Abstract
Tubulin is a heterodimer of α- and β-tubulin polypeptides. Assembly of the tubulin heterodimer in vitro requires the CCT chaperonin complex, and a set of five proteins referred to as the tubulin cofactors (Tian, F., Y. Huang, H. Rommelaere, J. Vandekerckhove, C. Ampe, and N.J. Cowan. 1996. Cell. 86:287–296; Tian, G., S.A. Lewis, B. Feierbach, T. Stearns, H. Rommelaere, C. Ampe, and N.J. Cowan. 1997. J. Cell Biol. 138:821–832). We report the characterization of Alf1p, the yeast ortholog of mammalian cofactor B. Alf1p interacts with α-tubulin in both two-hybrid and immunoprecipitation assays. Alf1p and cofactor B contain a single CLIP-170 domain, which is found in several microtubule-associated proteins. Mutation of the CLIP-170 domain in Alf1p disrupts the interaction with α-tubulin. Mutations in α-tubulin that disrupt the interaction with Alf1p map to a domain on the cytoplasmic face of α-tubulin; this domain is distinct from the region of interaction between α-tubulin and β-tubulin. Alf1p-green fluorescent protein (GFP) is able to associate with microtubules in vivo, and this localization is abolished either by mutation of the CLIP-170 domain in Alf1p, or by mutation of the Alf1p-binding domain in α-tubulin. Analysis of double mutants constructed between null alleles of ALF1 and PAC2, which encodes the other yeast α-tubulin cofactor, suggests that Alf1p and Pac2p act in the same pathway leading to functional α-tubulin. The phenotype of overexpression of ALF1 suggests that Alf1p can act to sequester α-tubulin from interaction with β-tubulin, raising the possibility that it plays a regulatory role in the formation of the tubulin heterodimer.
Keywords: tubulin, microtubule, Saccharomyces cerevisiae, chaperonin, CLIP-170
Microtubules are essential and ubiquitous cytoskeletal elements composed of heterodimers of α- and β-tubulin. Much of the work on microtubules has focused on the requirements for polymerization of the α-β heterodimer into microtubules, and the temporal and spatial regulation of that polymerization. Less well understood are the steps leading to formation of the α-β heterodimer in the cell, including regulation of the amounts of individual subunits, folding of the α- and β-tubulin monomeric subunits, and assembly of the monomers into heterodimers.
For many proteins, folding into the correct three-dimensional structure occurs spontaneously, driven by primary sequence determinants, as demonstrated for ribonuclease A by Anfinsen (Anfinsen, 1973). However the tubulins do not fold spontaneously and require the action of cytosolic chaperonin (c-cpn; also referred to as TRiC or Cct complex; Frydman et al., 1992; Yaffe et al., 1992; Gao et al., 1993; Stoldt et al., 1996), a multisubunit toroidal complex that generates potentially productive folding intermediates via multiple rounds of ATP-hydrolysis (for review, see Hendrick and Hartl, 1995). The known in vivo substrates of c-cpn include actin, and α-, β-, and γ-tubulin (Frydman et al., 1992; Gao et al., 1992; Melki et al., 1993; Sternlicht et al., 1993). In in vitro reactions that use denatured protein as the starting material, both actin and γ-tubulin are fully functional after interaction with the c-cpn (Gao et al., 1992; Melki et al., 1993), but α- and β-tubulin must interact with several additional proteins before they form a heterodimer that is competent for assembly into microtubules (Gao et al., 1993). This observation led to the purification and characterization of the mammalian cofactors (A through E) for tubulin formation (Tian et al., 1996, 1997). It has been proposed that the known cofactors interact with partially folded α- and β-tubulin and bring about their successful folding and subsequent joining to form the heterodimer. This would occur by a set of sequential interactions, the early steps of which are separate for α- and β-tubulin (Tian et al., 1996, 1997; also, see Fig. 8). Cofactors A and D are specific for β-tubulin, whereas cofactors B and E are specific for α-tubulin, and the order of action of the cofactors has been determined biochemically with purified proteins (Tian et al., 1997).
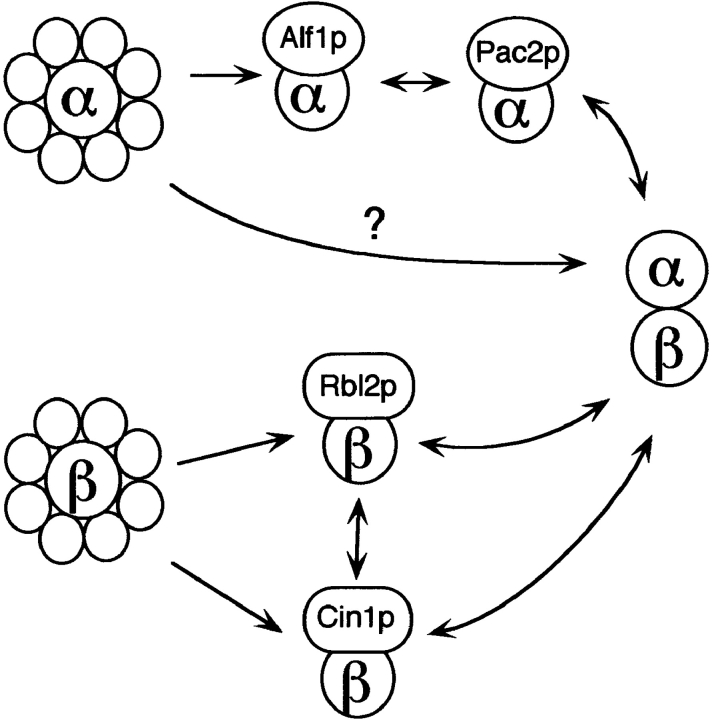
Figure 8.
Model of the cofactor pathway in yeast, adapted from Tian et al. (1997). In this model, α- and β-tubulin are released from chaperonin complexes and interact with monomer-specific cofactors before formation of the tubulin heterodimer. α-Tubulin can become functional either by complexing with Alf1p and Pac2p, or via a putative alternate pathway. β-Tubulin can become functional by interaction with either Rbl2p or Cin1p. The pathways converge at the point of α-β-heterodimer formation.
Cofactors A, B, D, and E each have a putative homologue in the Saccharomyces cerevisiae genome, and genetic experiments have helped to define the in vivo role of the cofactors. RBL2, the homologue of cofactor A, was identified as a gene that when overexpressed is able to compensate for the lethality of β-tubulin overexpression (Archer et al., 1995). ALF1, the homologue of cofactor B, was identified by sequence homology (Tian et al., 1997). CIN1, the homologue of cofactor D, was identified as a mutant with reduced fidelity of chromosome segregation and increased sensitivity to the anti-microtubule drug benomyl (Hoyt et al., 1990; Stearns et al., 1990). PAC2, the homologue of cofactor E, was identified as a mutation that is lethal in combination with a mutation in CIN8, which encodes a kinesin-like protein (Geiser et al., 1997). Although the mammalian cofactors are required for the in vitro folding of tubulin, null mutations in each of the yeast genes are viable. Nonetheless, the mutants all have phenotypes indicative of defects in microtubule function, such as supersensitivity to benomyl. Interestingly, the cofactor D homologue in Schizosaccharomyces pombe, alp1 +, is essential for viability (Hirata et al., 1998).
Although the mammalian cofactors have been shown to interact with tubulin, little is known regarding the molecular nature of that interaction. The only clue from the sequence of the cofactors is that both cofactor B/Alf1p and cofactor E/Pac2p have a single CLIP-170 domain, originally defined in the microtubule-associated protein CLIP-170 (Pierre et al., 1992; Hoyt et al., 1997; Tian et al., 1997). This domain is present in several other microtubule-associated proteins including the dynactin subunit p150Glued and the yeast microtubule-associated protein Bik1p (Pierre et al., 1992). The relevance of the CLIP-170 domain to cofactor function has not been established, indeed, CLIP-170, p150Glued, and Bik1p are thought to interact with microtubules, rather than monomeric tubulin.
With the hope of understanding the role of the cofactors in tubulin biogenesis, we have focused on Alf1p/cofactor B. Cofactor B was identified as a protein that greatly enhances the recovery of α- and β-tubulin heterodimer in an in vitro assembly assay (Tian et al., 1997). Cofactor B interacts with α-tubulin monomer and may serve as a reservoir for folding or dimerization intermediates (Tian et al., 1997). We previously presented an initial characterization of the yeast homologue of cofactor B, which we named ALF1 (α-tubulin formation 1; Tian et al., 1997). An alf1 null mutation is viable but results in supersensitivity to benomyl and lethality in combination with the α-tubulin allele, tub1-1, consistent with ALF1 being involved in microtubule function. The results here indicate that Alf1p is an α-tubulin monomer-binding protein. Residues involved in the Alf1p–α-tubulin interaction were mapped onto the structure of α-tubulin, defining a binding site for Alf1p. This interaction depends upon the CLIP-170 domain in Alf1p, suggesting that the CLIP-170 domain may specifically recognize α-tubulin.
Materials and Methods
Yeast Strains, Media, and Plasmids
The yeast strains used in this study are listed in Table I. Bacterial strains and plasmids are listed in Table II. Yeast extract/peptone (YEP)1, synthetic dextrose (SD) and sporulation media were as described (Adams et al., 1997). Strains requiring expression of a plasmid-borne PGAL1-ALF or -TUB3 construct were grown on minimal media + 2% galactose. Yeast molecular genetic methods were as described (Adams et al., 1997). Benomyl, 98.6% pure, was a generous gift from E.I. duPont de Nemours and Co., Inc., and was kept in a 10 mg/ml stock in DMSO at −20°C. Benomyl was added to media in final concentrations of 1, 2, 5, 10, 15, 20, 30, 50, and 80 μg/ml. Growth of strains on solid media was assayed by suspending cells in sterile water and spotting onto plates using a multipronged inoculating device.
Table I.
Yeast Strains
| Strain | Genotype | Source | ||
|---|---|---|---|---|
| TSY678 | MATa alf1::HIS3 ade2-101 his3Δ-200 ura3-52 lys2-801 leu2-3,-112 | This study | ||
| TSY679 | MATa alf1::HIS3 his3Δ-200 ura3-52 lys2-801 leu2-3,-112 | This study | ||
| TSY680 | MATa alf1::URA3 ade2-101 his3Δ-200 ura3-52 lys2-801 leu2-3,-112 | This study | ||
| TSY681 | MATa alf1::URA3 his3Δ-200 ura3-52 lys2-801 leu2-3,-112 | This study | ||
| TSY828 | MATa bik1::ADE2 TUB1:LEU2 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | Botstein lab | ||
| TSY979 | MATa alf1::URA3 bik1::ADE2 TUB1:LEU2 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | This study | ||
| TSY829 | MATa tub1-836:LEU2 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | Botstein lab | ||
| TSY980 | MATa alf1::URA3 tub1-836:LEU2 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | This study | ||
| TSY830 | MATa tub1-831:LEU2 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | Botstein lab | ||
| TSY981 | MATa alf1::URA3 tub1-831:LEU2 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | This study | ||
| TSY831 | MATa tub2-442:URA3 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | Huffaker lab | ||
| TSY982 | MATa alf1::HIS3 tub2-442:URA3 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | This study | ||
| TSY832 | MATa tub1-839:LEU2 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | Botstein lab | ||
| TSY983 | MATa alf1::URA3 tub1-839:LEU2 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | This study | ||
| TSY833 | MAT tub2-408:URA3 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | Huffaker lab | ||
| TSY984 | MATa alf1::HIS3 tub2-408:URA3 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | This study | ||
| TSY834 | MATa tub3::HIS3 TUB1:LEU2 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | Botstein lab | ||
| TSY985 | MATa alf1::URA3 tub3::HIS3 TUB1:LEU2 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | This study | ||
| TSY835 | MATa tub1-801:LEU2 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | Botstein lab | ||
| TSY837 | MATa tub2-428:URA3 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | Huffaker lab | ||
| TSY987 | MATa alf1::HIS3 tub2-428:URA3 his3-Δ200 ade2-101 leu2-Δ1 lys2-801 ura3-52 | This study | ||
| TSY839 | MATa bim1::URA3 ura3-52 leu2-Δ1 his3-Δ200 | Botstein lab | ||
| TSY989 | MATa alf1::HIS3 bim1::URA3 ura3-52 leu2-Δ1 his3-Δ200 | This study | ||
| TSY906 | MATa cin1::HIS3 his3-Δ200 lys2-801 leu2-3,-112 ura3-52 | This lab | ||
| TSY990 | MATa alf1::URA3 cin1::HIS3 his3-Δ200 lys2-801 leu2-3,-112 ura3-52 | This study | ||
| TSY907 | MATa cin2::LEU2 his3-Δ200 lys2-801 leu2-3,-112 ura3-52 | This lab | ||
| TSY991 | MATa alf1::URA3 cin2::LEU2 his3-Δ200 lys2-801 leu2-3,-112 ura3-52 | This study | ||
| TSY993 | MATa cin1::HIS3 cin2::LEU2 his3-Δ200 lys2-801 leu2-3,-112 ura3-52 | This lab | ||
| TSY994 | MATa alf1::URA3 cin1::HIS3 cin2::LEU2 his3-Δ200 lys2-801 leu2-3,-112 ura3-52 | This study | ||
| TSY790 | MATa pac2::URA3 his3-Δ200 lys2-801 leu2-3,-112 ura3-52 | This study | ||
| TSY789 | MATa alf1::HIS3 pac2::URA3 his3-Δ200 lys2-801 leu2-3,-112 ura3-52 | This study | ||
| FSY611 | MATa rbl2::URA3-hisG ura3-52 leu2-3,-112 lys2-801 his3-Δ200 | Solomon lab | ||
| TSY973 | MATa alf1::HIS3 rbl2::URA3-hisG ura3-52 leu2-3,-112 lys2-801 his3-Δ200 | This study | ||
| TSY908 | MATa tub1-1 his3-Δ200 lys2-801 leu2-3,-112 ura3-52 | This lab | ||
| Y190 | MATa gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3,-112 URA3::GAL→ lacZ LYS2::GAL→ HIS3 cyl | Elledge lab | ||
| TSY963 | MATa ALF1-myc:URA3 ura3-52 leu2-3,-112 lys2-801 his3-Δ200 | This study | ||
| TSY1066 | MATa ura3-52 leu2-3,-112 lys2-801 his3-Δ200 [pTS887] | This study | ||
| TSY986 | MATa alf1-1-myc:URA3 ura3-52 leu2-3,-112 lys2-801 his3-Δ200 | This study | ||
| TSY964 | MATα cin1::HIS3 ura3-52 leu2-3,-112 lys2-801 his3-Δ200 [pTS210] | This study | ||
| TSY965 | MATα cin1::HIS3 ura3-52 leu2-3,-112 lys2-801 his3-Δ200 [pTS863] | This study | ||
| TSY966 | MATa ura3-52 leu2-3,-112 lys2-801 his3-Δ200 [pTS210] | This study | ||
| TSY967 | MATa ura3-52 leu2-3,-112 lys2-801 his3-Δ200 [pTS863] | This study | ||
| TSY968 | MATα cin2::LEU2 ura3-52 leu2-3,-112 his3-Δ200 [pTS210] | This study | ||
| TSY969 | MATα cin2::LEU2 ura3-52 leu2-3,-112 his3-Δ200 [pTS863] | This study | ||
| TSY970 | MATa cin4::LEU2 ura3-52 leu2-3,-112 lys2-801 his3-Δ200 [pTS210] | This study | ||
| TSY971 | MATa cin4::LEU2 ura3-52 leu2-3,-112 lys2-801 his3-Δ200 [pTS863] | This study | ||
| TSY974 | MATα cin1::HIS3 ura3-52 leu2-3,-112 lys2-801 his3-Δ200 [pTS863, pTS249] | This study | ||
| TSY975 | MATα cin1::HIS3 ura3-52 leu2-3,-112 lys2-801 his3-Δ200 [pTS336, pTS210] | This study | ||
| TSY976 | MATα cin1::HIS3 ura3-52 leu2-3,-112 lys2-801 his3-Δ200 [pTS336, pTS863] | This study | ||
| TSY977 | MATα cin1::HIS3 ura3-52 leu2-3,-112 lys2-801 his3-Δ200 [pTS249, pTS210] | This study | ||
| TSY972 | MATa/α ura3-52/ura3-52 leu2-3,-112/leu2-3,-112 lys2-801/lys2-801 his3-Δ200/his3-Δ200 ade2-101/ADE2 [pTS864] | This study | ||
| TSY995 | MATa/α ura3-52/ura3-52 leu2-3,-112/leu2-3,-112 lys2-801/LYS2 his3-Δ200/his3-Δ200 ade2-101/ADE2 [pTS864] | This study | ||
| TSY996 | MATa/α ura3-52/ura3-52 leu2-3,-112/leu2-3,-112 lys2-801/LYS2 his3-Δ200/his3-Δ200 ade2-101/ADE2 [pTS868] | This study | ||
| TSY904 | MATa tub1-850:LEU2 tub3::HIS3 ura3-52 leu2-3,-112 lys2-801 ade2-101 [pTS864] | This study |
Table II.
Plasmid List
| Plasmid | Relevant Markers | Source | ||
|---|---|---|---|---|
| pTS210 | Pgal1; URA3, YCp | This lab | ||
| pTS249 | Pgal1; LEU2, YCp | This lab | ||
| pAS1 | Padh; GAL4 DNA-binding domain TRP1; YEp | Elledge lab | ||
| pACTII | Padh; GAL4 activation domain LEU2; YEp | Elledge lab | ||
| pDAb1 | Padh; GAL4 DNA-binding domain TRP1; YCp | Botstein lab | ||
| pTS869 | CIN1-pAS1 | This study | ||
| pTS870 | CIN1-pACTII | This study | ||
| pTS862 | PAC2-pACTII | This study | ||
| pTS860 | ALF1-pAS1 | This study | ||
| pTS861 | ALF1-pACTII | This study | ||
| pTS863 | Pgal1-ALF1, URA3,YCp | This study | ||
| pTS887 | Pgal1-ALF1-myc, URA3,YCp | This study | ||
| pTS864 | Pgal1-ALF1-GFP, URA3, YCp | This study | ||
| pTS865 | ALF1-myc, URA3, YIp | This study | ||
| pTS866 | alf1-1-pAS1 | This study | ||
| pTS867 | alf1-1-pACTII | This study | ||
| pTS868 | alf1-1-GFP, URA3, YCp | This study | ||
| pTS889 | alf1-1-myc, LEU2, YIp | This study | ||
| pTS336 | Pgal1-TUB3, LEU2, YCp | This study | ||
| pTS871 | TUB1, pACTII* | This study | ||
| pTS872 | CCT5-pACTII* | This study | ||
| pRB2514 | TUB1-pAS1 | Botstein lab | ||
| pRB2516 | TUB1-pACTII | Botstein lab |
Plasmids were isolated from a two-hybrid screen with Alf1p.
Indicates that the TUB2 allele used in the two-hybrid assay is not wild-type (see Results for description).
Indicates that the CIN2 allele used in the two-hybrid assay is not full-length (see Materials and Methods for more details).
Fluorescence and Immunological Techniques
Immunofluorescence was performed as described (Pringle et al., 1989) with the following changes. To visualize microtubules, cells were fixed by adding formaldehyde to a final concentration of 3.7% and incubating at room temperature for 2 h, followed by methanol/acetone treatment. Primary antibody YOL1/34 (Kilmartin et al., 1982) was detected with Texas red–conjugated donkey anti–rat antibodies (Jackson ImmunoResearch Laboratories, Inc.).
Green fluorescent protein (GFP) fluorescence was visualized in living cells using a fluorescein filter set (Hi-Q FITC; Chroma Technology Corp.) on a microscope (Axioskop; Carl Zeiss, Inc.) equipped with an HBO100 mercury lamp and ×100/1.3 objective lens. To visualize GFP in living cells, strains containing GFP plasmids under the control of the GAL1 promoter were grown overnight at 30°C in minimal media containing 2% galactose + 0.5% glucose; the addition of glucose reduces the expression from the GAL1 promoter, thus reducing background fluorescence (Marschall et al., 1996).
ALF1 Plasmid Constructs
To construct a PGAL1 -ALF1 plasmid, the ALF1 gene was generated by PCR with BamHI and XbaI ends using the following primers: YNL148.5 5′-GGCGGATTCCATAAATGGTTAGAGTTGTCA-3′ and YNL148.7 5′-GCCTCTAGATCAAATTTCATCATCGCTCTC-3′.
All PCR reactions used Vent polymerase (New England Biolabs) to amplify directly from genomic DNA unless otherwise specified. The resultant PCR product was cleaved with BamHI and XbaI and cloned into the same sites in pTS210, a centromere based PGAL1 vector with the ACT1 transcriptional terminator.
To construct an Alf1p-GFP fusion protein, a version of the ALF1 gene with BamHI and XbaI ends was generated by PCR using the following primers: YNL148.5 (described above) and YNL148.6 5′-GGCTCTAGAAATTTCATCATCGCTCTCCAC-3′. The resulting PCR product was cleaved with BamHI and XbaI and cloned into pTS586, a centromere based PGAL1-GFP vector with the ACT1 transcriptional terminator, containing the GFP mutant, GFPMUT3 (Cormack et al., 1996), creating the PGAL1-ALF1-GFP construct, pTS864. pTS864 was transformed into a wild-type diploid yeast strain to create TSY972.
Integrated myc-tagged ALF1 was created by cloning the ALF1 BamHI-XbaI fragment from pTS864 into pTS776, an integrating vector with the URA3 marker, to generate pTS865. pTS865 was cut with EcoRI, which cuts in the ALF1 gene, and transformed into a wild-type haploid strain (DBY4974) to create TSY963. PGAL1-ALF1-myc was constructed by PCR using the YNL148.5.6 primers (described above). The ALF1 gene was cloned into pTS466, a CEN-based vector with the GAL1 promoter and a 3′ myc tag, creating pTS887. pTS887 was transformed into a haploid yeast strain, creating TSY1066; Alf1p-myc was induced by growing in selective medium containing either 2% galactose, or 2% galactose and 0.5% glucose.
The Alf1p two-hybrid bait plasmid was constructed as follows. The ALF1 gene was generated by PCR with NcoI and BamHI ends using the following primers: YNL148.13 5′-GCCCCATGGTTAGAGTTGTCATA-3′ and YNL148.14 5′-GCCGGATCCAATTTATCATCGCTTCTC-3′. The resulting PCR product was digested with NcoI and BamHI and cloned into pAS1-CYH2 to create pTS860 (Durfee et al., 1993). This plasmid was transformed into the two-hybrid strain Y190 and tested for expression of the fusion protein by Western analysis. The resulting strain was named TSY1013.
Two-Hybrid Analyses
The two hybrid system used was that described in Durfee et al. (1993). In this system, both HIS3 and lacZ are used as reporters of interaction. The λYES cDNA library was titered and amplified in bacterial strain LE392 (Elledge et al., 1991). Plasmid inserts were excised from the λYES cDNA library by infection of strain BNN132 (Elledge et al., 1991). TSY1013 was transformed with library DNA by a lithium acetate protocol (Adams et al., 1997) and plated on SD + 10 mg/ml adenine + 25 mM 3-aminotriazole. The plates were incubated at 30°C for 9 d. Approximately 9 × 106 transformants were screened in this manner.
As a secondary test, His+ strains were assayed for β-galactosidase activity. Strains were patched onto SC-trp-leu media, grown overnight, lifted on nitrocellulose filters (BA85; Schleicher and Schuell, Keene, NH) and immersed in liquid nitrogen for 10 s. The filters were then placed on Whatman paper soaked with 3 ml of Z buffer (60 mM Na2HPO4, 40 mM NaH2P04, 10 mM KCl, 1 mM MgSO4, and 50 mM β-mercaptoethanol, pH 7.0) containing 0.05% X-Gal. Filters were incubated at 30°C for 6 h and scored for the development of blue color. Clones that passed the secondary test were tested for solo activation by growth on SC-leu media with 5 μg/ml cycloheximide. The strains were confirmed to have lost the TRP1-containing bait plasmid by failure to grow on SC-trp media. Library inserts were sequenced on one strand with a Thermo Sequenase dye terminator cycle sequencing kit (Amersham Life Science) with primer 2-H1 (TGATGAAGATACCCCACC) and identified by a BLAST search (Altschul et al., 1990) via the Saccharomyces Genome Database (http://genome-www.stanford.edu).
Yeast Protein Extract Preparation
Protein extracts for Western analysis were prepared by disrupting yeast cells with glass beads as described (Kaiser et al., 1987). Protein extracts for immunoprecipitations were made in the following manner. Yeast were grown in 20 ml of YPD overnight to late log phase, harvested, and washed. Cells were resuspended in 5 ml 20 mM Tris, pH 7.8, 0.1 M β-mercaptoethanol and 1 M sorbitol at 30°C for 10 min. Cells were centrifuged and resuspended in 5 ml 1 M sorbitol, 10 mM potassium phosphate solution, pH 6.0, in the presence of protease inhibitors (100 μg/ml PMSF, and 5 μg/ml each of pepstatin, leupeptin, and chemostatin) and 100 μg/ml zymolyase, followed by incubation at 30°C for 20–30 min. The resulting spheroplasts were centrifuged and resuspended in 200 μl HBS (50 mM Hepes, 150 mM NaCl, pH 7.4, and 0.5% Triton X-100) in the presence of protease inhibitors, and transferred to a 15-mm test tube. Acid-washed glass beads were added to the liquid meniscus and tubes were vortexed twice for 40 s each time. Extract was removed from the beads and centrifuged at 4°C for 10 min at 10,000 g. Bradford assays (Bio-Rad) were performed on the cleared extract.
Immunoprecipitations
Cell extracts were prepared as described above. Equal amounts of protein were added to two Eppendorf tubes, each containing protein A–Sepharose (Sigma Chemical Co.) and the monoclonal anti-myc antibody 9E10. In one tube a 10-fold molar excess of competing antigenic peptide was added. Immunoprecipitations were performed at 4°C for 1 h on a rotator. The immunoprecipitates were washed twice in lysis buffer, twice in 50 mM Hepes, 250 mM NaCl, pH 7.4, and 0.5% Triton X-100, and once again in lysis buffer. 60 μl of sample buffer was added to each tube and incubated for 5 min at room temperature, and 20 μl was used per lane on a protein gel. Incubation at room temperature was found to dissociate the antibodies from protein A but not to dissociate the two heavy chains of the antibody, resulting in the heavy chains migrating at a molecular weight that does not interfere with the migration and visualization of tubulin (Murphy et al., 1998).
Immunoblotting
Protein samples were separated on 10% SDS–polyacrylamide gels, electrophoretically transferred to nitrocellulose, and then blocked overnight in 4% nonfat dried milk in TBST (10 mM Tris, pH 8.0, 150 mM NaCl, 0.05% IGEPAL). The following primary antibodies were used for this study: 206, rabbit anti-β-tubulin (F. Solomon, MIT, Cambridge, MA); TAT-1, mouse anti-α-tubulin (K. Gull, University of Manchester, Manchester, England); 345, rabbit anti-α-tubulin (D. Botstein, Stanford University, Stanford, CA); and guinea pig anti-actin (D. Botstein). Primary antibody incubations were followed by incubation with HRP-conjugated anti–mouse, anti–rabbit, or anti–protein A antibodies (Jackson ImmunoResearch Laboratories, Inc.). Immunoreactive proteins were visualized by chemiluminescence (Amersham Pharmacia Biotech Inc.).
Construction of the ALF1 CLIP-170 Mutant
The conserved CLIP-170 motif GKNDG of Alf1p was mutated to AENDA as follows. Complementary oligonucleotides were designed to introduce into ALF1 both the desired amino acid changes and a PvuII site for diagnostic purposes:
ALF1.8 5′-AATTCCCGGAAGCCGCAGCTGAAAACGATGCTCGCATAAATGGGGTAACACTGTTTGGGCCTGTGG-3′ and ALF1.9 5′-CGCCACAGGCCCAAACAGTGTTACCCCATTTATGCGAGCATCGTTTTCAGCTGCTGCGGCTTCCGGG-3′. When hybridized, the ends of the resulting duplex form NarI and EcoRI overhangs for cloning. The oligos were hybridized and the ends were kinased with T4 polynucleotide kinase (New England Biolabs). pTS826, containing wild-type ALF1 under its own promoter, was cut with EcoRI and religated to make pBF87, deleting a 0.7-kb piece of ALF1. pBF87 was cut with EcoRI and NarI and ligated to ALF1.8.9 plus the 0.7-kb piece removed in the previous step, thereby replacing the wild-type CLIP-170 domain in ALF1, resulting in pTS878.
The mutant, alf1-1, was cloned into two-hybrid vectors by PCR using YNL148.13 and YNL148.14 primers and Vent polymerase, and the sequence of the resulting plasmid, pTS866 was confirmed by sequencing. This plasmid was transformed into the two-hybrid strain Y190 and tested for expression of the fusion protein by Western analysis.
Integrated myc-tagged alf1-1 was created in the following manner. alf1-1 was cloned by PCR into pTS776, an integrating vector with a 3′ myc-tag and a URA3 marker, to generate pTS886. alf1-1-myc was removed from pTS886 by digestion with BamHI and SalI, and subcloned into pRS305, a LEU2-based integration vector, creating pTS889. pTS889 was cut with MscI, which cuts 5′ to the mutation in alf1-1, and transformed into a wild-type haploid strain (DBY4974) to create TSY986.
Tubulin Mapping
A panel of alanine scan tub1 mutants generated by K. Richards (Richards, 1997) was cloned into pAS1 (Schwartz et al., 1997). Two PCR isolates of each tub1 mutant were transformed separately into Y190, containing Alf1p-pACTII, and tested for their ability to activate the reporter gene lacZ, in the X-gal filter lift assay. The Alf1p constructs and assay are described above.
The tub1 mutants that did not interact with Alf1p were mapped onto the predicted structure of yeast α-tubulin, as determined by homology modeling using the mammalian α-tubulin structure. Modeling was done on a Silicon Graphics Indigo workstation (Silicon Graphics), using Look 3.0 software (Molecular Applications Group).
Results
Alf1p Physically Interacts with α-Tubulin
Cofactor B was identified on the basis of its interaction with α-tubulin, and its ability to assist in the formation of the tubulin heterodimer. This role predicts a direct interaction with α-tubulin, and possibly with other members of the same pathway. To identify such interacting proteins we performed a yeast two-hybrid screen with the yeast cofactor B ortholog Alf1p. ALF1 was fused to the GAL4 DNA-binding domain and the construct was shown to express the expected 40-kD fusion protein. A library of yeast cDNAs fused to the GAL4 activation domain was screened for interaction with Alf1p. Among the positive clones, TUB1 and TUB3, the major and minor α-tubulin genes, and CCT5, a subunit of the Cct/TRiC complex, were represented by several independent clones (Fig. 1 A). The α-tubulin isolates were full-length clones, whereas the CCT5 isolates were not full-length, but each contained at least the last 600 bp of the ORF. Alf1p was also found to interact with TUB1 when tested directly in the two-hybrid assay (Table III).
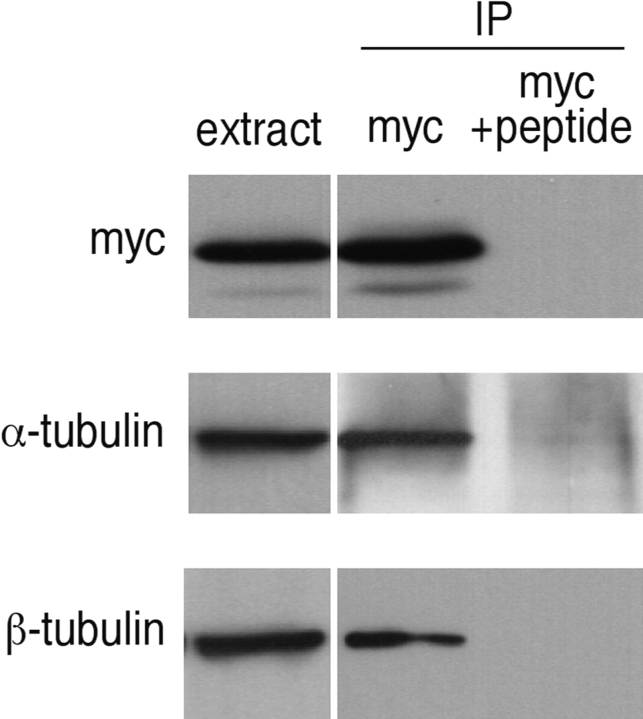
Figure 1.
Alf1p interactions. (A) TUB1 and CCT5 are isolated in a two-hybrid screen with Alf1p. The strains tested are indicated at the left of the figure; the gene on top was cloned into the plasmid pAS1, and the gene on the bottom was cloned into the plasmid pACTII. All strains can grow on permissive medium (+his). Only strains carrying ALF1 as well as TUB1 or CCT5, but not vector alone, can grow on restrictive medium (−his +3-AT). (B) Alf1p coimmunoprecipitates with α-tubulin, but not β-tubulin. Alf1p-myc was immunoprecipitated from extracts with anti-myc mAb in the absence (myc) or presence (myc + peptide) of competing antigenic peptide. Precipitates were immunoblotted with either anti-myc, anti-α-tubulin, or anti-β-tubulin antibodies, as indicated on the left-hand side of the figure.
Table III.
Summary of Two-Hybrid Interactions
| DNA-binding domain fusion (pAS1) | Activation domain fusion (pACTII) | Interaction | ||
|---|---|---|---|---|
| ALF1 | TUB1 | + | ||
| ALF1 | ALF1 | − | ||
| ALF1 | CIN1 | − | ||
| ALF1 | PAC2 | − | ||
| TUB1 | PAC2 | + | ||
| CIN1 | PAC2 | − | ||
| CIN1 | TUB1 | − |
Constructs were tested in the Y190 strain for their ability to activate the lacZ reporter gene in a colormetric assay (see Materials and Methods).
We were interested to know whether Alf1p interacted with Pac2p, since their mammalian counterparts both interact with α-tubulin, and are thought to act sequentially (Tian et al., 1996). Alf1p and Pac2p failed to interact with each other in the assay, but Pac2p did interact with α-tubulin (Table III), consistent with the biochemical results (Tian et al., 1996). Alf1p was also tested for interactions with Cin1p and itself, with negative results (Table III). Interaction between Alf1p, or the other cofactors, and β-tubulin could not be tested directly because overexpression of either the β-tubulin gene, TUB2, or the required two-hybrid fusion is lethal in yeast.
To determine whether the interaction of Alf1p with α-tubulin observed in the two-hybrid assay reflects a genuine in vivo association, we immunoprecipitated Alf1p and tested for the presence of α-tubulin in the precipitate. A single myc-tag was integrated at the 3′ end of the genomic copy of ALF1 in a haploid strain. This strain was phenotypically wild-type, indicating that the tag did not interfere with Alf1p function. Western blots of anti-myc immunoprecipitates from this strain were tested for the presence of Alf1p, α-tubulin, and β-tubulin. α-Tubulin coimmunoprecipitated with the epitope-tagged Alf1p but β-tubulin did not (Fig. 1 B). Under the conditions used, the sensitivity of detection of the anti-α-tubulin and anti-β-tubulin antibodies was comparable. These results indicate that Alf1p physically interacts with α-tubulin but not β-tubulin. In addition, this interaction appears to be specifically with the α-tubulin monomer rather than the tubulin heterodimer, because β-tubulin did not coimmunoprecipitate with the Alf1p–α-tubulin complex.
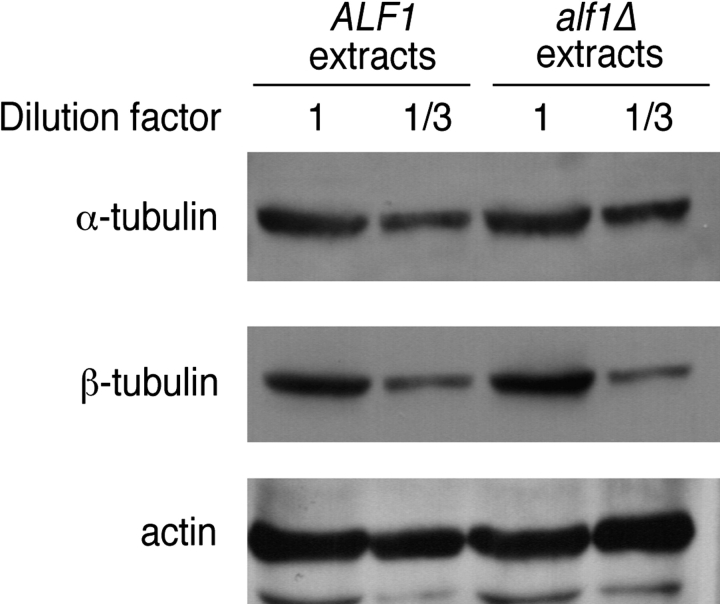
If Alf1p were involved with the folding or biogenesis of α-tubulin, we reasoned that alf1Δ strains might show reduced amounts of α-tubulin, due to the degradation of unfolded intermediates. This would, in part, explain the benomyl sensitivity of alf1 null strains, because strains with reduced amounts of α-tubulin (i.e., tub3Δ strains) are benomyl sensitive (Schatz et al., 1986). Extracts from alf1Δ and wild-type cells were tested by Western blotting and were found to have approximately the same amounts of both α- and β-tubulin (Fig. 2).
Figure 2.
alf1Δ cells have normal levels of α- and β-tubulin. Cytoplasmic extracts were prepared from ALF1 and alf1Δ cells and immunoblotted with anti-α-tubulin, anti-β-tubulin, and anti-actin antibodies. Actin was used as a control for loading. The amounts of α- and β-tubulin were quantitated by densitometry.
Interaction of Alf1p with a α-Tubulin requires the Alf1p CLIP-170 Domain
Alf1p and cofactor B contain one copy of the CLIP-170 microtubule-binding domain found in the mammalian CLIP-170 protein, the p150Glued subunit of dynactin, and yeast BIK1 (Pierre et al., 1992; Fig. 3 A). This domain is also found in cofactor E and its yeast ortholog Pac2p (Hoyt et al., 1997; Tian et al., 1997; Fig. 3 A). CLIP-170 and Bik1p have been shown to localize to microtubules in vivo (Berlin et al., 1990; Pierre et al., 1992). We wished to test whether the CLIP-170 domain was important for Alf1p interaction with α-tubulin, as well as for Alf1p function in general. This was tested by creating a mutant allele, alf1-1, in which the conserved CLIP-170 domain sequence GKNDG was changed to AENDA (Fig. 3 A); the same mutation was made previously in CLIP-170 and found to abolish microtubule binding (Pierre et al., 1994).
Figure 3.
The CLIP-170 domain of Alf1p is required for the interaction with α-tubulin. (A) Alf1p contains a CLIP-170 microtubule-binding domain. The CLIP-170 domains of the yeast proteins, Alf1p, Pac2p, and Bik1p, and the mammalian proteins, CLIP-170, cofactor B, cofactor E and p150Glued are aligned. Shaded regions indicate residues identical or similar to Alf1p. Site-directed mutations were constructed in the CLIP-170 domain of Alf1p, creating alf1-1. Arrows indicate the amino acids changed. (B) alf1-1 does not interact with α-tubulin in the two-hybrid assay. The strains tested are indicated at the left of the figure; the gene on top was cloned into the plasmid pAS1, and the gene on the bottom was cloned into the plasmid pACTII. All strains grow on permissive medium (+his). Strains bearing both ALF1 and TUB1 plasmids are able to grow on the restrictive condition (−his + 3-AT), whereas those bearing alf1-1 and TUB1 are not, indicating that alf1-1p does not interact with Tub1p as either bait or prey. (C) alf1-1p-myc does not coimmunoprecipitate with α-tubulin. alf1-1p-myc was immunoprecipitated from extracts with anti-myc mAb in the absence (myc) or presence (myc + peptide) of competing antigenic peptide. Precipitates were immunoblotted with either anti-myc, anti-α-tubulin, or anti-β-tubulin antibodies, as indicated on the left-hand side of the figure.
alf1-1 was cloned into vectors to test complementation of the alf1 null phenotype, and for the ability to interact with α-tubulin. Expressed at endogenous levels, alf1-1 did not complement the benomyl sensitivity of an alf1 null mutant. In the two-hybrid assay alf1-1 was unable to interact with TUB1; this result was obtained with alf1-1 as either bait or prey in the assay (Fig. 3 B). Consistent with the two-hybrid data, an epitope-tagged version of alf1-1p, alf1-1p-myc was unable to coimmunoprecipitate α-tubulin (Fig. 3 C). Although the alf1-1 mutant did not complement an alf1 null and did not interact with α-tubulin, it was expressed at a level similar to that of ALF1 (not shown), could be immunoprecipitated (Fig. 3 C), and was able to interact with CCT5 in the two-hybrid assay (not shown). These results suggest both that the interaction of Alf1p with α-tubulin depends on the CLIP-170 domain, and that the loss of this activity results in the benomyl supersensitivity in the alf1 null mutant.
Mapping the Alf1p Binding Site on α-Tubulin
The above experiments demonstrate that the CLIP-170 domain is necessary for Alf1p interaction with α-tubulin. Next we determined the residues of α-tubulin involved in interaction with Alf1p, taking advantage of a panel of α-tubulin alanine-scan mutants (Richards, 1997; Schwartz et al., 1997). In this set of mutants, clusters of charged residues in TUB1 were changed to alanines, with the rationale that such residues are likely to be on the surface of the molecule, and potentially involved in interaction with other proteins (Cunningham and Wells, 1989).
We used the two-hybrid system to assess the ability of these α-tubulin mutants to interact with Alf1p, as described previously for other α-tubulin ligands (Schwartz et al., 1997). 53 of the tub1 mutant alleles were fused to the GAL4 DNA-binding domain and tested for their ability to interact with Alf1p. Each combination was tested for its ability to activate the lacZ reporter gene (Table IV). By this assay, most of the α-tubulin mutants could still interact with Alf1p, however the following alleles failed to interact: tub1-814 (R106A, H108A), tub1-819 (T146A), tub1-822 (K167A, E169A), tub1-823 (E197A, H198A, D200A), tub1-824 (D206A,E208A), tub1-829 (R265A, H267A), tub1-834 (R321A, D323A), tub1-840 (D393A, R394A, K395A, D397A), tub1-842 (K395A, D397A), tub1-843 (K395A), tub1-844 (D397A), tub1-845 (K402A, R403A), and tub1-847 (E416A, E418A), tub1-848 (E421A, R423A, E424A, D425A), tub1-849 (E430A, R431A, D432A), tub1-850 (E435A, D439A), tub1-851 (E443A, E444A, E445A, E446A), tub1-852 (F447A), and tub1-853 (E183A). In addition, the following alleles displayed positive, but weak, interaction: tub1-818 (D128A, D131A), tub1-821 (E161A, K164A, K165A), tub1-830 (K279A, K281A), and tub1-838 (D373A, R374A).
Table IV.
Two-Hybrid Interaction of Alf1p with tub1 Mutants
| tub1 allele | Interacts in Assay | tub1 allele | Interacts in Assay | tub1 allele | Interacts in Assay | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| tub1−801 | + | −820 | d.l. | −837 | + | |||||
| −802 | + | −821 | +‡ | −838 | + | |||||
| −803 | + | −822 | − | −839 | + | |||||
| −804 | + | −823* | − | −840 | − | |||||
| −805 | + | −824 | − | −841 | + | |||||
| −808 | + | −825 | + | −842 | − | |||||
| −809 | + | −826 | + | −843 | − | |||||
| −810 | + | −827 | + | −844 | − | |||||
| −811 | + | −828 | d.l. | −845* | − | |||||
| −812 | + | −829 | − | −846 | + | |||||
| −813 | + | −830 | +‡ | −847 | − | |||||
| −814* | − | −831 | + | −848 | − | |||||
| −815 | + | −832 | + | −849 | − | |||||
| −816 | + | −833 | + | −850 | − | |||||
| −817 | + | −834* | − | −851 | − | |||||
| −818 | +‡ | −835 | + | −852 | − | |||||
| −819 | − | −836 | + | −853 | − |
Strains bearing both ALF1 and TUB1 (allele indicated) plasmids were patched onto selective media and after 1 d of growth were assayed for lacZ expression. "+" indicates that the tub1 allele tested positive in the assay with ALF1, and "−" indicates a negative result.
d.l., Dominant lethal allele.
*Alleles of tub1 that do not interact with any known tubulin-binding proteins, suggesting that the negative result may not be specific to Alf1p.
Indicates a very weak positive interaction.
To determine whether these differential interactions define a specific domain on the surface of α-tubulin, we mapped the mutations onto the structure of yeast Tub1p. Fig. 4 A shows the putative space-filling structure of yeast α-tubulin, modeled on the mammalian α-tubulin structure, and oriented with the GTP-binding site at the top of the figure (Nogales et al., 1998). Residues that either do not interfere with Alf1p binding when mutated or were not mutated are displayed in gray. The residues that abolish Alf1p interaction when mutated are displayed in yellow. Interestingly, these residues are clustered together on the outside face of α-tubulin. Note that gray residues within the yellow patches might be involved in interaction with Alf1p, but were not mutagenized because they are not part of charged clusters. The tub1-814, tub1-823, tub1-834, and tub1-845 alleles do not interact with Alf1p, but are not shown as yellow in the figure, because they do not interact with several other known α-tubulin interacting proteins in the two-hybrid assay, such as Bim1p and Bik1p (Schwartz et al., 1997). Most of these alleles map to regions buried within the α-tubulin structure, suggesting that the mutations might affect Alf1p binding in a nonspecific manner. The tub1-850, tub1-851, tub1-852, and tub1-853 alleles were also not included in the figure because they map to the α-tubulin COOH terminus, which is not resolved in the existing structure (Nogales et al., 1998). The interactions shown in Fig. 4 A were also mapped onto the α-tubulin α-carbon backbone, shown in stereo view in Fig. 4 B.
Figure 4.

Alf1p binding site on α-tubulin. The structure of the yeast TUB1 α-tubulin was determined by homology modeling using the mammalian α-tubulin structure (Richards, 1997). Both views of the α-tubulin structure are shown in the same orientation, with the GTP-binding site at the top. (A) Space-filling model of α-tubulin. The residues that disrupt the interaction with Alf1p when mutated are indicated in yellow. (B) Stereo view of the α-tubulin α-carbon backbone (magenta) in the context of a protofilament. The protofilament runs vertically and the surface facing the viewer corresponds to the outside of the microtubule. The residues that disrupt the interaction with Alf1p when mutated are indicated in yellow. Several of the helices in α-tubulin, as described in Nogales et al. (1998) are indicated for reference. A dimer is formed with the partial β-subunit at the top (violet), while the partial β-tubulin at the bottom (aquamarine) corresponds to another dimer.
Alf1p Localization to Microtubules In Vivo Depends on Its CLIP-170 Domain
We have shown that Alf1p binds to α-tubulin monomer and that the CLIP-170 domain is required for this interaction. Yet, the CLIP-170 domain was originally identified in proteins that associate with microtubule polymer. To determine whether Alf1p associates with microtubule polymer in vivo, an ALF1-GFP fusion was constructed. When expressed at endogenous levels, no specific localization of Alf1p-GFP was apparent. To increase the amount of Alf1p-GFP in the cell, the fusion protein was placed under the control of the GAL1 promoter. Under conditions of full induction, which resulted in 200-fold overexpression relative to wild-type Alf1p, Alf1p-GFP was present throughout the cytoplasm (not shown). However, when the level of expression from the GAL1 promoter was reduced to only several-fold above that of wild-type Alf1p, the fusion protein localized to both nuclear and cytoplasmic microtubules throughout the cell cycle (Fig. 5 A). The Alf1p-GFP fluorescence was brighter on the spindle than on cytoplasmic microtubules, consistent with the greater number of spindle microtubules relative to cytoplasmic microtubules (Byers and Goetsch, 1975; Winey et al., 1995). To determine whether localization of Alf1p-GFP to microtubules was dependent on the CLIP-170 domain, an alf1-1p-GFP fusion was created. When expressed under the same conditions of induction as in Fig. 5 A, alf1-1p-GFP did not localize to microtubules (Fig. 5 B). Taken together, these results suggest that Alf1p can localize to the microtubule cytoskeleton, and that this localization is dependent upon its CLIP-170 domain.
Figure 5.
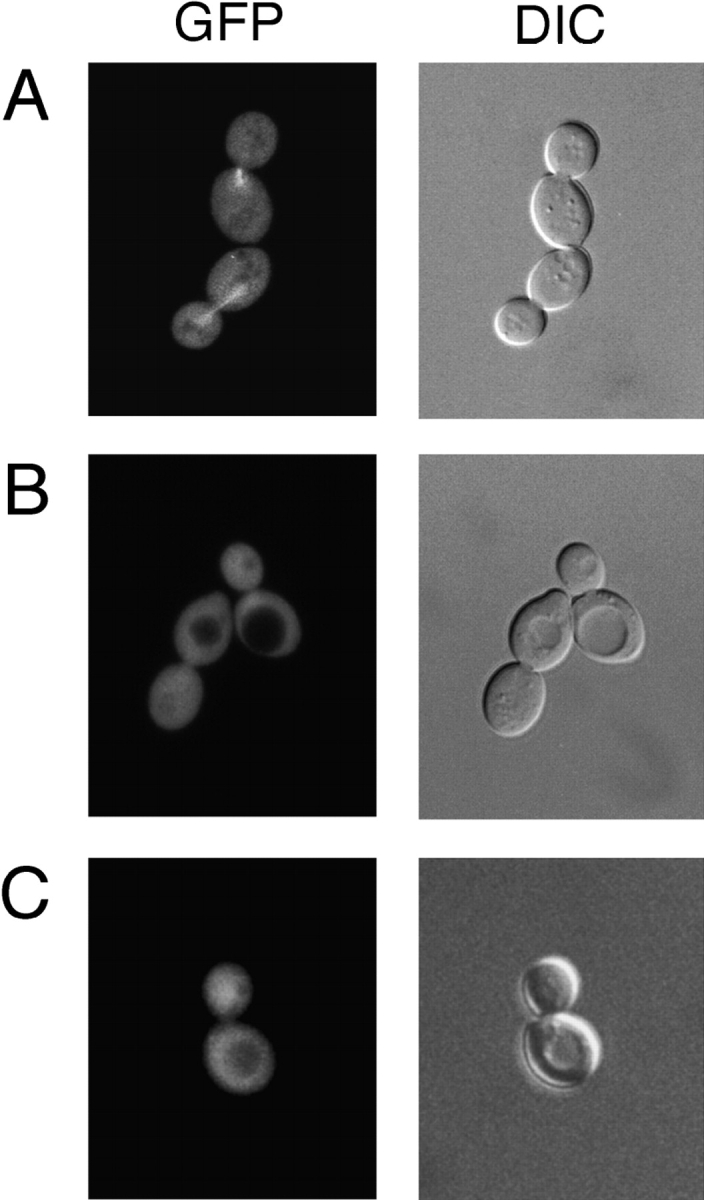
Localization of Alf1p-GFP to microtubules depends on both the Alf1p CLIP-170 domain and the domains of α-tubulin identified in the two-hybrid assay (Table V). (A) Localization of Alf1p-GFP in living cells. Wild-type diploid cells expressing Alf1p-GFP were examined by fluorescence microscopy. Alf1p is present on both spindle and cytoplasmic microtubules, but the cytoplasmic microtubules are very faint. Expression of Alf1p-GFP was under the control of the GAL1 promoter and is induced by growth in 2% galactose and 0.5% glucose. (B) alf1-1p-GFP does not localize to the microtubule cytoskeleton in wild-type cells. (C) Wild-type Alf1p-GFP does not localize to the microtubule cytoskeleton in tub1-850 cells. Cells were grown in minimal media containing 2% galactose and 0.5% glucose.
The Alf1p binding site on α-tubulin defined in the two-hybrid study above includes much of the cytoplasmic face of α-tubulin, as it is oriented in the microtubule. We wished to test whether Alf1p associated with microtubule polymer using this same binding site. Alf1p-GFP was tested for localization to microtubules in two of the tub1 mutants that failed to interact with Alf1p in the two-hybrid assay, but which have a morphologically normal microtubule cytoskeleton. When Alf1p-GFP was expressed in either tub1-850 or tub1-852 strains, Alf1p-GFP did not localize to microtubules (tub1-850 shown in Fig. 5 C). This result suggests that Alf1p interacts with the α-tubulin monomer and the microtubule polymer via the same site on α-tubulin.
Phenotypes of ALF1 Overexpression
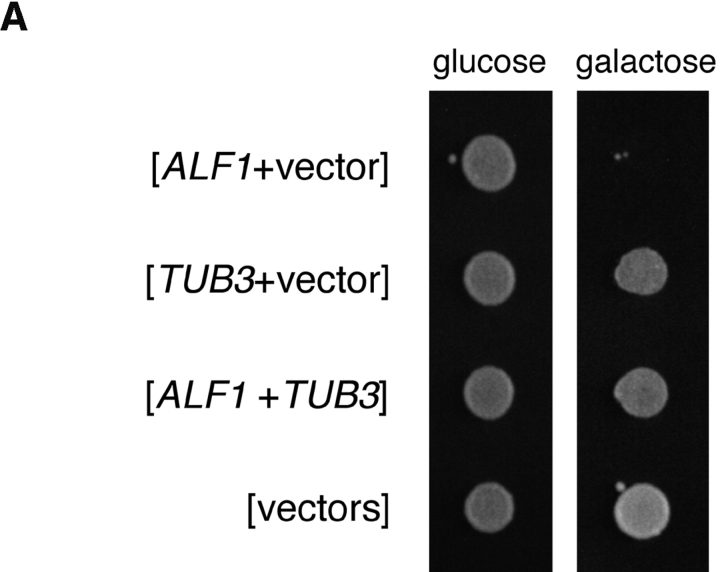
The association of Alf1p with α-tubulin monomer raises the possibility that Alf1p can act to sequester α-tubulin from interaction with β-tubulin. If this were the case, then overexpression of Alf1p might result in an imbalance in the ratio of α-tubulin to β-tubulin, which is detrimental to microtubule function in yeast (Burke et al., 1989; Katz et al., 1990; Weinstein and Solomon, 1990). To determine the effect of increased Alf1p levels, ALF1 was placed under the control of the inducible GAL1 promoter. Induction of PGAL1-ALF1 in wild-type cells had no effect on growth (Fig. 6 A). We next tested the phenotype of Alf1p overexpression in cin1 and rbl2 mutants, which function in the production of β-tubulin, with the rationale that compromising their function would exacerbate any effect of Alf1p overexpression. Induction of PGAL1-ALF1 in a rbl2 mutant strain caused a slight reduction growth rate (not shown), whereas induction in a cin1 mutant strain resulted in lethality (Fig. 6 A). This effect appears to be dependent upon the ability of Alf1p to interact with α-tubulin, since overexpression of alf1-1p is viable in a cin1Δ strain (not shown). If the lethality of overexpression of Alf1p were due to sequestration of α-tubulin, then it should be possible to overcome it by overexpressing α-tubulin with Alf1p. Consistent with this hypothesis, a cin1 strain expressing both PGAL1-TUB3 and PGAL1-ALF1 was viable (Fig. 6 A). We note that overexpression of Alf1p also results in association of Alf1p with tubulin heterodimer (see below), thus it is possible that the observed lethality is due to sequestration of the tubulin heterodimer rather than the α-tubulin monomer.
Figure 6.
Consequences of overexpressing Alf1p in cin1Δ cells. (A) Overexpression of ALF1 is lethal in cin1Δ cells. Cooverexpression of α-tubulin rescues the lethality of ALF1 overexpression in a cin1Δ strain. cin1Δ strains bearing the indicated plasmids were spotted onto selective media containing either 2% glucose or 2% galactose. The vectors were pTS210 (CEN; GAL1 promoter; URA3 marker) and pTS249 (CEN; GAL1 promoter; LEU2 marker). (B) cin1Δ cells overexpressing Alf1p exhibit a loss of microtubules. At time zero, cultures were resuspended at ∼2 × 106 cells/ml in selective medium containing 2% galactose. Samples were fixed at various time points by the addition of formaldehyde to 3.7%, and the fixed cells were stained with YOL1/34, an anti-α-tubulin mAb (the 9 h time point is shown).
ALF1 overexpression in cin1 cells caused a cell cycle arrest consistent with a defect in microtubule function. After 12 h of ALF1 induction, 75% of the cin1 cells had large buds, whereas cin1 cells with a control plasmid had a bud size distribution typical of exponentially growing cells (not shown). Large-budded arrest is typical of, but not specific to, defects in the microtubule cytoskeleton. To determine directly if ALF1 overexpression had an effect on the microtubule cytoskeleton in cin1 cells, microtubules were visualized by immunofluorescence at time points after ALF1 induction. After 9 h of ALF1 induction, virtually all of the cells lacked microtubule structures, although some retained dots of staining adjacent to the nucleus, presumably representing the spindle pole bodies (Fig. 6 B). In contrast, ∼95% of the cin1 cells bearing the control vector plasmid contained microtubules observable by immunofluorescence.
The amount of α-tubulin associated with Alf1p under overexpression conditions was determined by immunoprecipitation. A myc-tagged version of ALF1 was placed under control of the GAL1 promoter on a CEN-based plasmid. This protein was overexpressed in alf1 null cells and immunoprecipitated with the anti-myc antibody. α-Tubulin coimmunoprecipitated with Alf1p-myc, as expected. Although the total amount of α-tubulin in these cells was approximately the same as in wild-type cells, the amount of α-tubulin associated with the overexpressed Alf1p-myc was 12-fold greater than found with endogenous levels of Alf1p-myc (Fig. 7). In addition, β-tubulin coimmunoprecipitated with Alf1p-myc. This is most likely due to association of Alf1p-myc with the tubulin heterodimer, although we cannot rule out the possibility that high levels of Alf1p result in association with β-tubulin monomer, or in association with a complex containing the tubulins and other cofactors. The amount of β-tubulin that coimmunoprecipitated with Alf1p-myc was strictly dependent on the amount of overexpression. At the full level of induction (∼200-fold over wild-type), approximately twice as much α-tubulin than β-tubulin was precipitated, whereas at the reduced level of induction used in the localization experiments above, ∼20 times more α-tubulin than β-tubulin was precipitated (not shown). We interpret these results to mean that Alf1p preferentially binds to α-tubulin monomer, thus under conditions of overexpression, Alf1p will associate with the available α-tubulin monomer first, then with tubulin heterodimer as the amount of Alf1p increases.
Figure 7.
Overexpression of Alf1p results in the coimmunoprecipitation of both α- and β-tubulin. Alf1p-myc was expressed from the GAL1 promoter was immunoprecipitated from extracts with anti-myc mAb in the absence (myc) or presence (myc + peptide) of competing antigenic peptide. Precipitates were immunoblotted with either anti-myc, anti-α-tubulin, or anti-β-tubulin antibodies, as indicated on the left-hand side of the figure.
Genetic Interactions of alf1Δ with Tubulin and Cofactor Mutants
It was shown previously that alf1 null mutants are lethal in combination with tub1-1, a mutation in the major yeast α-tubulin gene (Stearns and Botstein, 1988; Tian et al., 1997). To further characterize alf1-tubulin genetic interactions, alf1 null strains were crossed to a set of tubulin mutants with a range of phenotypic defects. Among α-tubulin alleles, combination with alf1 null resulted in either lethality or increased sensitivity to benomyl in comparison to either of the parental single mutants (Table V). Most alf1Δ tub1 double mutants also exhibited increased sensitivity to cold (not shown), which, like benomyl sensitivity, is generally indicative of compromised microtubule function. Among β-tubulin alleles, combination with alf1 null resulted in increased sensitivity to benomyl with two of three tested alleles (Table V). An interesting exception to the above was the alf1Δ tub2-428 double mutant that was more resistant to benomyl than either parental single mutant. If Alf1p was required to fold, or otherwise aid in the biosynthesis, of α-tubulin, then this suppression of tub2-428 might be mimicked by a reduction in α-tubulin levels. One way of reducing α-tubulin levels is to delete the minor α-tubulin gene, TUB3 (Schatz et al., 1986). However, combination of tub3Δ with tub2-428 resulted in lethality rather than suppression (not shown).
Table V.
Synthetic Genetic Interactions with alf1Δ
| Strain | Benomyl Sensitivity | |||
|---|---|---|---|---|
| ALF1 | alf1Δ | |||
| μg/ml | ||||
| Wild-type | 15 | 5 | ||
| tub1-801 | 2 | SL | ||
| tub1-831 | 10 | 1 | ||
| tub1-839 | 30 | 2 | ||
| tub3Δ | 5 | 2 | ||
| tub2-428 | 2 | 10 | ||
| tub2-408 | 10 | 2 | ||
| tub2-442 | 80 | 20 | ||
| bik1Δ | 20 | 2 | ||
| cin1Δ | 1 | <1 | ||
| pac2Δ | 2 | 2 | ||
| rbl2Δ | 10 | 1 | ||
Double mutants were created by crosses. SL indicates that the double mutant is synthetically lethal. All strains were tested on plates containing either 0, 1, 2, 5, 10, 15, 20, 30, or 80 μg/ml of benomyl. The highest concentration which exhibited growth at 25°C is reported.
The alf1 null mutant was also tested for genetic interactions with the other yeast cofactor genes. As with the tubulin mutants, most of the double mutants were more sensitive to benomyl than either of the parental single mutants (Table V). An exception was the alf1Δ pac2Δ double mutant, which exhibited the same sensitivity to benomyl as the pac2Δ single mutant. This result is consistent with the biochemical determination that cofactor B/Alf1p and cofactor E/Pac2p act in the same pathway (Tian et al., 1997). To see whether the β-tubulin cofactors displayed a similar epistasis relationship, we crossed cin1 and rbl2 mutant strains. The putative double mutants from this cross were inviable; occasional spore microcolonies that did appear could not be propagated.
Discussion
Formation of the α- and β-tubulin heterodimer requires the action of several proteins and protein complexes in vitro. We have tested the in vivo function of one of these proteins, Alf1p, the yeast ortholog of cofactor B. Alf1p binds to α-tubulin, and can localize to microtubules in vivo. The interaction with α-tubulin monomer and microtubules is dependent upon both the single CLIP-170 microtubule-binding domain of Alf1p and a specific domain of α-tubulin. ALF1 displays genetic interactions with the tubulin genes and other genes involved in microtubule function. Here we interpret ALF1 function in the context of tubulin biogenesis and regulation.
Physical Interactions of Alf1p
We examined the interactions between several of the yeast cofactors and yeast tubulin, as well as potential interactions between the cofactors themselves. First, in a screen against the entire yeast genome, the two yeast α-tubulin genes, TUB1 and TUB3, were identified as interacting with ALF1. We confirmed this interaction by demonstrating that α-tubulin immunoprecipitates with Alf1p. Similarly, cofactor B has been shown biochemically to interact with α-tubulin (Tian et al., 1996; 1997). In the same two-hybrid screen, CCT5 was identified as interacting with ALF1. Cct5p is one of the eight Cct protein subunits of the chaperonin complex. It is proposed that the tubulin proteins interact with this complex before interacting with cofactors A-E (Gao et al., 1993), and cofactor B/Alf1p is thought to be the first protein to bind α-tubulin after the chaperonin complex. Thus this interaction provides a potential mechanism for transfer of the monomeric α-tubulin to the cofactor pathway.
Another important result from the two-hybrid analysis is the lack of detectable interaction between Alf1p and the other yeast cofactors in direct pairwise tests. Alf1p did not interact with either Cin1p or Pac2p; similarly, Cin1p and Pac2p did not interact. Although there are many possible reasons for the lack of an interaction in the yeast two-hybrid system, each of the fusion proteins involved was independently able to interact with other proteins. The lack of interactions among the cofactors is of interest because it leaves open the questions of how the monomeric tubulin subunits are transferred from one protein to another, and of how they are ultimately joined to form the heterodimer.
Finally, the results we present here, in combination with the work of others, show that the interactions of the yeast cofactors with the tubulin proteins are the same as those of the mammalian cofactors. Alf1p and Pac2p associate with α-tubulin, and not β-tubulin (see Fig. 1 B; Vega et al., 1998), whereas Rbl2p binds to β-tubulin, and not α-tubulin (Archer et al., 1995, 1998). We have also found that Cin1p interacts with β-tubulin, and not α-tubulin (Feierbach, B., and T. Stearns, unpublished results) The similarity of these interactions with those of the mammalian proteins suggests that similar pathways are operating in both yeast and mammalian cells.
Nature of the Alf1p–α-Tubulin Interaction
The above evidence indicates that Alf1p binds to monomeric α-tubulin. We have identified the domains required for this interaction on both Alf1p and α-tubulin. Alf1p contains a single copy of the CLIP-170 domain, and mutation of this domain abolishes interaction with α-tubulin. Cofactor E/Pac2p also contains a CLIP-170 domain, and also binds to α-tubulin (Table III; Tian et al., 1996, 1998). Since both Pac2p and Alf1p specifically interact with α-tubulin, it seems likely that the CLIP-170 domain specifically recognizes α-tubulin. In addition, other proteins that have a CLIP-170 domain, such as CLIP-170 and Bik1p, are known to bind to the microtubule polymer and it is possible that these proteins are interacting with the polymer by specifically interacting with α-tubulin. We note that the known polymer-binding CLIP-170 proteins have not, to our knowledge, been tested for interaction with monomeric tubulin, and it remains possible that they are similar to Alf1p in their preference for monomer vs. polymer.
We used a panel of defined α-tubulin mutants to map the Alf1p binding site on α-tubulin. The approach used was similar to that previously used for actin and actin binding proteins (Wertman et al., 1992; Holtzman et al., 1994; Amberg et al., 1995), and was made possible by the availability of a complete set of clustered charged-to-alanine mutations in the yeast TUB1 α-tubulin gene (Richards, 1997; Schwartz et al., 1997). We reasoned that α-tubulin mutations that disrupted the interaction with Alf1p in the two-hybrid assay would define the side chains making up the interacting surface. By modeling the yeast TUB1 α-tubulin sequence onto the mammalian α-tubulin three-dimensional structure, we determined that the mutations that disrupt the interaction fall, for the most part, on one face of the α-tubulin three-dimensional structure. This face would be exposed to the cytoplasm in the proposed structure of the microtubule (Nogales et al., 1998), and is different from that determined for the binding of β-tubulin in either the heterodimer or the microtubule (Nogales et al., 1998). Thus, this domain would be accessible to both monomer and polymer binding proteins, consistent with the ability of CLIP-170 domain proteins in general to bind to the polymer. Given the proposed sequential action of Alf1p and Pac2p, it will be interesting to determine whether Pac2p recognizes the same site on α-tubulin.
Although Alf1p interacts only with α-tubulin monomer under normal conditions, we found that overexpression of Alf1p results in the coimmunoprecipitation of both α-tubulin and β-tubulin. This experiment was done under conditions of both mild (about fivefold, not shown) and extreme (200-fold; Fig. 7) overexpression; in both cases there was detectable association with β-tubulin, although much less relative to α-tubulin under mild Alf1p overexpression conditions. This result could be either direct, by interaction of Alf1p with β-tubulin, or indirect by interaction of Alf1p with α-tubulin in the heterodimer. The latter seems most likely given the other evidence for the specificity of the Alf1p-α-tubulin interaction, and that the Alf1p binding site on α-tubulin would be accessible in both the monomer and heterodimer.
The Yeast Cofactor Pathway
Tian et al. (1997) proposed a model of the cofactor pathway in which the tubulin monomers interact sequentially with the cofactors after release from the cytosolic chaperonin complex (c-cpn/TriC/Cct complex). In this model, cofactors B/Alf1p and E/Pac2p interact with α-tubulin monomer, and cofactors A/Rbl2p and D/Cin1p interact with β-tubulin to promote their assembly into the heterodimer. The interactions described here between the yeast cofactors and specific tubulin monomers are entirely consistent with this model. The alf1Δ pac2Δ double mutant is viable and the phenotype is no more severe than the pac2Δ single mutant. In contrast, the phenotypes of the alf1Δ cin1Δ and alf1Δ rbl2Δ double mutants are more severe than either of the single mutants. These results are consistent with Alf1p and Pac2p acting in the same pathway and Cin1p and Rbl2p acting in a separate pathway. However, other genetic results suggest two modifications to the original model: (a) because α-tubulin is essential for viability, and an alf1Δ pac2Δ double mutant is viable, there must be another path to functional α-tubulin; (b) because a cin1Δ rbl2Δ strain is inviable whereas the single mutants are viable, there are likely to be two independent pathways to functional β-tubulin, one dependent on Cin1p, and the other on Rbl2p. Both of these changes are incorporated in a revised model of the yeast cofactor pathway, shown in Fig. 8.
Alf1p Function
The mammalian tubulin cofactor proteins have been proposed to be tubulin monomer binding proteins that facilitate the formation of the tubulin heterodimer (Tian et al., 1997). Our results with ALF1 are consistent with this hypothesis: Alf1p binds to monomeric α-tubulin, the alf1 null mutant is supersensitive to benomyl, as expected for a reduction in the amount of functional tubulin heterodimer, and the ALF1 overexpression phenotype suggests that Alf1p can sequester α-tubulin monomer. One of the issues regarding cofactor function is whether the proteins are required for the folding of the tubulin monomers, or acting as assembly factors, bringing together two fully folded tubulin monomers to make the heterodimer. Several of our results bear on this issue. First, the level of α-tubulin protein is the same in alf1 and wild-type cells. If there were significant amounts of partially folded α-tubulin, one might expect to observe decreased α-tubulin levels due to protein turnover. Similarly, the alf1Δ phenotype with respect to genetic interactions with tub2-428 is distinct from that of deletion of the minor α-tubulin gene TUB3, suggesting that the afl1Δ defect is not equivalent to a reduction of α-tubulin levels. Second, Alf1p can bind to microtubule polymer, which must represent native tubulin. In addition, overexpressed Alf1p can immunoprecipitate both α- and β-tubulin; this seems most likely to be due to association with the heterodimer, again suggesting that Alf1p can interact with native α-tubulin. The ability to associate with native tubulin might be common to all of the cofactors. For example, cofactors B and C (no known homologue in yeast) and cofactor E/Pac2p have been shown to behave as microtubule-associated proteins in vitro (Tian et al., 1996), and the cofactor D/Cin1p homologue in Schizosaccharomyces pombe, alp1 +, also localizes to microtubules in vivo (Hirata et al., 1998).
Acknowledgments
We thank Kristy Richards, Katja Schwartz, Jon Mulholland, and Kirk Anders of the Botstein lab for freely sharing strains, plasmids, antibodies and unpublished results. We thank Jan Carminati, Laura Marschall, and Kristy Richards for useful discussions on the manuscript. We also thank Nick Cowan for first identifying ALF1, and David Pellman for helpful advice on CLIP-170 domain mutations.
This work was supported by grants from the American Cancer Society and the Searle Scholars Program to T. Stearns. B. Feierbach was supported by a Cell and Molecular Biology NIH Predoctoral Training Grant (5T32 GM07276-22).
Abbreviations used in this paper
- c-cpn
cytosolic chaperonin
- GFP
green fluorescent protein
- SD
synthetic dextrose
- YEP
yeast extract/peptone
- SD
synthetic dextrose
Footnotes
Address correspondence to Tim Stearns, Department of Biological Sciences, Stanford University, Stanford, CA 94305-5020. Tel.: (650) 725-6934. Fax: (650) 725-8309. E-mail: stearns@stanford.edu
References
- Adams, A., D.E. Gottschling, C.A. Kaiser, and T. Stearns. 1997. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press. 177.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amberg DC, Basart E, Botstein D. Defining protein interactions with yeast actin in vivo. Nat Struct Biol. 1995;2:28–35. doi: 10.1038/nsb0195-28. [DOI] [PubMed] [Google Scholar]
- Anfinsen DC. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Archer JE, Magendantz M, Vega LR, Solomon F. Formation and function of the Rbl2p-beta-tubulin complex. Mol Cell Biol. 1998;18:1757–1762. doi: 10.1128/mcb.18.3.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer JE, Vega LR, Solomon F. Rbl2p, a yeast protein that binds to beta-tubulin and participates in microtubule function in vivo. Cell. 1995;82:425–434. doi: 10.1016/0092-8674(95)90431-x. [DOI] [PubMed] [Google Scholar]
- Berlin V, Styles CA, Fink GR. BIK1, a protein required for microtubule function during mating and mitosis in Saccharomyces cerevisiae, colocalizes with tubulin. J Cell Biol. 1990;111:2573–2586. doi: 10.1083/jcb.111.6.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D, Gasdaska P, Hartwell L. Dominant effects of tubulin overexpression in Saccharomyces cerevisiae. . Mol Cell Biol. 1989;9:1049–1059. doi: 10.1128/mcb.9.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. . J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Valdiria RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A, Lee W-H, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Mulligan JT, Ramer SW, Spottswood M, Davis RW. λYES: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia colimutations. Proc Natl Acad Sci USA. 1991;88:1731–1735. doi: 10.1073/pnas.88.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO (Eur Mol Biol Organ) J. 1992;11:4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Thomas JO, Chow RL, Lee GH, Cowan NJ. A cytoplasmic chaperonin that catalyzes beta-actin folding. Cell. 1992;69:1043–1050. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
- Gao Y, Vainberg IE, Chow RL, Cowan NJ. Two cofactors and cytoplasmic chaperonin are required for the folding of alpha- and beta-tubulin. Mol Cell Biol. 1993;13:2478–2485. doi: 10.1128/mcb.13.4.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, Schott EJ, Kingsbury TJ, Cole NB, Totis LJ, Bhattacharya G, He L, Hoyt MA. Saccharomyces cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol Biol Cell. 1997;8:1035–1050. doi: 10.1091/mbc.8.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrick JP, Hartl FU. The role of molecular chaperones in protein folding. FASEB J. 1995;9:1559–1569. doi: 10.1096/fasebj.9.15.8529835. [DOI] [PubMed] [Google Scholar]
- Hirata D, Masuda H, Eddison M, Toda T. Essential role of tubulin-folding cofactor D in microtubule assembly and its association with microtubules in fission yeast. EMBO (Eur Mol Biol Organ) J. 1998;17:658–666. doi: 10.1093/emboj/17.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DA, Wertman KF, Drubin DG. Mapping actin surfaces required for functional interactions in vivo. J Cell Biol. 1994;126:423–432. doi: 10.1083/jcb.126.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Macke JP, Roberts BT, Geiser JR. Saccharomyces cerevisiaePAC2 functions with CIN1, 2 and 4 in a pathway leading to normal microtubule stability. Genetics. 1997;146:849–857. doi: 10.1093/genetics/146.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Stearns T, Botstein D. Chromosome instability mutants of Saccharomyces cerevisiaethat are defective in microtubule-mediated processes. Mol Cell Biol. 1990;10:223–234. doi: 10.1128/mcb.10.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser CA, Preuss D, Grisafi P, Botstein D. Many random sequences functionally replace the secretion signal sequence of yeast invertase. Science. 1987;235:312–317. doi: 10.1126/science.3541205. [DOI] [PubMed] [Google Scholar]
- Katz W, Weinstein B, Solomon F. Regulation of tubulin levels and microtubule assembly in Saccharomyces cerevisiae: consequences of altered tubulin gene copy number. Mol Cell Biol. 1990;10:5286–5294. doi: 10.1128/mcb.10.10.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J, Wright B, Milstein C. Rat monoclonal anti-tubulin antibodies derived by using a new nonsecreting rat cell line. J Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall L, Jeng R, Mulholland J, Stearns T. Analysis of Tub4p, a yeast γ-tubulin-like protein: implications for microtubule organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melki R, Vainberg IE, Chow RL, Cowan NJ. Chaperonin-mediated folding of vertebrate actin-related protein and gamma-tubulin. J Cell Biol. 1993;122:1301–1310. doi: 10.1083/jcb.122.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SM, Urbani L, Stearns T. The mammalian gamma-tubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J Cell Biol. 1998;141:663–674. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Pierre P, Pepperkok R, Kreis TE. Molecular characterization of two functional domains of CLIP-170 in vivo. J Cell Sci. 1994;107:1909–1920. doi: 10.1242/jcs.107.7.1909. [DOI] [PubMed] [Google Scholar]
- Pierre P, Scheel J, Rickard JE, Kreis TE. CLIP-170 links endocytic vesicles to microtubules. Cell. 1992;70:887–900. doi: 10.1016/0092-8674(92)90240-d. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AEM, Stearns T, Drubin DG, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Richards, K. 1997. Genetic analysis of the TUB1 gene in Saccharomyces cerevisiae. Ph.D. Thesis, Stanford University.
- Schatz PJ, Pillus L, Grisafi P, Solomon F, Botstein D. Two functional α-tubulin genes of the yeast Saccharomyces cerevisiaeencode divergent proteins. Mol Cell Biol. 1986;6:3711–3721. doi: 10.1128/mcb.6.11.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz PJ, Solomon F, Botstein D. Genetically essential and non-essential α-tubulin genes specify functionally interchangeable proteins. Mol Cell Biol. 1986;6:3722–3733. doi: 10.1128/mcb.6.11.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz K, Richards K, Botstein D. BIM1 encodes a microtubule-binding protein in yeast. Mol Biol Cell. 1997;8:2677–2691. doi: 10.1091/mbc.8.12.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Botstein D. Unlinked noncomplementation: isolation of new conditional-lethal mutations in each of the tubulin genes of Saccharomyces cerevisiae. . Genetics. 1988;119:249–260. doi: 10.1093/genetics/119.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Hoyt MA, Botstein D. Yeast mutants sensitive to antimicrotubule drugs define three genes that affect microtubule function. Genetics. 1990;124:251–262. doi: 10.1093/genetics/124.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht HG, Farr GW, Sterlicht M, Driscoll KJ, Willison K, Yaffe M. The t-complex polypeptide 1 complex is a chaperonin for tubulin and actin in vivo. . Proc Natl Acad Sci USA. 1993;90:9422–9426. doi: 10.1073/pnas.90.20.9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoldt V, Rademacher F, Kehren V, Ernst JF, Pearce DA, Sherman F. Review: the Cct eukaryotic chaperonin subunits of Saccharomyces cerevisiaeand other yeasts. Yeast. 1996;12:523–529. doi: 10.1002/(SICI)1097-0061(199605)12:6%3C523::AID-YEA962%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Tian G, Huang Y, Rommelaere H, Vandekerckhove J, Ampe C, Cowan NJ. Pathway leading to correctly folded beta-tubulin. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- Tian G, Lewis SA, Feierbach B, Stearns T, Rommelaere H, Ampe C, Cowan NJ. Tubulin subunits exist in an activated conformational state generated and maintained by protein cofactors. J Cell Biol. 1997;138:821–832. doi: 10.1083/jcb.138.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega LR, Fleming J, Solomon F. An alpha-tubulin mutant destabilizes the heterodimer: phenotypic consequences and interactions with tubulin-binding proteins. Mol Biol Cell. 1998;9:2349–2360. doi: 10.1091/mbc.9.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein B, Solomon F. Phenotypic consequences of tubulin overproduction in Saccharomyces cerevisiae: differences between alpha-tubulin and beta-tubulin. Mol Cell Biol. 1990;10:5295–5304. doi: 10.1128/mcb.10.10.5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertman KF, Drubin DG, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiaemitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Farr GW, Miklos D, Horwich AL, Sternlicht ML, Sternlicht H. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature. 1992;358:245–248. doi: 10.1038/358245a0. [DOI] [PubMed] [Google Scholar]