Abstract
To identify septin-interacting proteins in Saccharomyces cerevisiae, we screened for mutations that are synthetically lethal with a cdc12 septin mutation. One of the genes identified was GIN4, which encodes a protein kinase related to Hsl1p/Nik1p and Ycl024Wp in S. cerevisiae and to Nim1p/Cdr1p and Cdr2p in Schizosaccharomyces pombe. The Gin4p kinase domain displayed a two-hybrid interaction with the COOH-terminal portion of the Cdc3p septin, and Gin4p colocalized with the septins at the mother–bud neck. This localization depended on the septins and on the COOH-terminal (nonkinase) region of Gin4p, and overproduction of this COOH-terminal region led to a loss of septin organization and associated morphogenetic defects. We detected no effect of deleting YCL024W, either alone or in combination with deletion of GIN4. Deletion of GIN4 was not lethal but led to a striking reorganization of the septins accompanied by morphogenetic abnormalities and a defect in cell separation; however, remarkably, cytokinesis appeared to occur efficiently. Two other proteins that localize to the neck in a septin-dependent manner showed similar reorganizations and also appeared to remain largely functional. The septin organization observed in gin4Δ vegetative cells resembles that seen normally in cells responding to mating pheromone, and no Gin4p was detected in association with the septins in such cells. The organization of the septins observed in gin4Δ cells and in cells responding to pheromone appears to support some aspects of the model for septin organization suggested previously by Field et al. (Field, C.M., O. Al-Awar, J. Rosenblatt, M.L. Wong, B. Alberts, and T.J. Mitchison. 1996. J. Cell Biol. 133:605–616).
Keywords: cytokinesis, Gin4p, protein kinase, Saccharomyces cerevisiae, septins
The septins are a family of proteins that were first identified in the budding yeast Saccharomyces cerevisiae but have since been identified also in a variety of other fungi and animals (for review see Cooper and Kiehart, 1996; Longtine et al., 1996; Longtine and Pringle, 1998). The septins localize to the cleavage site and are essential for cytokinesis in both yeast and animal cells, but their precise role(s) in this process are unknown. In addition, it appears that the septins also have roles in processes other than cytokinesis. For example, in Drosophila, septins are highly concentrated in the basolateral surfaces of ovarian follicle cells and in neurons of the embryonic central nervous system (Neufeld and Rubin, 1994; Fares et al., 1995); there are as yet no good clues to the roles of the septins in these polarized, nondividing cells. In S. cerevisiae, two of the seven septins are expressed only in sporulating cells and appear (from protein-localization data and mutant phenotypes) to be involved in the poorly understood process by which the meiotic nuclei are packaged into spores (De Virgilio et al., 1996; Fares et al., 1996).
At least some septin functions appear to involve a role in organizing other proteins at the cell surface. For example, the septins are involved in the selection of axial budding sites in haploid cells (Flescher et al., 1993; Chant et al., 1995); this function reflects the role of the septins in the localization of Bud3p and Bud4p, two proteins that are constituents of a transient mark at the division site that serves to direct the next budding event to an adjacent location (Chant et al., 1995; Sanders and Herskowitz, 1996). In addition, the septins are necessary for the proper localization of Chs3p (the catalytic subunit of chitin synthase III), and hence for the normal localization of chitin deposition to a ring at the base of the bud (DeMarini et al., 1997). Localization of Chs3p involves at least two other proteins, Chs4p, which appears to interact directly with Chs3p, and Bni4p, which appears to interact directly both with Chs4p and with one of the septins.
Another interesting class of questions about the septins concerns the relationship between the function of these proteins and their assembly into higher-order structures. In S. cerevisiae, the vegetatively expressed septins appear to be constituents of a set of filaments found on the cytoplasmic face of the plasma membrane in the mother–bud neck (Byers and Goetsch, 1976; Byers, 1981; Longtine et al., 1996; Frazier et al., 1998). However, the relationship between the function of the septins and their assembly into these filaments remains obscure, as do the mechanisms controlling the assembly and disassembly of the filaments. In addition, although some progress has been made in analyzing septin complexes (including short filaments) isolated from Drosophila (Field et al., 1996), it is not yet clear whether the septins of Drosophila or other organisms are organized in vivo into filaments like those seen in S. cerevisiae.
In this paper, we report the results of a genetic screen in S. cerevisiae that identified a protein kinase, Gin4p, that appears to coassemble with the septins at the mother–bud neck and play an important role in their assembly. Interestingly, analysis of gin4 mutant strains suggests that the septins and septin-associated proteins can remain largely functional after a seemingly drastic change in their organization.
Materials and Methods
Strains, Growth Conditions, and Genetic and DNA Methods
The S. cerevisiae strains used in this study are described in Table I. Yeast media (YM-P rich liquid medium, YPD rich solid medium, synthetic minimal [SD]1 medium, synthetic complete [SC] medium lacking specific nutrients, and sporulation medium) have been described previously (Lillie and Pringle, 1980; Guthrie and Fink, 1991). S. cerevisiae strains were grown at 30°C except where noted. Cultures were treated with α mating pheromone (Sigma Chemical Co., St. Louis, MO) as described by Evangelista et al. (1997). Escherichia coli strain DH12S (Life Technologies, Gaithersburg, MD) and standard media and methods (Ausubel et al., 1995) were used for plasmid manipulations. Transformation of yeast was performed as described by Gietz et al. (1992), and other yeast genetic manipulations used standard procedures (Guthrie and Fink, 1991).
Table I.
S. cerevisiae Strains Used in This Study
| Name | Relevant genotype* | Source | ||
|---|---|---|---|---|
| JFY54A | α cdc12-5 “sl54” ade2 ade3 his7 leu2 ura3 [pB12G] | This study‡ | ||
| M-100 | α cdc12-5 “sl54” ade2 ade3 his7 leu2 ura3 [YEp24(CDC12)N] | This study‡ | ||
| DL454 | a mpk1Δ::TRP1 can1r his4 leu2-3,112 ura3-52 | Lee et al., 1993 | ||
| EGY48J | a his3 leu2::pLEU2-lexAop6 trp1 ura3-52 [pJK103] | This study§ | ||
| YEF473 | a/α his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 lys2-801/lys2-801 trp1-Δ63/trp1-Δ63 ura3-52/ura3-52 | Bi and Pringle, 1996 | ||
| YEF473A | a his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 | Segregant from YEF473 | ||
| YEF473B | α his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 | Segregant from YEF473 | ||
| SY2625 | a sst1Δ ade2-1 can1-100 his3Δ::FUS1-HIS3 leu2-3,112 mfa2Δ::FUS1-lacZ trp1-1 ura3-1 | Adames et al., 1995 | ||
| M-238 | α cdc12-6 his3 leu2 trp1-Δ63 ura3 | This study‖ | ||
| M-261 | as YEF473 except GIN4/gin4-Δ9 | This study ¶ | ||
| M-267 | as YEF473A except gin4-Δ9 | Segregant from M-261 | ||
| M-272 | as YEF473 except gin4-Δ9/gin4-Δ9 | This study** | ||
| M-283 | as YEF473 except YCL024W/ycl024W-Δ2 | This study ¶ | ||
| M-286 | as YEF473 except ycl024W-Δ2/ycl024W-Δ2 | This study** | ||
| M-292 | as YEF473 except GIN4/gin4-Δ9 YCL024W/ycl024W-Δ2 | This study** | ||
| M-326 | a cdc12-6 gin4-Δ9 his3 leu2 lys2-801 trp1-Δ63 ura3 | This study†† | ||
| M-441 | as YEF473A except gin4-Δ9 ura3::URA3:GIN4-GST | This study§§ | ||
| M-450 | a cdc12-6 gin4-Δ9 ura3::URA3:GIN4-GST his3 leu2 lys2-801 trp1-Δ63 ura3 | This study‖‖ | ||
| M-460 | as YEF473 except gin4-Δ9/gin4-Δ9 ycl024W-Δ2/ycl024W-Δ2 | This study ¶¶ | ||
| M-623 | as YEF473 except GIN4/GIN4-3HA:kan | This study** | ||
| M-692 | as YEF473 except gin4K48A-3HA:kan/gin4K48A-3HA:kan | This study†† ‡ |
Plasmids are indicated in brackets and are described in Table II.
“sl54” refers to the mutations producing synthetic lethality in combination with cdc12-5 at 23°C. Strain JFY54A is a segregant from a cross of the original synthetic-lethal mutant to strain JF1.2 Strain M-100 was derived from JFY54A as described in the text.
Strain EGY48 (Zervos et al., 1993) containing lacZ-reporter plasmid pJK103 (Estojak et al., 1995).
Derived by several crosses from a segregant from strain JPTA1493-HO1 (Adams and Pringle, 1984).
YEF473 transformed with the appropriate PCR product (see Materials and Methods). For gin4-Δ9, the selectable marker was TRP1; for ycl024W-Δ2, the selectable marker was HIS3.
Constructed by mating segregants from M-261 and/or M-283.
Segregant from a cross of M-238 with a segregant from M-261.
Derived by transforming a segregant from M-261 with YIp/GIN4-GST (see Materials and Methods).
Segregant from a cross of M-238 to M-441.
Constructed by mating segregants from M-292.
YEF473 transformed with the PCR product obtained using primers ML136 and ML137 (Table III) and plasmid pFA6a-3HA-kanMX6 (Longtine et al., 1998) as template. Correct integration of the 3HA-kan DNA cassette at the 3′ end of GIN4 was confirmed by PCR using genomic DNA from strain M-623 as template and primers ML136 and MLP54-5.
‡‡
‡ a and α GIN4-3HA:kan segregants were obtained from M-623, and GIN4 was replaced by gin4K48A (from plasmid pDK49) in each strain as described by Altman and Kellogg (1997). The resulting strains were mated to generate strain M-692.
Standard methods (Ausubel et al., 1995) were used for DNA manipulations except where noted. DNA–DNA blot hybridization was performed as described previously (Ausubel et al., 1995) and site-specific mutagenesis was carried out using the pALTER system as described by the manufacturer (Promega, Madison, WI). The oligonucleotides used in this study are described in Table III and were obtained from Integrated DNA Technologies (Coralville, IA). Vent DNA polymerase (New England Biolabs, Beverly, MA) was used for the PCR synthesis of DNA fragments for cloning; Taq DNA polymerase (Promega) was used in other PCR applications. Primer pairs ML38 and ML39 and ML40 and ML41 were used to generate DNA cassettes for replacement of GIN4 and YCL024W with TRP1 and HIS3, respectively, using the PCR method of Baudin et al. (1993) and plasmids pRS304 and pRS303 (Sikorski and Hieter, 1989) as templates. The success of the gene replacements was verified by PCR using yeast genomic DNAs as templates, primers ML38 and MLP54-5 for GIN4, and two different pairs of primers (ML18 and ML41; ML195 and ML196) for YCL024W.
Table III.
Oligonucleotides Used in This Study
| Name | Sequence | |
|---|---|---|
| ML38* | 5′-ATCAAGAAAGATATTCGCAGCACAATACAATAATAACATTCCTCCTTACGCATCTGTGCGG-3′ | |
| ML39* | 5′-CAAAACGAAGGAGACAAAACATGATTGCATTACATTAGCAGTACAATCTTGATCCGGAGC-3′ | |
| MLP54-5‡ | 5′-GGTAAATATAATAGTACCTCGAGTCCAGTACCTCC-3′ | |
| ML40* | 5′-TGATAATAGTGCTGACGTAAATTACCAGAACTACTGCAGTGCCTCGTTCAGAATGACACG-3′ | |
| ML41* | 5′-GTGTGGGCTCTGCGCTTTTTGATATTGGAGATTGGTTTGGCTCTTGGCCTCCTCTAGTAC-3′ | |
| ML18‡ | 5′-GAGTGCGACCTCAAGTCATCAAAAGGC-3′ | |
| ML195‡ | 5′-AGAAAACGCGTAACGCAGTCTTAG-3′ | |
| ML196‡ | 5′-TCAGGATTTGCGCCTTTGGATGAG-3′ | |
| ML62§ | 5′-GGACAAGAGGCGGCAGTTATGGTAATATCAAAAGCAGTA-3′ | |
| ML63‖ | 5′-GAAGGCGTTCTACAAAAAGGCGGCCGCTAGTGCTAATGTAATGCA-3′ | |
| ML67‖ | 5′-AAAACTCGAGACTTGGATATTTTTGTAATA-3′ | |
| ML69‖ | 5′-AAAACCATGGATAAGAGATTCTAAAAGT-3′ | |
| ML70‖ | 5′-AAAACTCGAGCTATTTTTGTAGAACGCCT-3′ | |
| ML72‖ | 5′-AAAAGAATTCATGGCAATCAATGGTAACAG-3′ | |
| ML73‖ | 5′-AAAAAGATCTATGGCAATCAATGGTAACAG-3′ | |
| ML102‖ | 5′-AAAAAAAGGATCCGCACATCCAAAGATTAAATCG-3′ | |
| ML103‖ | 5′-AAAAAAGAATTCAAGCTTAGCACTATTTTTGTAGAAC-3′ | |
| ML136 ¶ | 5′-AGTTGAGAACGTCCTGAATAAGGAAGGCGTTCTACAAAAACGGATCCCCGGGTTAATTAA-3′ | |
| ML137 ¶ | 5′-AAATATTATGGCAGAACAAAACGAAGGAGACAAAACATGAGAATTCGAGCTCGTTTAAAC-3′ |
Primers used to generate deletion cassettes for replacement of GIN4 (ML38 and ML39) and YCL024W (ML40 and ML41) with TRP1 and HIS3, respectively. Underlined nucleotides correspond to those flanking the gene to be deleted; the remaining nucleotides correspond to regions upstream and downstream of the selectable marker genes.
Primers used to verify deletion of GIN4 (MLP54-5) and of YCL024W (ML18, ML195, and ML196) (see Materials and Methods).
Primer containing altered nucleotide (double underline) used to alter codon 48 (AAG to ATG) to create the gin4K48M mutation.
Primers used in plasmid constructions (see Materials and Methods). Underlining indicates restriction enzyme sites incorporated into the primers: GCGGCCGC, NotI; CTCGAG, XhoI; CCATGG, NcoI; GAATTC, EcoRI; AGATCT, BglII; GGATCC, BamHI; AAGCTT, HindIII.
Primers used to generate a cassette for tagging the 3′ end of GIN4 with 3HA:kan (see Table I, strain M-623). Underlined nucleotides correspond to those just upstream of the GIN4 stop codon (ML136) and downstream of the GIN4 ORF (ML137).
Plasmid Constructions
Most plasmids used in this study are listed in Table II; others are described where appropriate below. YCpGALGST was constructed by inserting an ∼1.8-kb StuI/HindIII fragment containing the GAL1/10 promoter from pEG(KT) (Mitchell et al., 1993) into SmaI/HindIII-digested YCplac111. YCp/CDC12 was constructed by subcloning an ∼2-kb XbaI/ PstI fragment containing CDC12 from YEp24(CDC12)N into XbaI/PstI-digested YCplac111. p54-6 was a primary isolate from the plasmid library that contained MPK1/SLT2, and p54-15 was a primary isolate from the plasmid library that contained GIN4 (see below). YCp/GIN4, YEp/GIN4, and pALTER/GIN4 were constructed by subcloning an ∼7-kb XbaI fragment from p54-15 into XbaI-digested YCplac111, YEplac181, and pALTER, respectively.
Table II.
Plasmids Used in This Study
| Name | Description* | Source | ||
|---|---|---|---|---|
| YCplac111 | CEN, LEU2 | Gietz and Sugino, 1988 | ||
| YEplac181 | 2 μ, LEU2 | Gietz and Sugino, 1988 | ||
| YIplac204 | URA3 | Gietz and Sugino, 1988 | ||
| pEG202 | 2 μ, HIS3, lexA DNA-binding domain (DBD) | Ausubel et al., 1995 | ||
| pJG4-5PL | 2 μ, TRP1, transcriptional activation domain (AD) | DeMarini et al., 1997 | ||
| pB12G | 2 μ, LEU2, CDC12, ADE3 | Fares et al.2 | ||
| YEp24(CDC12)N | 2 μ, URA3, CDC12 | Fares et al.2 | ||
| YCp50[MPK1] | CEN, LEU2, MPK1 | Lee et al., 1993 | ||
| YCpGALGST | CEN, LEU2, GST under GAL1/10 promoter control | See text | ||
| YCp/CDC12 | CEN, LEU2, CDC12 | See text | ||
| P54-6 | CEN, LEU2, ∼12 kb of chromosome VIII DNA including MPK1 (SLT2) | See text | ||
| p54-15 | CEN, LEU2, ∼12 kb of chromosome IV DNA including GIN4 | See text | ||
| YCp/GIN4 | CEN, LEU2, GIN4 | See text and Fig. 1 | ||
| YEp/GIN4 | 2 μ, LEU2, GIN4 | See text | ||
| pDK49 | URA3, gin4K48A | Altman and Kellogg, 1997 | ||
| pDK50 | CEN, URA3, gin4K48A | D. Kellogg, University of California, Santa Cruz, CA | ||
| YCp/gin4K48M | CEN, LEU2, gin4K48M | See text | ||
| YEp/gin4K48M | 2 μ, LEU2, gin4K48M | See text | ||
| YCp/GIN4-3HA | CEN, LEU2, GIN4-3HA | See text | ||
| YCp/gin4K48M-3HA | CEN, LEU2, gin4K48M-3HA | See text | ||
| YIp/GIN4-GST | URA3, GIN4-GST | See text | ||
| YCpGALGST/GIN4N | CEN, LEU2, GAL1/10-GST-GIN4 (codons 1–315) | See text | ||
| YCpGALGST/GIN4C | CEN, LEU2, GAL1/10-GST-GIN4 (codons 295–1,142) | See text | ||
| pEG202/GIN4N | 2 μ, HIS3, DBD-GIN4 (codons 1–294) | See text | ||
| pEG202/GIN4C | 2 μ, HIS3, DBD-GIN4 (codons 295–1,142) | See text | ||
| pEG202/GIN4NFL | 2 μ, HIS3, DBD-GIN4 (codons 18–1,142) | See text |
CEN plasmids are low copy; 2 μ plasmids are high copy; other plasmids do not have a yeast replication origin and can transform only by chromosomal integration.
To construct plasmids containing tagged or mutated alleles of GIN4, single-stranded DNA from plasmid pALTER/GIN4 was used with primer ML62 (which mutates the lysine codon at position 48 to a methionine codon), primer ML63 (which introduces a NotI site just upstream of the GIN4 stop codon), or both, to create plasmids pALTER/gin4K48M, pALTER/ GIN4NotI, and pALTER/gin4K48MNotI. NotI fragments containing sequences encoding either a triple hemagglutinin tag (3HA) (Tyers et al., 1993) or glutathione-S-transferase (GST) (provided by D. DeMarini, University of North Carolina, Chapel Hill, NC) were cloned into NotI-digested pALTER/GIN4NotI and pALTER/gin4K48MNotI to yield plasmids pALTER/GIN4-3HA, pALTER/GIN4-GST, and pALTER/gin4K48M-3HA. The appropriate XbaI fragments from the pALTER derivatives were then cloned into XbaI-digested YCplac111, YEplac181, or YIplac204 to yield plasmids YCp/gin4K48M, YEp/gin4K48M, YCp/GIN4-3HA, YCp/gin4K48M-3HA, and YIp/GIN4-GST. Before transformation into yeast, YIp/GIN4-GST was digested with ApaI to target integration to the ura3 locus.
To construct plasmid YCpGALGST/GIN4N, which contains the kinase region of GIN4 (codons 1–315) fused to GST-encoding sequences under the control of the GAL1/10 promoter, the PCR product obtained using primers ML67 and ML73 and pALTER/GIN4 as template was digested with BglII and XhoI and cloned into BamHI/SalI-digested YCpGALGST. The GIN4 ORF in YCpGALGST/GIN4N includes eight COOH-terminal non-GIN4-encoded amino acids (LDSSSSLA) followed by a stop codon. To construct plasmid YCpGALGST/GIN4C, which contains the COOH-terminal nonkinase region of GIN4 (codons 295–1,142) fused to GST-encoding sequences under the control of the GAL1/10 promoter, an ∼2.2-kb SalI fragment from pEG202/GIN4C (see below) was cloned into SalI-digested YCpGALGST.
To construct plasmids for two-hybrid analyses, the kinase (codons 1–294) and nonkinase (codons 295–1,142) regions of GIN4 were amplified by PCR using primers ML67 and ML72 and ML69 and ML70, respectively, and plasmid pALTER/GIN4 as template. The products were digested with EcoRI and XhoI or with NcoI and XhoI, respectively, and cloned into EcoRI/XhoI- or NcoI/XhoI-digested pEG202 to yield plasmids pEG202/GIN4N and pEG202/GIN4C, respectively. pEG202/ GIN4NFL, which contains nearly full-length GIN4 (codon 18 to past the stop codon), was constructed by cloning an NcoI-XhoI fragment from plasmid p54-15 into NcoI/XhoI-digested pEG202.
Cloning and Sequencing of GIN4
A plasmid shuffle was performed in mutant isolate JFY54A (see Results) to replace plasmid pB12G with the URA3-marked plasmid YEp24 (CDC12)N, creating strain M-100. M-100 was then transformed with a library of S. cerevisiae genomic DNA fragments in the low-copy plasmid YCp50-LEU2 (provided by P. Hieter, University of British Columbia, Vancouver, British Columbia, Canada), and ∼9,000 transformants were replica-plated at 23°C to SC-Leu medium containing 0.1% 5-fluoroorotic acid (5-FOA) to select against cells containing plasmid YEp24(CDC12)N. Nine clones were able to grow, indicating that they contained plasmids that rescued the synthetic lethal phenotype. Six of these clones were also able to grow at 37°C and thus were presumed to harbor CDC12-containing plasmids. Plasmids from the remaining three clones (p54-4, p54-6, and p54-15) were recovered into E. coli and confirmed to rescue the 5-FOA sensitivity of strain M-100. Restriction enzyme and DNA–DNA blot hybridization analyses indicated that plasmids p54-4 and p54-15 contained identical ∼12-kb inserts of genomic DNA (data not shown); thus, only p54-15 was used in further studies.
An ∼7-kb XbaI fragment from p54-15 was ligated in both orientations into XbaI-digested pRS315 (Sikorski and Hieter, 1989); both of the resulting plasmids (p54-110 and p54-170) rescued the 5-FOA sensitivity of strain M-100. Plasmid pΔSacI was created by digesting p54-110 with SacI, which cuts once in GIN4 and once in the plasmid polylinker, and then religating. Other deletions in the XbaI inserts of plasmids p54-110 and p54-170 were made using an exonuclease III/mung-bean nuclease deletion method as described previously (Ausubel et al., 1995), with ApaI used as the blocker enzyme and HindIII used to allow digestion by exonuclease III. The resulting deletion plasmids (including pΔ110-1, pΔ110-3, pΔ170-20, and pΔ170-10; see Fig. 1 A) were assayed for the ability to confer 5-FOA resistance to strain M-100 and were used as substrates for double-stranded DNA sequencing using T3 or T7 primers (Sikorski and Hieter, 1989) (Stratagene, La Jolla, CA) or custom primers where necessary to complete the DNA sequence. Approximately 4 kb were sequenced (from 109 nucleotides upstream of the GIN4 start codon to 454 nucleotides downstream of the stop codon); the sequence was identical to that subsequently released by the genome project.
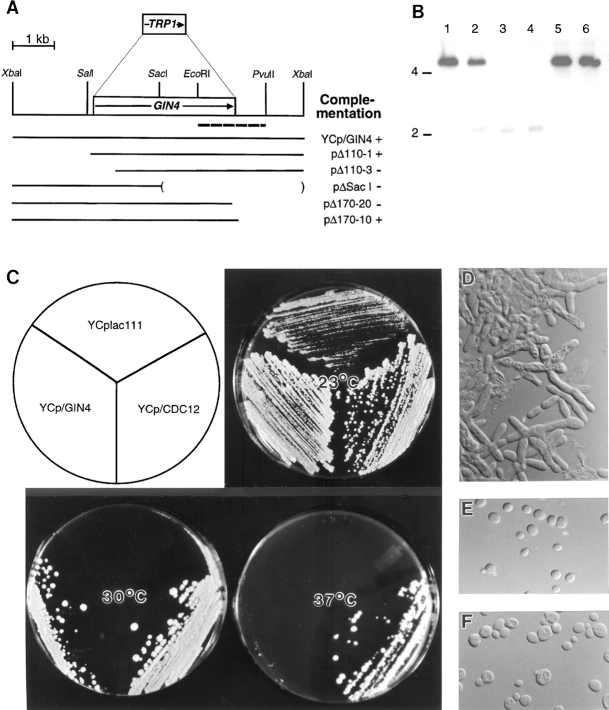
Figure 1.
(A) Structures of the GIN4 region and of the gin4-Δ9 allele. Open boxes, ORFs; arrows, directions of transcription. Dashed line, the fragment used as probe for blot hybridization (B). Solid lines, DNA segments assayed (in the indicated plasmids; see Materials and Methods) for complementation of the synthetic-lethal phenotype of strain M-100. Plasmids pΔ110-1 and pΔ110-3 contain DNAs beginning at nucleotides −109 and +517, respectively, relative to the beginning of the 3426-nucleotide GIN4 ORF. Plasmids pΔSacI, pΔ170-20, and pΔ170-10 contain DNAs ending at nucleotides +1619, +3382, and +3477, respectively. To construct gin4-Δ9, the GIN4 ORF was precisely replaced by TRP1 using a PCR-generated deletion cassette (see Materials and Methods). (B) DNA–DNA blot hybridization analysis confirming the deletion of GIN4 in the gin4-Δ9 strains. Genomic DNAs were digested with SalI and PvuII and the blot was hybridized to an EcoRI-PvuII fragment (see A) isolated from YCp/ GIN4. GIN4 and gin4-Δ9 are predicted to yield fragments of 4,253 and 1,980 bp, respectively. Lane 1, YEF473 (wild type); lane 2, M-261 (GIN4/ gin4-Δ9); lanes 3–6, Trp+ (lanes 3 and 4) and Trp− (lanes 5 and 6) segregants from a complete tetrad from M-261. The positions of 2- and 4-kb molecular size markers are indicated. (C–F) Synthetic interaction between gin4-Δ9 and cdc12-6 alleles. (C) Strain M-326 (cdc12-6 gin4-Δ9) transformed with YCplac111, YCp/GIN4, or YCp/CDC12, as indicated, was streaked on SC-Leu plates and incubated for 2 d at the indicated temperature. (D–F) Cells from the plate incubated at 23°C (C) were examined by DIC microscopy. Cells contained plasmid YCplac111 (D), YCp/GIN4 (E), or YCp/CDC12 (F).
Two-hybrid Analysis
Two-hybrid analyses (Fields and Sternglanz, 1994) were conducted using the system described by Brent and coworkers (Gyuris et al., 1993; Ausubel et al., 1995). Strain EGY48J was cotransformed with DNA-binding domain (DBD) and activation domain (AD) plasmids and transformants were selected on solid SD + Leu medium. Quantitative assays for β-galactosidase activity were then performed in triplicate as described by De Virgilio et al. (1996). In brief, at least three independent transformants were grown for 16 h at 30°C in SD + Leu medium containing 1% raffinose and 2% galactose instead of glucose, and the levels of β-galactosidase activity were determined. pJG4-5PL-derived plasmids encoding AD fusions of full-length Cdc10p and full-length, NH2-terminal, and COOH-terminal regions of Cdc3p, Cdc11p, and Cdc12p have been described previously (De Virgilio et al., 1996).
Antibodies, Protein Procedures, and Microscopy
To prepare Gin4p-specific antibodies, the COOH-terminal region of GIN4 from codon 936 to 16 nucleotides past the stop codon was amplified by PCR using primers ML102 and ML103 and pALTER/GIN4 as template. The PCR product was digested with BamHI and EcoRI or with BamHI and HindIII and ligated into BamHI/EcoRI-digested pMAL-C2 (New England Biolabs) or BamHI/HindIII-digested pGEX-2T (Pharmacia Biotech, Piscataway, NJ) to create plasmids pMAL-C2/GIN4C and pGEX-2T/GIN4C, respectively. Fusion proteins were expressed from these plasmids and purified by batch methods according to standard protocols (Ausubel et al., 1995), then injected into rabbits using standard procedures (Cocalico Biologicals, Reamstown, PA). The antibodies raised against GST-Gin4p were then affinity purified using MalE-Gin4p on nitrocellulose blots (Pringle et al., 1991). Antibodies to Cdc3p, to Cdc11p, to Bni4p, and to Bud4p have been described previously (Ford and Pringle, 1991; Kim et al., 1991; Sanders and Herskowitz, 1996; DeMarini et al., 1997). Polyclonal antibodies to GST (Pharmacia Biotech) were purified as described by Bi and Pringle (1996). Monoclonal anti-HA antibody (HA.11) and monoclonal anti-tubulin antibody (YOL1/34) were purchased from Berkeley Antibody Co. (Richmond, CA) and Accurate Chemical and Scientific Company (Westbury, NY), respectively. FITC- or rhodamine-labeled secondary antibodies and rhodamine-phalloidin were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA) and Molecular Probes (Eugene, OR), respectively.
Yeast proteins were isolated for Western blot analysis by resuspending whole cells in 2× Laemmli sample buffer (Laemmli, 1970) and boiling for 5 min. Western blot analysis was performed as described by Ausubel et al. (1995), using the enhanced chemiluminescence system (Amersham, Arlington Heights, IL) for detection.
Differential interference contrast (DIC) and fluorescence microscopy were performed using a Nikon Microphot SA microscope (Tokyo, Japan). Cells were stained with Calcofluor to visualize chitin, stained with rhodamine-phalloidin to visualize F-actin, or prepared for immunofluorescence as described previously (Pringle et al., 1989, 1991), using bisBenzamide (Sigma Chemical Co.) in the mounting medium to stain DNA. To determine whether the cells in clumps had completed cytokinesis, cells were either sonicated or fixed and then treated with cell wall-digesting enzymes without sonication (Pringle and Mor, 1975). To evaluate axial or nonaxial budding, ≥200 Calcofluor-stained cells with three or more bud scars were examined; cells were scored as budding axially if all bud scars were in a single chain.
Results
Identification of MPK1/SLT2 and GIN4 by Synthetic Lethal Interaction with a cdc12 Septin Mutation
To identify proteins that interact with the S. cerevisiae septins, we screened for mutations that are lethal in combination with a cdc12-5 temperature-sensitive mutation. This screen identified several genes including BNI1 and CHS4 (DeMarini et al., 1997).2 Another complementation group, defined by strain JFY54A, displayed a segregation pattern suggesting that two mutations were required for synthetic lethality with cdc12-5 at 23°C. To identify the corresponding genes, a library was screened for plasmids that could rescue the synthetic lethality of strain M-100 (a derivative of JFY54A), yielding plasmids p54-6 and p54-15 (see Materials and Methods). Cells of strain M-100 containing p54-15 in place of the CDC12 plasmid grew at a rate comparable to that of strain M-100 itself, but cells containing only p54-6 grew more slowly.
Deletion and partial sequence analysis of p54-6 (data not shown) indicated that the complementing open reading frame (ORF) was MPK1/SLT2, which encodes one of several S. cerevisiae MAP kinase homologues (for review see Levin and Errede, 1995); mutation of MPK1 results in temperature-dependent osmotic sensitivity (Torres et al., 1991; Lee et al., 1993). A deletion of MPK1, in the absence of other known mutations, was synthetically lethal with the cdc12-6 temperature-sensitive allele at 30°C; that is, in 23 tetrads dissected from a cross of strains M-238 and DL454 (Table I), no segregants were recovered that were Trp+ (indicating the presence of mpk1Δ) and unable to grow at 37°C on medium containing 1 M sorbitol (indicating the presence of cdc12-6), yet able to grow at 30°C on medium lacking sorbitol (a condition normally permissive for both cdc12-6 and mpk1Δ single-mutant strains). Low-copy plasmids containing either CDC12 (YCp/CDC12) or MPK1 (YCp50[MPK1]) could rescue the synthetic lethality at 30°C. In the absence of a rescuing plasmid, cdc12-6 mpk1Δ double-mutant cells incubated at 30°C underwent several cell divisions before arresting growth with cells of nearly normal morphology that appeared to be in various stages of the cell cycle (data not shown).
An ∼3.6-kb region of p54-15 responsible for rescue of the synthetic lethality of strain M-100 was identified by subcloning and deletion analysis (Fig. 1 A). Sequencing of this region revealed a single long ORF encoding a predicted protein of 1,142 amino acids (129,778 kD). The sequence of codons 497–535 of this ORF had been deposited previously in GenBank; the identified ORF was named GIN4 because it was isolated in a screen for sequences whose overexpression resulted in a growth inhibitory phenotype (Akada et al., 1997). Subsequent release of the sequence of chromosome IV by the Saccharomyces genome project identified GIN4 as ORF YDR507C.
Homology of Gin4p to Protein Kinases
Analysis of the predicted Gin4p sequence revealed that it contains an NH2-terminal protein kinase domain and a long COOH-terminal nonkinase region (Fig. 2 A), and Gin4p has been shown to have protein kinase activity in vitro (Altman and Kellogg, 1997). The nonkinase region contains no obvious motifs except for a region predicted (Lupas, 1996) to form a coiled coil (Fig. 2 A). Gin4p is most similar to the protein encoded by S. cerevisiae ORF YCL024W, identified by the genome project; Ycl024Wp displays 45% overall identity to Gin4p, with 76% identity in the kinase domain and 35% identity in the nonkinase region (Fig. 2 A). Although the Gin4p and Ycl024Wp nonkinase regions are not significantly homologous to any other proteins currently in the databases, their kinase domains are particularly similar to the corresponding domains of four other kinases, S. cerevisiae Hsl1p/Nik1p (62% identity), Schizosaccharomyces pombe Nim1p/ Cdr1p (49% identity) and Cdr2p (52% identity), and a Caenorhabditis elegans kinase identified during genomic sequencing (51% identity) (Fig. 2 B). In addition, phylogenetic analysis (performed by S. Hanks, Vanderbilt University, Nashville, TN) suggests that Gin4p, Ycl024Wp, Hsl1p/Nik1p, Nim1p/Cdr1p, Cdr2p, and (less closely) the C. elegans kinase constitute a distinct family (Fig. 2 B).
Figure 2.

Analysis of Gin4p sequence. (A) Comparison of Gin4p (GenBank/EMBL/DDBJ accession number U33140) and Ycl024Wp (see corrected sequence at http://www.mips.biochem. mpg.de/htbin/search_code/YCL024W). Sequences were aligned using the GCG GAP program. Black boxes, identical amino acids; gray boxes, similar amino acids (I, L, V; S, T; D, E; Q, N; and K, R). Dots, gaps introduced by the program to maximize alignment. Roman numerals, conserved kinase domains as described by Hanks and Hunter (1995); *, the conserved lysine in Gin4p that was mutated to create Gin4pK48M and Gin4pK48A; #, the last residue of the kinase domains (residue 289 in Gin4p). Underline, regions predicted by the Coils program (Lupas, 1996) to form coiled-coil structures with a probability ≥0.88. (B) Phylogenetic analysis of kinase domains of putative Nim1p family members (provided by S. Hanks). The kinase domains of S. cerevisiae Gin4p, Ycl024Wp, and Hsl1p/Nik1p (GenBank/EMBL/DDBJ accession number P34244), of Schizosaccharomyces pombe Nim1p/ Cdr1p (GenBank/EMBL/DDBJ accession number X57549) and Cdr2p (gene 02 on cosmid c57A10; GenBank/EMBL/DDBJ accession number Z94684), and of a Caenorhabditis elegans kinase (ORF 6 of cosmid F15A2; GenBank/ EMBL/DDBJ accession number Z70207) were identified as described by Hanks and Hunter (1995). Phylogenetic analysis using the PAUP program with these kinase domains and a data set of 37 other kinase domains indicates that Gin4p, Ycl024Wp, Hsl1p/Nik1p, Nim1p/Cdr1p, Cdr2p, and (less strongly) the F15A2.6-encoded kinase constitute a family of related kinases contained within one branch of the CaMK family (see Fig. 3 C of Hanks and Hunter [1995] for an extended phylogenetic tree; note that Ycl24 in this figure is Ycl024Wp). Numbers, amino acid changes needed to reach hypothetical common ancestors at internal nodes.
Deletion of GIN4 and Genetic Interactions between GIN4 and CDC12
The GIN4 ORF was precisely replaced with TRP1 as described in Materials and Methods (Fig. 1, A and B). Tetrads from strain M-261 (GIN4/gin4-Δ9) segregated 2+:2− for TRP1 (gin4-Δ9) and 4+:0− for viability at temperatures from 18° to 39°C, indicating that GIN4 is not essential for viability.
Because of the complicated genetics of the original synthetic lethal mutant, we asked if gin4-Δ9 and cdc12 mutations were synthetically lethal in the absence of other known mutations. Strains M-238 (cdc12-6 GIN4) and M-267 (CDC12 gin4-Δ9) were crossed, and 24 tetrads were dissected onto YPD plates containing 1 M sorbitol at 23°C. 94 out of 96 spores were viable. The segregants were scored for gin4-Δ9 (Trp+) and cdc12-6 (inviability at 37°C) and streaked on YPD plates to assay viability at 30°C. As expected, all 22 wild-type segregants and all 25 gin4-Δ9 single-mutant segregants were viable at 30°C, and cdc12-6 single-mutant segregants were mostly viable at this temperature (22 viable out of 24 total). In contrast, all 23 cdc12-6 gin4-Δ9 double-mutant segregants were inviable at 30°C. The synthetic lethality was confirmed by showing that a low-copy plasmid containing either GIN4 or CDC12, but not a control plasmid, could rescue the viability of cdc12-6 gin4-Δ9 segregants at 30°C (Fig. 1 C). cdc12-6 gin4-Δ9 double-mutant strains grew slowly even at 23°C (Fig. 1 C) and displayed a morphology very similar to that of a cdc12-6 single-mutant strain at restrictive temperature (Fig. 1 D) (Hartwell, 1971; Adams and Pringle, 1984); as expected from their abnormal morphology, the cdc12-6 gin4-Δ9 cells grown at 23°C also lacked detectable septin localization to the mother–bud necks (data not shown). A low-copy plasmid containing either GIN4 or CDC12 restored both normal cell morphology (Fig. 1, E and F) and septin localization to the necks (data not shown). These data suggest that deletion of GIN4 exacerbates the functional defect of the mutant Cdc12p and hence that Gin4p normally has a positive role in septin function.
To test this hypothesis further, we asked if overexpression of GIN4 could rescue the temperature-sensitive lethality of a cdc12-6 strain. As expected, cells of strain M-238 containing a control plasmid were viable at 23°C but inviable at 32° or 37°C, and cells containing a CDC12 plasmid were viable at all three temperatures (Fig. 3 A). In contrast, cells containing either a low-copy or a high-copy GIN4 plasmid were able to grow at 32°C although not at 37°C (Fig. 3 A), and the low-copy GIN4 plasmid even restored nearly normal morphology to the cells grown at 32°C (Fig. 3, B–D). (Because a high-copy GIN4 plasmid itself causes morphological defects [see below], it could not be tested for rescue of the cdc12-6 morphological defect.)
Figure 3.
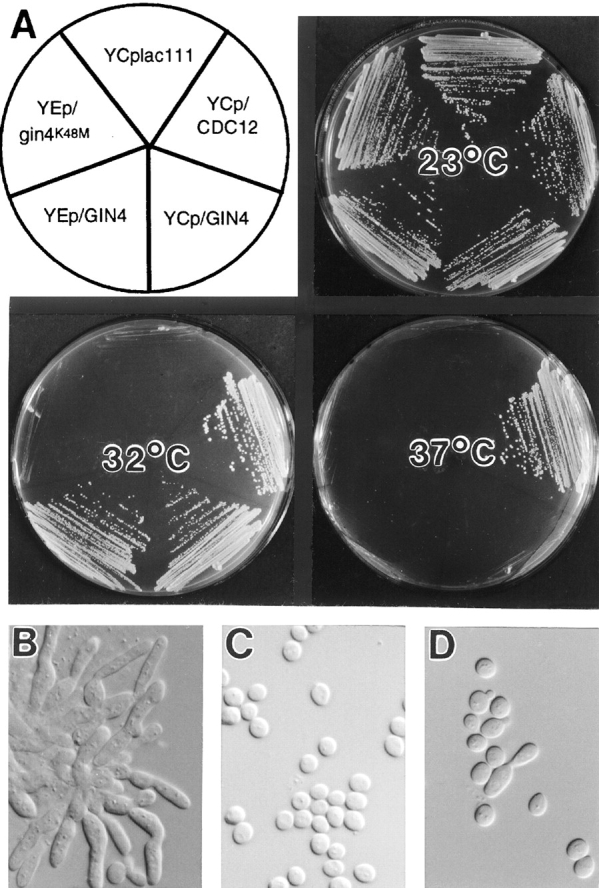
Partial suppression of cdc12-6 by overexpression of GIN4 but not of gin4K48M. (A) Strain M-238 (cdc12-6) transformed with YCplac111, YCp/CDC12, YCp/GIN4, YEp/GIN4, or YEp/gin4K48M, as indicated, was streaked on SC-Leu plates and incubated for 3 d at the indicated temperature. (B–D) Cells from the plate incubated at 32°C (A) were examined by DIC microscopy. Cells contained plasmid YCplac111 (B), YCp/CDC12 (C), or YCp/GIN4 (D).
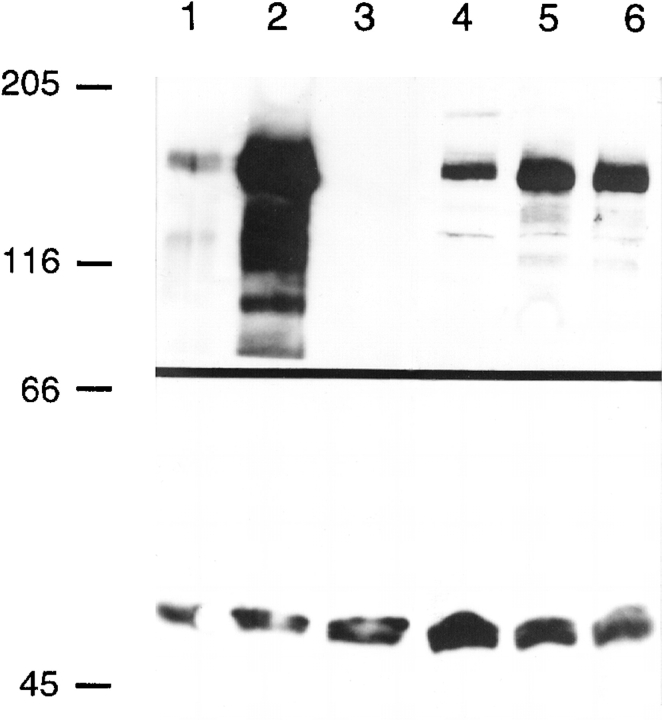
To ask if the protein kinase activity of Gin4p is necessary for its positive role in septin function, we tested the suppression ability of the gin4K48M and gin4K48A alleles, in which an invariant lysine of the kinase domain is altered (Fig. 2 A). Mutation of this lysine eliminates or greatly reduces the catalytic ability of a variety of kinases (for review see Bossemeyer, 1995; Hanks and Hunter, 1995; Taylor et al., 1995), and Gin4pK48A has been shown to have little or no kinase activity in vitro (Altman and Kellogg, 1997). In contrast to the corresponding GIN4 plasmids, a low-copy gin4K48M plasmid, a low-copy gin4K48A plasmid, and a high-copy gin4K48M plasmid all failed to rescue either the synthetic lethality of a cdc12-6 gin4-Δ9 strain or the viability of a cdc12-6 strain at 32°C (Fig. 3 A, and data not shown). Moreover, low-copy gin4K48M and gin4K48A plasmids also failed to rescue the morphology of cdc12-6 cells (strain M-238) grown at 32°C (data not shown). The lack of rescue by gin4K48M and gin4K48A was not due to a lack of protein, as these alleles are expressed at levels similar to those of wild-type Gin4p (Fig. 4, lanes 4 and 6) (Altman and Kellogg, 1997). Thus, the positive role of Gin4p in septin function appears to involve its kinase activity (see also below).
Figure 4.
Western blot analysis of Gin4p. Proteins from exponential-phase cells of the indicated strains were stained with antibodies to Gin4p (top) and to Cdc11p (bottom; to confirm equal protein loading). Lanes 1 and 2, strain YEF473 (wild-type) containing no plasmid (lane 1) or plasmid YEp/GIN4 (lane 2); lanes 3–6, strain M-272 (gin4-Δ9/gin4-Δ9) containing no plasmid (lane 3), plasmid YCp/GIN4 (lane 4), plasmid YCp/GIN4-3HA (lane 5), or plasmid YCp/gin4K48M (lane 6). Sizes of molecular weight markers (in kD) are indicated.
Two-hybrid Interaction between Gin4p and Cdc3p
Gin4p might affect septin function directly or indirectly. As one approach to this question, we tested for Gin4p– septin interactions using the two-hybrid system. Interaction was detected between the Gin4p kinase domain and either full-length Cdc3p or a fragment comprising the 101 COOH-terminal amino acids of Cdc3p (Table IV). Other interactions were not detected; in particular, a construct containing all but the first 17 codons of Gin4p did not show a detectable interaction with Cdc3p (Table IV; see Discussion). Thus, Gin4p may influence septin function, at least in part, by a direct interaction between its kinase domain and the COOH-terminal portion of Cdc3p.
Table IV.
Two-hybrid Interaction between the Gin4p Kinase Domain and Cdc3p
| DBD fusion | AD fusion | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cdc3p | Cdc3p-N | Cdc3p-C | Cdc10p | Cdc11p | Cdc12p | pJG4-5PL | ||||||||
| Gin4p-N | 242 | 11 | 727 | 7 | 9 | 9 | 22 | |||||||
| Gin4p-C | 9 | 12 | 11 | 12 | 14 | 11 | — | |||||||
| Gin4p-NFL | 7 | 9 | 8 | 9 | 11 | 11 | — | |||||||
Plasmids encoding the indicated DBD and AD fusions are described in Materials and Methods. Shown for each pair of plasmids are the units of β-galactosidase activity, measured as described in Materials and Methods. NH2-terminal and COOH-terminal fragments of Cdc11p and Cdc12p (De Virgilio et al., 1996) were also tested and showed no interaction with Gin4p.
Localization of Gin4p in Wild-type and Septin-mutant Strains
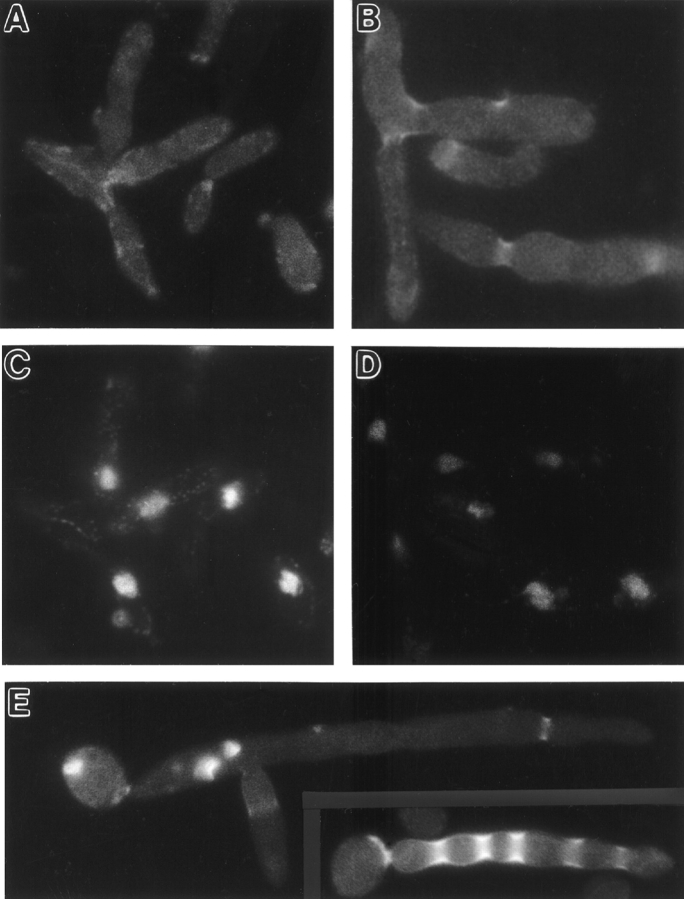
To explore further the apparent Gin4p–septin interaction, we investigated the intracellular localization of Gin4p using three approaches. First, we raised and affinity purified antibodies against the COOH-terminal portion of Gin4p (see Materials and Methods). In extracts of wild-type cells, the purified antibodies recognized a polypeptide of ∼140 kD, close to the predicted size of Gin4p (Fig. 4, lane 1). This polypeptide was absent in extracts from a gin4-Δ9 strain (Fig. 4, lane 3) and more abundant in extracts from a strain containing a high-copy GIN4 plasmid (Fig. 4, lane 2), confirming that the purified antibodies are specific for Gin4p. Second, we introduced a 3HA tag just upstream of the GIN4 stop codon to create GIN4-3HA (see Materials and Methods). gin4-Δ9 cells harboring this construct in a low-copy plasmid expressed Gin4p-3HA at levels similar to those of the wild-type protein (Fig. 4, compare lanes 1, 4, and 5), and this plasmid fully complemented the morphological defects of a gin4-Δ9 strain (see below, and data not shown). Third, a bacterial GST gene was introduced just upstream of the GIN4 stop codon to create GIN4-GST (see Materials and Methods); after integration into the chromosome, GIN4-GST fully rescued the morphological defects of a gin4-Δ9 strain (see below, and data not shown). In immunofluorescence experiments, we obtained essentially identical results using antibodies to Gin4p to localize Gin4p (Fig. 5 A) or antibodies to HA or GST to localize Gin4p-3HA (Fig. 5, B and E) or Gin4p-GST (Fig. 6 A), respectively.
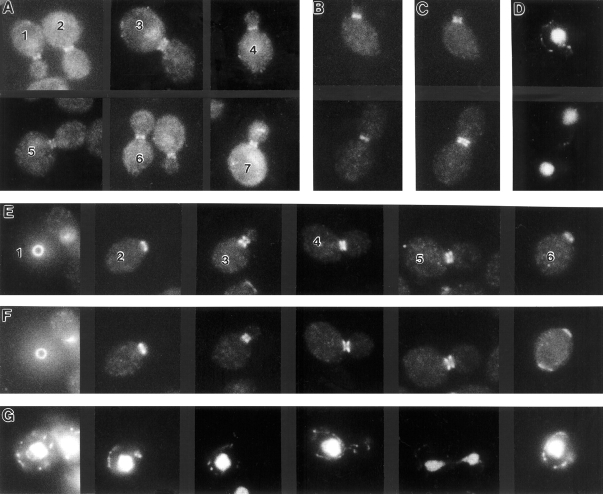
Figure 5.
Localization of Gin4p and septins during the cell cycle. Exponential-phase cultures of wild-type strain YEF473 (A) and of gin4-Δ9/gin4-Δ9 strain M-272 containing plasmid YCp/GIN4-3HA (B–G) were examined by immunofluorescence microscopy. Cells were stained with antibodies to Gin4p (A ) or double stained with antibodies to the HA epitope (B and E) and to Cdc11p (C and F) to localize Gin4p-3HA and Cdc11p, respectively. (D and G) DNA staining of cells shown in B and C and in E and F, respectively. Some cells are numbered for reference in the text.
Figure 6.
Dependence of Gin4p localization on the septins and on the Gin4p COOH-terminal (nonkinase) region. (A–D) Strain M-450 (cdc12-6 gin4-Δ9 URA3:GIN4-GST) containing plasmid YCp/CDC12 (A and B) or YCplac111 (C and D) was grown at 23°C to exponential phase, then shifted to 37°C for 45 min before examination by immunofluorescence using antibodies to GST (A and C) and to Cdc3p (B and D). (E–G) Strain M-272 (gin4-Δ9/gin4-Δ9) carrying galactose-inducible plasmids expressing GST (E; plasmid YCpGALGST) or GST fused to the NH2-terminal (kinase) region (F; plasmid YCpGALGST/GIN4N) or the COOH-terminal nonkinase region (G; plasmid YCpGALGST/GIN4C) of Gin4p were grown at 23°C to exponential phase in SC-Leu medium containing 1% raffinose. Galactose was then added to 2%, and cells were cultured for 2.5 h before examination by immunofluorescence with antibodies to GST.
In many unbudded cells, Gin4p was visualized as a single ring in the cell cortex (Fig. 5 E, cells 1, 2, and 6). These rings appear to correspond to incipient bud sites, as they were always located at a cell pole with polarized F-actin (as detected by staining with rhodamine phalloidin; data not shown), and they corresponded precisely to rings of septin proteins (Fig. 5, E and F, cells 1, 2, and 6) that had the appearance (diameter and brightness) of septin rings marking incipient bud sites rather than of those marking previous division sites (Kim et al., 1991; Ford and Pringle, 1991). Consistent with this interpretation, when such cells had two rings of septin proteins, the Gin4p ring always (40 out of 40 cells observed) corresponded to the septin ring that appeared to be at the incipient bud site (Fig. 5, E and F, cell 6); unbudded cells with two rings of Gin4p were not observed. Thus, unlike the septins, which typically remain visible at the division site for some time after cytokinesis (Kim et al., 1991; Ford and Pringle, 1991), Gin4p seems to disappear from the neck/division site at about the time of cytokinesis. Gin4p appears to arrive at the incipient bud site in late G1, coincident with the arrival of the septins. Every unbudded cell identified as having a ring of Gin4p-3HA had a corresponding ring of Cdc11p (50 out of 50 cells observed), and nearly every unbudded cell identified as having a ring of Cdc11p at the incipient bud site had a corresponding ring of Gin4p-3HA (48 out of 50 cells observed). (The cells with a ring of Cdc11p at the incipient bud site but no corresponding ring of Gin4p-3HA might have lost the GIN4-3HA plasmid before immunofluorescence staining.)
Like the septins, Gin4p was localized to the mother–bud neck throughout the budded phase of the cell cycle (Fig. 5, A, B, and E). However, in budded cells, the septins are consistently visualized on both sides of the neck as an apparent double ring (Haarer and Pringle, 1987; Kim et al., 1991; Ford and Pringle, 1991) (Fig. 5 C; Fig. 5 F, cells 3–5), whereas Gin4p was visualized as either an apparent single ring, (Fig. 5 A, cells 1, 2, 4, and 6; Fig. 5 B) or an apparent double ring (Fig. 5 A, cells 3, 5, and 7; Fig. 5 E, cells 3–5). The “single rings” of Gin4p were located in the middle or on the mother side of the neck, within the region occupied by the septins, and the “double rings” precisely colocalized with the septins. Careful focusing up and down revealed that Gin4p in double rings was, like the septins, in fact present as a continuous band throughout the neck region, at least until very late in the cell cycle; thus, the single ring versus double ring appearance seems simply to reflect differences in the width of the band of Gin4p along the mother–bud axis rather than a discrete difference in protein organization. Both “single rings” and “double rings” of Gin4p were observed at all stages in the cell cycle, but there was a sharp increase in the frequency of cells with “double rings” at about the time of the G2/M transition (data not shown).
The overlapping localization of Gin4p and the septins suggested that Gin4p localization may be septin dependent. To test this, a temperature-sensitive cdc12 septin mutant expressing Gin4p-GST and harboring either a control plasmid or a low-copy CDC12 plasmid was grown to exponential phase at 23°C, shifted to 37°C for 45 min, and stained with antibodies to GST or antibodies to Cdc3p. Both Gin4p-GST and Cdc3p remained detectable at the neck in cells containing the CDC12 plasmid (Fig 6, A and B), but both proteins were lost from the neck in cells containing the control plasmid (Fig. 6, C and D). Thus, Gin4p localization to the neck indeed appears to be septin dependent.
To investigate further the determinants of Gin4p localization to the neck, we created plasmids YCpGALGST, YCpGALGST/GIN4N, and YCpGALGST/GIN4C, which express GST or fusions of GST to the kinase domain or the nonkinase region of Gin4p, respectively, under control of the GAL1/10 promoter. When the proteins were expressed in wild type (strain YEF473) or gin4-Δ9 mutant (strain M-272) cells and localized by immunofluorescence using antibodies to GST, GST localized throughout the cytoplasm in a partially punctate pattern (Fig. 6 E), and GST-Gin4pN localized diffusely throughout the cytoplasm (Fig. 6 F). In contrast, GST-Gin4pC localized to the neck (Fig. 6 G), like full-length Gin4p. Thus, the nonkinase region of Gin4p appears to be responsible for localization to the mother–bud neck. Consistent with this hypothesis, Gin4p kinase activity does not appear to be required for localization of the full-length protein: when expressed from a low-copy plasmid (YCp/gin4K48M-3HA) in wild-type or gin4-Δ9 cells or from an integrated single copy (strain M-692) at room temperature, Gin4pK48M-3HA and Gin4pK48A-3HA both localized indistinguishably from normal Gin4p or Gin4p-3HA (data not shown).
Effects of Gin4p Overexpression
No obvious effects were seen upon long-term overexpression of GST or of the fusion of GST to the Gin4p kinase domain (data not shown). However, long-term overexpression of the fusion of GST to the nonkinase region of Gin4p in wild-type cells, although not lethal, produced striking morphological abnormalities. Cells frequently had multiple, elongated buds (Fig. 7, A, B, and E) containing multiple nuclei that appeared to have segregated efficiently (Fig. 7, C and D). This phenotype was similar to that of temperature-sensitive septin mutants incubated at restrictive temperature (Hartwell, 1971; Adams and Pringle, 1984), suggesting that septin localization or assembly might be defective in these cells. Indeed, septin staining was usually weak and diffuse compared with that in wild-type cells (Fig. 7 A; compare Fig. 5, C and F), and many cells lacked detectable septin staining. The localization of GST-Gin4pC resembled that of the septins (Fig. 7 B). As expected from the abnormal septin organization (DeMarini et al., 1997) (see Introduction), cells overexpressing GST-Gin4pC also displayed abnormal patterns of chitin deposition (Fig 7 E; compare with Fig. 9 G). Similar results were obtained when GST-Gin4pC was overexpressed in gin4-Δ9 cells or (in a smaller proportion of the cells) when wild-type or gin4-Δ9 cells contained a high-copy GIN4 or gin4K48M plasmid (data not shown). Thus, overexpression of the nonkinase region of Gin4p, catalytically inactive Gin4p, or normal Gin4p appears to interfere with septin localization or assembly.
Figure 7.
Aberrant cell morphology, Gin4p localization, septin localization, and chitin deposition resulting from overexpression of the COOH-terminal (nonkinase) region of Gin4p. Strain YEF473 carrying plasmid YCpGALGST/GIN4C was grown to exponential phase in SC-Leu medium containing 1% raffinose and 2% galactose. (A and B) Cdc11p and GST-Gin4pC were localized using antibodies to Cdc11p (A) and to GST (B), respectively. The exposure time for photomicroscopy of this septin staining was the same as that for the septin staining shown in Figs. 5 and 8. (C and D) DNA staining of cells shown in A and B, respectively. (E) Chitin was detected by staining with Calcofluor.
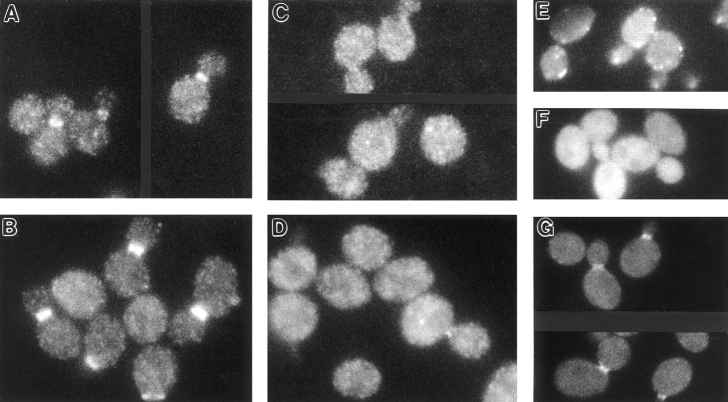
Figure 9.
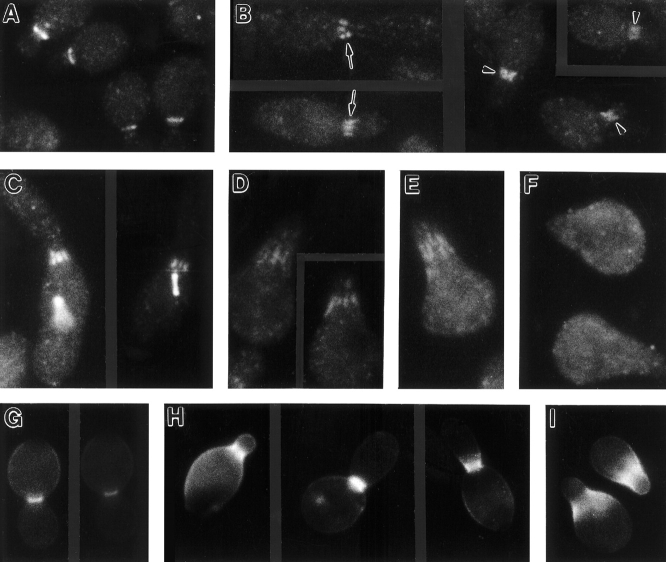
Localization of Bni4p, Bud4p, and chitin in wild-type and gin4-Δ9 vegetative cells, and localization of septins, Gin4p, and chitin in shmooing cells. (A–C, G, and H) Wild-type strain YEF473 (A and G) and gin4-Δ9/gin4-Δ9 strain M-272 (B, C, and H) were grown at 30°C to exponential phase and observed by immunofluorescence using antibodies to Bni4p (A and B) or Bud4p (C) or stained with Calcofluor to visualize chitin (G and H). This preparation of anti-Bud4p antibodies stains the spindle microtubules (out of focus in the cell on the left) as well as Bud4p (Sanders and Herskowitz, 1996; DeMarini, D.J., and M.S. Longtine, unpublished results). (D–F and I) Strain SY2625 was treated with α-factor to induce shmoo formation and examined by immunofluorescence using antibodies to Cdc11p (D), Cdc3p (E), or Gin4p (F), or stained with Calcofluor to visualize chitin (I). Arrows and arrowheads, features described in the text.
Abnormal Morphology and Septin Organization in gin4 Mutants
To characterize further the function of Gin4p, we observed the effects of GIN4 deletion and of mutations that reduce or eliminate Gin4p kinase activity (see above) on cell morphology, septin organization, and cytokinesis. In comparison to wild-type cells (Fig. 8, A and B), most gin4-Δ9 cells grown at 23°C were moderately elongated and clumped (Fig. 8 C), and many cells had enlarged bud necks (Fig. 8 C, arrows). These abnormalities were more pronounced in gin4-Δ9 cells grown at 37°C (Fig. 8 D); fewer than 20% of such cells had a wild-type morphology. Cell clumping at both 30° and 37°C appeared to be due mostly to a defect in cell separation rather than a defect in cytokinesis, because either sonication or fixation and cell wall digestion (see Materials and Methods) yielded predominantly single-budded and unbudded cells (data not shown). Similar abnormalities, but less pronounced, were observed in strains expressing only a kinase-dead GIN4 allele (data not shown).
Figure 8.
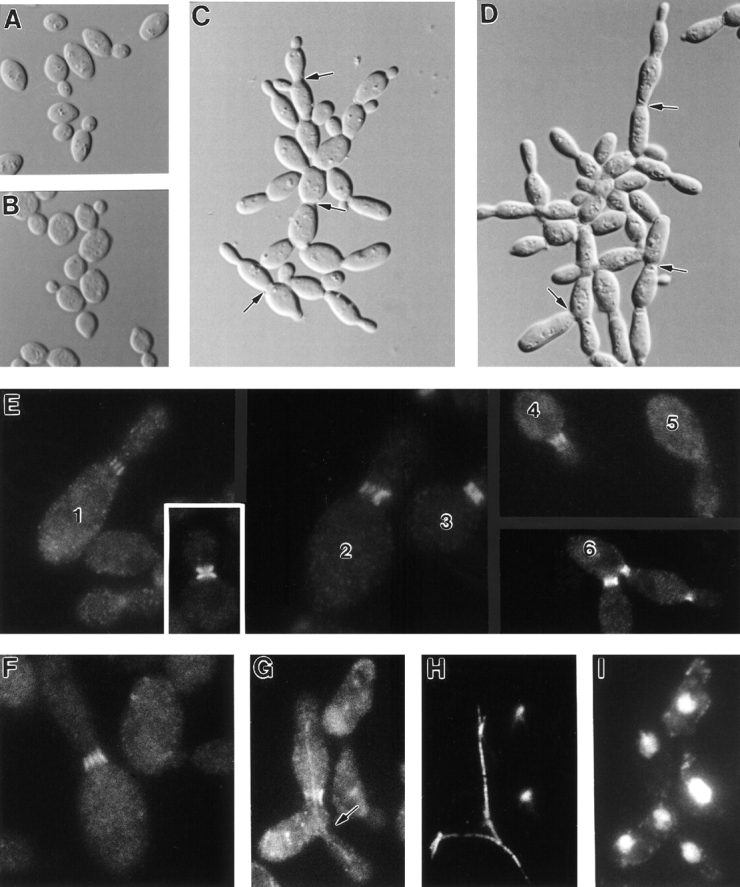
Abnormal morphology and septin organization in gin4-Δ9 cells. Wild-type strain YEF473 (A, B, and inset in E) and gin4-Δ9/ gin4-Δ9 strain M-272 (C–I) were grown to exponential phase at 23°C (A and C), 30°C (E and F), or 37°C (B, D, and G–I) and examined by DIC microscopy (A–D) or by immunofluorescence using antibodies to Cdc11p (E and G ) or Cdc3p (F). Cells in G were also stained with antibodies to tubulin (H) and with bisBenzamide to visualize DNA (I). Arrows in C and D, enlarged bud necks; arrow in G, a neck without septin staining. Some cells in E are numbered for reference in the text.
Immunofluorescence staining revealed that gin4-Δ9 cells grown at various temperatures were also frequently abnormal in septin organization. In both diploid and haploid mutant strains, a fraction of cells (ranging from ∼61% at 23°C to ∼10% at 37°C) displayed approximately normal septin staining (Fig. 8 E, cell 3; compare to the wild-type cells in the inset and in Fig. 5, C and F) (Table V). However, other cells displayed one of several aberrant staining patterns. First, and most strikingly, many cells displayed a set of five to eight parallel “bars” of septin staining running through the neck along the mother–bud axis (Fig. 8 E, cells 1 and 2; Fig. 8 F) (Table V). Second, many cells displayed a band of septin staining in the neck whose endpoints were less well defined (“fuzzier”) than those in wild-type cells (Fig. 8 E, cell 4) (Table V). Often (as in Fig. 8 E, cell 4), it appeared that fuzzy bands of septin staining might actually be poorly resolved sets of bars. (Similarly, the higher ratio of fuzzy bands to bars scored in haploid cells, relative to diploid cells, may reflect, at least in part, a lower resolution of structures in the smaller necks of haploid cells.) Both fuzzy and bar septin staining patterns were observed both in morphologically abnormal cells and in cells of relatively normal morphology, suggesting that the abnormalities in septin organization are not simply a consequence of abnormal cell morphology. Necks with either fuzzy or bar septin staining appeared to contain amounts of septins comparable to those in wild-type necks, as judged by the intensity of immunofluorescence staining; thus, the absence of Gin4p appears to affect the organization of septins at the neck rather than their ability to localize to that part of the cell. Third, in some cells, septins were not detectable at the neck (Fig. 8, E, cell 5, and G, arrow); such cells were rare at 23°C but more common at higher temperatures (Table V), and their overall morphology typically resembled that of temperature-sensitive septin mutants incubated at restrictive temperature (Hartwell, 1971; Adams and Pringle, 1984). The absence of detectable septin staining indeed appeared to correlate with a defect in cytokinesis: the cytoplasm appeared to be continuous through necks that were devoid of septin staining, and microtubules could span them (Fig. 8 H); moreover, the two or more nuclei in such cells were typically synchronized in the cell cycle (Fig. 8 I and data not shown). Occasionally, even necks with septin staining appeared to fail in cytokinesis or cell separation (Fig. 8 E, cell 6); in these cells, septin staining persisted at the neck(s) as if septin disassembly had been abnormally delayed.
Table V.
Localization of Cdc11p, Bni4p, and Bud4p in gin4-Δ9 Strains
| Strain | Protein | Temperature | Localization pattern | Cells with multiple nuclei | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild-type | “Fuzzy” | “Bars” | Absent | |||||||||||
| (°C) | (%) | (%) | (%) | (%) | (%) | |||||||||
| M-272 | Cdc11p | 23 | 61 | 17 | 21 | <1 | <1 | |||||||
| 30 | 45 | 14 | 32 | 9 | 4 | |||||||||
| 37 | 10 | 20 | 41 | 29 | 18 | |||||||||
| 30* | 86 | 11 | 2 | 1 | — | |||||||||
| 30‡ | 86 | 12 | 2 | 0 | — | |||||||||
| 30§ | 67 | 20 | 11 | 1 | — | |||||||||
| M-272 | Bni4p | 23 | 7 | 73 | 20 | <1 | — | |||||||
| 30 | 4 | 59 | 37 | <1 | — | |||||||||
| 37 | 2 | 8 | 11 | 79 | — | |||||||||
| M-268 | Cdc11p | 30 | 61 | 27 | 8 | 4 | — | |||||||
| 34 | 33 | 42 | 21 | 4 | — | |||||||||
| M-268 | Bud4p | 30 | 64 | 26 | 10 | —‖ | — | |||||||
| 34 | 39 | 32 | 29 | —‖ | — | |||||||||
Strains M-272 (gin4-Δ9/gin4-Δ9) and M-268 (gin4-Δ9) were grown to exponential phase at the temperature indicated, and cells were stained with bisBenzamide and with antibodies to Cdc11p, Bni4p, or Bud4p. For each sample, 200 budded cells were scored for the pattern of Cdc11p staining, 200 small- or medium-budded cells were scored for the pattern of Bni4p staining, and 200 medium- or large-budded cells were scored for the pattern of Bud4p staining. For Cdc11p and Bud4p, the wild-type pattern is symmetric localization through the neck with well defined endpoints (see Fig. 8 E, inset, for an example of Cdc11p staining in a wild-type cell); for Bni4p, the wild-type pattern is asymmetric localization to the mother side of the neck (see Fig. 9 A for examples). Aberrant staining patterns observed were as follows. “Fuzzy” for Cdc11p and Bud4p, localization throughout the neck but with endpoints abnormally ill-defined and fuzzy (see Fig. 8 E, cell 4, for an example of “fuzzy” Cdc11p staining); for Bni4p, localization symmetric on both sides of the neck (see Fig 9 B, arrowheads, for examples). “Bars” for all three proteins, localization symmetrically through the neck as several bars or lines (see Fig. 8 E, cells 1 and 2; Fig. 8 F; Fig. 9 B, arrows; and Fig. 9 C for examples). Absent, no localized staining was detected. In control experiments with wild-type strains YEF473 and YEF473A grown at 30° or 37°C, >97% of cells showed the wild-type staining patterns for Cdc11p, Bni4p, and Bud4p (data not shown).
Strain M-272 transformed with plasmid YCp/GIN4.
Strain M-272 transformed with plasmid YCp/GIN4-3HA.
Strain M-272 transformed with plasmid YCp/gin4K48M-3HA.
Because Bud4p is present at the neck only at certain times in the cell cycle, it was difficult to know whether the absence of detectable Bud4p in a given cell reflected an abnormal failure of localization or simply that cell's position in the cell cycle. Thus, the Bud4p localization pattern was only scored in cells in which a Bud4p signal was visible at the neck.
Remarkably, however, in most cases, gin4-Δ9 bud necks with fuzzy or bar septin staining appeared to be competent to undergo cytokinesis. In gin4-Δ9/gin4-Δ9 diploid cells, at each temperature examined, the fraction of multibudded and/or multinucleate cells (indicative of failures of cytokinesis) was much less than the fraction of cells with fuzzy or bar septin staining (Table V).
To ask if the role of Gin4p in promoting normal septin organization at the neck depends on the Gin4p kinase activity, we examined cells of a gin4Δ strain that had been transformed with plasmids expressing either Gin4p-3HA or Gin4pK48M-3HA. Although the former plasmid restored septin organization as effectively as did a plasmid expressing normal Gin4p, the plasmid expressing Gin4pK48M-3HA only partially restored septin organization (Table V). Similar results were obtained with a strain (M-692) expressing only Gin4pK48A-3HA. Thus, the Gin4p kinase activity appears to promote, but not to be absolutely essential for, normal septin organization at the neck.
To investigate further the functional properties of the abnormally organized septins, we examined the localization and function of Bni4p and Bud4p, two proteins that localize to the neck in a septin-dependent manner (see Introduction). In wild-type cells, Bni4p is visualized as a sharp band on the mother-cell side of the neck (DeMarini et al., 1997) (Fig. 9 A). In contrast, in gin4-Δ9 cells, Bni4p was typically visualized as a fuzzy band spanning the neck (Fig. 9 B, arrowheads) (Table V) or as a set of bars similar to the bars of septin staining (Fig. 9 B, arrows) (Table V); moreover, in cells grown at 37°C, Bni4p was often undetectable (Table V). At all temperatures tested, aberrant Bni4p organization was detected more frequently than was aberrant septin organization (Table V), suggesting that subtle alterations of septin organization could have more pronounced (and/or more easily detected) effects on Bni4p localization or organization. Remarkably, the abnormally organized Bni4p appeared to be functional in assembling components of the chitin synthase III complex: at 23° or 30°C, most gin4-Δ9 cells displayed either fuzzy or bar Bni4p staining (Table V), but nearly all cells deposited chitin primarily at the neck (Fig. 9 H) (Table VI). Strikingly, like Bni4p itself, the chitin was typically present on both sides of the neck (Fig. 9 H) (Table VI), rather than being restricted to the mother-cell side of the neck as in wild-type cells (Fig. 9 G) (Table VI).
Table VI.
Chitin Localization in Wild-type and gin4-Δ9 Strains
| Strain | Temperature | Chitin localization | ||||||
|---|---|---|---|---|---|---|---|---|
| Asymmetric | Symmetric | Delocalized | ||||||
| (°C) | (%) | (%) | (%) | |||||
| YEF473 | 30 | 100 | 0 | 0 | ||||
| M-272 | 23 | 13 | 87 | 0 | ||||
| 30 | 4 | 96 | 0 | |||||
| 37 | 2 | 19 | 79 | |||||
Strains YEF743 (wild-type) and M-272 (gin4-Δ9/gin4-Δ9) were grown to exponential phase at the temperature indicated and stained with Calcofluor. For each sample, 100 budded cells were scored for their patterns of chitin staining. Asymmetric, chitin localized to the mother side of the mother–bud neck (see Fig. 9 G for examples). Symmetric, chitin localized to the neck region but with apparently equal staining intensity on both mother and bud sides of the neck (see Fig. 9 H for examples). Delocalized, chitin dispersed more or less uniformly around the cell periphery. Chitin localization in YEF473 cells grown at 23° or 37°C was indistinguishable from that of YEF473 cells grown at 30°C.
Similar results were obtained with Bud4p. In wild-type cells, Bud4p localizes to the neck from G2 until the end of the cell cycle in a tight band that is symmetric on the mother and bud sides of the neck (Sanders and Herskowitz, 1996). In contrast, in haploid and diploid gin4-Δ9 cells, Bud4p was often visualized in fuzzy or bar patterns (Fig. 9 C and data not shown) whose frequencies were similar to those observed for Cdc11p (Table V). The abnormally organized Bud4p appeared to be largely functional in axial bud-site selection: at 30°C, ∼36% of the cells displayed Bud4p in a fuzzy or bar pattern (Table V), whereas only ∼6% of the cells budded in nonaxial patterns as judged by Calcofluor staining of bud scars (see Materials and Methods). Similarly, at 34°C, ∼61% of the cells displayed Bud4p in a fuzzy or bar pattern (Table V), whereas only ∼15% of the cells budded in nonaxial patterns.
Interestingly, in cells undergoing polarized morphogenesis (shmooing) in response to mating pheromone, the septins are also typically visualized either as a fuzzy band (Kim et al., 1991; Ford and Pringle, 1991) or as a set of bars parallel to the projection axis (Fig. 9, D and E). This septin localization corresponds to that of the broad and somewhat fuzzy band of chitin deposited at the bases of the shmoo projections (Schekman and Brawley, 1979; Kim et al., 1991; Konopka et al., 1995) (Fig. 9 I), suggesting that in shmooing cells, as in vegetative cells (DeMarini et al., 1997), the septins serve as a template for the localization and/or assembly of Chs3p (Santos and Snyder, 1997) and the other components of the chitin synthase III complex. These observations raise the intriguing possibility that the absence of Gin4p in mutant vegetative cells might result in an altered organization of the septins that mimics what occurs normally in shmooing cells. Consistent with this hypothesis, Gin4p was not detectable by immunofluorescence in shmooing cells (Fig. 9 F), although the protein could be detected by Western blotting in such cells (Altman and Kellogg, 1997).
We did not detect any other defects in gin4-Δ9 strains. Haploid gin4-Δ9 cells mated efficiently with both wild-type and gin4-Δ9 partners, and a diploid gin4-Δ9/gin4-Δ9 strain sporulated with a frequency similar to that of an otherwise isogenic wild-type strain and produced ascospores of normal viability.
Apparent Lack of Redundancy between Gin4p and Ycl024Wp
To investigate the function of the Gin4p homologue Ycl024Wp, its ORF was precisely replaced with HIS3 (see Materials and Methods). Haploid and homozygous diploid ycl024W-Δ2 strains were viable and displayed no readily detectable defects in mating, cell morphology, chitin deposition, bud-site selection, sporulation, spore germination, growth on YPD plates at temperatures ranging from 18° to 39°C, or growth on plates containing 1 M sorbitol, 0.4 M KCl, or 0.9 M KCl (data not shown). Moreover, gin4-Δ9 ycl024W-Δ2 haploid and homozygous diploid double-mutant strains were indistinguishable from the gin4-Δ9 single- mutant strains; they displayed neither enhanced nor novel phenotypes. Thus, somewhat surprisingly, Ycl024Wp does not appear to be redundant, or to share an overlapping function, with Gin4p.
Discussion
Identification of Septin-interacting Proteins by a Synthetic-lethal Screen
A screen for mutations lethal in combination with the cdc12-5 septin mutation has identified several genes whose products interact with the septins (DeMarini et al., 1997; this study).2 In the work reported here, we found that loss of function mutations in either of two protein kinase genes, GIN4 and MPK1/SLT2, are synthetically lethal with cdc12 mutations. As discussed below, abundant evidence demonstrates an intimate interaction between Gin4p and the septins. In contrast, it remains unclear how directly Mpk1p and the septins interact. mpk1Δ cdc12-6 double mutants arrest growth with cells at various stages in the cell cycle and do not resemble a septin loss of function mutant in morphology; thus, it does not appear that the absence of Mpk1p simply compromises septin function. As both Mpk1p (Cid et al., 1995; Levin and Errede, 1995; Banuett, 1998) and the septins (Hartwell, 1971; Adams and Pringle, 1984; Longtine et al., 1996; DeMarini et al., 1997) are involved in the control of cell-surface growth, the synthetic lethality may reflect a synergistic effect on the maintenance of cell integrity.
Interaction of Gin4p and the Septins
Many lines of evidence show that Gin4p interacts intimately with the septins and plays a positive role in septin function. First, these proteins colocalize through most of the cell cycle. Both Gin4p and the septins assemble into a ring at the presumptive bud site ∼15 min before bud emergence (this study; Ford and Pringle, 1991; Kim et al., 1991), and both do so in an actin-independent manner (Ayscough et al., 1997). The proteins then remain concentrated in a band at the mother–bud neck throughout the period of bud growth (this study; Okuzaki et al., 1997). Interestingly, the band of Gin4p often occupies only a part of the region defined by the band of septins, and, unlike the septins (Ford and Pringle, 1991; Kim et al., 1991) (Fig. 5 F, cell 6), Gin4p disappears from the neck at about the time of cell division. These observations suggest that Gin4p localization might depend on the septins but not vice versa. Consistent with this hypothesis, Gin4p localization is lost when a septin mutant is shifted to restrictive temperature, whereas the septins still generally localize to the neck (albeit in abnormal patterns; see below) in the absence of Gin4p. Second, interaction between Gin4p and the septins has been observed both in the two-hybrid system (Table IV) and by affinity chromatography (Carroll et al., 1998). Third, deletion of GIN4 in a wild-type background, although not lethal, results both in a dramatically altered reorganization of the septins (as discussed more fully below) and in a mutant cell morphology that suggests that septin function is compromised. Fourth, gin4 and cdc12 mutations are synthetically lethal, and the morphology of the double mutants suggests that septin function is lost in the absence of Gin4p. Fifth, mild overexpression of GIN4 can rescue both the viability and the morphology of a cdc12 temperature-sensitive mutant at intermediate temperatures. Finally, more extreme overexpression of full-length Gin4p or (especially) of a COOH-terminal Gin4p fragment causes disorganization of the septins and an abnormal cell morphology that resembles that of septin mutants.
The protein kinase activity of Gin4p appears to be important for its positive role in septin function, as kinase-dead mutants of Gin4p can rescue neither the cdc12 single mutant at intermediate temperatures nor the synthetic lethality of the gin4 cdc12 double mutant, and strains expressing only a kinase-dead allele have many cells with conspicuously abnormal septin organization (Table V). Moreover, the two-hybrid interaction observed was between the COOH-terminal region of Cdc3p and the kinase domain of Gin4p; this interaction appeared to depend on the presence of an intact kinase domain, as a construct lacking only the NH2-terminal 17 amino acids of Gin4p failed to interact (Table IV; note the caveat that negative results in two-hybrid assays can have a variety of causes). As two-hybrid interactions have been detected between other protein kinases and their substrates (Yang et al., 1992; Staudinger et al., 1995; Cook et al., 1996) and Cdc3p appears to be phosphorylated in vivo (Healy, A.M., and J.R. Pringle, unpublished results; Reed, S.I., personal communication), it is possible that the COOH-terminal region of Cdc3p is a substrate of Gin4p; phosphorylation of the predicted coiled-coil domain in this region of Cdc3p might regulate possible homotypic interactions, interactions with other septins, or interactions with septin-associated proteins. However, further studies will be required to test these possibilities and to identify other possible substrates of Gin4p.
In addition, it is also clear that the interaction of Gin4p with the septins does not only involve the kinase domain of Gin4p. In contrast to the kinase domain, the nonkinase region of Gin4p is both necessary (this study; Okuzaki et al., 1997) and sufficient (this study) for colocalization with the septins at the mother–bud neck, and expression of the nonkinase region alone perturbs septin organization. It is possible that the nonkinase region plays a structural role in septin organization (see below); this possibility is supported by the observation that strains expressing only a kinase-dead Gin4p have milder defects in septin organization than do cells containing no Gin4p (Table V).
Possible Redundancy of Gin4p Function
If Gin4p plays an important role in septin organization and function, why is it nonessential for growth? It seemed possible that Gin4p was redundant in function with one of the structurally related protein kinases. Ycl024Wp, the only protein to share significant sequence similarity with Gin4p outside of the kinase domain, was the most obvious candidate, but we could detect no effect of deleting YCL024W, either alone or in combination with deletion of GIN4. Similarly, although Hsl1p/Nik1p and Gin4p may be partially redundant with respect to their roles in cell-cycle control (Okuzaki et al., 1997; see below), we saw no obvious effect of deleting HSL1/NIK1 (either alone or in combination with deletion of GIN4) on septin organization or the structure of the neck, and even the gin4 hsl1 ycl024W triple mutant appeared to grow as well as the gin4 single mutant (Longtine, M.S., and J.R. Pringle, unpublished results). Thus, although it remains possible that Gin4p is redundant in function with some less closely related protein, we favor the hypothesis that the role of Gin4p in septin organization is helpful but not strictly necessary for septin function (see also below).
Roles of Gin4p and Related Kinases in Cell Cycle Control
While our studies of Gin4p and its interaction with the septins were in progress, at least two other groups identified GIN4 during studies of the control of mitotic events by the Cdc28p cyclin-dependent kinase. In particular, Kellogg and coworkers have presented multiple lines of evidence that Gin4p functions together with the protein Nap1p in the control of mitotic events (the G2/M transition and the switch from apical to isotropic bud growth) by the Cdc28p/Clb2p complex (Altman and Kellogg, 1997; Carroll et al., 1998). Independently, Okuzaki et al. (1997) focused on Gin4p because of its similarity to Hsl1p/Nik1p, a previously identified regulator of the Swe1p kinase and hence of Cdc28p (Ma et al., 1996; Tanaka and Nojima, 1996), and also presented evidence for a role of Gin4p in association with Cdc28p in mitotic control. Such a role for Gin4p would also be consistent with the similarity between its kinase domain and that of S. pombe Nim1p/ Cdr1p, which has a well-established role in mitotic control (MacNeill and Nurse, 1997).
However, several lines of evidence suggest that Gin4p does not function only at the G2/M transition. First, consistent with the presence in its upstream region of apparent MCB (at −310 to −303 and −287 to −280) and SCB (at −249 to −243) sequences (Koch and Nasmyth, 1994; Breeden, 1996), GIN4 appears to be transcribed differentially early in the cell cycle (Okuzaki et al., 1997), like other genes whose products function in late G1 or S phase. (In this regard, however, we also note that a plasmid-borne GIN4 that has only 109 bp of upstream sequence, and hence lacks the MCB and SCB sequences, is able to provide GIN4 function; see plasmid pΔ110-1, Fig. 1 A.) Second, GIN4 was also identified on the basis of the synthetic lethality of gin4 cln1 cln2 cells (Benton et al., 1997), suggesting a role for Gin4p early in the cell cycle when the Cdc28p/G1 cyclin complexes are active. Finally, Gin4p is already colocalized with the septins before bud emergence, and the abnormal organization of the septins in gin4Δ cells is evident even in cells with tiny buds, implying that Gin4p is involved in septin organization from the beginning of the cell cycle.
Thus, Gin4p may have two discrete times of action in the cell cycle. However, at this time it also seems possible that the observed delays in the G2/M transition and in the switch from apical to isotropic bud growth in gin4 mutant strains represent checkpoint-mediated responses to abnormal morphogenesis that occurs earlier in the cell cycle when Gin4p is absent. Such an interpretation might explain the otherwise puzzling observation that a protein implicated in mitotic control is colocalized with the septins in the cell cortex. Further studies (e.g., determination of whether the mitotic effects depend on Swe1p and the tyrosine phosphorylation of Cdc28p; for review see Lew and Reed, 1995; Sia et al., 1996) will be necessary to resolve these issues.
Relationship between Septin Organization and Septin Function
The results presented in this study appear to provide important insights into the mode of septin action and the relationship between septin organization and septin function. Previous studies have suggested that a major role for the septins is to serve as a scaffold or template for other proteins that must assemble at specific sites on the cell surface (Chant et al., 1995; Sanders and Herskowitz, 1996; DeMarini et al., 1997; Giot and Konopka, 1997; Bi et al., 1998). The results presented here provide strong support for this model. First, Gin4p provides another example of a protein that assembles at the mother–bud neck in a septin-dependent manner. As in the other cases studied to date, the relationship is not reciprocal, as the septins can still localize to the neck in the absence of Gin4p. Second, the altered septin organization observed in the absence of Gin4p (a set of bars running through the neck rather than the normal smooth band around the neck) is paralleled by an essentially identical reorganization of at least two other proteins (Bni4p and Bud4p) that normally localize to a band at the neck in a septin-dependent manner, supporting the hypothesis that the septins provide a scaffold or template for these other proteins. In this regard, it is interesting that the failure of Gin4p to associate with the septins in the mating projections of wild-type cells is paralleled by an organization of the septins (and, at least as judged by the pattern of chitin deposition, of septin-associated proteins) that appears similar to that observed in gin4Δ vegetative cells.
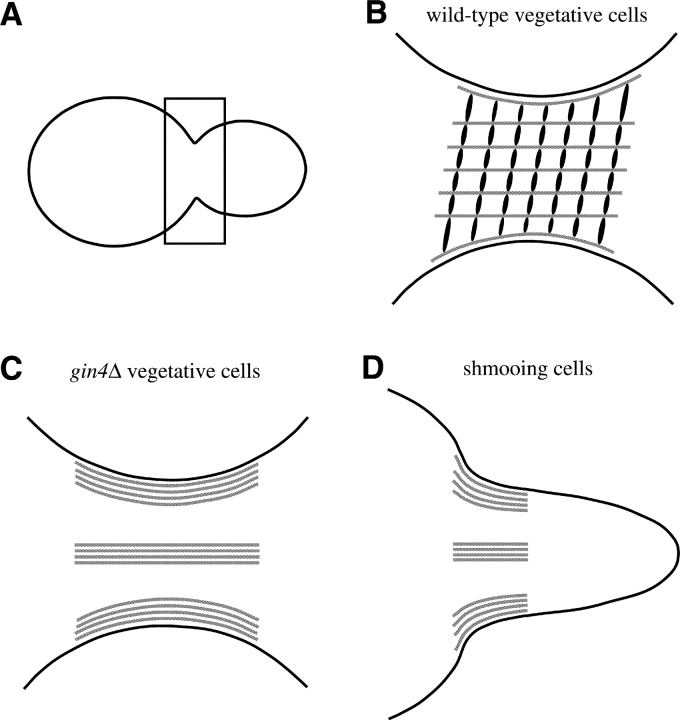
Remarkably, despite the seemingly very different organization of the septins in the absence of Gin4p, the septins themselves and the proteins assembled onto them are apparently able to function almost normally, as judged by the efficient occurrence of cytokinesis, of axial bud-site selection, and of localized chitin deposition (both in gin4Δ vegetative cells and in wild-type cells responding to mating pheromone). (Other evidence also indicates that the septins function during the morphogenetic response to pheromone [Giot and Konopka, 1997].) These observations suggest two important conclusions. First, it seems that the basic requirement for septin function may simply be the localization of these proteins to the correct region of the cell (so that they can recruit and/or anchor other proteins to the same region) and that the details of septin organization (as mediated by Gin4p) may play only a fine-tuning role (e.g., note the loss of mother-daughter asymmetry in the pattern of chitin deposition in the gin4Δ mutant). Second, it seems likely that the alteration of septin organization that occurs in the absence of Gin4p is actually less drastic than it appears, an interpretation that would support one important aspect of the novel model for septin organization proposed by Field et al. (1996). Based largely on measurements of isolated septin complexes from Drosophila, these investigators suggested that the septins in yeast might be arranged in filaments that run through the neck along the mother–bud axis rather than in the helically arranged filament(s) suggested by electron microscopic studies of the neck region (Byers and Goetsch, 1976; Byers, 1981). In the model of Field et al. (1996), the apparent helical filaments observed in the electron micrographs were suggested to represent a periodic density along the actual septin filaments. Modifying this model slightly, we suggest that the apparent filaments seen in the electron micrographs may in fact correspond to the periodic distribution of a protein(s) that normally links and spaces the septin filaments (Fig. 10 B). The linker protein(s) might be Gin4p itself and/or another protein(s) whose assembly or function depends on the Gin4p kinase. Thus, in the absence of Gin4p and hence of functional spacer protein(s), the septin filaments would coalesce into laterally associated bundles (Fig. 10, C and D). The coalescence into bundles would not, however, drastically affect the ability of the septin filaments to recruit and anchor other proteins to the neck, where they would function more or less normally.
Figure 10.
A model for septin organization and the role of Gin4p. Following the suggestion by Field et al. (1996), the model proposes that the vegetatively expressed septins are organized into filaments that run through the neck along the mother–bud axis at the face of the plasma membane. (The model does not address the question of how the septins are assembled into the filaments, which seems likely to be more complicated than envisioned in the original model of Field et al. [1996]; see Frazier et al. [1998].) (A) Diagram of a vegetative cell; the region depicted in panels B and C is boxed. (B) In wild-type vegetative cells, the septin filaments (gray lines) are proposed to be spaced by a linker protein(s) (black bars) that might be Gin4p itself and/or a protein(s) whose assembly or function depends on Gin4p. The regular spacing of this linker protein along the septin filaments and its relatively heavy staining during preparation for electron microscopy would result in the appearance of a helical filament(s) in electron micrographs of the neck region. (C) In the absence of Gin4p (and hence of functional linker protein) in a gin4Δ mutant, the septin filaments would coalesce into bundles but otherwise retain their organization and function. (D) Although Gin4p is present in wild-type cells responding to mating pheromone (Altman and Kellogg, 1997), it appears to have no kinase activity (Altman and Kellogg, 1997) and does not colocalize with the septins (this study). Thus, the septin filaments would also form bundles like those in gin4Δ vegetative cells, while retaining their ability to function.
An attractive feature of this view of septin organization and function is that it may help to explain two otherwise puzzling observations on the septins and functionally related proteins. First, despite the wide conservation of the septin family of proteins in other fungi and animal cells, “filaments” like those seen S. cerevisiae have not been observed by electron microscopy in other types of cells (except for the rather closely related Candida albicans: Soll and Mitchell, 1983). Second, although other protein kinases have kinase domains resembling that of Gin4p, no full-length homologues of Gin4p have yet been identified (except for Ycl024Wp in S. cerevisiae itself). Perhaps in other organisms the septins assemble at appropriate locations (perhaps even in filaments like those pictured in Fig. 10) and fulfill their putative scaffold or template role without assuming the particular higher-order structure (giving the appearance of filaments in electron micrographs) that would be provided by a Gin4p-like protein.
Acknowledgments
We thank D. Kellogg for plasmids and valuable discussions; J. Frazier (University of California, San Francisco, CA), C. Field (Harvard University Medical School, Boston, MA), H. Nojima, S. Tanaka, and D. Okuzaki (all three from Osaka University, Osaka, Japan) for valuable discussions; S. Hanks for valuable discussions and for performing the phylogenetic analysis shown in Fig. 2; D. Levin (Johns Hopkins University, Baltimore, MD) for strains, plasmids, and helpful comments; E. Bi (University of Pennsylvania, Philadelphia, PA), D. DeMarini, and C. De Virgilio (Botanisches Institut, Universität Basel, Basel, Switzerland) for antibodies, plasmids, and valuable discussions; P. Hieter for a plasmid library; S. Sanders (Massachusetts Institute of Technology, Cambridge, MA) for antibodies; other members of our laboratory for encouragement and valuable discussions; and S. Whitfield (University of North Carolina, Chapel Hill, NC) for expert assistance with the illustrations.
This work was supported by a National Institutes of Health (NIH) grant to J.R. Pringle (GM-31006), by funds from the RJEG Trust, and by an NIH postdoctoral fellowship to M.S. Longtine (GM-15766).
Abbreviations used in this paper
- AD
activation domain
- DBD
DNA-binding domain
- DIC
differential interference contrast
- 5-FOA
5-fluoroorotic acid
- GST
glutathione-S-transferase
- 3HA
triple hemagglutinin epitope
- ORF
open reading frame
- SC
synthetic complete medium
- SD
synthetic minimal medium
Footnotes
H. Fares's present address is Department of Biochemistry, Columbia University, New York, NY 10032.
Address all correspondence to J.R. Pringle, Department of Biology, CB#3280, Coker Hall, University of North Carolina, Chapel Hill, NC 27599-3280. Tel.: (919) 962-2293. Fax: (919) 962-0320. E-mail: jpringle@email.unc.edu
2. Fares, H., M.S. Longtine, and J.R. Pringle, manuscript submitted for publication.
References
- Adames N, Blundell K, Ashby MN, Boone C. Role of yeast insulin-degrading enzyme homologs in propheromone processing and bud site selection. Science. 1995;270:464–467. doi: 10.1126/science.270.5235.464. [DOI] [PubMed] [Google Scholar]
- Adams AEM, Pringle JR. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. . J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akada R, Yamamoto J, Yamashita I. Screening and identification of yeast sequences that cause growth inhibition when overexpressed. Mol Gen Genet. 1997;254:267–274. doi: 10.1007/s004380050415. [DOI] [PubMed] [Google Scholar]
- Altman R, Kellogg D. Control of mitotic events by Nap1 and the Gin4 kinase. J Cell Biol. 1997;138:119–130. doi: 10.1083/jcb.138.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, editors. 1995. Current Protocols in Molecular Biology. John Wiley and Sons, New York.
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett F. Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol Mol Biol Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. . Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton BK, Tinkelenberg A, Gonzalez I, Cross FR. Cla4p, a Saccharomyces cerevisiaeCdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol Cell Biol. 1997;17:5067–5076. doi: 10.1128/mcb.17.9.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Pringle JR. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. . Mol Cell Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, Pringle JR. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiaecytokinesis. J Cell Biol. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossemeyer D. Protein kinases—structure and function. FEBS (Fed Eur Biochem Soc) Lett. 1995;369:57–61. doi: 10.1016/0014-5793(95)00580-3. [DOI] [PubMed] [Google Scholar]
- Breeden L. Start-specific transcription in yeast. Curr Top Microbiol Immunol. 1996;208:95–127. doi: 10.1007/978-3-642-79910-5_5. [DOI] [PubMed] [Google Scholar]
- Byers, B. 1981. Cytology of the yeast life cycle. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 59–96.
- Byers B, Goetsch L. A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol. 1976;69:717–721. doi: 10.1083/jcb.69.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Altman R, Schieltz D, Yates J, Kellogg D. The septins are required for the mitosis-specific regulation of the Gin4 kinase. J Cell Biol. 1998;143:709–717. doi: 10.1083/jcb.143.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J, Mischke M, Mitchell E, Herskowitz I, Pringle JR. Role of Bud3p in producing the axial budding pattern of yeast. J Cell Biol. 1995;129:767–778. doi: 10.1083/jcb.129.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid VJ, Durán A, del Rey F, Snyder MP, Nombela C, Sánchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. . Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JG, Bardwell L, Kron SJ, Thorner J. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. . Genes Dev. 1996;10:2831–2848. doi: 10.1101/gad.10.22.2831. [DOI] [PubMed] [Google Scholar]
- Cooper JA, Kiehart DP. Septins may form a ubiquitous family of cytoskeletal elements. J Cell Biol. 1996;134:1345–1348. doi: 10.1083/jcb.134.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini DJ, Adams AEM, Fares H, De Virgilio C, Valle G, Chuang JS, Pringle JR. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiaecell wall. J Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C, DeMarini DJ, Pringle JR. SPR28, a sixth member of the septin gene family in Saccharomyces cerevisiaethat is expressed specifically in sporulating cells. Microbiology. 1996;142:2897–2905. doi: 10.1099/13500872-142-10-2897. [DOI] [PubMed] [Google Scholar]
- Estojak J, Brent R, Golemis EA. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Fares H, Goetsch L, Pringle JR. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. . J Cell Biol. 1996;132:399–411. doi: 10.1083/jcb.132.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H, Peifer M, Pringle JR. Localization and possible functions of Drosophilaseptins. Mol Biol Cell. 1995;6:1843–1859. doi: 10.1091/mbc.6.12.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CM, Al-Awar O, Rosenblatt J, Wong ML, Alberts B, Mitchison TJ. A purified Drosophilaseptin complex forms filaments and exhibits GTPase activity. J Cell Biol. 1996;133:605–616. doi: 10.1083/jcb.133.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- Flescher EG, Madden K, Snyder M. Components required for cytokinesis are important for bud site selection in yeast. J Cell Biol. 1993;122:373–386. doi: 10.1083/jcb.122.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford SK, Pringle JR. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC11gene product and the timing of events at the budding site. Dev Genet. 1991;12:281–292. doi: 10.1002/dvg.1020120405. [DOI] [PubMed] [Google Scholar]
- Frazier JA, Wong ML, Longtine MS, Pringle JR, Mann M, Mitchison TJ, Field C. Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J Cell Biol. 1998;143:737–749. doi: 10.1083/jcb.143.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia colishuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Giot L, Konopka JB. Functional analysis of the interaction between Afr1p and the Cdc12p septin, two proteins involved in pheromone-induced morphogenesis. Mol Biol Cell. 1997;8:987–998. doi: 10.1091/mbc.8.6.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G.R. Fink, editors. 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194:1–933. [PubMed]
- Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- Haarer BK, Pringle JR. Immunofluorescence localization of the Saccharomyces cerevisiae CDC12gene product to the vicinity of the 10-nm filaments in the mother-bud neck. Mol Cell Biol. 1987;7:3678–3687. doi: 10.1128/mcb.7.10.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB (Fed Am Soc Exp Biol) J. 1995;9:576–596. [PubMed] [Google Scholar]
- Hartwell LH. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- Kim HB, Haarer BK, Pringle JR. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3gene product and the timing of events at the budding site. J Cell Biol. 1991;112:535–544. doi: 10.1083/jcb.112.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Nasmyth K. Cell cycle regulated transcription in yeast. Curr Opin Cell Biol. 1994;6:451–459. doi: 10.1016/0955-0674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Konopka JB, DeMattei C, Davis C. AFR1 promotes polarized apical morphogenesis in Saccharomyces cerevisiae. . Mol Cell Biol. 1995;15:723–730. doi: 10.1128/mcb.15.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee KS, Irie K, Gotoh Y, Watanabe Y, Araki H, Nishida E, Matsumoto K, Levin DE. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol Cell Biol. 1993;13:3067–3075. doi: 10.1128/mcb.13.5.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE, Errede B. The proliferation of MAP kinase signaling pathways in yeast. Curr Opin Cell Biol. 1995;7:197–202. doi: 10.1016/0955-0674(95)80028-x. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J Cell Biol. 1995;129:739–749. doi: 10.1083/jcb.129.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie SH, Pringle JR. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: response to nutrient limitation. J Bacteriol. 1980;143:1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., and J.R. Pringle. 1998. Septins. In Guidebook to the Cytoskeletal and Motor Proteins. T. Kreis and R. Vale, editors. Oxford University Press, New York. In press.
- Longtine MS, DeMarini DJ, Valencik ML, Al-Awar OS, Fares H, De Virgilio C, Pringle JR. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, DeMarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. . Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Ma X-J, Lu Q, Grunstein M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. . Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- MacNeill, S.A., and P. Nurse. 1997. Cell cycle control in fission yeast. In The Molecular and Cellular Biology of the Yeast Saccharomyces. Cell Cycle and Cell Biology. J.R. Pringle, J.R. Broach, and E.W. Jones, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 697–763.
- Mitchell DA, Marshall TK, Deschenes RJ. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–723. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- Neufeld TP, Rubin GM. The Drosophila peanutgene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell. 1994;77:371–379. doi: 10.1016/0092-8674(94)90152-x. [DOI] [PubMed] [Google Scholar]
- Okuzaki D, Tanaka S, Kanazawa H, Nojima H. Gin4 of S. cerevisiaeis a bud neck protein that interacts with the Cdc28 complex. Genes Cells. 1997;2:753–770. doi: 10.1046/j.1365-2443.1997.1590358.x. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Mor J-R. Methods for monitoring the growth of yeast cultures and for dealing with the clumping problem. Methods Cell Biol. 1975;11:131–168. doi: 10.1016/s0091-679x(08)60320-9. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Adams AEM, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AEM, Stearns T, Drubin DG, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Sanders SL, Herskowitz I. The Bud4 protein of yeast, required for axial budding, is localized to the mother/bud neck in a cell cycle-dependent manner. J Cell Biol. 1996;134:413–427. doi: 10.1083/jcb.134.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R, Brawley V. Localized deposition of chitin on the yeast cell surface in response to mating pheromone. Proc Natl Acad Sci USA. 1979;76:645–649. doi: 10.1073/pnas.76.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia RAL, Herald HA, Lew DJ. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol Biol Cell. 1996;7:1657–1666. doi: 10.1091/mbc.7.11.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll DR, Mitchell LH. Filament ring formation in the dimorphic yeast Candida albicans. . J Cell Biol. 1983;96:486–493. doi: 10.1083/jcb.96.2.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger J, Zhou J, Burgess R, Elledge SJ, Olson EN. PICK1: a perinuclear binding protein and substrate for protein kinase C isolated by the yeast two-hybrid system. J Cell Biol. 1995;128:263–271. doi: 10.1083/jcb.128.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Nojima H. Nik1: a Nim1-like protein kinase of S. cerevisiaeinteracts with the Cdc28 complex and regulates cell cycle progression. Genes Cells. 1996;1:905–921. doi: 10.1046/j.1365-2443.1996.d01-213.x. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Radzio-Andzelm E, Hunter T. How do protein kinases discriminate between serine/threonine and tyrosine? Structural insights from the insulin receptor protein-tyrosine kinase. FASEB (Fed Am Soc Exp Biol) J. 1995;9:1255–1266. doi: 10.1096/fasebj.9.13.7557015. [DOI] [PubMed] [Google Scholar]
- Torres L, Martin H, García-Saez MI, Arroyo J, Molina M, Sánchez M, Nombela C. A protein kinase gene complements the lytic phenotype of Saccharomyces cerevisiae lyt2mutants. Mol Microbiol. 1991;5:2845–2854. doi: 10.1111/j.1365-2958.1991.tb01993.x. [DOI] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiaeG1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO (Eur Mol Biol Organ) J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Hubbard EJA, Carlson M. A protein kinase substrate identified by the two-hybrid system. Science. 1992;257:680–682. doi: 10.1126/science.1496382. [DOI] [PubMed] [Google Scholar]
- Zervos AS, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]