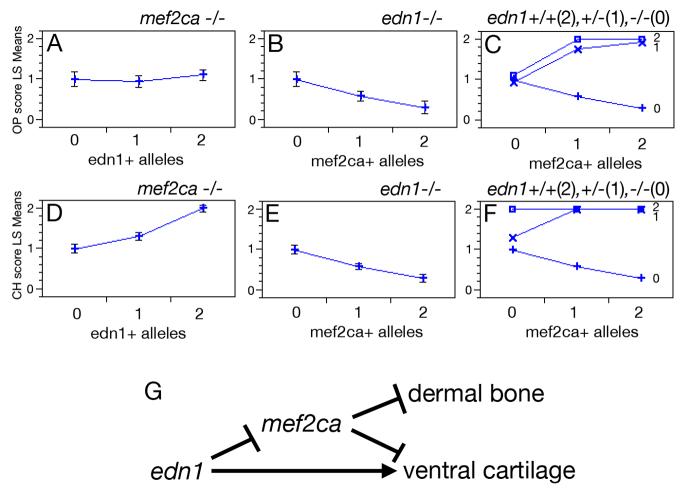
Fig. 4.
Epistatic interactions between mef2ca and edn1. Hyoid bone (opercle, OP) (A-C) and cartilage (ceratohyal, CH) (D-F) phenotypes as a function of: edn1+ alleles in homozygous mef2ca mutants (A,D), mef2ca+ alleles in homozygous edn1 mutants (B, E), and genotypes at both mef2ca and edn1 (C,F). In C and F, edn1 genotype is denoted by the number of edn1+ alleles: open box = 2 edn1+ alleles; “X” = 1 edn1+ allele, “+” = 0 edn1+ alleles. Phenotypes were scored on individual sides of PCR-genotyped larvae (see Methods). OP bone scoring scale: strong edn1 loss-of-function phenotype (loss of bone) = 0, weak edn1 loss-of-function phenotype (expanded bone) = 1, wild-type = 2. CH cartilage scoring scale: strong edn1 loss-of-function phenotype (complete loss) = 0, weak edn1 loss-of-function phenotype (partial loss) = 1, wild-type = 2. See Kimmel et al. (2003) and Miller and Kimmel (2001) for strong and weak edn1 loss-of-function bone and cartilage mutant phenotypes. Phenotypes for each genotypic category are shown as least square means (LS means). Error bars represent 95% confidence intervals from ANOVA. (G) Genetic model for zebrafish hyoid cartilage and dermal bone development. mef2ca represses bone and cartilage formation. edn1 represses mef2ca, but also activates a mef2ca-independent pathway for ventral cartilage development.
In contrast to this rescue (or suppression) of the edn1−/− phenotype, we see a striking enhancement of the phenotype of the edn1+/− heterozygote with loss of a single wild-type allele of mef2ca (i.e., in the double heterozygote, edn1+/−; mef2ca+/−, Fig. 3E). In the example shown dermal bone is dramatically enlarged, and the DV joint is lost (*) on one side of the fish, whereas the phenotype on the other side of this animal appears wild type, the usual case for the single heterozygote, edn1+/−; mef2ca+/+.
To examine these interactions quantitatively, we scored both the dermal bone (OP) and cartilage (CH) phenotypes according to severity in fish from all nine possible genotypic combinations of mef2ca and edn1 mutant alleles (shown in Fig. 3). For the bone we see a mild edn1 loss-of-function phenotype in the mef2ca−/−; edn1−/− double homozygous mutant, and no effect of adding 1 or 2 edn1+ alleles in the absence of any mef2ca+ alleles (Fig. 4A). In contrast, adding mef2ca+ alleles in the absence of edn1+ alleles yields more severe bone phenotypes in a dose-dependent manner (Fig. 4B). The straight-forward interpretation is that mef2ca is acting as a repressor. The function of edn1 in dermal bone development becomes clear when wild-type alleles of the two genes are combined (Fig. 4C); even only a single edn1+ allele strongly reverses the mef2ca repression. Hence edn1 acts hierarchically upstream of mef2ca, serving to repress the repressor (Fig. 4G).
Similar analysis of the CH phenotype shows a variation on this theme. Whereas mef2ca again shows itself to be repressing skeletal development (Fig. 4E) and edn1 again reverses this repression (Fig. 4F), a separate, additional function of Edn1 is apparent when examined in the mef2ca−/− background (Fig. 4E), namely a mef2ca-independent activation of CH development (Fig. 4D). Hence, whereas edn1 may be regulating dermal bone entirely through repressing mef2ca, it regulates ventral cartilage size in both a mef2ca-dependent and mef2ca-independent fashion (Fig. 4G).
mef2ca is expressed in early postmigratory pharyngeal arch CNC and later in pharyngeal arch muscle cores
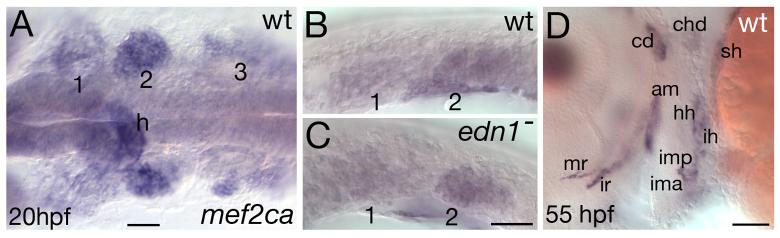
If, as our epistasis evidence suggests, Mef2ca functions downstream of Edn1, then we predict mef2ca to be expressed in CNC-derived arch mesenchyme, the target of Edn1 signaling. In the mouse, mef2c expression during embryonic development is detected in complex and dynamic patterns in CNC, cardiac and skeletal muscle, the vasculature, and the CNS (De Val et al., 2004; Dodou et al., 2004; Edmondson et al., 1994; Lyons et al., 1995). Consistent with the mouse expression pattern, we find transient expression of mef2ca in pharyngeal arch CNC. At 20 hpf, we detect mef2ca expression in mandibular, hyoid and branchial CNC (Fig. 5A,B). This CNC expression domain does not appear to require edn1 signaling, as expression appears unchanged in edn1 mutants (Fig. 5C). Reciprocally, the expression of edn1 in arch epithelia and mesodermal cores appears unaffected in mef2ca mutants (data not shown). mef2ca CNC expression is dynamic and seems to sweep in an anterior-posterior fashion in that expression at 24 hpf is strongest in the third arch, at 30 hpf is strongest in the fourth arch, and at 32 hpf is strongest in the fifth arch (data not shown).
Fig. 5.
mef2ca expression in early CNC and late head muscles. In situ hybridization showing mef2ca expression in wild-type (A,B,D) and edn1 mutant (C) embryos at 20 hpf (A-C) and 55 hpf (D). (A-B) Dorsal (A) and lateral (B) views of mef2ca expression in all three migrating streams of CNC at 20 hpf. (C) CNC expression in edn1 mutants appears unaffected. (D) Lateral view of mef2ca expression in head muscles at 55 hpf. For A-C, arches are numbered. Pharyngeal pouches are outlined in B-C. For description of the larval head muscle pattern see (Lin et al., 2006; Schilling and Kimmel, 1997). am adductor mandibulae; cd, constrictor dorsalis; chd, constrictor hyoideus dorsalis; hh, hyohyoideus; ih, interhyoideus; ima, intermandibularis anterior; imp, intermandibularis posterior; ir, inferior rectus; mr, medial rectus; sh, sternohyoideus.
Beginning around 30 hpf, we detect mef2ca expression in pharyngeal arch muscle cores, first appearing in the adductor mandibulae core in the first arch (data not shown). By 55 hpf, all cranial muscle cores and differentiating head muscles express mef2ca (Fig. 5D). While the mef2ca head mesoderm expression is detectable after stages known to be critical for transmission of the Edn1 signal, the CNC expression domain is in precisely the right place and at the right time to transduce the Edn1 signal (Miller et al., 2000).
mef2ca is autonomously required in cranial neural crest
If mef2ca functions downstream of edn1 in CNC, mef2ca should function autonomously in CNC to regulate head skeletal development, a hypothesis we can directly test by mosaic analysis. We transplanted wild-type CNC unilaterally into mef2ca mutant hosts (Fig. 6A,B). We found that wild-type CNC can rescue skeletal patterning on the transplanted side, while the non-transplanted control side exhibits the characteristic mef2ca mutant phenotypes (Fig. 6C). In every instance (n=7), wild-type CNC cells rescued the cartilage morphology of mutant hosts on the transplanted side. In contrast, the control side exhibited the mutant phenotype 100% of the time. Additionally, in reciprocal transplants, mef2ca mutant CNC conferred a mutant cartilage phenotype in 88% (n=7/8) of experimental sides . Thus mef2ca functions autonomously in CNC for pharyngeal cartilage morphogenesis. Together our epistasis and mosaic analyses indicate that mef2ca functions as a downstream effector of edn1 in CNC cells.
Fig. 6.
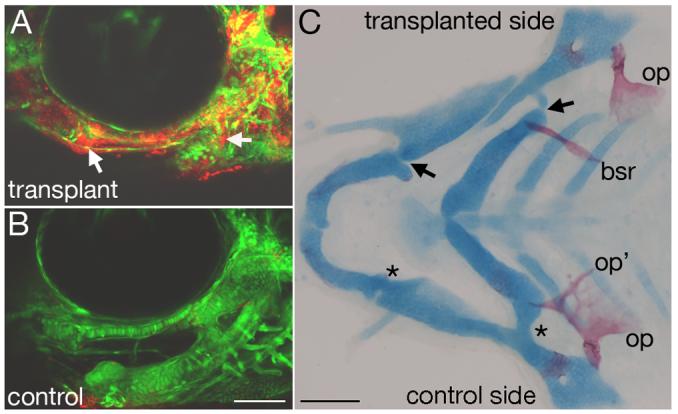
mef2ca is autonomously required in cranial neural crest for skeletal patterning. Wild-type rhodamine-labeled fli1:EGFP CNC cells were unilaterally transplanted into mef2ca mutant hosts. (A-B) Confocal micrographs of mef2ca mutant with wild-type CNC mosaic fish from the tranplanted side (A) and control side (B). On the transplanted side, lineage tracer is detected throughout dorsal and ventral cartilages, as well as joint regions (arrows). On the control side, only mef2ca mutant host fli1:GFP CNC derivatives are seen. (C) Flatmounted pharyngeal skeleton, double stained for cartilage in blue and bone in red. Dorsal/ventral cartilage joints have been rescued on the transplanted side (arrows) but not on the control side (asterisks). Opercle and branchiostegal ray morphology is also completely rescued on the transplanted side, but not the control side. bsr, branchiostegal ray; op, opercle.
mef2ca is required for expression of edn1-dependent genes
We next asked where mef2ca functions in the cascade of known effectors of edn1 signaling in CNC. We previously identified two transcription factors that partially mediate the edn1-dependent processes of ventral cartilage and joint formation. The bHLH transcription factor Hand2, whose ventrally-restricted expression is dramatically reduced in edn1 mutants, is also required for ventral pharyngeal cartilage formation (Miller et al., 2003). Hand2 is also known to physically interact with Mef2c (Zang et al., 2004). A second effector of Edn1 signaling is the homeobox transcription factor Bapx1, which is required for jaw joint formation in the mandibular arch (Miller et al., 2003). bapx1 is expressed in an intermediate first arch expression domain in wild types, but this domain is absent in edn1 mutants. As mef2ca mutants typically lack jaw joints, and variably have ventral cartilage reduction, we asked whether mef2ca functions upstream of these two edn1 effectors by examining the expression of hand2 and bapx1 in mef2ca mutants. At 30 hpf, the ventral arch expression of hand2 is downregulated in mef2ca mutants (Fig. 7A,B). However, by 55 hpf, ventral arch hand2 expression has recovered in mef2ca mutants (Fig. 7C-F), similar to furinA mutants, but unlike edn1 mutants (Walker et al., 2006a). At 48 hpf, the intermediate first arch domain of bapx1 is downregulated in mef2ca mutants (Fig. 7G,H).
Fig. 7.
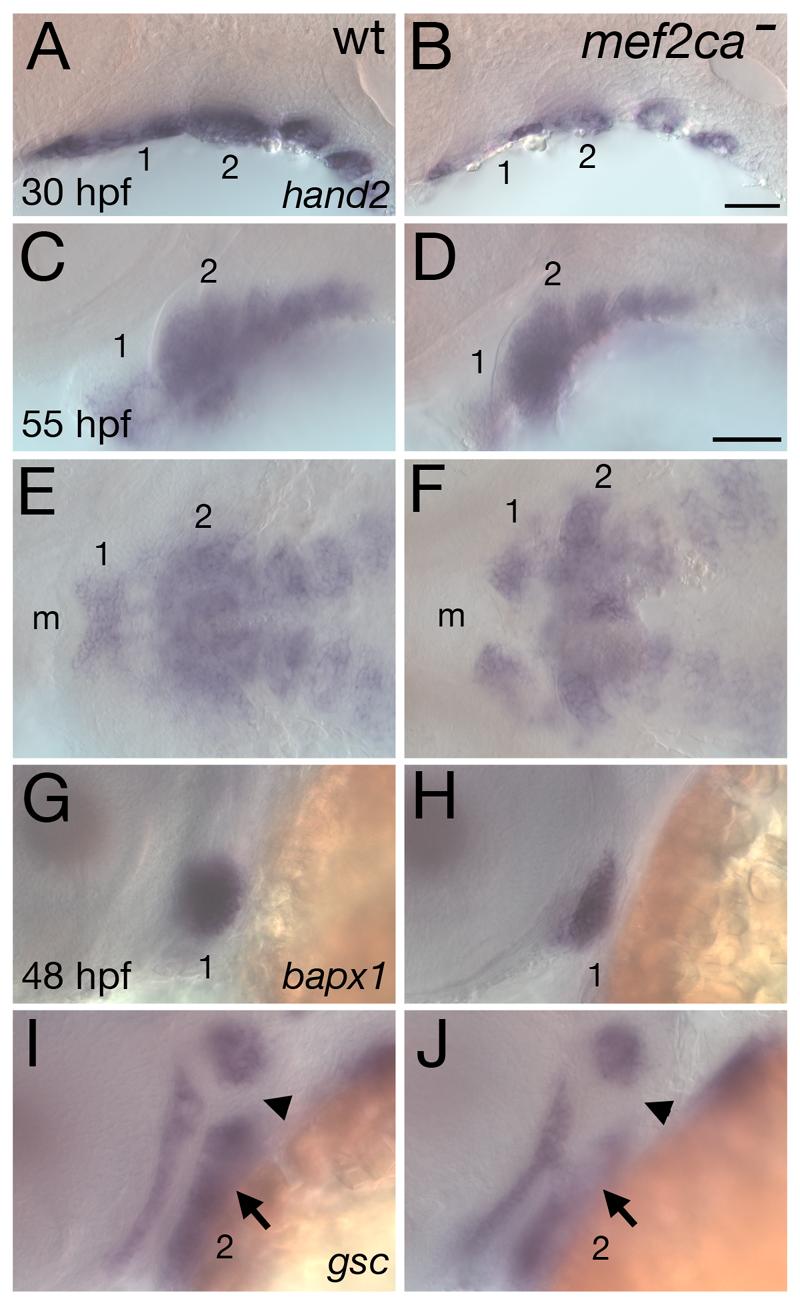
mef2ca is required for proper expression of hand2, bapx1, and gsc in postmigratory CNC. Lateral (A-D, G-J) and ventral (E,F) views of expression of hand2 (A-F) at 30 hpf (A,B) and 55 hpf (C-F), and bapx1 (G,H) and gsc (I,J) at 48 hpf in wild-type (A,C,E,G,I) and mef2ca mutants (B,D,F,H,J). (A-F) Ventrally restricted expression of hand2 is reduced in mef2ca mutants at 30 hpf, but recovers by 55 hpf. (G,H) bapx1 expression prefiguring the jaw joint is reduced in mef2ca mutants. (I,J) Ventral hyoid expression of gsc (arrow) is downregulated in mef2ca mutants. The hyoid joint region (arrowhead) remains devoid of gsc expression. Arches are numbered. m, mouth.
In the second (hyoid) arch of wild types, gsc is expressed in dorsal and ventral domains, but not in the intermediate domain, which includes the presumptive joint (Crump et al., 2006; Miller et al., 2000; Walker et al., 2006a). In edn1 mutant zebrafish, ventral arch two expression of goosecoid (gsc) is missing, while dorsal arch two gsc expression ectopically expands into the intermediate domain (Miller et al., 2000; Walker et al., 2006a; Walker et al., 2006b). Like edn1 mutants, mef2ca mutants have reduced ventral arch two expression of gsc. However, unlike edn1 mutants, and also unlike plcβ3 and furinA mutants, mef2ca mutants have no detectable ectopic expression of gsc in the intermediate hyoid domain (Fig. 7I,J). Thus, consistent with the phenotypic similarities of mef2ca and edn1 mutants, we find mef2ca also regulates these three edn1-dependent genes. Paralleling the less severe skeletal phenotype of mef2ca mutants, expression of hand2, bapx1, and gsc are affected less severely in mef2ca mutants than in edn1 mutants.
mef2ca activates dlx5a, dlx6a, and dlx4b, but represses dlx3b
Critical homeotic effectors of edn1 signaling are dlx genes, which in mice function at least in part upstream of hand2 (Depew et al., 2002; Ozeki et al., 2004; Ruest et al., 2004). We previously showed that in edn1 mutants, the ventrally-restricted pharyngeal arch CNC expression of dlx5a, dlx6a and dlx3b are extremely reduced, while the widespread expression of dlx2a was only effected in the ventral-most CNC (Miller et al., 2000; Walker et al., 2006a). Furthermore, we showed that reduction of dlx3b and dlx5a function causes phenotypes similar to the mef2ca mutant phenotype (Walker et al., 2006a). Given these functions and edn1-dependent expression domains of dlx genes, we tested the hypothesis that mef2ca functions as an upstream activator of dlx genes by assaying dlx expression in arch primordia of mef2ca mutants. Like in edn1 mutants, the broad CNC expression of dlx2a appears largely unaffected in mef2ca mutants (Fig. 8A,B). As expected and like hand2 and bapx1, dlx5a and dlx6a are downstream of mef2ca, as pharyngeal arch CNC expression of both genes is downregulated in mef2ca mutants (Fig. 8C-F). Contrary to our prediction, the edn1- dependent ventrally-restricted dlx3b expression expands dorsally in mef2ca mutants (Fig. 8G,H). Thus, mef2ca does function upstream of dlx3b, but represses dlx3b expression in more intermediate domains. Conversely, expression of the convergently transcribed linked dlx gene, dlx4b, is severely downregulated in mef2ca mutants (Fig. 8I,J). Thus, mef2ca plays a crucial role in effecting the Edn1 signal to establish the correctly nested expression patterns of dlx genes.
Fig. 8.
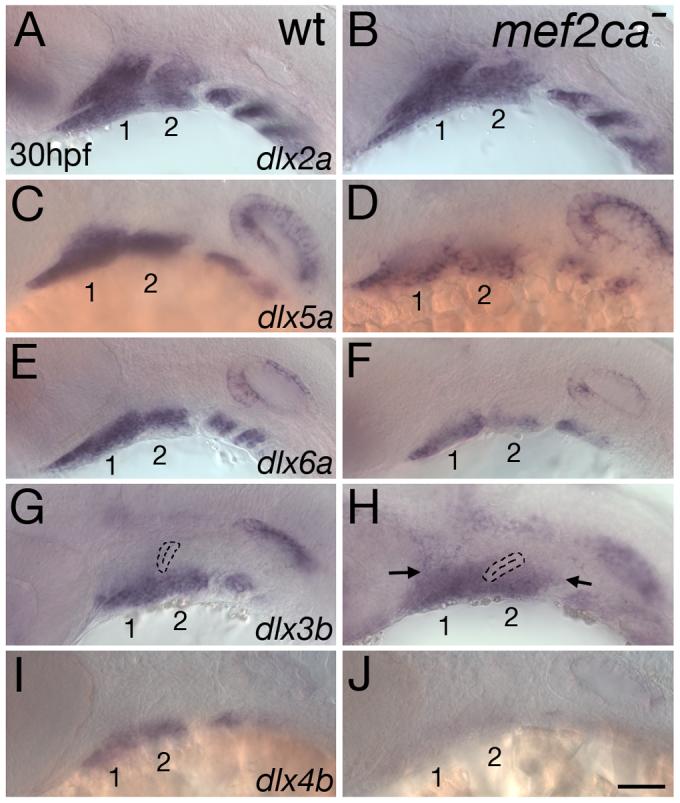
mef2ca activates dlx5a, dlx6a, and dlx4b but represses dlx3b. Lateral views of expression of dlx2a (A,B), dlx5a (C,D), dlx6a (E,F), dlx3b (G,H) and dlx4b (I,J) in wild-type (A,C,E,G,I) and mef2ca mutant (B,D,F,H,J) heads of 30 hpf zebrafish embryos. (A,B) mef2ca mutants have no detectable alteration in dlx2a expression, which is expressed throughout pharyngeal arch CNC. (C-F) Expression of both dlx5a and dlx6a, in ventral and intermediate CNC in wild-types, is reduced in mef2ca mutants. (G,H) dlx3b expression is ectopically expressed in dorsal arch CNC of mef2ca mutants (arrows). (I,J) In contrast, dlx4b expression, restricted to ventral arch CNC, is largely undetectable in mef2ca mutants. The first two arches are numbered.
Discussion
We present fine mapping, sequencing, and morpholino phenocopy results that together reveal a critical role for mef2ca in patterning the pharyngeal skeleton. The skeletal and gene expression similarities of mef2ca mutants and animals with reduced edn1 function support a model in which mef2ca functions in the same pathway as edn1. Epistasis analyses order mef2ca function downstream of edn1. Our expression and mosaic analyses support this model, and demonstrate mef2ca to be required in CNC, the tissue which receives the Edn1 signal from the surrounding epithelial and mesodermal environment. While mef2ca expression in CNC does not require Edn1 signaling, mef2ca is required for aspects of the edn1-dependent activation of hand2, bapx1, gsc, dlx5a, dlx6a, and dlx4b, and represses edn1-dependent dlx3b expression.
A zebrafish mef2c gene is required in CNC for Edn1 signal transduction
In mice, Mef2c plays critical roles in patterning mesodermal tissue in the heart and vasculature. Mice deficient for Mef2c die around e9.5, lacking a right ventricle and displaying complex vascular defects (Bi et al., 1999; Lin et al., 1998; Lin et al., 1997). While cardiac and vasculature expression domains of Mef2c likely underlie these early phenotypes, Mef2c is expressed in many other domains (Dodou et al., 2004; Edmondson et al., 1994). For example, in the embryonic pharyngeal arches, Mef2c is expressed in postmigratory CNC (Dodou et al., 2004).
Here we show a critical role for a zebrafish mef2c gene, mef2ca, in patterning the CNC-derived head skeleton. mef2ca mutant zebrafish have pharyngeal skeletal defects resembling defects seen upon partial reduction of edn1 function. The more mild skeletal phenotypes seen in mef2ca mutants compared to edn1 mutants might be explained by the fact that there are other mef2 genes in zebrafish, any one or more of which could act redundantly with mef2ca in the CNC. If so, the condition would be similar to furin genes (Walker et al., 2006a), where duplicate copies in fish allow roles in later developmental processes to be uncovered and studied.
Similarly, the less severe phenotype of zebrafish mef2ca mutants compared to the early embryonic lethality of mouse mef2c mutants could be explained in part by redundant roles of mef2ca and a second mef2c gene in zebrafish, mef2cb. Consistent with this model, we detect expression of both mef2ca and mef2cb during heart development (M.E.S. and M.B.W., unpublished results). Future studies can test whether mef2ca and mef2cb are functionally redundant in zebrafish, as we predict.
The genetic alleles of mef2ca appear to represent an allelic series. The weakest mef2ca allele, b631, is clearly a hypomorphic allele, as it arose on and was maintained on the same inbred genetic background as b1086, but results in lower penetrance and expressivity. The phenotypically intermediate allele, tn213, arose on and has been maintained on a different genetic background as the other two alleles, which could account for some or all of the differences in it's penetrance and/or expressivity from the other two alleles. The phenotypically strongest mef2ca allele, b1086, is predicted to result in a protein lacking the MEF domain and carboxyl terminus, which are required for Mef2c function (Molkentin et al., 1996a). Furthermore expression of mef2ca in b1086 mutants appears severely reduced by in situ hybridization (M.E.S., unpublished data). Thus, b1086 is clearly at least a strong loss-of-function allele, and likely a null.
Together our phenotypic, epistasis, mosaic, and gene expression results indicate that mef2ca functions within CNC downstream of Edn1 signaling. Unlike other previously identified downstream targets of Edn1 signaling, transcription of mef2ca in CNC cells does not require Edn1 signaling. Thus, the requirement of mef2ca for edn1- dependent gene expression must be a mechanism other than a sequential transcriptional hierarchy. During myogenesis, class II histone deacetylases (HDACs) regulate MEF2 protein activity by binding to MEF2 and repressing MEF2's ability to transactivate (reviewed in McKinsey et al., 2001). Chang et al., (2005) showed that signaling from G-protein coupled receptors, including EdnrA, leads to phosphorylation of class II HDACs which has been shown to cause dissociation from MEF2 (Chang et al., 2005; Lu et al., 2000). Perhaps in CNC cells, activation of EdnrA by Edn1 signaling signals to Mef2ca through HDAC regulation. Alternatively, Edn1 signaling could effect Mef2c more directly and involve one or more of the known post-transcriptional modifications to Mef2 proteins, such as phosphorylation or sumoylation, or an effect on the nuclear or subnuclear localization (De Angelis et al., 1998; Gregoire et al., 2006; Gregoire and Yang, 2005; Han et al., 1997; Kato et al., 2000; Lazaro et al., 2002; Molkentin et al., 1996b). Future research can determine how Mef2ca is activated in CNC by Edn1 signaling.
mef2a is required for proper expression patterns of dlx genes
The primacy of the master regulator Edn1 for lower jaw development has been demonstrated in the mouse. Both Edn1 and EdnrA mutant mice display similar homeotic transformations as those first reported for the Dlx5; Dlx6 double mutant mice (Depew et al., 2002; Ozeki et al., 2004; Ruest et al., 2004). These phenotypic similarites between Edn1 pathway mutants and Dlx5; Dlx6 double mutants, together with the findings that in mice and fish CNC expression of dlx5 and dlx6 requires edn1 signaling (Ruest et al., 2004; Walker et al., 2006a), identify Dlx transcription factors as critical effectors of the Edn1-mediated program of intermediate and ventral pharyngeal arch development. We find that CNC expression of both dlx5a and dlx6a also depends on mef2ca. Thus mef2ca functions in CNC to transduce the Edn1 signal to dlx genes.
The ventral to dorsal homeotic transformation seen in Dlx5; Dlx6 mutant mice provides strong support for the model of a Dlx code where dorsal-ventral fates in pharyngeal arches are assigned according to the combination of Dlx genes expressed. Functional analysis of other Dlx genes supports this idea of a Dlx code (Depew et al., 1999; Depew et al., 2002; Depew et al., 2005; Qiu et al., 1995). Although mice homozygous for Dlx3 mutations die during early embryonic development due to placental defects, precluding analysis of the head skeleton (Morasso et al., 1999), Depew et al. demonstrated a genetic interaction between Dlx3 and Dlx5. Dlx3 heterozygosity results in distinctive additional craniofacial phenotypes occuring on a Dlx5-/- mutant background (Depew et al., 2005). In zebrafish, dlx3 and dlx5 also interact, as reducing dlx3b and dlx5a function with morpholino oligonucleotides results in craniofacial phenotypes similar to mef2ca mutants (Walker et al., 2006a). It is interesting that in these same experiments, reduction of dlx5a and dlx6a did not produce dramatic ventral-to-dorsal homeotic transformations as seen in mice. This could either reflect incomplete knockdown of these genes by morpholinos, or reveal differences in dlx gene function between mice and fish. However, these experiments establish that dlx3b and dlx5a both participate in zebrafish craniofacial development.
Despite this fundamental importance of dlx genes in patterning the face, little is known about the molecular mechanisms that establish their nested expression patterns. Our finding that mef2ca mutants display ectopic intermediate domain expression of dlx3b suggests that the establishment of the nested dlx expression domains involves repression of ventrally-restricted dlx genes from more dorsal domains, with mef2ca mediating this dorsal repression of dlx3b. Thus, mef2ca plays multiple roles in establishing a Dlx code. In addition to being required for the expression of dlx4b, dlx5a, and dlx6a in intermediate and ventral domains, mef2ca restricts dlx3b expression from more dorsal domains. This dorsal repression of dlx3b was unpredicted, as dlx3b requires edn1 signaling (Miller et al., 2000; Walker et al., 2006a). Although both edn1 and mef2ca function upstream of dlx3b, they exert opposite regulatory influences. We propose that the edn1 activation of dlx3b expression occurs through mef2ca-independent (see below).
Molecular genetic analyses of dlx cis-regulation have identified conserved enhancers driving pharyngeal arch expression. A conserved enhancer between dlx3 and dlx4 is both necessary and sufficient for pharyngeal arch expression in mice (Sumiyama et al., 2003). Another conserved enhancer between dlx5 and dlx6 is also sufficient to drive expression in arch CNC in mice, and is dependent upon Edn signaling (Ghanem et al., 2003; Park et al., 2004). Whether mef2ca directly regulates dlx expression and how mef2ca exerts opposite regulation of dlx3b and dlx4b are interesting questions for future research.
A genetic model for the Edn1 network of craniofacial development
Our identification of mef2ca as a critical effector of Edn1 signaling further elaborates a model of the genetic circuitry controlling craniofacial development in zebrafish. Upstream of edn1 in arch epithelia and mesoderm, tbx1 and wdr68 positively regulate edn1 expression (Nissen et al., 2003; Piotrowski et al., 1996). Furin cleaves Edn1, followed by ECE1 cleavage, to produce the biologically active mature Edn1 ligand (Denault et al., 1995; Walker et al., 2006a; Xu et al., 1994). The mature secreted Edn1 ligand binds to, and activates, EdnrA receptors, which are required for Edn1 signaling in both mice and fish (Clouthier et al., 1998; Nair et al., 2007). Next, G-protein activation by EdnrA would signal to Plc (Ivey et al., 2003; Walker et al., 2006a). Our mosaic and gene expression analyses of mef2ca mutants place mef2ca as the most upstream known transcription factor that transduces the Edn1 signal in zebrafish. mef2ca transcription in CNC cells is apparently unaltered in edn1 mutants, thus we propose that some posttranscriptional alteration to Mef2ca occurs upon Edn1 signaling (e.g. dissociation of a MEF2/HDAC complex by HDAC phosphorylation upon EdnrA activation, see above and Chang et al., 2005). Mef2ca then either directly or indirectly transactivates the CNC expression of dlx5a, dlx6a, hand2, and bapx1. As dlx5a, hand2, and bapx1 are all required in some context for proper joints and/or ventral pharyngeal cartilage development, the failure to properly express these downstream genes in mef2ca mutants likely partially underlies the mef2ca mutant head skeletal phenotypes (Miller et al., 2003; Walker et al., 2006a).
Our data suggests that additional mef2ca-independent pathways likely transduce other branches of the Edn1 signal. For instance, while Edn1 is required for most ventral arch early and late hand2 expression and all jaw joint bapx1 expression, mef2ca only contributes to early hand2 expression and is only required for some bapx1 expression. Furthermore, the expression of dlx3b is repressed by mef2ca but activated by edn1. The expression of gsc is also ectopically activated in the intermediate hyoid arch in edn1 and furinA mutants, but not mef2ca mutants. We propose that other mef2 genes play roles in at least some of these mef2ca-independent processes.
That mef2ca mutations partially rescue or suppress the edn1 mutant bone and cartilage phenotypes provides strong evidence that mef2a and edn1 interact genetically, which we also infer from the similar gene expression and skeletal phenotypes seen in the two single mutants. One potential explanation for these results, supported by our epistasis analyses, is that mef2ca represses skeletal development and that edn1 represses mef2ca function (see Fig. 4G). Consistent with this model, emphasizing repression, all four of the typical mef2ca mutant phenotypes (joint loss, ectopic cartilage nodules, ectopic processes emanating off of the upper jaw cartilage, enlarged opercle bone) represent inappropriate overdevelopment of skeletal structures. However, at the level of target genes we see a positive, not negative, function in mef2ca regulation of the edn1-dependent transcription of hand2, a gene required for ventral cartilage development, and, as well, for correct patterning of some features of the intermediate domain. Thus, mef2ca is clearly not simply repressing the Edn1-dependent transcriptional cassette. dlx3b stands out as the only gene we have found to be repressed by mef2ca. We hypothesize that the repression of dlx3b contributes to the rescue of the edn1 mutant phenotypes by mef2ca mutations.
We summarize our major findings, and put them in context of previous work, with a model (Fig. 9). Secreted from ventral arch epithelia and mesoderm, the secreted Edn1 forms a gradient, with high levels ventrally, and low levels dorsally. The Edn1 signal is received by early postmigratory CNC cells, which interpret the gradient to specify fates along the DV axis. mef2ca is broadly expressed in CNC and exerts opposite effects on downstream effectors. In part via dlx genes, which mef2ca itself regulates, mef2ca mediates transduction of the edn1 signal to bapx1 and hand2 in intermediate and ventral cells.
Fig. 9.
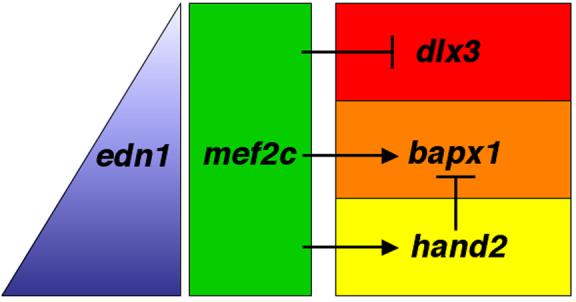
The edn1-effector mef2c regulates dorsal/ventral patterning in pharyngeal arch CNC. Edn1 secreted from ventral pharyngeal epithelia and mesoderm forms a gradient of Edn1 (blue triangle), with high levels ventrally (bottom of triangle) and low levels dorsally (top of triangle). mef2c, broadly expressed in early arch postmigratory CNC (green rectangle) exerts differential effects on downstream target genes to help specify domains along the dorsal-ventral axis. In the dorsal arch (red), mef2c represses dlx3 expression. In the intermediate (orange) and ventral (yellow) domains, mef2c activates transcription of bapx1 and hand2, two effectors of intermediate and ventral skeletal fates, respectively. hand2 helps delimit the intermediate domain by repressing bapx1 expression from the ventral domain (Miller et al., 2003). The red, orange, and yellow domains are in part defined by the nested expression patterns of dlx genes and hand2. The dorsal red domain expresses dlx2a, but not dlx3b or dlx6a. The orange and yellow domains express dlx2a, dlx3b, dlx5a, and dlx6a. The expression of hand2 defines the ventral yellow compartment. See Walker et al. (2006a) for a more detailed spatiotemporal model of dlx and hand2 regulation by Edn1.
The downregulation of hand2 in mef2ca mutants argues that mef2ca functions upstream of hand2 transcription in ventral CNC cells (Fig. 9). Since we find dlx5a and dlx6a to be downregulated in ventral CNC cells of mef2ca mutants, and dlx5 and dlx6 function upstream of hand2 expression in ventral CNC in mice (Depew et al., 2002), we propose that the positive regulation of hand2 by mef2ca occurs via dlx genes including dlx5a and dlx6a. The epistatic relationship of mef2ca and hand2 can be tested by analyzing animals with mutant alleles for both mef2ca and hand2, as we have done here for mef2ca and edn1.
The interaction between mef2ca and hand2 is of particular interest because of our previous findings that at the level of target gene regulation in the pharyngeal arch mesenchyme, hand2 itself is prominently, or entirely, functioning as a repressor (Miller et al., 2003). Repression of ventrally expressed genes, such as the edn1-dependent msxE and msxB, was unexpected since hand2 mutants, like edn1 mutants, lack nearly all ventral cartilage. In a manner that we don't yet understand, repression of hand2 target gene function must be transposed into activation of skeletal development by the same cells. Since hand2 expression is only transiently downregulated in mef2ca mutants, it is likely that the important time of hand2 function for ventral cartilage to be made is relatively late in the patterning and morphogenesis of pharyngeal arch mesenchyme. This finding is just as we showed previously for the function of furinA, and we presented a model that the intermediate domain and ventral domain are dynamically segregating during the critical period when hand2 must function (Walker et al. 2006a). Our new findings, especially the repression of dlx3b by mef2ca that we report in this paper, reveal the genetic circuitry of the downstream response to edn1 in CNC to be intricate and show frequent repression. The identification of additional molecules involved in transducing the Edn1 signal, as forward genetics in zebrafish has allowed, provides key progress toward a molecular genetic understanding of how the face is patterned.
Acknowledgments
We thank Will Talbot and Heather Stickney for mapping the b1086 allele, current and former members of the Kimmel laboratory, particularly Gage Crump, Tom Schilling, and Bruce Draper for useful discussions and suggestions, UO zebrafish researchers for their contributions to the ENU mutagenesis screens, and John Dowd for fish husbandry. Research support was provided by NIH grants DE13834, HD22486, an NRSA to J.K.E., and NIH training grant GM07413.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akimenko MA, et al. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J Neurosci. 1994;14:3475–86. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo S, et al. Conservation of sequence and expression of Xenopus and zebrafish dHAND during cardiac, branchial arch and lateral mesoderm development. Mech Dev. 2000;95:231–7. doi: 10.1016/s0925-4773(00)00334-8. [DOI] [PubMed] [Google Scholar]
- Bi W, et al. The transcription factor MEF2C-null mouse exhibits complex vascular malformations and reduced cardiac expression of angiopoietin 1 and VEGF. Dev Biol. 1999;211:255–67. doi: 10.1006/dbio.1999.9307. [DOI] [PubMed] [Google Scholar]
- Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–96. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- Chang S, et al. An expression screen reveals modulators of class II histone deacetylase phosphorylation. Proc Natl Acad Sci U S A. 2005;102:8120–5. doi: 10.1073/pnas.0503275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charite J, et al. Role of Dlx6 in regulation of an endothelin-1-dependent, dHAND branchial arch enhancer. Genes Dev. 2001;15:3039–49. doi: 10.1101/gad.931701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier DE, et al. Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development. 1998;125:813–24. doi: 10.1242/dev.125.5.813. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Schilling TF. Understanding endothelin-1 function during craniofacial development in the mouse and zebrafish. Birth Defects Res C Embryo Today. 2004;72:190–9. doi: 10.1002/bdrc.20007. [DOI] [PubMed] [Google Scholar]
- Crump JG, et al. Moz-dependent Hox expression controls segment-specific fate maps of skeletal precursors in the face. Development. 2006;133:2661–9. doi: 10.1242/dev.02435. [DOI] [PubMed] [Google Scholar]
- Crump JG, et al. An integrin-dependent role of pouch endoderm in hyoid cartilage development. PLoS Biol. 2004;2:E244. doi: 10.1371/journal.pbio.0020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis L, et al. Inhibition of myogenesis by transforming growth factor beta is density-dependent and related to the translocation of transcription factor MEF2 to the cytoplasm. Proc Natl Acad Sci U S A. 1998;95:12358–63. doi: 10.1073/pnas.95.21.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val S, et al. Mef2c is activated directly by Ets transcription factors through an evolutionarily conserved endothelial cell-specific enhancer. Dev Biol. 2004;275:424–34. doi: 10.1016/j.ydbio.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Denault JB, et al. Processing of proendothelin-1 by human furin convertase. FEBS Lett. 1995;362:276–80. doi: 10.1016/0014-5793(95)00249-9. [DOI] [PubMed] [Google Scholar]
- Depew MJ, et al. Dlx5 regulates regional development of the branchial arches and sensory capsules. Development. 1999;126:3831–46. doi: 10.1242/dev.126.17.3831. [DOI] [PubMed] [Google Scholar]
- Depew MJ, et al. Specification of jaw subdivisions by Dlx genes. Science. 2002;298:381–5. doi: 10.1126/science.1075703. [DOI] [PubMed] [Google Scholar]
- Depew MJ, et al. Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. J Anat. 2005;207:501–61. doi: 10.1111/j.1469-7580.2005.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E, et al. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–42. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- Edmondson DG, et al. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994;120:1251–63. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- Ghanem N, et al. Regulatory roles of conserved intergenic domains in vertebrate Dlx bigene clusters. Genome Res. 2003;13:533–43. doi: 10.1101/gr.716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire S, et al. Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J Biol Chem. 2006;281:4423–33. doi: 10.1074/jbc.M509471200. [DOI] [PubMed] [Google Scholar]
- Gregoire S, Yang XJ. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol Cell Biol. 2005;25:2273–87. doi: 10.1128/MCB.25.6.2273-2287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, et al. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–9. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- Ivey K, et al. Galphaq and Galpha11 proteins mediate endothelin-1 signaling in neural crest-derived pharyngeal arch mesenchyme. Dev Biol. 2003;255:230–7. doi: 10.1016/s0012-1606(02)00097-0. [DOI] [PubMed] [Google Scholar]
- Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–91. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- Kato Y, et al. Big mitogen-activated kinase regulates multiple members of the MEF2 protein family. J Biol Chem. 2000;275:18534–40. doi: 10.1074/jbc.M001573200. [DOI] [PubMed] [Google Scholar]
- Kelly PD, et al. Genetic linkage mapping of zebrafish genes and ESTs. Genome Res. 2000;10:558–67. doi: 10.1101/gr.10.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf H, et al. Pharmacological inactivation of the endothelin type A receptor in the early chick embryo: a model of mispatterning of the branchial arch derivatives. Development. 1998;125:4931–41. doi: 10.1242/dev.125.24.4931. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, et al. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, et al. The shaping of pharyngeal cartilages during early development of the zebrafish. Dev Biol. 1998;203:245–63. doi: 10.1006/dbio.1998.9016. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, et al. Endothelin 1-mediated regulation of pharyngeal bone development in zebrafish. Development. 2003;130:1339–51. doi: 10.1242/dev.00338. [DOI] [PubMed] [Google Scholar]
- Knapik EW, et al. A microsatellite genetic linkage map for zebrafish (Danio rerio) Nat Genet. 1998;18:338–43. doi: 10.1038/ng0498-338. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, et al. Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature. 1994;368:703–10. doi: 10.1038/368703a0. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–18. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Lazaro JB, et al. Cyclin D-cdk4 activity modulates the subnuclear localization and interaction of MEF2 with SRC-family coactivators during skeletal muscle differentiation. Genes Dev. 2002;16:1792–805. doi: 10.1101/gad.U-9988R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, et al. Myogenic regulatory factors Myf5 and Myod function distinctly during craniofacial myogenesis of zebrafish. Dev Biol. 2006;299:594–608. doi: 10.1016/j.ydbio.2006.08.042. [DOI] [PubMed] [Google Scholar]
- Lin Q, et al. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development. 1998;125:4565–74. doi: 10.1242/dev.125.22.4565. [DOI] [PubMed] [Google Scholar]
- Lin Q, et al. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–7. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay EA, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- Liu D, et al. Fgf3 and Fgf8 dependent and independent transcription factors are required for otic placode specification. Development. 2003;130:2213–24. doi: 10.1242/dev.00445. [DOI] [PubMed] [Google Scholar]
- Lu J, et al. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci U S A. 2000;97:4070–5. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons GE, et al. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J Neurosci. 1995;15:5727–38. doi: 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, et al. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- Merscher S, et al. TBX1 is responsible for cardiovascular defects in velo-cardiofacial/DiGeorge syndrome. Cell. 2001;104:619–29. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- Miller CT, Kimmel CB. Morpholino phenocopies of endothelin 1 (sucker) and other anterior arch class mutations. Genesis. 2001;30:186–7. doi: 10.1002/gene.1061. [DOI] [PubMed] [Google Scholar]
- Miller CT, et al. moz regulates Hox expression and pharyngeal segmental identity in zebrafish. Development. 2004;131:2443–61. doi: 10.1242/dev.01134. [DOI] [PubMed] [Google Scholar]
- Miller CT, et al. sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–28. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- Miller CT, et al. Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development. 2003;130:1353–65. doi: 10.1242/dev.00339. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, et al. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol Cell Biol. 1996a;16:2627–36. doi: 10.1128/mcb.16.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, et al. Phosphorylation of the MADS-Box transcription factor MEF2C enhances its DNA binding activity. J Biol Chem. 1996b;271:17199–204. doi: 10.1074/jbc.271.29.17199. [DOI] [PubMed] [Google Scholar]
- Morasso MI, et al. Placental failure in mice lacking the homeobox gene Dlx3. Proc Natl Acad Sci U S A. 1999;96:162–7. doi: 10.1073/pnas.96.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, et al. Requirements for Endothelin type-A receptors and Endothelin-1 signaling in the facial ectoderm for the patterning of skeletogenic neural crest cells in zebrafish. Development. 2007;134:335–45. doi: 10.1242/dev.02704. [DOI] [PubMed] [Google Scholar]
- Neff MM, et al. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 1998;14:387–92. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- Neuhauss SC, et al. Mutations affecting craniofacial development in zebrafish. Development. 1996;123:357–67. doi: 10.1242/dev.123.1.357. [DOI] [PubMed] [Google Scholar]
- Nissen RM, et al. Zebrafish foxi one modulates cellular responses to Fgf signaling required for the integrity of ear and jaw patterning. Development. 2003;130:2543–54. doi: 10.1242/dev.00455. [DOI] [PubMed] [Google Scholar]
- Ozeki H, et al. Endothelin-1 regulates the dorsoventral branchial arch patterning in mice. Mech Dev. 2004;121:387–95. doi: 10.1016/j.mod.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Park BK, et al. Intergenic enhancers with distinct activities regulate Dlx gene expression in the mesenchyme of the branchial arches. Dev Biol. 2004;268:532–45. doi: 10.1016/j.ydbio.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Piotrowski T, et al. The zebrafish van gogh mutation disrupts tbx1, which is involved in the DiGeorge deletion syndrome in humans. Development. 2003;130:5043–52. doi: 10.1242/dev.00704. [DOI] [PubMed] [Google Scholar]
- Piotrowski T, et al. Jaw and branchial arch mutants in zebrafish II: anterior arches and cartilage differentiation. Development. 1996;123:345–56. doi: 10.1242/dev.123.1.345. [DOI] [PubMed] [Google Scholar]
- Qiu M, et al. Null mutation of Dlx-2 results in abnormal morphogenesis of proximal first and second branchial arch derivatives and abnormal differentiation in the forebrain. Genes Dev. 1995;9:2523–38. doi: 10.1101/gad.9.20.2523. [DOI] [PubMed] [Google Scholar]
- Ruest LB, et al. Endothelin-A receptor-dependent and -independent signaling pathways in establishing mandibular identity. Development. 2004;131:4413–23. doi: 10.1242/dev.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling TF, Kimmel CB. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development. 1997;124:2945–60. doi: 10.1242/dev.124.15.2945. [DOI] [PubMed] [Google Scholar]
- Schilling TF, et al. Jaw and branchial arch mutants in zebrafish I: branchial arches. Development. 1996;123:329–44. doi: 10.1242/dev.123.1.329. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, et al. Expression of zebrafish goosecoid and no tail gene products in wild-type and mutant no tail embryos. Development. 1994;120:843–52. doi: 10.1242/dev.120.4.843. [DOI] [PubMed] [Google Scholar]
- Sumiyama K, et al. The role of gene duplication in the evolution and function of the vertebrate Dlx/distal-less bigene clusters. J Struct Funct Genomics. 2003;3:151–9. [PubMed] [Google Scholar]
- Thomas T, et al. A signaling cascade involving endothelin-1, dHAND and msx1 regulates development of neural-crest-derived branchial arch mesenchyme. Development. 1998;125:3005–14. doi: 10.1242/dev.125.16.3005. [DOI] [PubMed] [Google Scholar]
- Ticho BS, et al. Three zebrafish MEF2 genes delineate somitic and cardiac muscle development in wild-type and mutant embryos. Mech Dev. 1996;59:205–18. doi: 10.1016/0925-4773(96)00601-6. [DOI] [PubMed] [Google Scholar]
- Walker MB, et al. Zebrafish furin mutants reveal intricacies in regulating Endothelin1 signaling in craniofacial patterning. Dev Biol. 2006a;295:194–205. doi: 10.1016/j.ydbio.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Walker MB, et al. phospholipase C, beta 3 is required for Endothelin1 regulation of pharyngeal arch patterning in zebrafish. Dev Biol. 2006b doi: 10.1016/j.ydbio.2006.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 82 doi: 10.1080/10520290701333558. in press. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish book : a guide for the laboratory use of zebrafish (Brachydanio rerio) University of Oregon Press; Eugene. Or.: 1993. [Google Scholar]
- Xu D, et al. ECE-1: a membrane-bound metalloprotease that catalyzes the proteolytic activation of big endothelin-1. Cell. 1994;78:473–85. doi: 10.1016/0092-8674(94)90425-1. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, et al. Targeted deletion of a branchial arch-specific enhancer reveals a role of dHAND in craniofacial development. Development. 2003;130:1069–78. doi: 10.1242/dev.00337. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, et al. Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development. 1998;125:825–36. doi: 10.1242/dev.125.5.825. [DOI] [PubMed] [Google Scholar]
- Zang MX, et al. Cooperative activation of atrial naturetic peptide promoter by dHAND and MEF2C. J Cell Biochem. 2004;93:1255–66. doi: 10.1002/jcb.20225. [DOI] [PubMed] [Google Scholar]