Abstract
We describe here the molecular and functional characterization of the Caenorhabditis elegans unc-97 gene, whose gene product constitutes a novel component of muscular adherens junctions. UNC-97 and homologues from several other species define the PINCH family, a family of LIM proteins whose modular composition of five LIM domains implicates them as potential adapter molecules. unc-97 expression is restricted to tissue types that attach to the hypodermis, specifically body wall muscles, vulval muscles, and mechanosensory neurons. In body wall muscles, the UNC-97 protein colocalizes with the β-integrin PAT-3 to the focal adhesion-like attachment sites of muscles. Partial and complete loss-of-function studies demonstrate that UNC-97 affects the structural integrity of the integrin containing muscle adherens junctions and contributes to the mechanosensory functions of touch neurons. The expression of a Drosophila homologue of unc-97 in two integrin containing cell types, muscles, and muscle-attached epidermal cells, suggests that unc-97 function in adherens junction assembly and stability has been conserved across phylogeny. In addition to its localization to adherens junctions UNC-97 can also be detected in the nucleus, suggesting multiple functions for this LIM domain protein.
Keywords: LIM domains, UNC-97, muscle development, adherens junction, touch neuron
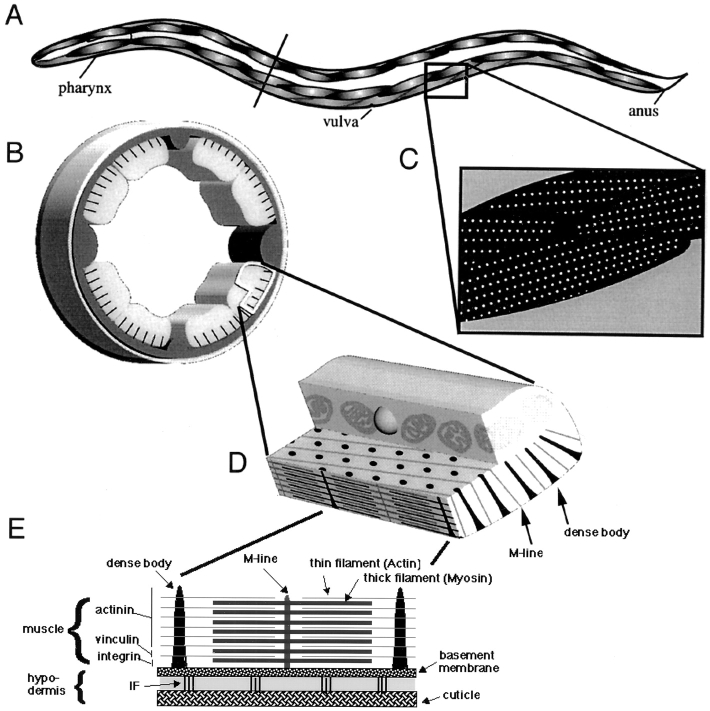
The nematode Caenorhabditis elegans has proven to be a useful system to study the development of muscle (Moerman and Fire, 1997; Waterston, 1988). Myoblasts arise after the end of gastrulation (at ∼290 min of embryonic development) and are defined by the accumulation of structural components such as myosin, vinculin, and integrin (Epstein et al., 1993; Coutu-Hresko et al., 1994). Myoblasts then migrate to their final positions, polarize, and flatten. In mid-embryogenesis (450 min), the muscle components organize into sarcomeres and attachment structures (Coutu-Hresko et al., 1994; see Fig. 1). These attachment structures, also termed dense bodies, are adherens junctions that provide a physical linkage between muscles and the hypodermis and are of critical importance to translate the mechanical movement of myofibrillar components to motion of the whole animal. These adherens junctions contain integrin heterodimers as central structural units that anchor cytoskeletal components to the extracellular matrix (Gettner et al., 1995). They also contain cytoskeletal adapter proteins such as vinculin, α-actinin, and talin (see Fig. 1) (Francis and Waterston, 1985; Barstead and Waterston, 1991; Moulder et al., 1996); thus, dense bodies are reminiscent of the structural composition of focal adhesions in tissue culture cells (Clark and Brugge, 1995). The regulatory steps that coordinate the assembly of adherens junction components into functional attachment structures that are capable of enduring and transmitting mechanical stress are largely undefined. We describe here a new LIM domain protein, UNC-97, that is required for assembly and stability of focal adhesion-like muscle attachment structures in C. elegans.
Figure 1.
Schematic view of nematode muscle organization. (A) Schematic view of C. elegans body wall muscles. (B) Cross-section showing muscle quadrants. (C) Lateral view onto muscle quadrants. White dots, focal-adhesion like muscle attachment sites (dense bodies); these represent a top view of the black lines in B. (D) Muscle cell. (E) Structural composition of sarcomeres (actin-myosin based contractile units) and their anchorage to the hypodermis. See text for references.
LIM domains are defined by the presence of two zinc coordinating, Cys/His-containing motifs and represent protein–protein interaction domains (Schmeichel and Beckerle, 1994; Feuerstein et al., 1994; Dawid et al., 1998). Some LIM proteins (LIM-only) are comprised almost exclusively of LIM domains whereas other LIM proteins (LIM-plus) display LIM domains linked to other functional domains including homeodomains, kinase domains or other protein–protein interaction domains (Dawid et al., 1998). LIM domain proteins display considerable specificity in their sites of subcellular localization. LIM homeodomain proteins and the LIM-only proteins LMO1, -2, and -3 are transcriptional regulatory proteins that function in the nucleus (Dawid et al., 1998). On the other hand, the LIM-only proteins zyxin and paxillin localize to actin stress fibers and focal adhesions, CRP family members localize to actin filaments and to the Z lines of myofibers in vertebrates (Turner et al., 1990; Crawford and Beckerle, 1991; Arber et al., 1997; Louis et al., 1997) and the Drosophila CRP family members, MLP60A and MLP84B, distribute along muscle fibers and to sites of muscle attachment (Stronach et al., 1996). Several of these cytoskeletal LIM proteins have been shown to display a remarkable dual subcellular localization; apart from its cytoskeletal localization, CRP3/MLP protein has also been reported to localize to the nucleus (Arber and Caroni, 1996), where it may participate in MyoD dependent transcriptional regulation (Kong et al., 1997). Moreover, although the LIM protein zyxin appears to be restricted to focal adhesions at steady-state levels, recent experiments have shown that it shuttles between focal adhesions and the nucleus (Nix and Beckerle, 1997).
The functional role of LIM proteins has mostly been addressed for those LIM proteins involved in transcriptional control. Mutations in LIM homeobox genes revealed their participation in lineage determination and neural differentiation in vertebrates and invertebrates (Dawid et al., 1998). Disruption of the LMO transcriptional regulator LMO1 causes haematopoietic lineage defects (Warren et al., 1994). In spite of their well-characterized biochemical properties, such as binding partners and subcellular localization, CRP3/MLP is the only cytoskeletal LIM protein for which a loss-of-function phenotypes has been reported so far; analysis of knockout mice revealed a requirement for CRP3/MLP in the organization of myofibrillar structures in cardiomyocytes (Arber et al., 1997). No physiological role can as yet be ascribed to other LIM-only protein families, such as the SLIM, zyxin, or paxillin families.
The C. elegans UNC-97 protein that we describe here defines a novel family of LIM-only proteins whose members have been conserved throughout evolution. We find UNC-97 to be expressed in a tissue restricted manner. It is present in touch neurons, sex muscles, and body wall muscles and colocalizes in muscle with β-integrin to sites of attachment of muscle to the underlying hypodermis. We show that disruption of UNC-97 function leads to the disruption of these focal adhesion-like structures as well as to functional defects of the mechanosensory neurons. The developmental expression profile of UNC-97, as well as its embryonic lethal null phenotype, is consistent with UNC-97 playing a role late in embryonic development in the process of terminal differentiation of muscles and mechanosensory neurons.
Materials and Methods
Strains and Genetics
The strains used in this study are: HE110 [unc-97(su110)], mec-3(u298), mec-3(e1338), dpy-20(e1282), and N2 wild-type strain. Double mutant mec-3(u298); unc-97(su110) animals were constructed by first marking unc-97(su110)X with dpy-20(e1282)IV and then repelling dpy-20(e1282) with the closely linked mec-3(u298).
Polarized Light Microscopy
Polarized light images were obtained essentially as described in Waterston et al. (1980). In brief, single live adult hermaphrodites were placed in a drop of M9 buffer on a slide and then a coverslip was carefully lowered onto the animals. Excess liquid was wicked away until the animals were immobilized and then animals were rolled gently until a good view of body wall muscle was obtained. Animals were viewed using a Zeiss Axiophot microscope (Carl Zeiss) equipped with polarizing optics.
Sequence Analysis
The unc-97(su110) molecular lesion was identified by PCR amplification of the F14D12.2 gene from unc-97(su110) mutant animals and subsequent DNA sequencing. BLAST searches identified unc-97 cDNAs in Y. Kohara's (Mishima, Japan) cDNA collection (yk460d6, yk457c11, yk455g5, yk457c10, yk403d5, yk267h9, yk114h11, yk184e5). yk184e5 was completely sequenced and found to encode a full-length clone. The nucleotide and protein coding sequence have been submitted to GenBank/EMBL/ DDBJ under accession number AF035583. UNC-97 homologous proteins were identified using the BLAST search algorithm. The cDNA representing the Drosophila expressed sequence tag (EST) sequence was obtained from Genome Systems. The complete nucleotide and protein coding sequence for d-pinch have been submitted to GenBank/EMBL/ DDBJ under accession number AF078907. Sequence alignments and dendograms were constructed using the pileup algorithm in the GCG software package.
Fusion Gene Constructs
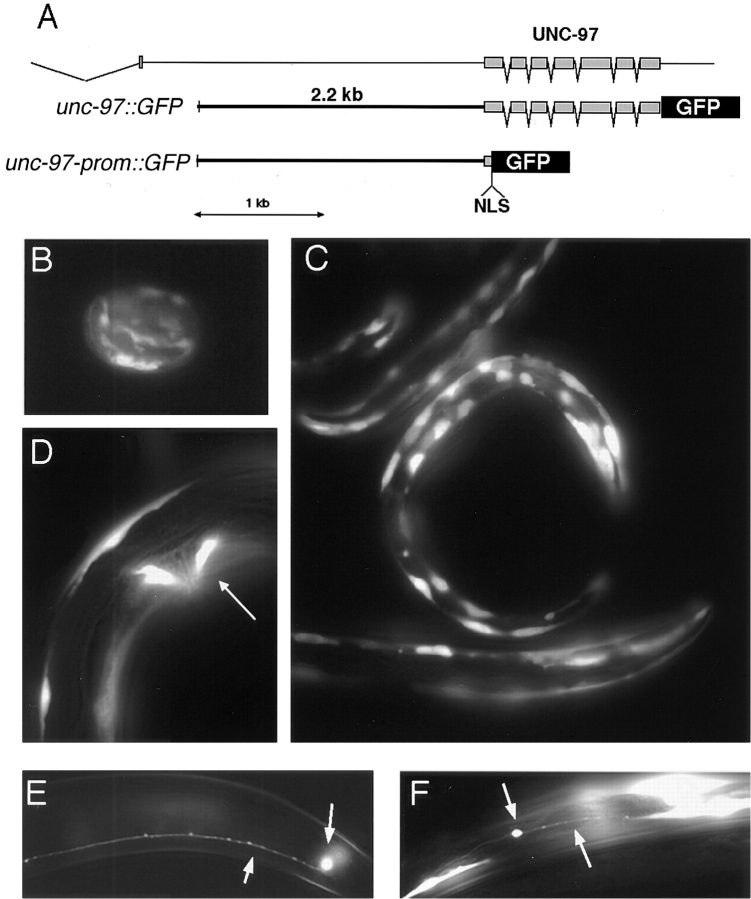
All three reporter gene constructs were constructed using a PCR fusion approach. First, two independent PCRs were performed, one amplifying the promoter and coding region (in the case of unc-97-prom::GFP just the promoter) from genomic DNA. The 3′ primer contained sequences overlapping with part of the green fluorescent protein (GFP)1 sequences on pPD95.75; the other PCR amplified GFP and the unc-54-3′-UTR from the pPD95.75 vector. The 5′ primer contained sequences overlapping with the genomic region targeted for fusion. The two PCR products were then fused by combining 1 μl of each PCR reaction and subsequent amplification with a set of nested primers, one at the beginning of the promoter, the other at the end of the unc-54-3′-UTR. For all the PCR reactions a mixture of Taq-polymerase and proofreading PwoI polymerase (Boehringer Mannheim, Indianapolis, IN) was used to ensure fidelity of the reaction. The fusion PCR product was injected at ∼100 ng/μl into wild-type worms. The promoter sequence used for both unc-97 reporter constructs starts at −2168 5′ of the predicted ATG start codon (reaching almost to the preceding predicted gene). unc-97-prom::GFP is a translational fusion encompassing the first 21 amino acids of UNC-97. unc-97::GFP is a translational fusion encompassing the complete genomic coding sequence of unc-97 (see Fig. 4). Note that both reporter gene constructs reveal a similar set of expressing cells, arguing that none of the introns contain regulatory sites. The promoter sequence used for pin-2::GFP starts at −2038 5′ of the predicted ATG start codon (reaching almost to the preceding predicted gene) and contains its complete, predicted genomic coding sequence.
Figure 4.
UNC-97 is expressed in muscles and neurons. (A) Schematic representation of reporter gene constructs used. Both constructs contain regulatory regions up to the mec-2 gene (last exon is shown), which precedes unc-97 and reveal a similar expression pattern. The unc-97-prom::GFP reporter gene constructs (shown below in B–F) uniformly labels the whole cell, whereas the unc-97::GFP fusion gene product reflects the subcellular localization of the UNC-97 protein shown in Figs. 6–8. (B) Expression of the unc-97-prom::GFP reporter gene in embryos at the comma stage (∼400 min). (C) Expression of unc-97-prom:: GFP in L1 larvae muscle cells. (D) Expression in vulval muscles (arrow). (E and F) Expression in the touch neurons PLM (E) and ALM (F). Bottom arrows, axonal processes; top arrows, cell bodies. Expression in the other touch neurons PVM and AVM is as strong but not shown here. The expression pattern described can be observed with an chromosomally integrated unc-97-prom:: GFP reporter gene.
Transgenic Animals and Rescue Analysis
Transgenic strains were constructed by injecting GFP reporter gene constructs as PCR products into N2 wild-type animals. No coinjection marker was used; transgenic animals were identified by their green fluorescence. The extrachromosomal GFP reporter gene arrays were integrated using a Stratalinker 1800 UV light source (Stratagene) at 300 J/m2. The transgenic strains used are: N2 Ex[unc-97::GFP], N2 Ex[unc-97-prom::GFP]; mgIs25 (integrated Ex[unc-97::GFP]), mgIs26 (integrated Ex[unc-97-prom:: GFP]); N2 Ex[pin-2::GFP, rol-6]. Rescue of unc-97(su110) was performed by making use of (a) the X-linkage of unc-97 and (b) an autosomally integrated, full-length unc-97::GFP copy: mgIs25 heterozygous males were mated with unc-97(su110) hermaphrodites. There are two types of F1 male cross progeny from this cross. One type contains a chromosome from the father that has no integrated unc-97::GFP. This type of F1 male cross progeny is not fluorescent and display the uncoordinated (unc) phenotype, since it is hemizygous for unc-97(su110). The other type received a chromosome from the father that does contain an integrated unc-97:: GFP (identified as fluorescent animals). If unc-97::GFP rescues, these fluorescent males should be non-unc. Indeed we found male animals of both phenotypes: nonfluorescent unc as well as fluorescent non-unc, thus demonstrating rescue specific for the animals that carry the unc97::GFP containing array.
Antibody Staining and GFP Expression Analysis
Fixation of animals for antibody staining was performed as described by Finney and Ruvkun (1990). In brief, animals were freeze-cracked by two rounds of freezing and thawing in dry-ice/ethanol and fixed in 3% paraformaldehyde, 80 mM KCl, 20 mM NaCl, 10 mM EGTA, 5 mM spermidine, 15 mM Pipes, pH 7.4, 25% methanol for 3 h at 4°C. Animals were washed twice in Tris/Triton buffer (100 mM Tris/HCl, pH 7.4; 1% Triton X-100, 1 mM EDTA), antigens reduced in Tris/Triton buffer + 1% β-mercaptoethanol for 1 h at 37°C. After washing once in borate buffer (33.3 mM boric acid, 16.6 mM NaOH, pH 9.5), antigens were oxidized in borate buffer + 0.3% hydrogen peroxide for 15 min at room temperature. After washing once in borate buffer, samples were blocked in antibody buffer (1× PBS, pH 7.4, 1% BSA [fraction V, 96–99%; Sigma, St. Louis, MO], 0.5% Triton X-100, 0.05% sodium azide, 1 mM EDTA) for at least 15 min. Antibodies used were: monoclonal mouse anti–pat-3 antibody MH25 (Francis and Waterston, 1985), monoclonal mouse anti-vinculin antibody MH24 (Francis and Waterston, 1985), and mouse polyclonal anti–unc-52 antibody GM1 (Moerman et al., 1996). MH24 and MH25 were used at 1:250, GM1 at 1:50, and incubated overnight at 4°C. Secondary antibodies (FITC- or TRITC-coupled) were incubated for 6–12 hours at 4°C. DNA was stained using 4′,6-diamidino-2-phenylindole (DAPI) at 10 μg/ml.
The expression of the unc-97::GFP reporter gene was either monitored in live animals or by fixing animals as described below for antibody staining. The fixation procedure used does not interfere with the fluorescent signal of GFP. We found that in several instances subcellular structures (such as the nuclear staining) could be better observed in fixed animals, although they were also clearly visible in the live animals, thus excluding fixation artifacts.
RNA Interference
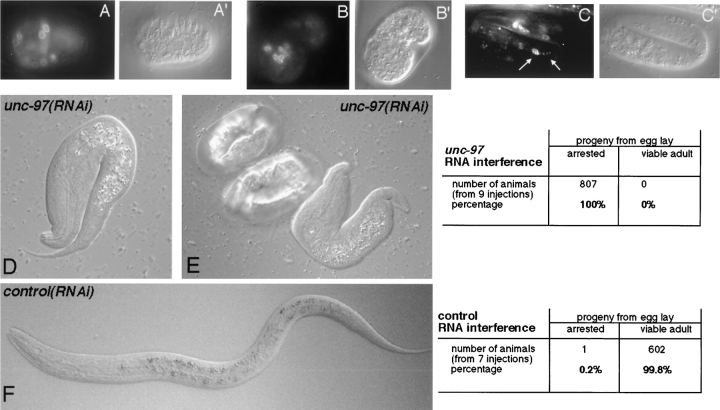
The probable null phenotype for the unc-97 gene was generated using RNA interference as described by Fire et al. (1998). The template for the sense and antisense unc-97 RNA transcripts was the complete unc-97 cDNA, subcloned into the Bluescript vector, which was amplified using Bluescript specific primers flanking the T3 and T7 primer sites. Sense and antisense RNA was produced from this PCR product in separate transcription reactions using T3 and T7 primers and commercially available RNA synthesis kits. The RNA was resuspended in water, the antisense and sense templates were mixed to a final concentration of ∼1–5 μg/μl and injected into the gonad. Scoring of the phenotype was done by allowing single injected animals to lay eggs at 20°C from hour 24 to hour 48 postinjection (see figure). The progeny derived from the hour 0 to hour 24 postinjection egg lay (which usually has not received as much RNA as the later progeny) also showed lethality; however, we occasionally observed the presence of entirely immobile and uncoordinated animals that grow to adulthood. The control RNA was derived from the COOH-terminal half of the cDNA of a predicted, nonessential homeobox gene on the CO4F1 cosmid.
Mechanosensory Assays
For each genotype 30–100 individual animals were scored by touching the animals with a thin hair 10 times, five times at the anterior and five times at the posterior. Normally, animals respond by backing up away from the touch. The responsiveness of every single animal was recorded so that, for example, six responses out of 10 touches were recorded as 60% responsiveness. Note that although unc-97(su110) display an uncoordinated phenotype associated with an almost immobile appearance, those mutant animals are still capable of responding to touch by backing up.
Whole Mount In Situ Hybridization of Drosophila Embryos
w1118 embryos were collected over a 17-h period and then prepared for whole mount in situ hybridization as described in Lehmann and Tautz (1993). The hybridization reaction proceeded for 12 h at 65°C in the following buffer, pH 4.5, 50% formamide (UltraPure; GIBCO BRL, Gaithersburg, MD), 5× SSC, 100 μg/ml tRNA, 100 ug/ml sonicated salmon-sperm DNA (denatured), 50 μg/ml heparin (Sigma), and 0.1% Tween-20, plus the appropriate riboprobe at a concentration of 0.25 ng/ml. After hybridization, samples were washed several times over a 2-h period at 65°C. For detection of probe, samples were incubated with a 1:5,000 dilution of anti-digoxigenin Fab fragments (Boehringer Mannheim), presorbed against fixed Drosophila embryos, either overnight at 4°C or 2 h at room temperature, and then processed according to the manufacturer's instructions.
Full-length digoxigenin-labeled RNA probes, corresponding to the sense and antisense strands of the d-pinch cDNA, were prepared using the Boehringer Mannheim Genius kit according to the manufacturer's instructions. The full-length probes were hydrolyzed to a size of ∼100 bp by treatment with carbonate buffer (60 mM Na2CO3, 40 mM NaHCO3, pH 10.2) at 60°C for 1 h.
Results
unc-97 Is Required for the Maintenance of C. elegans Adherens Junctions
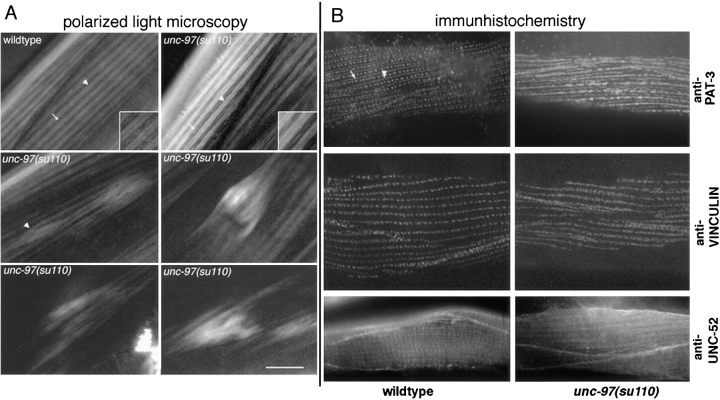
The X-linked unc-97 gene was identified by Zengel and Epstein (1980) and is defined by a single recessive allele, su110. Adult animals homozygous for the su110 mutation are limp, egg laying–defective, and show movement defects that vary from slow moving to paralyzed animals. This probably reflects the degree of disorganization of the vulval and body wall muscles. Zengel and Epstein (1980) reported that unc-97 mutants have shallow, easily disrupted muscle sarcomeres. Using polarized light microscopy we also find that mutant muscle cells are fragile, but if animals are handled carefully intact sarcomeres can be observed (Fig. 2 A). As can be seen in Fig. 2 A (top left panel), when handled gently, unc-97(su110) muscle largely resembles wild-type muscle. Sarcomere dimensions and overall organization are similar between unc-97(su110) and wild-type muscle, but the dense bodies in su110 animals are never as clearly defined as in wild-type animals. Fig. 2 A (bottom four panels) illustrates the range of defects than can be observed within individual muscle cells of su110 animals. These panels are all of cells where the sarcomeres had already become disorganized. The combination of coverslip pressure and slight rolling of an unc-97(su110) animal will also lead to the collapse of the sarcomere structure. Under similar conditions sarcomeres within wild-type muscle cells do not collapse. Anchorage of the sarcomere complex to the plasma membrane is therefore weak and easily disrupted in unc-97(su110) animals.
Figure 2.
Analysis of muscle structure in unc-97 mutant animals. (A) High magnification views of individual body wall muscle cells in N2 wild-type and unc-97(su110) living adult animals using polarized light microscopy. In N2 wild-type dense bodies and M lines are easily detected. In the first unc-97(su110) panel, although the overall structure of the muscle cell is similar to that observed in wild type, individual dense bodies are not as easily detected and the edges of the A bands are often ragged. Arrowheads, A bands; arrows, individual dense bodies (adhesion plaques). Bottom four unc-97(su110) panels, varying levels of disorganization that can be observed in su110 muscle cells. Relatively mild disorganization with several sarcomeres are just beginning to detach can be observed (arrowhead). Note how the birefringent material accumulates at the ends of cells in the bottom two panels. (B) Immunohistochemical analysis of dense body structure in wild-type and unc-97(su110) with the monoclonal β-integrin/PAT-3 antibody MH25, the monoclonal vinculin antibody MH24, and the polyclonal anti–UNC-52 antibody. White arrow, dense bodies; white triangle; M line. Note the occasional fusion of diffuse stripes staining with anti-vinculin antibodies in unc-97(su110) animals. Bar, 10 μm.
The lability of the sarcomeres as observed by polarized light microscopy prompted us to analyze the structural integrity of the muscle attachment structures in more detail. To this end, we determined the subcellular localization of two defined components of these muscle–hypodermal adherens junctions. Immunhistochemistry with a β-integrin/ PAT-3 monoclonal antibody (Gettner et al., 1995) revealed that the dense bodies do not for the most part show up as rows of discrete spots in unc-97 mutants, but instead appear primarily as diffuse stripes running parallel to the M line (Fig. 2 B). Similarly, vinculin, which normally localizes exclusively to dense bodies (Barstead and Waterston, 1991), shows a diffuse stripe-like appearance in unc-97 mutants (Fig. 2 B); the diffuse stripes do not always run in parallel, but occasionally fuse to one another. Due to the presence of M lines in the anti–PAT-3–stained animals, the fusion of these stripes can more easily be observed using anti-vinculin stained antibodies. These observations are consistent with UNC-97 playing a role in determining the structural integrity of adherens junctions, perhaps through clustering integrin or other dense body-associated proteins.
The C. elegans proteoglycan perlecan, encoded by the unc-52 gene, localizes to the basement membrane underlying the muscle (Francis and Waterston, 1991; Rogalski et al., 1993). Staining with anti–UNC-52 antibodies reveals staining at periodicities corresponding to the sites of the dense bodies and M-line structures (Fig. 2 B) (Francis and Waterston, 1991). In unc-97(su110) mutant animals, the periodicity of the staining is abrogated; instead, UNC-52 appears more diffuse (Fig. 2 B). The effect of the intracellular UNC-97 protein on the localization of the extracellular UNC-52 protein demonstrates that the correct localization of extracellular matrix components depends on the structural integrity of the muscular integrin complexes.
unc-97 Defines a New Family of LIM Domain Proteins
unc-97(su110) was mapped to linkage group X in the lon-2 dpy-7 interval by Zengel and Epstein (1980). The cosmid F14D12 maps to this interval and contains a predicted LIM domain protein (F14D12.2; Fig. 3, A and B). Sequencing of this predicted gene from the mutant strain revealed that unc-97(su110) harbors a G→ A splice site mutation at the last intron–exon boundary, which is predicted to disrupt the structural integrity of the last LIM domain of the UNC-97 protein (Fig. 3 A). A genomic PCR fragment containing the predicted unc-97 gene rescues the uncoordinated phenotype of unc-97(su110) (Materials and Methods). Taken together, these results demonstrate that the unc-97 gene is encoded by the predicted LIM domain containing protein F14D12.2.
Figure 3.
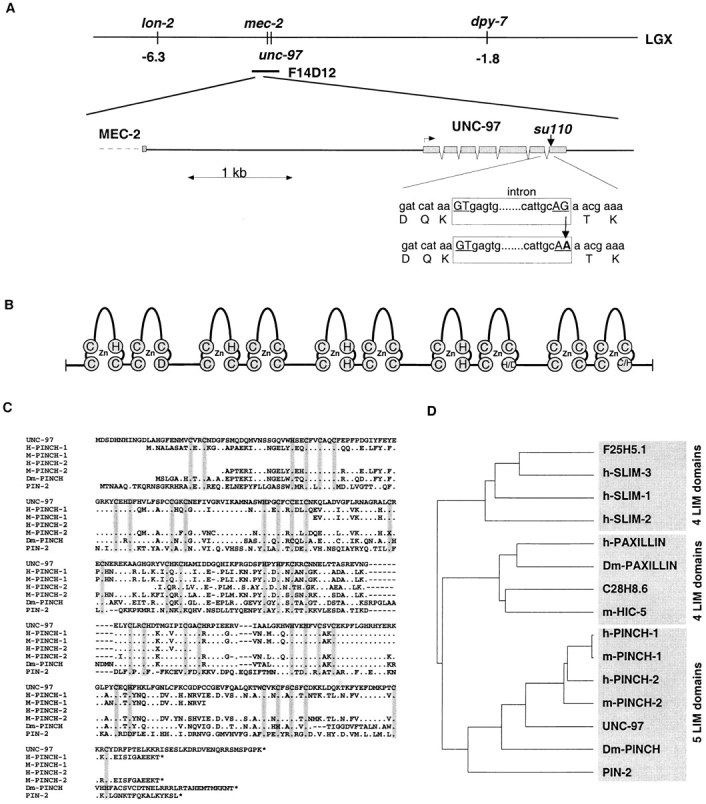
UNC-97 and the PINCH family of LIM proteins. (A) Chromosomal localization of unc-97. The unc-97(su110) mutation is a G→ A transition in a highly conserved splice acceptor site. Due to the invariance of G at splice sites, these sites represent common targets for the EMS mutagen in C. elegans. (B) Schematic domain structure of UNC-97, D-PINCH, and other PINCH family members. (C) Alignment of the PINCH family. PIN-2 corresponds to F07C6.1. We propose the names h-PINCH-1 for the first original PINCH gene described by Rearden (1994), h-PINCH-2 for the incomplete human EST clone c-0qd01 (GenBank/EMBL/ DDBJ accession number Z42656), m-PINCH-1 for the mouse EST clone vc92b05.r1 (GenBank/EMBL/DDBJ accession number AA289280), and m-PINCH-2 for the mouse EST clones vf78h08.r1 (GenBank/EMBL/DDBJ accession number AA450826) and vg99h07.r1 (GenBank/ EMBL/DDBJ accession number AA471768). All of the ESTs are incomplete and were corrected for several obvious errors causing reading frame shifts. Black arrowhead, location of the splice site mutation that is predicted to disrupt the structural integrity of the last LIM domain in UNC-97. X, ambiguities in the EST sequences. (D) The relationship of the UNC-97 family to one another and to other LIM domain subfamilies that also consist of multiple LIM domains is shown in this dendrogram. Note that this dendrogram created by the pileup program represents a clustering based on similarity and is not representative of evolutionary distance. SLIM family members are characterized by the presence of four LIM domains preceded by an incomplete fifth LIM domain. Paxillin family members contain four LIM domains. Vertebrate paxillins, but not the C. elegans homologue C28H8.6 contain an NH2-terminal extension. Note that all subfamilies contain at least one C. elegans homologue. Database accession numbers (incl. references) are: h-PAXILLIN: Swiss prot P49023; m-HIC-5: GenBank/EMBL/DDBJ L22482; C28H8.6: Swiss prot Q09476; Dm-PAXILLIN (EST GM04891.5): GenBank/EMBL/DDBJ AA696001; h-SLIM-3 (identical to h-DRAL): Swiss prot Q14192; h-SLIM-2: Swiss prot Q13643; h-SLIM-1 (the human orthologue of the mouse KyoT gene): Swiss prot Q13642; F25H5.1: GenBank/EMBL/DDBJ Z81068.
Eight independent unc-97 cDNA clones were present in Y. Kohara's EST collection library. We completely sequenced the largest cDNA clone and found it to represent a likely full-length clone containing a poly-A tail and an in-frame stop codon 5′ of the putative ATG start codon. The open reading frame (Fig. 3 C) was similar to the predicted gene structure; no evidence for alternative splice forms was detected in the other 7 cDNA clones. The UNC-97 protein encoded by the open reading frame (GenBank/EMBL/DDBJ accession number AF035583) consists entirely of 5 LIM domains (Fig. 3, B and C). Database searches revealed the presence of highly related proteins from other species, including the human PINCH protein (Rearden, 1994), another predicted C. elegans protein, which we termed pin-2 (for PINCH-related gene 2) and several ESTs from mice, humans, and Drosophila (Fig. 4 D). To address cross-species conservation in more detail, we obtained the cDNA corresponding to the Drosophila EST, and after fully sequencing the transcript, found it to contain the entire open reading frame (GenBank/EMBL/ DDBJ accession number AF078907). Like the other family members, Drosophila PINCH is a LIM-only protein comprised of five LIM domains and contains a significant sequence identity (on the order of 60%) to UNC-97 in each of its LIM modules (Fig. 3 C).
We propose to term this new family of LIM proteins the PINCH family after its founding member, PINCH (Rearden, 1994; renamed to h-PINCH-1 in Fig. 3). The PINCH family can be clearly distinguished from other multiple LIM domain-containing subfamilies (Fig. 3 D) and is defined by the following features: (a) all sequenced members consist exclusively of five LIM domains; (b) the spacing between the Zn-coordinating amino acids, a variable parameter in different LIM domain proteins, is with two exceptions invariant in the PINCH family; and (c) the Zn-coordinating amino acids in each LIM domain are with two exceptions invariant. Most notably, the first Zn finger of the last LIM domain of all PINCH family members is characterized by the highly unusual replacement of a His in the Cys-Cys-His-Cys LIM domain consensus motif by a Cys residue.
Expression of C. elegans unc-97 and Its Drosophila Homologue d-pinch in Muscle
To monitor the expression pattern of UNC-97, we fused the transcriptional regulatory regions of the unc-97 gene to GFP and analyzed its expression in transgenic C. elegans. A translational fusion protein, encoding all exons and introns of unc-97 reveals a similar set of expressing cells (see below). In both larvae and adult animals, unc-97 is expressed in two different cell types, muscles and neurons (Fig. 4). Strong expression is detectable in body wall muscle cells and vulval muscle cells, whereas no expression can be detected in pharyngeal muscles, intestinal muscle, or the anal depressor muscle. Weak expression can be observed in the anal sphincter muscle (data not shown). In the nervous system, unc-97 is expressed in the six mechanosensory receptor neurons ALML/R, PLML/R, AVM, and PVM (Fig. 4), which are responsible for sensing light touch (Chalfie et al., 1985). Intriguingly, a common theme of all the unc-97–expressing cell types is their attachment via the extracellular matrix to the hypodermis. We next examined unc-97 expression at earlier developmental stages to determine its onset of expression in development. We found that in embryos, unc-97 expression can first be observed at mid-embryogenesis at ∼300 min of development (Fig. 4).
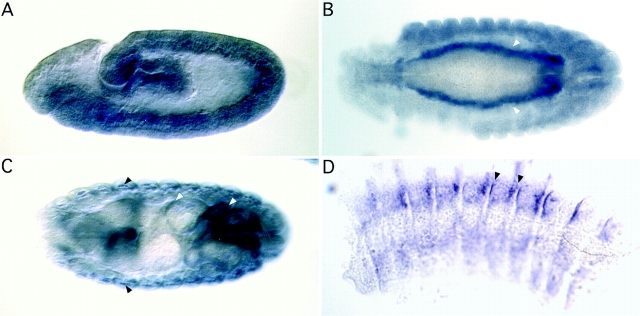
In an effort to deduce some conserved function among PINCH family members, we characterized the expression pattern of the Drosophila homologue of UNC-97, d-pinch. We generated a developmental profile and expression pattern for d-pinch by whole mount RNA in situ hybridization on 0–17-h embryos (Fig. 5). D-pinch transcripts were first detected in stage 10 embryos, where it is expressed in the visceral and body wall muscle. By stage 13, the expression has increased, and some pharyngeal muscle staining is also detected. Of particular interest is the intense expression of the transcripts at the sites of gut constriction. In late-stage 16 embryos, when the myotendinous junction is just beginning to form, we detect d-pinch transcripts in the epidermal tendon cells, as well as the aforementioned muscle lineages (Fig. 5). At this time of development, the heart musculature has differentiated as well; however, no d-pinch expression is seen in this muscle lineage.
Figure 5.
Distribution of d-pinch RNA during Drosophila development. (A) Lateral view of a stage 9 embryo, hybridized with antisense d-pinch probe; oriented with anterior left, dorsal side up. The transcript is concentrated in the developing mesoderm, whereas there is no specific staining seen with the negative control (data not shown). (B) Dorsal view of an early stage 13 embryo, hybridized with antisense d-pinch probe. D-pinch transcript is detected strongly in the visceral mesoderm flanking the developing gut (arrowheads), as well as in the somatic musculature. (C) Dorsal view of a late stage 16 embryo, hybridized with antisense d-pinch probe. The d-pinch transcript is still strongly expressed in the visceral mesoderm (white arrowheads) that now completely envelopes the gut and the somatic body wall muscles (black arrowheads). Pharyngeal muscle and epidermal tendon cell staining is also present at this stage; however, these tissues are not discernible in this plane of focus. (D) Isolated epidermis from a stage 16 embryo, hybridized with d-pinch probe. Note the strong hybridization at the segment boundaries where the somatic muscle cells attach to the epidermis. Arrowheads, two attachment sites.
Like unc-97–expressing cells, d-pinch–expressing cells are attached to extracellular matrix components via integrin receptors. The timing of d-pinch expression in these cells is consistent with the expression of integrin subunits as well. A very striking example of coexpression is the temporal pattern of d-pinch and the integrin subunit αPS1 in tendon cells. By late stage 16, high levels of αPS1 are detected in the tendon cells with no other epidermal expression of the transcript (Wehrli et al., 1993); it is at this embryonic stage that we first detect d-pinch transcript in tendon cells.
Adherens Junction and Nuclear Localization of UNC-97 in Muscle
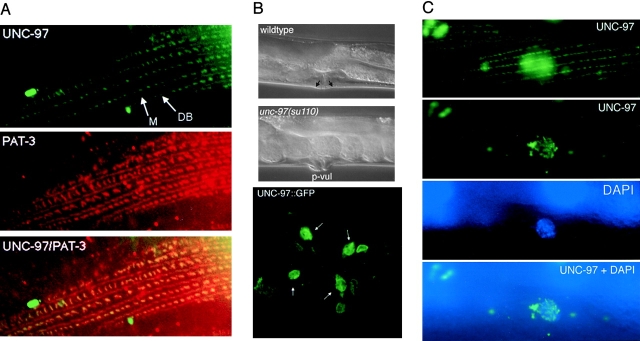
The complete coding sequence of unc-97 was fused to GFP to reveal the subcellular localization of UNC-97. The unc-97::GFP fusion gene rescues the unc-97(su110) mutant phenotype (Materials and Methods) and is thus likely to reveal a functionally relevant subcellular localization. In adult body wall muscles UNC-97 is located in discrete spots and lines along the body wall muscle (Fig. 6). Double labeling with an antibody directed against the C. elegans β-integrin PAT-3 (Gettner et al., 1995) revealed a colocalization of UNC-97 with PAT-3 at dense bodies and along the M lines (Fig. 6, see also Fig. 1). Costaining with anti-vinculin antibodies, which also stain dense bodies, confirms that UNC-97 is part of the integrin complex at muscle-hypodermal attachment sites (data not shown). Taken together with the requirement of unc-97 for the integrity of dense bodies shown in Fig. 2, the localization of UNC-97 to dense bodies suggests that UNC-97 acts at the dense bodies to ensure the integrity of this type of adherens junction.
Figure 6.
Subcellular localization of UNC-97. (A) Top, UNC-97::GFP localization to dense bodies (DB) and M lines; middle, PAT-3 localization visualized with the anti-MH25 antibody; bottom, overlay of the first and second panel showing the overlap between green (UNC-97::GFP) and red (PAT-3) in yellow. (B) Top two panels show lateral Nomarski images of wild-type and unc-97(su110) animals, the latter animal displaying a protruding vulva phenotype (p-vul). The p-vul phenotype displays a 56% penetrance (n = 32). Black arrows in top panel, approximate attachment sites of the vulval muscles. The shape of the vulval muscles can also be seen in Fig. 4 D. Bottom panel, ventral view of UNC-97::GFP localization in wild-type adult animals. UNC-97::GFP localizes to the attachment sites (white arrows) of the vulval muscles to the hypodermis (for a schematic drawing of the vulval muscles see White, 1988). (C) Top, UNC-97:: GFP localization in the muscle sarcomere. Thin stripes, M line structures; dots, dense body structures (for a schematic description of these structures see Fig. 1). In the center of the panel, fuzzy nuclear staining can be observed which is out of focus. Second panel, the same cell as in the first panel, but in a different plane of focus showing nuclear localization of UNC-97 in dots. Third panel, DAPI staining of the same nucleus. Bottom, overlay between the second and third panel. The animal shown in this series of micrographs has been fixed with formaldehyde to stabilize sarcomeres and nuclear localization of UNC-97.
In the vulval muscles UNC-97 also localizes at sites of muscle attachment to the hypodermis (Fig. 6). 56% (n = 32) of the unc-97(su110) animals display a protruding vulva (p-vul) phenotype (Fig. 7), which could be ascribed to defects of these attachment sites in the absence of full unc-97 function. Structural defects of the vulval muscle attachments could explain the egg laying–defective phenotype of unc-97(su110), which is manifested by an accumulation of eggs in the gonad (data not shown).
Figure 7.
unc-97 functions in muscle development. (A–C): UNC-97::GFP localization in live worms at embryonic stage ∼300 min (A), ∼400 min (comma stage) (B), and ∼500 min (threefold “pretzel” stage) (C). (A′–C′) Corresponding DIC micrographs. White arrows in C, a muscle nucleus and dense bodies, respectively. Note UNC-97::GFP localization to the periphery of nuclei in A–C. (D–F): Loss-of-function phenotype of unc-97 as determined by RNAi. D and E show the progeny of animals subjected to unc-97 RNAi. Embryos either arrest and die at the twofold stage or die shortly after hatching (both categories were scored as arrested). Note that the pharynx develops relatively normally but that elongation of the rest of the animal is arrested. All these phenotypes are hallmarks of the pat phenotype (Williams and Waterston, 1994). Bottom, progeny of animals subject to RNAi with a control ds RNA derived from a new homeobox gene (Hobert, O., and G. Ruvkun, unpublished data), which displays no lethality.
Surprisingly, we also found UNC-97 to localize to the nuclei of muscle cells (Fig. 6). UNC-97 appears in discrete dots in muscle nuclei. We have not analyzed these dots in detail, however, their subnuclear localization appears to be somewhat variable. In many cases, these spots do not appear to be confined to submembraneous structures of the nuclear envelope, but appear to localize throughout the nucleoplasm (Fig. 6). In younger animals, however, particularly in embryos, we observe these subnuclear dots to be more concentrated towards the periphery of the nucleus (Fig. 7, A–C).
RNA Interference of unc-97 Causes an Embryonic Arrest Phenotype
Using the full-length UNC-97::GFP fusion protein, we analyzed the developmental progression of UNC-97 expression and localization. In accordance with results from the promoter fusion (see above), we observe UNC-97 expression from ∼300 min of embryonic development onward (Fig. 7, A–C). This is about the time when myoblasts adopt their final identity, as judged by the onset of expression of structural muscle components, but is before the positioning of muscles into quadrants (Epstein et al., 1993; Coutu-Hresko et al., 1994). Upon formation of the muscle quadrant and the onset of muscle elongation, UNC-97 expression becomes stronger and its localization into discrete nuclear spots becomes visible (350–420 min; comma stage). This is about the stage when muscle components are first assembled into sarcomeres. Due to the strong punctate nuclear pattern it is hard to discern whether UNC-97 localizes to the newly formed dense bodies at this early stage. Discrete punctate UNC-97::GFP localization to dense bodies, which is clearly discernible from the punctate nuclear localization, can be observed around the time of hatching. Although most of the embryonic UNC-97::GFP signals can be accounted for by body wall muscle localization, some additional signals appear in the head and tail, which presumably are touch neurons that form late in embryogenesis (Sulston et al., 1983).
The early embryonic expression of UNC-97 in muscle prompted us to study its involvement in embryonic muscle development. Early defects in muscle development caused by the loss of the dense body components β-integrin/pat-3 or vinculin/deb-1 function have been shown to cause a specific developmental arrest phenotype, termed Pat phenotype (paralyzed, arrested elongation at twofold) (Barstead and Waterston, 1991; Williams and Waterston, 1994). unc-97(su110) mutant animals do not display such a phenotype; however, the molecular nature of the (su110) allele (Fig. 3) strongly suggests that this mutation does not lead to a complete loss of unc-97 function. To address this issue, we thus determined the probable loss-of-function phenotype of unc-97 using RNA interference (RNAi). This technique has been shown to effectively decrease the expression of both maternally and zygotically expressed genes and, in many cases, has been shown to phenocopy the null phenotype of a gene (Fire et al., 1998). We found that unc-97 RNAi causes a characteristic developmental arrest phenotype, which shows all the hallmarks of the Pat phenotype (Fig. 7, D and E). Embryos grow to the twofold stage, but arrest body elongation. The pharynx occasionally develops, but the embryo is paralyzed. Several embryos hatch, but are misshapen and then die (Fig. 7, D and E). None of these phenotypes could be observed upon injection of control double-stranded (ds) RNA. The Pat phenotype of unc-97 RNAi is similar to the loss-of-function phenotype of β-integrin/pat-3 and vinculin/deb-1, both factors which colocalize with UNC-97 at adherens junctions. Taken together, these data indicate that unc-97 is essential for early stages of muscle development.
UNC-97 expression is maintained in muscles throughout the postembryonic life of the animal (data not shown). Also, its dual subcellular localization in muscle nuclei and muscle attachment sites is not altered in development. These observations suggest that UNC-97 is continuously required to maintain the structural integrity of muscle attachment sites during growth and in the adult. The sarcomere fragility of unc-97(su110) described above clearly supports this notion. Conditional alleles of unc-97 will be required to address this issue in more detail.
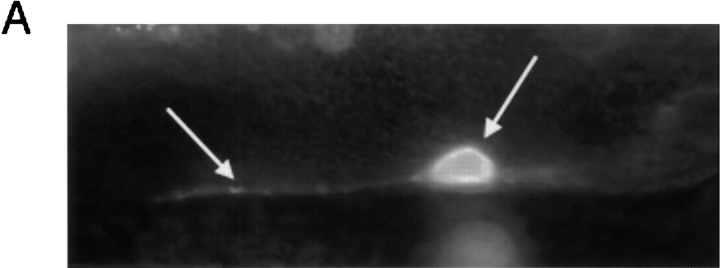
unc-97 Functions in Mechanosensation
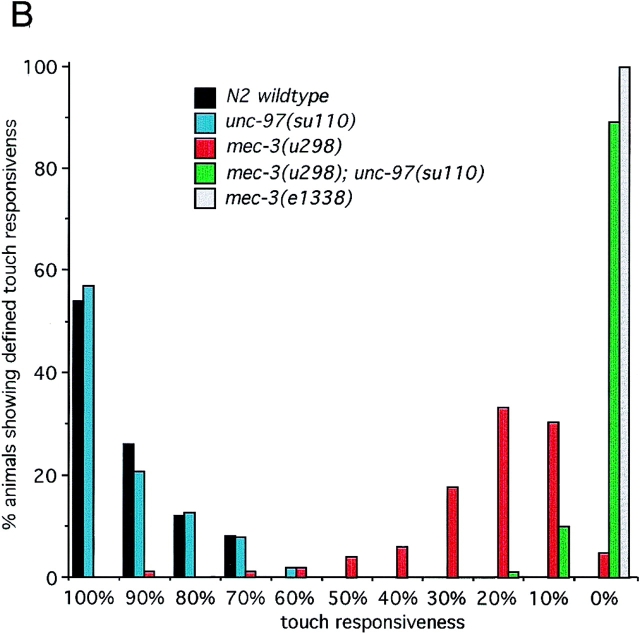
The subcellular localization of UNC-97::GFP in touch neurons is more diffuse than in muscles (Fig. 8 A). Although UNC-97 appears to be concentrated in submembraneous regions, it can be found throughout the cytosol as well as throughout the nucleus (Fig. 8 A). Occasionally we observe a concentration of UNC-97::GFP in the periphery of the touch neuron nuclei in what appear to be ring like structures (data not shown). The nuclear staining profile of UNC-97::GFP in neurons thus appears to be distinct from that in muscles, suggesting that UNC-97 associates with cell type–specific and nonubiquitous subnuclear domains. We investigated whether unc-97 is required for the function of mechanosensory neurons. In behavioral touch assays we found that the partial loss-of-function allele unc-97(su110) displays no mechanosensory touch defectiveness on its own (Fig. 8 B). However, we find that this partial loss of unc-97 function strongly enhances the mechanosensory defect of a partial loss-of-function mutation in mec-3, a LIM homeobox gene required for mechanosensory function (Way and Chalfie, 1988). Although the strong mec-3 allele e1338 is completely touch insensitive, mec-3(u298) mutant animals retain some responsiveness to touch (Fig. 8 B). Examining a unc-97(su110); mec-3(u298) double mutant, we found that this partial responsiveness is almost completely abolished by the partial reduction unc-97 gene activity (Fig. 8 B). This experiment reveals that unc-97 contributes to the function of mechanosensory neurons. Due to the arrested lethal phenotype of a complete loss of unc-97 function as determined by RNAi (Fig. 7), we could not address the null phenotype of unc-97 in touch neurons. Genetic mosaic experiments will address this issue in the future.
Figure 8.
unc-97 genetically interacts with the mec-3 gene. (A) UNC-97::GFP expression in the ALM touch neurons. Left arrow, axonal process; right arrow, cell body. (B) Genetic interaction of unc-97 and mec-3. Mechanosensory assays were performed on the individual genotypes as described in Materials and Methods. The responsiveness of an individual animal is defined by how often an animal can respond to a total of ten touches on the anterior and posterior half of the animal (e.g., two responses from ten touches is recorded as a 20% responsiveness). Animals tested: unc-97(su110) n = 102; mec-3(u298) n = 102; unc-97(su110), mec-3(u298) n = 100; mec-3(e1338) n = 30; N2 wild-type n = 30. The fraction of the tested animals that show any of the ten different touch responses is plotted against the touch responsiveness.
The UNC-97–related LIM protein PIN-2 Is Expressed in Neurons and Intestine
The now completely sequenced genome of C. elegans reveals the presence of a single other PINCH family member in C. elegans, which we termed PIN-2 (Fig. 3). Although it represents the most diverged member of the PINCH family, it clearly shares all the features of this family including the presence of five LIM domains and a characteristic signature of Zn-coordinating residues of the LIM domains (Fig. 4; PIN-2 shows 55.7% amino acid similarity to UNC-97 and 58.5% amino acid similarity to h-PINCH-1). pin-2 maps on LGIV, ∼7 kb to the left of the cloned unc-31 gene; no candidate mutations have been mapped to this region.
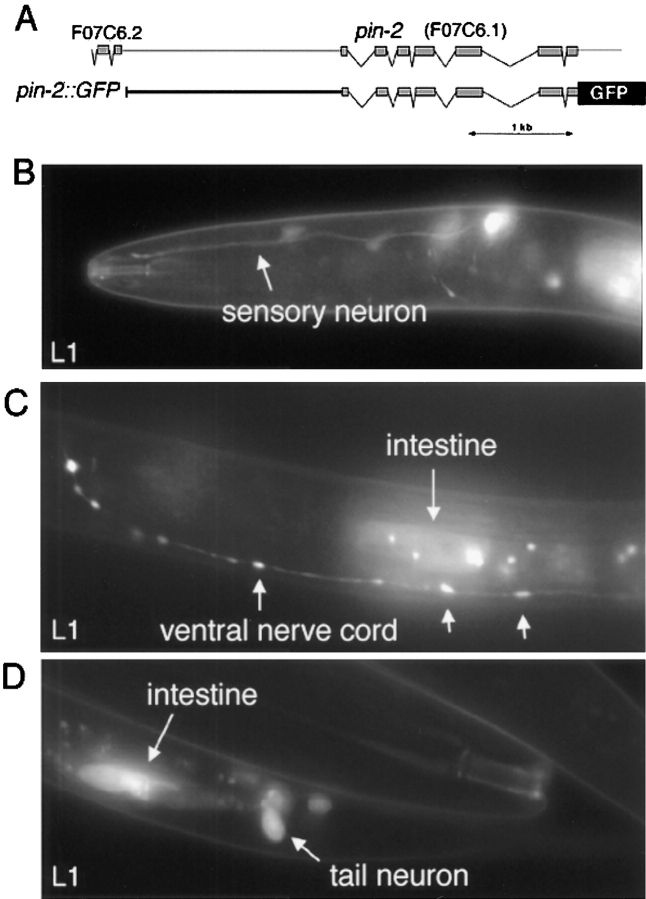
We were motivated to look for the expression and subcellular localization of PIN-2 to address whether PIN-2 might perform a role in muscle development similar to UNC-97 or whether this diverged PINCH family member might be used in distinct cellular processes. A pin-2::GFP fusion gene construct (Fig. 9), which contains the full coding region of pin-2 and presumably reflects the authentic subcellular localization of PIN-2, is not expressed during early stages of embryogenesis; expression is first detectable in embryos shortly before hatching (data not shown). In early larval stages, PIN-2::GFP is expressed in two major tissue types, neurons and intestinal cells (Fig. 9). It does not reveal any overlap of expression with UNC-97. Notably, however, PIN-2 expression in the intestine might be somewhat related to the expression of Drosophila d-pinch in the visceral mesoderm that envelopes the gut. Neural expression of PIN-2::GFP is restricted to few neurons, arguing for a cell type–specific role of PIN-2. Moreover, whereas PIN-2::GFP expression fades in intestinal cells and is almost undetectable in adults, its expression is maintained in neurons throughout adulthood (data not shown), suggesting a role for PIN-2 in neural maintenance. PIN-2::GFP localizes uniformly throughout the cytoplasm and nucleus. In neurons, it localizes uniformly along axonal processes. PIN-2 appears to label variscosities of the ventral cord axon where it is expressed. The functional significance of this observation is unclear at the moment.
Figure 9.
Expression of the UNC-97–related LIM protein PIN-2. (A) The GFP reporter gene construct is depicted schematically. The last exons of the predicted gene F07C6.2 that precedes PIN-2/F07C6.1 are shown to demonstrate that virtually the whole 5′ upstream regulatory region of PIN-2 has been incorporated into the reporter gene construct. (B–D) Expression of the PIN-2:: GFP reporter construct. PIN-2::GFP expression in neurons is highly penetrant; however, the exact number of PIN-2::GFP– expressing cells varies slightly from animal to animal. We have not observed more than 10 head neurons and three tail neurons labeled in any one individual. The strongest expressing tail neuron (D) has a size and location consistent with the PVT interneuron, and sends a process into the nerve ring (C). At least one of the head neurons is an amphid sensory neuron (B). Arrows in C, variscosities in the ventral nerve cord. Those can be also seen in unanaesthetized animals, ruling out artefactual induction of variscosities as often observed, for example, with sodium azide.
In summary, both the tissue type expression and the subcellular distribution of the only two PINCH family members in C. elegans, UNC-97 and PIN-2 is strikingly different, thus arguing for a divergence in function of the genes.
Discussion
PINCH Family Members across Phylogeny
The invertebrate PINCH-like genes described here as well as several as yet uncharacterized vertebrate PINCH genes define a new family of LIM proteins that have been highly conserved across phylogeny. No other LIM domain family contains the same LIM domain architecture as the PINCH family. Considering the involvement of LIM domains in protein–protein interactions (see introduction), we propose that PINCH proteins represent adapter proteins capable of assembling multiprotein complexes. Within C. elegans, we identified two PINCH-like genes, unc-97 and pin-2. unc-97 is more closely related to the vertebrate PINCH genes. The function of unc-97 at focal adhesion sites appears to be conserved as well, since vertebrate PINCH-1 colocalizes with integrins to sites of cell–matrix contact (Tu et al., 1999).
Additional evidence that PINCH family members may function at focal adhesion sites comes from the analysis of d-pinch expression during Drosophila embryogenesis. D-pinch transcripts are expressed in both the body wall muscles and epidermal tendon cells, coincident with integrin subunit expression in those tissues (Bogaert et al., 1987; Leptin et al., 1989; Wehrli et al., 1993). The timing and tissue-specific expression of d-pinch suggests that it is involved in the terminal differentiation of muscles and tendon cells; one common feature of these cell types is their formation of adherens junctions. Integrin complexes are crucial for the stability of the junctions, as demonstrated by the dramatic muscle detachment phenotype seen in myospheroid/βPS-integrin mutants (Wright, 1960; Leptin et al., 1989; Volk et al., 1990). Based on the observation that the null unc-97 phenotype is very severe and phenocopies mutations in the pat-3/β-integrin gene, we predict that loss of d-pinch will also result in a severe disruption of muscle function.
The PIN-2 protein, which displays a high degree of similarity to UNC-97 in architectural composition, as well as primary sequence, is expressed in entirely different tissue types and, unlike UNC-97, displays a uniform subcellular distribution. Nevertheless, it is conceivable that, although deployed in different tissue types, distinct PINCH gene paralogues (such as UNC-97 and PIN-2) might fulfill a similar function. For example, integrin complexes in C. elegans are not only present in muscle cells but also in neurons, where they do not appear to localize in discrete spots (Baum and Garriga, 1997). It is possible that PIN-2 might be involved in neural integrin complex assembly and/or signaling. The expression of PIN-2 in the intestine could potentially indicate a similarity to the expression of Drosophila d-pinch in the visceral mesoderm that envelopes the gut.
UNC-97 and Muscle Development
The structural components that anchor myofibers to the extracellular matrix, such as integrin, vinculin, talin, and α-actinin are remarkably conserved in C. elegans and resemble the protein composition of adherens junctions in other systems, such as focal adhesions in tissue culture cells (Waterston et al., 1980; Francis and Waterston, 1985; Waterston, 1988; Moerman and Fire, 1997). It is thus likely that the fundamental mechanisms required to assemble these structure are conserved as well. Based on our functional analysis of UNC-97 and the presence of highly related homologous molecules in flies and vertebrates, we suggest that members of the PINCH family of LIM-only proteins are a critical component of muscle attachment and adherens junction assemblies across phylogeny. Indeed, the mouse UNC-97 homologue PINCH-1 has recently been shown to colocalize with β1-integrins at focal adhesions (Tu et al., 1999). This interaction appears to be mediated by the ankyrin-repeat containing serine/threonine kinase ILK, which by direct association with both β1-integrins (Hannagan et al., 1996) and the mouse UNC-97 homologue PINCH-1 (Wu, C., personal communication) appears to serve as a bridging molecule.
Cellular attachment sites contain several LIM domain proteins, such as zyxin, paxillin, CRP proteins, and others (Sadler et al., 1992; Turner et al., 1990; Crawford et al., 1994; Arber and Caroni, 1996). In muscles, the CRP3/ MLP and ALP proteins localize to Z-discs, attachment points of myofibers in vertebrates (Arber et al., 1997; Xia et al., 1997). Drosophila CRP homologues also localize to muscle attachment sites (Stronach et al., 1996). However, the functional requirement for any LIM containing protein at these subcellular sites has until now only been reported for the CRP3/MLP protein, whose loss-of-function causes disorganization of cardiac myofibers (Arber et al., 1997). We demonstrated here a similar requirement for the LIM-only protein UNC-97 in body wall muscles in C. elegans. Since the C. elegans genome contains a gene highly related to the CRP proteins (T04C9.4; data not shown), we speculate that other LIM proteins besides UNC-97 will also be involved in adherens junction assembly.
The su110 allele indicates that UNC-97 has an important role in maintaining sarcomere organization in growing and adult animals. Moreover, the RNAi (probable loss-of-function) experiment indicates an important regulatory role for UNC-97 during embryonic muscle development. However, it is unclear so far as to whether the embryonic defects caused by unc-97 RNA interference reflect a requirement for UNC-97 in the initial stages of assembly of the adherens junctions or reflects a requirement for UNC-97 in stabilizing adherens junctions after they are formed to resist the mechanical stress they are exposed to upon elongation of the animal. Since UNC-97 expression clearly coincides with the onset of expression of adherens junction components and their assembly into these structures, we favor a model in which UNC-97 is involved in the initial assembly of the adherens junction components and keeps residing in these structures to ensure their stability. We will address these issues in more detail in the future by isolating deletion alleles of unc-97, which will provide a better source for a detailed characterization of the embryonic phenotype than the dsRNA injected animals described here.
Although it is unclear how UNC-97 cooperates with β-integrins and other focal adhesion components such as vinculin to determine adherens junction integrity, the modular organization of UNC-97 into five LIM domains implicates UNC-97 in binding to multiple proteins. UNC-97 might be a scaffold or adapter protein onto which various different components assemble to form a functional muscle attachment site. Since defective myofibrils cause as a secondary consequence the destabilization of muscle attachment sites (Epstein, 1986), it is for example possible that UNC-97 serves as an anchor between myofibrils and membrane-anchored integrin components. In this model, dense body defects in unc-97(su110) mutant animals would not arise from direct defects in the dense body structure per se, but would represent a secondary consequence of the myofibril disorganization.
A Nuclear Function for UNC-97?
Our analysis of UNC-97 localization in a living, nonfixed animal, using a rescuing, i.e., functionally intact unc-97:: GFP reporter gene, lends support for an authentic in vivo dual subcellular localization of this LIM protein. However, at this point it is unclear whether the nuclear localization of UNC-97 is indeed functionally significant. In contrast to the localization of UNC-97 to adherens junctions, which correspond to the site of action of UNC-97 as inferred from the unc-97 mutant phenotype, no such clear correlation exists to a possible nuclear function of UNC-97. However, our demonstration of a genetic interaction of unc-97 with the LIM homeodomain transcription factor mec-3 could be explained on the basis of a physical interaction between these proteins and could thus reflect a functional requirement for UNC-97 in the nucleus. LIM– LIM interaction have been previously described (Sadler et al., 1992) and the direct interaction of UNC-97 with MEC-3 could affect the transcriptional activity of the MEC-3 transcription factor. Alternatively, it is also entirely possible that the genetic interaction of unc-97 and mec-3 reflects an independent requirement for these genes in mechanosensory processes.
Although further experiments will need to address the physiological significance of UNC-97 in the nucleus, there are several attractive hypotheses regarding a potential nuclear function of UNC-97. As mentioned above, UNC-97 could be directly involved in gene regulatory events by interacting with specific transcription factors. The vertebrate LIM-only proteins CRP3/MLP and SLIM1/KyoT have been directly implicated in gene regulation via interaction with the transcription factors MyoD and RBP-J, respectively (Kong et al., 1997; Tanigushi et al., 1998), whereas the LIM-only protein LMO2 assembles higher order transcriptional activation complexes by bridging other transcription factors that are directly involved in DNA binding (Wadman et al., 1997). Alternatively, but not necessarily mutually exclusive, UNC-97 could represent a structural component of specific subnuclear domains. The localization of UNC-97 to discrete dots in muscle nuclei supports this hypothesis, although the nature of these dots is entirely unclear. Subnuclear domains of different types, such as speckles, coiled bodies, gems, and Kr bodies/nuclear domains have been described in various systems. Factors localizing to these domains are involved in distinct nuclear processes such transcriptional silencing and RNA processing (for review see Lamond and Earnshaw, 1998).
Lastly, in regard to the dual subcellular localization of UNC-97 it is also tempting to speculate that UNC-97 transmits a signal from attachment sites to the nucleus. Although we have not addressed the question whether UNC-97 dynamically shuttles between attachment sites and the nucleus, focal adhesion–nuclear shuttling has been recently demonstrated for the LIM protein zyxin (Nix and Beckerle, 1997). Recently, the mouse UNC-97 homologue PINCH-1 was shown to interact specifically with, and might serve as a substrate of, the integrin-linked kinase ILK (Tu et al., 1999), a serine-threonine kinase implicated in integrin signal transduction (Hannagan et al., 1996). This observation might point to a potential role for UNC-97 in integrin-mediated signal transduction.
Acknowledgments
We wish to thank Y. Liu for expert technical assistance in DNA and RNA microinjection, Y. Kohara for providing cDNA clones, R. Waterston (Washington University, St. Louis, MO) for his gift of MH24 and MH25 antibodies, H. Epstein (Baylor College of Medicine, Houston, TX) for helpful discussions on the unc-97 phenotype, the Caenorhabditis Genetics Center (funded by the National Institutes of Health Center for Research Resources) for providing strains, B. Reinhart for critical reading of the manuscript, and C. Wu (University of Alabama, Birmingham, AL) for sharing his results on PINCH function in vertebrates.
We acknowledge support from Hoechst AG to G. Ruvkun, the National Institutes of Health to M.C. Beckerle (R01 HL60591), and National Science and Engineering Research Council (963226), and Medical Research Council of Canada (962565) grants to D. Moerman. M.C. Beckerle is a recipient of a faculty research award from the American Cancer Society. O. Hobert was supported by a postdoctoral fellowship from the Human Frontiers in Science Organization.
Abbreviations used in this paper
- EST
expressed sequence tag
- GFP
green fluorescent protein
- LIM
LIN-11, ISL-1, MEC-3
- MEC
mechanosensory
- PAT
paralyzed and arrested at twofold stage
- RNAi
RNA interference
- UNC
uncoordinated
References
- Arber S, Caroni P. Specificity of single LIM motifs in targeting and LIM/LIM interactions in situ. Genes Dev. 1996;10:289–300. doi: 10.1101/gad.10.3.289. [DOI] [PubMed] [Google Scholar]
- Arber S, Hunter JJ, Ross J, Jr, Hongo M, Sansig G, Borg J, Perriard JC, Chien KR, Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- Barstead RJ, Waterston RH. Vinculin is essential for muscle function in the nematode. J Cell Biol. 1991;114:715–724. doi: 10.1083/jcb.114.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum PD, Garriga G. Neuronal migrations and axon fasciculation are disrupted in ina-1 integrin mutants. Neuron. 1997;19:51–62. doi: 10.1016/s0896-6273(00)80347-5. [DOI] [PubMed] [Google Scholar]
- Bogaert T, Brown N, Wilcox M. The DrosophilaPS2 antigen is an invertebrate integrin that, like the fibronectin receptor, becomes localized to muscle attachments. Cell. 1987;51:929–940. doi: 10.1016/0092-8674(87)90580-0. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN, Brenner S. The neural circuit for touch sensitivity in C. elegans. . J Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark EA, Brugge JS. Integrins and signal transduction pathways: the road taken. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- Coutu-Hresko M, Williams BD, Waterston RH. Assembly of body wall muscle and muscle cell attachment structures in C. elegans. . J Cell Biol. 1994;124:491–506. doi: 10.1083/jcb.124.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford AW, Pino JD, Beckerle MC. Biochemical and molecular characterization of the chicken cysteine-rich protein, a developmentally regulated LIM-domain protein that is associated with the actin cytoskeleton. J Cell Biol. 1994;124:117–127. doi: 10.1083/jcb.124.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid IB, Breen JJ, Toyama R. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- Epstein, H.F. 1986. Different roles of myosin isoforms in filament assembly. In Molecular Biology of Muscle Development. Vol. 29. C. Emerson, D. Fischman, B. Nadal-Ginard, and M.A.Q. Siddiqui, editors. Liss, New York. 653– 666.
- Epstein HF, Casey DL, Ortiz I. Myosin and paramyosin of Caenorhabditis elegansembryos assemble into nascent structures distinct from thick filaments and multifilament assemblages. J Cell Biol. 1993;122:845–858. doi: 10.1083/jcb.122.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerstein R, Wang X, Song D, Cooke NE, Liebhaber SA. The LIM/double zinc-finger motif functions as a protein dimerization domain. Proc Natl Acad Sci USA. 1994;91:10655–10659. doi: 10.1073/pnas.91.22.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. . Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Francis GR, Waterston RH. Muscle organization in C. elegans: Localization of proteins implicated in thin filament attachment and I-band organization. J Cell Biol. 1985;101:1532–1549. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis GR, Waterston RH. Muscle cell attachment in Caenorhabditis elegans. . J Cell Biol. 1991;114:465–479. doi: 10.1083/jcb.114.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gettner SN, Kenyon C, Reichardt LF. Characterization of β pat-3 heterodimers, a family of essential integrin receptors in C. elegans. . J Cell Biol. 1995;129:1127–1141. doi: 10.1083/jcb.129.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannigan GE, Leung-Hagesteijn C, Fitz-Gibbon L, Coppolino MG, Radeva G, Filmus J, Bell JC, Dedhaer S. Regulation of cell adhesion and anchorage-dependent growth by a new β1-integrin-linked protein kinase. Nature. 1996;379:91–96. doi: 10.1038/379091a0. [DOI] [PubMed] [Google Scholar]
- Kong Y, Flick MJ, Kudla AJ, Konieczny SF. Muscle LIM protein promotes myogenesis by enhancing the activity of MyoD. Mol Cell Biol. 1997;17:4750–4760. doi: 10.1128/mcb.17.8.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lehmann, R., and D. Tautz. 1993. In situ hybridization to RNA. In Drosophila melanogaster: Practical Uses in Cell and Molecular Biology. L.S.B. Goldstein and E.A. Fyrberg, editors. Academic Press, San Diego, CA. 575–598.
- Leptin M, Bogaert T, Lehmann R, Wilcox M. The function of PS integrins during Drosophiladevelopment. Cell. 1989;56:401–408. doi: 10.1016/0092-8674(89)90243-2. [DOI] [PubMed] [Google Scholar]
- Louis HA, Pino JD, Schmeichel KL, Pomies P, Beckerle MC. Comparison of three members of the cysteine-rich protein family reveals functional conservation and divergent patterns of gene expression. J Biol Chem. 1997;272:27484–27491. doi: 10.1074/jbc.272.43.27484. [DOI] [PubMed] [Google Scholar]
- Moerman, D.G., and A. Fire. 1997. Muscle: structure, function and development. In C. elegans II. D.L. Riddle, T. Blumenthal, B.J. Meyer, and J.R. Priess, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 417–470. [PubMed]
- Moerman DG, Hutter H, Mullen GP, Schnabel R. Cell autonomous expression of perlecan and plasticity of cell shape in embryonic muscle of Caenorhabditis elegans. . Dev Biol. 1996;173:228–242. doi: 10.1006/dbio.1996.0019. [DOI] [PubMed] [Google Scholar]
- Moulder GL, Huan MM, Waterston RH, Barstead RJ. Talin requires β-integrin but not vinculin for its assembly into focal adhesion-like structures in the nematode C. elegans. . Mol Biol Cell. 1996;7:1181–1193. doi: 10.1091/mbc.7.8.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix DA, Beckerle MC. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rearden A. A new LIM protein containing an autoepitope homologous to “senescent cell antigen.” . Biochem Biophys Res Commun. 1994;1201:1124–1131. doi: 10.1006/bbrc.1994.1822. [DOI] [PubMed] [Google Scholar]
- Rogalski TM, Williams BD, Mullen GP, Moerman DG. Products of the unc-52 gene in Caenorhabditis elegansare homologous to the core protein of the mammalian basement membrane heparan sulfate proteoglycan. Genes Dev. 1993;7:1471–1484. doi: 10.1101/gad.7.8.1471. [DOI] [PubMed] [Google Scholar]
- Sadler I, Crawford AW, Michelsen JW, Beckerle MC. Zyxin and cCRP: two interactive LIM domain proteins associated with the cytoskeleton. J Cell Biol. 1992;119:1573–1587. doi: 10.1083/jcb.119.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeichel KL, Beckerle MC. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Stronach BE, Siegrist SE, Beckerle MC. Two muscle-specific LIM proteins in Drosophila. . J Cell Biol. 1996;134:1179–1195. doi: 10.1083/jcb.134.5.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenber E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. . Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Furukawa T, Tun T, Han H, Honjo T. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol Cell Biol. 1998;18:644–654. doi: 10.1128/mcb.18.1.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Y., F. Li, S. Goicoechea, and C. Wu. 1999. The LIM-only protein PINCH directly interacts with the integrin-linked kinase (ILK) and is recruited to integrin-rich sites in spreading cells. Mol. Cell. Biol. In press. [DOI] [PMC free article] [PubMed]
- Turner CE, Glenney JR, Jr, Burridge K. Paxillin: a new vinculin-binding protein present in focal adhesions. J Cell Biol. 1990;111:1059–1068. doi: 10.1083/jcb.111.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk T, Fessler LI, Fessler JH. A role for integrin in the formation of sarcomeric architecture. Cell. 1990;63:525–536. doi: 10.1016/0092-8674(90)90449-o. [DOI] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grütz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid DNA-binding complex which includes TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO (Eur Mol Biol Organ) J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren AJ, Colledge WH, Carlton MB, Evans MJ, Smith AJ, Rabbitts TH. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erthroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H. 1988. Muscles. In The Nematode Caenorhabditis elegans. W.B. Wood, editor. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 281–336.
- Waterston RH, Thomson JN, Brenner S. Mutants with altered muscle structure in C. elegans. . Dev Biol. 1980;77:271–302. doi: 10.1016/0012-1606(80)90475-3. [DOI] [PubMed] [Google Scholar]
- Way JC, Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. . Cell. 1988;54:5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- Wehrli M, DiAntonio A, Fearnley IM, Smith RJ, Wilcox M. Cloning and characterization of αPS1, a novel Drosophila melanogasterintegrin. Mech Dev. 1993;43:21–36. doi: 10.1016/0925-4773(93)90020-x. [DOI] [PubMed] [Google Scholar]
- White, J. 1988. The anatomy. In The Nematode Caenorhabditis elegans. W.B. Wood, editor. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 81–122.
- Williams BD, Waterston RH. Genes critical for muscle development and function in Caenorhabditis elegansidentified through lethal mutations. J Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright TRF. The phenogenetics of the embryonic mutant, lethal myospheroid in Drosophila melanogaster. . J Exp Zool. 1960;143:77–99. doi: 10.1002/jez.1401430107. [DOI] [PubMed] [Google Scholar]
- Xia H, Winokur ST, Kuo WL, Altherr MR, Bredt DS. Actinin-associated LIM protein: identification of a domain interaction between PDZ and spectrin-like repeat motifs. J Cell Biol. 1997;139:507–515. doi: 10.1083/jcb.139.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel JM, Epstein HF. Identification of genetic elements associated with muscle structure in the nematode Caenorhabditis elegans. . Cell Motil. 1980;1:73–97. doi: 10.1002/cm.970010107. [DOI] [PubMed] [Google Scholar]