Abstract
The subcellular distribution and posttranslational modification of human chromatin assembly factor 1 (CAF-1) have been investigated after UV irradiation of HeLa cells. In an asynchronous cell population only a subfraction of the two large CAF-1 subunits, p150 and p60, were found to exist in a chromatin-associated fraction. This fraction is most abundant during S phase in nonirradiated cells and is much reduced in G2 cells. After UV irradiation, the chromatin-associated form of CAF-1 dramatically increased in all cells irrespective of their position in the cell cycle. Such chromatin recruitment resembles that seen for PCNA, a DNA replication and repair factor. The chromatin-associated fraction of p60 was predominantly hypophosphorylated in nonirradiated G2 cells. UV irradiation resulted in the rapid recruitment to chromatin of phosphorylated forms of the p60 subunit. Furthermore, the amount of the p60 and p150 subunits of CAF-1 associated with chromatin was a function of the dose of UV irradiation. Consistent with these in vivo observations, we found that the amount of CAF-1 required to stimulate nucleosome assembly during the repair of UV photoproducts in vitro depended upon both the number of lesions and the phosphorylation state of CAF-1. The recruitment of CAF-1 to chromatin in response to UV irradiation of human cells described here supports a physiological role for CAF-1 in linking chromatin assembly to DNA repair.
Keywords: assembly, CAF-1, chromatin, repair, UV irradiation
In the eukaryotic cell, genetic information is present in the form of chromatin (van Holde, 1988; Wolffe, 1995). The assembly and maintenance of this nucleoprotein complex is important to ensure regulated DNA metabolism. In proliferating cells, the bulk of histone synthesis and chromatin assembly occurs during DNA replication in S phase (Krude, 1995b ; Sogo and Laskey, 1995; Kaufman, 1996). However, reassembly of chromatin may also be important after destabilizing events that may arise during transcription (Tsukiyama and Wu, 1997), recombination (Lichten and Goldman, 1995), and repair (Smerdon, 1991).
Biochemical approaches have allowed the proteins involved in assembly of the basic repeating unit of chromatin to be identified. This unit, the nucleosome core, consists of ∼1.8 superhelical turns of DNA wrapped around a histone octamer (Luger et al., 1997). Nucleosome cores can be reconstituted in vitro simply by mixing the four core histones with DNA. This is based on the self-assembly properties of histones to form octamers and their transfer onto DNA which can be facilitated by salt gradient dialysis or the presence of polyanions (Laskey and Earnshaw, 1980). Several histone-binding proteins including nucleoplasmin, N1/N2, nucleosome assembly protein 1, DF 31, and Spt6p have the ability to facilitate regulated deposition of histones at physiological ionic strength. These proteins seem to act as molecular chaperones, i.e., they facilitate the formation of nucleosome cores without being a component of the final reaction product. Chromatin assembly factor 1 (CAF-1)1 is also thought to function as a molecular chaperone but, unlike the other proteins mentioned above, CAF-1 promotes assembly of nucleosomes preferentially onto replicating DNA (Smith and Stillman, 1989) and is associated with newly synthesized and modified histones H3 and H4 in cell extracts (Kaufman et al., 1995; Verreault et al., 1996). After the deposition of histones onto DNA, the formation of long arrays of regularly spaced nucleosomes was shown to be facilitated by two additional factors, recently purified from Drosophila embryo extracts, chromatin accessibility complex (CHRAC) (Varga-Weisz et al., 1997) and ATP-utilizing chromatin assembly and remodeling factor (ACF) (Ito et al., 1997). Since all these factors were isolated using in vitro biochemical approaches, additional cell biology studies should prove useful to determine the cellular events in which these proteins are involved in vivo.
The three-subunit CAF-1 protein was initially purified from human cells based upon its ability to promote chromatin assembly onto replicating DNA in the SV-40 replication system (Stillman, 1986; Smith and Stillman, 1989). The small subunit of CAF-1, known as RbAp48, or p48, is part of multiple complexes involved in different aspects of histone metabolism (Roth and Allis, 1996; Verreault et al., 1998). In contrast, the two large subunits, p150 and p60, are specific to the CAF-1 protein. The immunolocalization of the two largest subunits of CAF-1 in an asynchronous population of cells revealed that these subunits are predominantly nuclear (Smith and Stillman, 1991). In addition, analysis of S phase nuclei demonstrated colocalization of CAF-1 and replication foci (Krude, 1995a ), consistent with a role for CAF-1 as an assembly factor during DNA replication. Complementation of nucleosome assembly in the human cell-free DNA replication system further allowed the identification of CAF-1 orthologues from other species, such as Drosophila, Xenopus, and Saccharomyces cerevisiae (Gaillard et al., 1996; Kamakaka et al., 1996; Kaufman et al., 1997). Although CAF-1 is not essential for S. cerevisiae viability (Enomoto et al., 1997; Kaufman et al., 1997), yeast strains lacking CAF-1 exhibit silencing defects and are sensitive to UV irradiation (Enomoto et al., 1997; Kaufman et al., 1997; Monson et al., 1997; Enomoto and Berman, 1998).
The cellular response to UV irradiation is a complex process which involves the translation of the presence of the toxic agent into cellular signaling, part of which is the detection and processing of DNA lesions (Herrlich et al., 1994). Indeed, ultraviolet C (UV-C), as a genotoxic agent (Pfeifer, 1997), produces mutagenic lesions in DNA, including cyclobutane pyrimidine dimers and 6-4 photoproducts, which are mainly repaired by nucleotide excision repair (NER) (Sancar, 1995; Wood, 1997). Importantly, studies monitoring the nuclease sensitivity of chromatin after UV irradiation revealed that NER is accompanied by nucleosomal rearrangements (Smerdon and Lieberman, 1978). Insights into the coordination between NER and chromatin assembly were obtained by in vitro studies using human cell extracts (Gaillard et al., 1996); these studies suggested a possible role for CAF-1 in linking these two events. The activity of CAF-1 during NER in vitro may account for the UV-sensitive phenotype that results from disruption of the genes encoding the CAF-1 subunits in S. cerevisiae.
To investigate the role of CAF-1 in vivo in human cells, we analyzed the subcellular distribution and posttranslational modification of CAF-1 after UV irradiation of HeLa cells. The association of the p150 and p60 subunits of CAF-1 with chromatin was found to increase substantially in irradiated cells. This occurred in parallel with the recruitment to chromatin of the DNA replication and repair factor proliferating cell nuclear antigen (PCNA). Furthermore, biochemical analysis of the chromatin-associated form of p60 revealed that it was mostly phosphorylated after UV irradiation. Association of a phosphorylated form of p60 with chromatin occurred rapidly and in a dose- dependent manner after UV irradiation. Consistent with these in vivo observations, we found that the amount of CAF-1 required to stimulate nucleosome assembly during the repair of UV photoproducts in vitro depended upon both the number of DNA lesions and the phosphorylation state of CAF-1. This parallel between the UV response in vivo and the in vitro assay are consistent with a response mediated through DNA damage processing rather than being a mere consequence of other cellular UV responses. This paper constitutes the first report of CAF-1 recruitment to chromatin in response to UV irradiation of human cells and supports a physiological role for CAF-1 in linking chromatin assembly to DNA repair.
Materials and Methods
Cell Culture, Synchronization, and UV Irradiation
HeLa cells were grown in Petri dishes (Falcon Plastics, Cockeysville, MD) in Dulbecco's modified Eagle's medium (DME) supplemented with 10% fetal calf serum, 10 mg/ml antibiotics (penicillin and streptomycin), and 2 mM l-glutamine (GIBCO BRL, Gaithersburg, MD) at 37°C under an atmosphere of 5% CO2. For synchronization, cells grown to ∼50% confluence were arrested in early S phase with 2 mM hydroxyurea (Sigma Chemical Co., St. Louis, MO) for 16 h then released by several washes in phosphate-buffered saline (PBS) before a final wash in fresh medium as described (Todorov et al., 1995). FACScan™ analysis revealed that these cells remained in S phase for 10 h before entering G2 phase (3 h), mitosis (1 h), and finally G1 phase ∼14 h after release. We routinely collected early G1 cells 15.5 h after release. The proportion of cells in the various phases of the cell cycle was determined by flow cytometry of fixed cells. For flow cytometry analysis, cells washed in PBS were collected by centrifugation after trypsinization, fixed in ethanol, washed with PBS, and then pelleted by centrifugation at 600 g to be treated with 100 mg/ml RNase A (Boehringer Mannheim, Mannheim, Germany) and 10 mg/ml propidium iodide (Sigma Chemical Co.) in PBS with 0.1% Tween. Data were collected using a FACScan™ flow cytometer (Becton Dickinson, San Jose, CA). For UV treatment, HeLa cells grown in a glass Petri dish at a density of 5 × 104 cells/cm2 (50% confluent), were rinsed with PBS twice, PBS was removed, and then the cells were exposed to UV-C light (254 nm) from a low-pressure mercury lamp. The distance between the lamp and the dish was adjusted to obtain a fluence of 1 J/m2 per second measured using a Latarjet Dosimeter (Institut Curie, Paris, France). The irradiation time was varied to obtain doses from 10 to 30 J/m2. After irradiation, growth medium was added and the cells were allowed to recover for the indicated times. Cell cycle arrest after UV irradiation was verified by flow cytometry by comparison with nonirradiated and mock-treated cells synchronized in parallel.
Cell Extractions
Extractions were performed by a method adapted from Todorov et al. (1995). For the immunofluorescence analysis, cells grown on coverslips were washed first with PBS+ which contains 0.5 mM MgCl2 and 0.5 mM CaCl2 (and then with cytoskeleton [CSK] buffer [10 mM Pipes-KOH, pH 7.0, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2 {Fey et al., 1986}]). At this stage, cells were either directly fixed with 4% paraformaldehyde in PBS+ for 20 min at 24°C or subjected to various treatments: (a) for the Triton extraction, cells were incubated in CSK buffer supplemented with 0.5% Triton X-100, 0.5 mM PMSF (Sigma Chemical Co.), and 10 mg/ml leupeptin (Boehringer Mannheim) for 5 min at 20°C; (b) for the DNase I digestion, 0.1 mg/ml DNase I (Grade II; Boehringer Mannheim) was added to the same extraction buffer and incubated 5 min at 20°C. After two washes in CSK buffer, the cells were fixed in 4% paraformaldehyde in PBS1. For biochemical analysis, the cells were trypsinized, pelleted (600 g for 10 min), and then washed with PBS+ and CSK buffer. Using the conditions described above, the cells in suspension were subjected to various extraction procedures. The Triton-extracted and DNase I–treated cells were harvested after addition of an excess of CSK buffer rapidly followed by centrifugation to collect the pellets. After a wash in CSK buffer, the pellets were resuspended in CSK buffer at a final dilution corresponding to 2.5 × 104 cells/ml. An equal volume of 2× Laemmli buffer was added to the samples before boiling for 10 min, loading on a SDS-polyacrylamide gel, and Western blotting.
Preparation of Cell-free Extracts
The procedure for preparing cytosolic extracts uses hypotonic buffer (Li and Kelly, 1984) and the properties of these extracts depends on the physiological state of the cells. In brief, the cytosolic extract was prepared from HeLa cells grown in dishes essentially as described (Krude et al., 1997). After rinsing twice in PBS buffer, cells were allowed to swell for 10 min in 20 ml of ice-cold extraction buffer E (20 mM Hepes-KOH, pH 7.8, 5 mM potassium acetate, 0.5 mM MgCl2, and 0.5 mM DTT) per dish and excess buffer was removed. All subsequent steps were carried out at 4°C. Mitotic cells that detached from the dish under the hypotonic conditions were discarded and interphase cells were then scraped off the plates and disrupted them with 25 strokes in a Dounce homogenizer (1-ml Dounce tissue grinder; Wheaton, Millville, NJ) using a loose-fitting pestle. Nuclei were pelleted at 1,500 g for 3 min and the supernatant was recentrifuged at 14,000 g for 20 min at 4°C. The cytosolic extract was then aliquoted and frozen in liquid nitrogen. For nuclear extract, pelleted nuclei were resuspended at 1.5 × 108 nuclei/ml in buffer E supplemented with NaCl to reach a final concentration of 0.6 M and incubated for 90 min at 4°C. Nuclear material was pelleted by centrifugation at 14,000 g for 20 min and the supernatant corresponding to the interphasic nuclear extract was collected. Aliquots were frozen in drops in liquid nitrogen. Mitotic extracts were prepared as previously described (Marheineke and Krude, 1998).
Antibodies
Monoclonal antibodies against the two large subunits of human CAF-1, p150 (mAb1), and p60 (mAb96) (Smith and Stillman, 1989) were kindly provided by B. Stillman (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). The rabbit polyclonal antibody, pAb1, directed against the p60 subunit was used to reveal both phosphorylated and nonphosphorylated forms of the p60 subunit as described (Marheineke and Krude, 1998). The specificity of pAb1 and mAb96 was verified by Western blotting (see Fig. 5) as well as by a double immunofluorescence labeling of fixed HeLa cells (data not shown). Double immunofluorescence assays were also carried out using pAb1 and mAb1 (Marheineke and Krude, 1998) (data not shown). The other monoclonal antibodies used were: PC10 (Dako, Carpinteria, CA) specific for PCNA, mAb 414 (Berkeley Antibody Co., Richmond, CA) detecting three nucleoporins nup62, nup175, and nup270 (Berkeley Antibody Co.) and an anti-NuMa antibody (Calbiochem-Novabiochem, La Jolla, CA) detecting the nuclear mitotic apparatus also called nuclear matrix-associated protein (NuMA).
Figure 5.
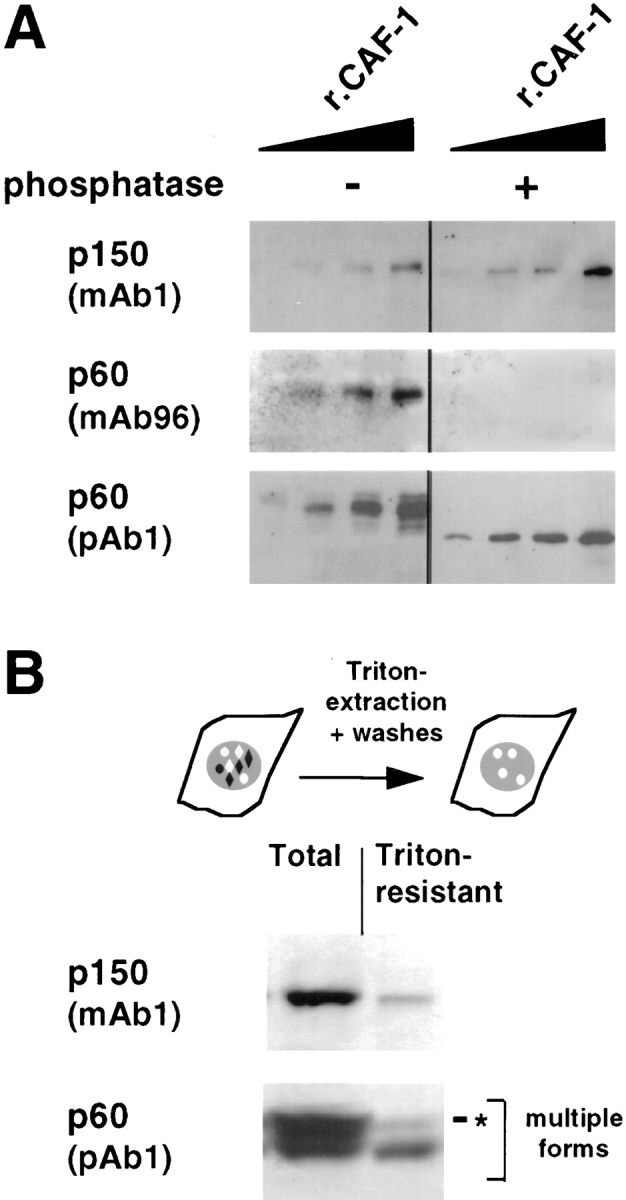
In G2 cells, the phosphorylated form of p60 is underrepresented in the Triton-resistant fraction. (A) Antibody specificity was determined by Western blotting using recombinant CAF-1 protein (rCAF-1) protein from baculovirus-infected insect cells. CAF-1, either treated (+) or not treated (−) with Lambda phosphatase, was loaded in various quantities (10, 20, 40, and 80 ng). The Western blots were probed with monoclonal antibodies against p150 (p150, mAb1) and p60 (p60, mAb96) as well as with a polyclonal antibody against p60 (p60, pAb1). (B) G2 cells were obtained as described in Fig. 4. Proteins from either whole cells (Total) or Triton-extracted cells (Triton-resistant) were analyzed by Western blotting. In each case, a lysate corresponding to 2.5 × 105 cells was loaded. The monoclonal antibody against p150 (p150, mAb1) and the polyclonal antibody against p60 (p60, pAb1) were used to probe the same blot. Multiple forms, multiple forms of the p60 subunit of CAF-1; asterisk, the slowest migration form.
Immunofluorescence Microscopy
Paraformaldehyde-fixed cells were first permeabilized with 0.2% Triton X-100 in PBS for 5 min and blocked in 5% BSA, 0.1% Tween in PBS (blocking buffer) for 10 min. Specific antibodies were added at the appropriate dilution in blocking buffer and incubated for 1 h at room temperature. For the double staining with PCNA, a second fixation step in methanol at −20°C for 10 min was included before addition of the anti-PCNA antibody to optimize recognition as reported earlier (Aboussekhra and Wood, 1995). To visualize the primary antibodies, fluorescein- or Texas red–conjugated goat anti–rabbit or anti–mouse IgG antibodies were used (Jackson ImmunoResearch Laboratories, West Grove, PA) and 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) (Sigma Chemical Co.) was added for DNA staining. The coverslips mounted in Vectashield (Vector Laboratories, Burlingame, CA) were observed using an epifluorescence photomicroscope (model DMR; Leica, Deerfield, IL) equipped with an HBO100 mercury lamp (Osram, Germany), 63× objective lenses, and a chilled charge-coupled device camera (Hamamatsu Phototonics, Hamamatsu City, Japan). Confocal analysis was carried out using an inverted microscope (Leica), and scanning was performed with an optical confocal scanner LEICA TCS 4. The images were produced using the Adobe Photoshop 4 software program (San Jose, CA).
Repair/Chromatin Assembly Reaction In Vitro
pBS plasmid (pBluescript KS; Stratagene, La Jolla, CA) isolated using a plasmid purification kit (QIAGEN, Santa Clarita, CA) was irradiated on ice with UV-C light (254 nm) from a low-pressure mercury lamp at various fluences (Wood et al., 1995). According to Wood et al. (1995), a fluence of 100 J/m2 induces roughly one photoproduct in 1,000 bp (∼0.75 cyclobutane pyrimidine dimer and 0.25 [6-4] photoproduct). It can thus be estimated for our plasmids that on average approximately four photoproducts will be present for 150 J/m2 and eight for 300 J/m2. The cytosolic extracts were used in 25 μl DNA repair reactions at 37°C essentially as described (Gaillard et al., 1996). In brief, reactions contained 100 μg of proteins contributed by the extract, 150 ng of nontreated or UV-C–irradiated pBS plasmid, 5 mM MgCl2, 40 mM Hepes-KOH, pH 7.8, 0.5 mM DTT, 4 mM ATP, 20 μM each of dGTP, dATP, and dTTP, 8 μM dCTP and 2 μCi [α-32P] dCTP (3,000 Ci/mmol), 40 mM phosphocreatine, 2 μg creatine phosphokinase (Type 1; Sigma Chemical Co.), and various sources of CAF-1. Reactions were stopped by the addition of EDTA to 15 mM, SDS to 0.35%, and 100 μg/ml RNase A (Boehringer Mannheim). After treatment with 200 μg/ml proteinase K (Boehringer Mannheim), DNA was isolated and subjected to electrophoresis (1.2% agarose, in TAE buffer) for DNA topology analysis. Ethidium bromide staining of the gel allowed verification of DNA recovery. The autoradiographs revealed the migration pattern of the plasmids labeled during the reaction. Quantification was carried out using a PhosphorImager and ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Phosphorylation Analysis
20–400 U of Lambda protein phosphatase (New England Biolabs, Beverly, MA) were used to treat 100 ng of recombinant human CAF-1 protein purified from baculovirus-infected Sf9 cells (Kaufman et al., 1995). When indicated, as a control to limit the phosphatase reaction and produce intermediate phosphorylated forms of recombinant CAF-1, 25 mM β-glycerophosphate (Sigma Chemical Co.) was added. Mock-treated samples were incubated similarly, but without Lambda phosphatase. Phosphorylation was monitored by Western blotting as shown in Fig. 8. All treated samples were adjusted to 25 mM β-glycerophosphate (Sigma Chemical Co.) and frozen before use. We reached in the repair/chromatin assembly reaction in vitro a 10 mM final concentration of β-glycerophosphate.
Figure 8.
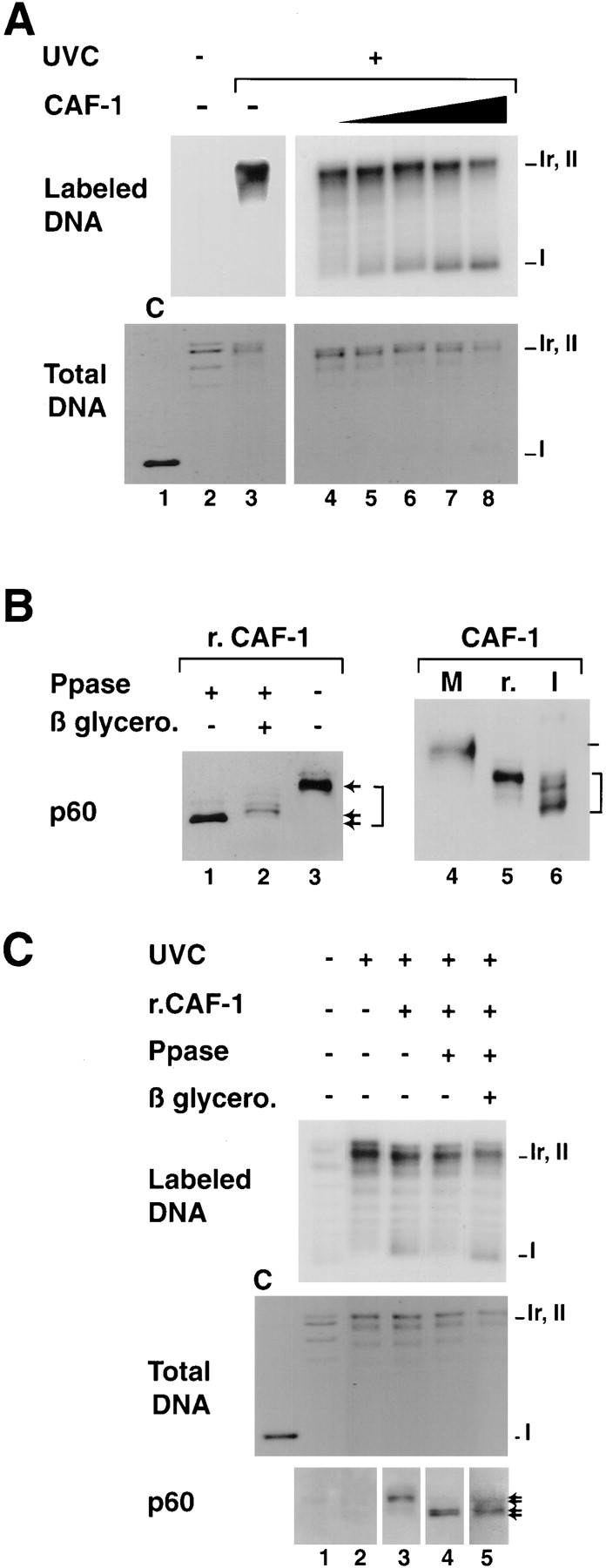
Titration conditions for recombinant CAF-1 to support nucleosome assembly during UV-induced synthesis and competence of various phosphorylated forms in the assay. (A) Plasmid DNA, either treated with UV-C at 500 J/m2 (+) or not treated (−) were incubated in a cytosolic extract in the presence of [α-32P] dCTP. Complementation in the supercoiling assay was achieved with increasing amounts of recombinant CAF-1 (solid triangle), 0 ng (lanes 2, 3, and 4), 5 ng (lane 5), 10 ng (lane 6), 20 ng (lane 7), or 40 ng (lane 8). Deproteinized DNA was purified and analyzed by agarose gel electrophoresis. The incorporation of radiolabel due to the UV-dependent DNA synthesis was visualized by autoradiography (Labeled DNA) and the total population of DNA molecules by ethidium bromide staining of the gel (Total DNA). Plasmids were processed as indicated in Materials and Methods. The positions of the supercoiled (form I), nicked (form Ir), and closed circular (form II) forms of plasmid DNA are indicated. Control supercoiled DNA was run in parallel (lane 1). (B) Recombinant CAF-1 was treated with Lambda phosphatase alone (lane 1), in the presence of β-glycerophosphate (lane 2) or mock treated (lanes 3 and 5), mitotic extract (M) and nuclear interphasic extract (I) were prepared as described (see Materials and Methods). All samples were processed for Western blot analysis and detection with the polyclonal anti–p60 antibody (p60). Bracket, various phosphorylation forms. (C) The three sources of recombinant CAF-1 (r.CAF-1): treated with Lambda phosphatase alone (lane 4), in the presence of β-glycerophosphate (lane 5), or mock treated (lane 3) (shown in B, left) were used in the assay as described above. The amount added per reaction was 10 ng (defined as the critical amount in A). The presence and amount of the phosphorylated p60 subunit of CAF-1 (exogenous and endogenous) at the end of each reaction was assessed by Western blotting using the polyclonal anti-p60 antibody (p60). Asterisk, slowest migrating phosphorylated form of p60.
Western Analysis
Protein samples in 1× Laemmli buffer were separated in SDS–7% polyacrylamide gels (Laemmli, 1970) to resolve both the p150 and the various phosphorylated forms of p60. Protein transfer to nitrocellulose membrane (0.45 μm; Schleicher & Schuell, Keene, NH) was achieved in 48 mM Tris, 39 mM glycine, 0.03% SDS, and 20% ethanol using a semi-dry electroblotting system (Bio-Rad, Hercules, CA). Ponceau S (Rhone-Poulenc, Lyon, France) staining allowed to verify protein recovery. After blocking in 5% non-fat dry milk in Tris-buffered saline (TBS; 136 mM NaCl, 2.7 mM KCl, 25 mM Tris, pH 8), antibodies were added in blocking buffer plus 0.1% Tween 20, incubated for 1 h, and then washed in 0.1% Tween 20 in TBS. The primary antibodies were detected with horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies (Jackson ImmunoResearch Laboratories), and the Supersignal substrate detection kit (Pierce Chemical Co., Rockford, IL). For semiquantitative analysis, various amounts of recombinant CAF-1 were processed in parallel and linearity of signal was verified.
Results
UV Irradiation Induces an Increase in the Triton-resistant Nuclear Pool of the Two Large Subunits of CAF-1
An asynchronous population of HeLa cells was subjected to a short pulse of UV-C irradiation (30 J/m2 at 254 nm, a dose previously used to detect UV response in HeLa cells) (Carty et al., 1994). After a 2-h recovery period, the subcellular localization of the two large subunits of CAF-1 was monitored using either monoclonal antibodies (mAb1 for p150 and mAb96 for p60; Smith and Stillman, 1991) or polyclonal antibodies (pAb1 for p60; Marheineke and Krude, 1998). Consistent with a previous report (Smith and Stillman, 1991), control cells before irradiation displayed a nuclear staining whose intensity varied in individual cells. After UV irradiation, the staining became reproducibly more intense in all nuclei irrespective of their position in the cell cycle (Fig. 1 A). However, this effect was not dramatic.
Figure 1.
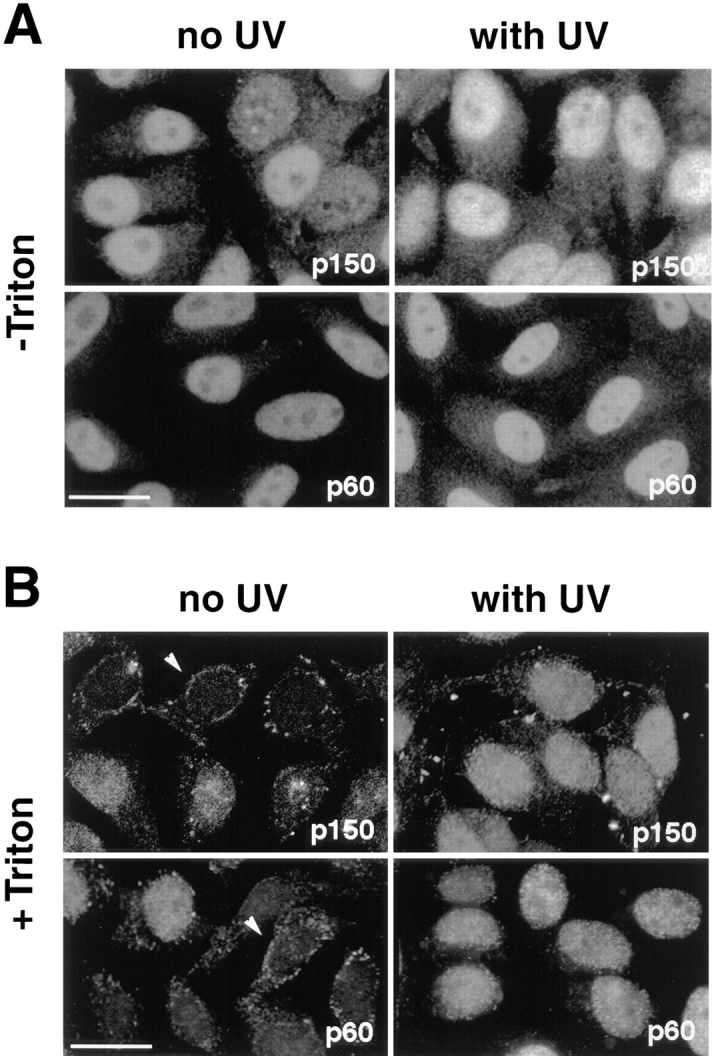
UV irradiation of HeLa cells induces an increase in the immunofluorescence staining and the Triton-resistance of p150 and p60 subunits of CAF-1. Asynchronous HeLa cells irradiated at 30 J/m2 (with UV) were compared with untreated cells (no UV). UV-irradiated cells were allowed to recover in growth medium for 2 h before immunofluorescence analysis. (A) Cells fixed directly with formaldehyde (−Triton) or (B) cells extracted with Triton X-100 (+Triton) before fixation. The p150 and p60 subunits of CAF-1 were visualized using the monoclonal antibodies mAb1 and mAb96. White arrows, cells with a low staining intensity. Bars, 10 μm.
Some DNA repair proteins such as the PCNA (see below) become resistant to extraction with the nonionic detergent Triton X-100 after UV irradiation of cells, a phenomenon that is thought to reflect the association of the repair proteins with insoluble nuclear structures at the sites of DNA repair (Cox, 1997). This prompted us to find out whether CAF-1 also became resistant to extraction by nonionic detergent during DNA repair. Control and UV-irradiated cells grown on coverslips were permeabilized with Triton X-100 before formaldehyde fixation and analysis by immunofluorescence (see Materials and Methods). Strikingly, in control cells, the signal for both the p150 and p60 subunits of CAF-1 showed a considerable variation in intensity among individual cells (Fig. 1 B, left). The most extreme situation under these extraction conditions was seen in a few cells which showed hardly any residual intranuclear staining, except for a rim at the periphery of the nucleus (Fig. 1 B, arrowheads). Remarkably, after UV irradiation, the entire cell population displayed a strong nuclear staining that was resistant to Triton extraction (Fig. 1 B). Thus, detergent extraction revealed the existence of a Triton-resistant nuclear pool of CAF-1 which increased in every cell upon UV irradiation.
A Parallel Behavior for the p60 Subunit of CAF-1 and PCNA
For the p60 subunit of CAF-1, either the monoclonal mAb96 (Fig. 1, p60) or the polyclonal antibody pAb1 (Fig. 2, p60*) allowed us to visualize a Triton-resistant nuclear pool of CAF-1 that increased in every cell upon UV irradiation. It was thus possible to perform a double staining with a monoclonal antibody raised against PCNA and the polyclonal antibody (pAb1) directed against the p60 subunit of CAF-1 (see Materials and Methods). The detection of PCNA allowed us to identify S phase cells in an asynchronous population of control cells and to ensure that the irradiated cells responded properly to UV irradiation (Bravo, 1987; Toschi, 1988).
Figure 2.
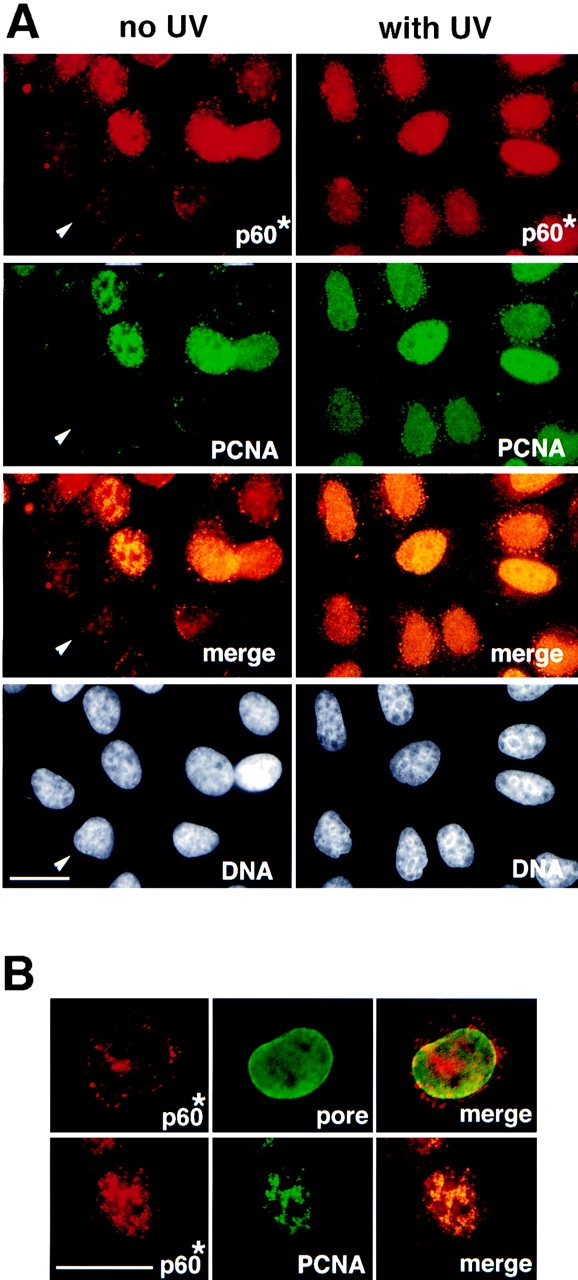
The p60 subunit of CAF-1 and PCNA similarly become Triton-resistant in response to UV irradiation. (A) Asynchronous HeLa cells, either untreated (no UV) or irradiated with UV (with UV), were extracted with Triton X-100 before fixation. The p60 subunit of CAF-1 (p60*) was visualized in red using a polyclonal antibody (pAb1) and PCNA labeled in green in the same cells using monoclonal antibody PC10. DAPI staining in grey (DNA) locates all the nuclei, including those with no signal detected for the immunolabeling (white arrow). The merge of the PCNA and p60 signals in yellow allows identification of positive cells for both markers. (B) A higher magnification control for the integrity of nuclei following the Triton extraction procedure by a double immunostaining using a polyclonal antibody against p60 (p60*, red) and a monoclonal antibody against nucleoporins (pore, green) performed with nonirradiated cells extracted with Triton. A cell displaying a typical weak staining for the p60 marker is presented in the top panel (see also Fig. 1 B, white arrow). The bottom panel shows at the same magnification a typical cell displaying a positive staining for both p60 (p60*, red) and PCNA (green). Bars, 10 μm.
In an asynchronous population of control cells, only the S phase cells exhibit high levels of Triton-resistant p60 subunit of CAF-1 and PCNA (Fig. 2 A, no UV, p60* and PCNA). In some cells where the number of PCNA foci were not very numerous, the p60 subunit of CAF-1 and PCNA clearly colocalized (Fig 2 B, p60* and PCNA). Double-staining with an antibody that detects three nucleoporins (mAb414) allowed us to verify that the nuclear envelope was preserved in cells that displayed a weak p60 staining following Triton extraction (Fig. 2 B, p60* and pore). In striking contrast, all cells in an asynchronous population exhibited intense p60 and PCNA staining after UV irradiation. These data suggest that CAF-1, like PCNA, is mobilized for a function required during both DNA replication and nucleotide excision repair.
The Triton-resistant Nuclear Fraction of CAF-1 p150 and p60 Is Associated with Chromatin
We then investigated the nature of the nuclear retention of the p150 and p60 subunits of CAF-1. Cells were treated with DNase I in the presence of detergent. Disappearance of the p150 and p60 signals paralleled degradation of DNA as shown by DAPI staining (Fig. 3, compare p150, p60*, and DNA). The disappearance of the staining cannot be attributed to a disruption of the nuclear envelope during the DNase I treatment since staining of the nuclear pores in the envelope with a nucleoporin-specific antibody was preserved (Fig. 3 A, pore). These data demonstrate that the Triton-resistant p150 and p60 subunits of CAF-1 are associated with chromatin.
Figure 3.
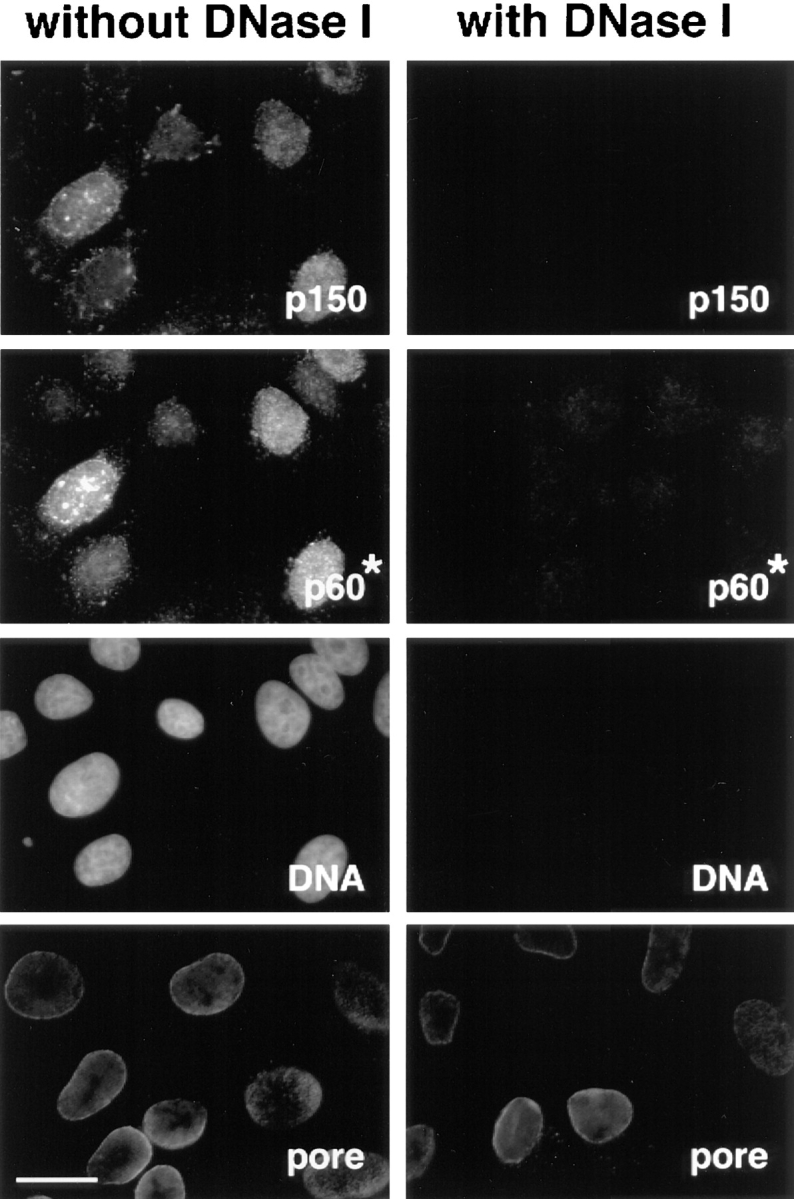
The immunofluorescence staining corresponding to the Triton-resistant fraction of the p150 and p60 subunits disappears following DNase I treatment. Asynchronous cells were extracted with Triton in the absence (without DNase I) or in the presence of 0.1 mg/ml DNase I (with DNase I). After fixation, cells were stained with a monoclonal anti-p150 antibody (p150) and polyclonal anti-p60 antibody (p60*). DAPI staining (DNA) confirmed the disappearance of DNA after DNase I digestion. Monoclonal antinucleoporins antibody (pore) was used to verify the integrity of the nuclear envelope. Bar, 10 μm.
The Chromatin-associated Fraction of the p60 Subunit of CAF-1 Increased Dramatically after UV Irradiation of G2 Cells
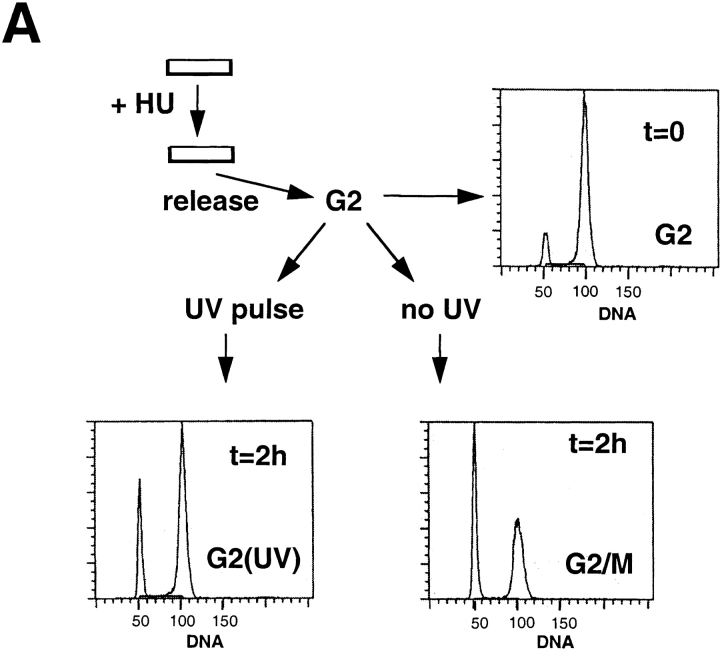
To further analyze the role of CAF-1 in the response to UV-induced DNA damage, HeLa cells were synchronized in the G2 phase of the cell cycle (Fig. 4 A, FACScan™ G2). After UV irradiation, we observed that cells were delayed in their cell cycle progression (Fig. 4 A, FACScans™ G2[UV] and G2/M) consistent with the activation of the G2-DNA damage checkpoint (for review see Nurse, 1997). Triton extraction and immunofluorescence analysis were then carried out to compare control G2 cells (Fig. 4 A, FACScan™ G2) and G2 cells irradiated with UV (Fig. 4 A, FACScans™ G2[UV]). The Triton-extracted G2 cells displayed a very weak nuclear staining for both p150 and p60 with either monoclonal (mAb1 and mAb96) or polyclonal antibodies (pAb1 for p60) (Fig. 4 B, left). This pattern was similar to the one observed for the Triton- extracted cells indicated by the arrows in Fig. 1. Therefore, our synchronization procedure did not promote the recruitment of p60 and p150 to Triton-resistant nuclear structures. After UV-C irradiation (30-J/m2 pulse, 2-h recovery), a strong nuclear staining could be detected in all cells (Fig. 4 B, right). This strong nuclear staining in response to UV irradiation, reflected numerous discrete sites as revealed by confocal microscopy analysis (Fig. 4 B, p60*) which were clearly distinct in size and number from the replication sites described for late S phase nuclei (O'Keefe et al., 1992). Since indirect immunofluorescence is strongly dependent upon the conformation and accessibility of the epitopes in the cell, we also investigated biochemically whether changes in total concentration and/or posttranslational modification of CAF-1 occurred in response to UV irradiation.
Figure 4.
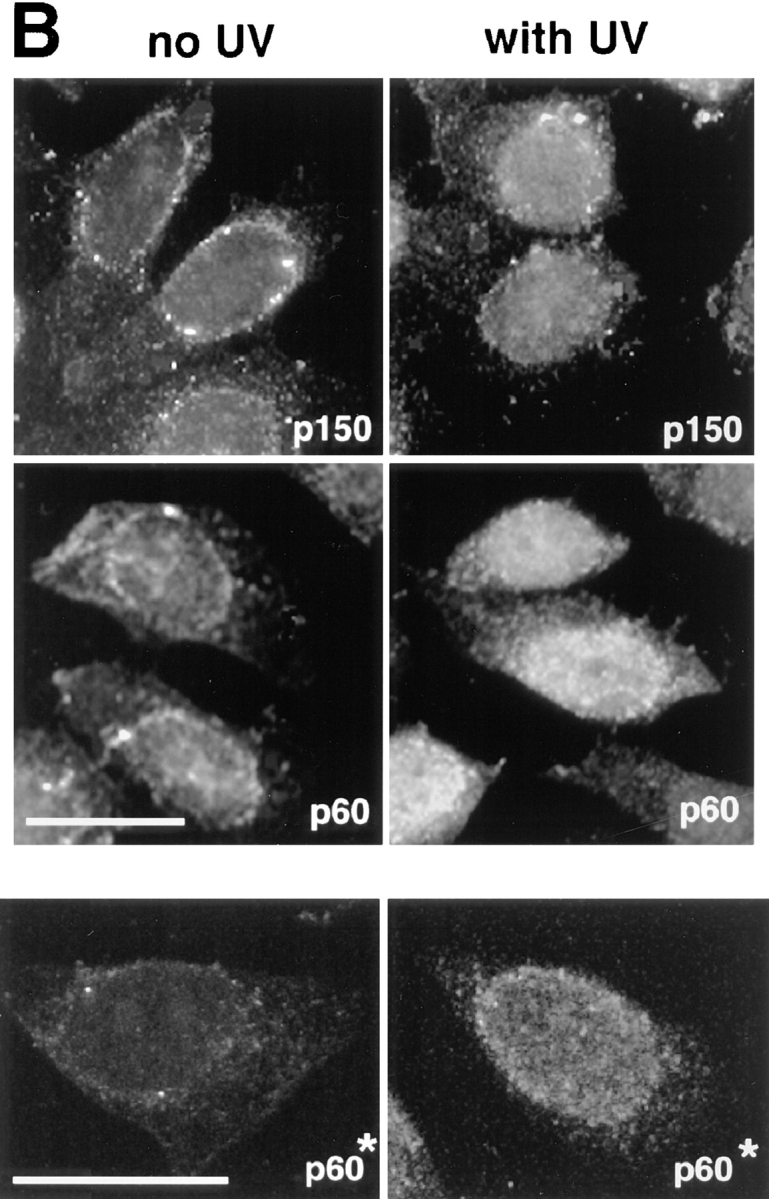
UV irradiation of G2 cells induces a significant increase of the immunofluorescence staining corresponding to the Triton-resistant fraction of p150 and p60 subunits of CAF-1. (A) Cells were synchronized by addition of hydroxyurea (HU). After release, G2 cells (FACScan™ analysis, t = 0, G2) were either immediately UV irradiated at 30 J/m2 (UV pulse) or not irradiated (no UV) and harvested after a 2-h recovery time period. FACScan™ analysis revealed that the majority of UV-irradiated cells were arrested in G2 (t = 2 h, G2 [UV]), whereas a significant proportion of nonirradiated cells were able to reach the G2/M transition (t = 2 h, G2/M). The x and y axis in the FACScans™ are in arbitrary units. (B) A comparison between cells corresponding to the FACScan™ analysis presented in A: t = 0, G2 (left, no UV) and t = 2 h, G2 (UV) (right, with UV). After Triton extraction, cells were fixed and stained with monoclonal antibodies against p150 (p150), monoclonal antibodies against p60 (p60), or a polyclonal antibody against p60 (p60*). Top (p150 and p60) represents data collected using a a standard epifluorescence microscope whereas the bottom panel (p60*) shows a confocal analysis accumulating four optical sections of 1 μm. Bar, 10 μm.
Phosphorylated Forms of the p60 Subunit Are Enriched in the Triton-resistant Fraction of CAF-1 from UV-irradiated Cells
The p60 subunit of CAF-1 consists of multiple phosphorylated forms that can be resolved by SDS-PAGE (Smith and Stillman, 1991). These various forms can be detected with the polyclonal antibody (pAb1, Marheineke and Krude, 1998) in recombinant CAF-1 purified from baculovirus-infected insect cells (Fig. 5 A). In contrast, when used on the same filter, a monoclonal antibody raised against the p60 subunit of CAF-1 purified from human cells (mAb96), detected only slow migrating forms of the p60 subunit (Fig. 5 A, mAb96). These forms correspond to phosphorylated forms of p60 as revealed by phosphatase treatment (Fig. 5 A, compare − and +). Thus, mAb96 is specific for phosphorylated form(s) of the p60 subunit in Western blots. In contrast, the monoclonal antibody against the p150 subunit of CAF-1 used in this study (mAb1) detected p150 irrespective of the phosphatase treatment used (Fig. 5 A and data not shown).
No major cell cycle-dependent changes in p60 phosphorylation could be detected in total cell lysates either from asynchronous cells or cells enriched in various stages of the cell cycle (Fig. 6 A) (data not shown). In G2 cells, a single band was detected for p150 and multiple bands in equal proportion for p60 (Fig. 5 B, Total). In these control G2 cells, Triton extracted most of the p150 and p60 molecules (Fig. 5 B, Triton-resistant) which is consistent with our immunofluorescence data (Fig. 4 B, no UV). Strikingly though, the various phosphorylated forms of p60 were preferentially extracted by Triton in G2 control cells leading to drastic removal of the phosphorylated forms (Fig. 5 B, p60, compare Total with Triton-resistant).
Figure 6.

UV irradiation of both G1 and G2 cells promotes the chromatin association of a phosphorylated form of p60. (A) Cells synchronized in early G1 or G2 phase were either immediately harvested (−) or subjected (+) to UV irradiation at 30 J/m2 followed by a 2-h recovery period. Total lysates were processed for Western blotting and detection was achieved using both the polyclonal anti-p60 antibody (p60, pAb1) and monoclonal anti-NuMA antibody (NuMA) as a control for extraction and protein loading. (B) The cells were Triton-extracted and an equivalent of 2.5 × 105 cells for G2 cells or 5 × 105 cells for G1 cells, corresponding to the postmitotic doubling of the cell population were lysed and processed for Western blotting. Detection was achieved with monoclonal anti-p150 (p150, mAb1), monoclonal anti-p60 (p60, mAb96), polyclonal anti-p60 (p60, pAb1) and monoclonal anti-PCNA (PCNA) antibodies. (C) DNase I digestion was performed in parallel with G1 or G2 cells in the absence of UV irradiation (−UV) and with UV-irradiated G2 cells (+UV) at 30 J/m2. The cells were Triton-extracted in the absence (−) or presence (+) of 0.1 mg/ml DNase I. Lysates equivalent to 2.5 × 105 cells were analyzed by Western blotting using both the polyclonal anti-p60 antibody (p60, pAb1) and monoclonal anti-NuMA antibody (NuMA) as a control for extraction and protein loading. Asterisk, slowest migration position of the p60 subunit.
To determine the fate of CAF-1 subunits during the cellular response to UV irradiation, cell populations enriched in G1 or G2 cells were treated with UV and, after a 2-h recovery period, the cells were processed to estimate first whether any change could be detected in the phosphorylation state of p60 in a total lysate (Fig. 6 A). For both G1 and G2 cells a comparable distribution of various forms of p60 could be detected without major change after UV irradiation. We then analyzed on parallel samples, the fraction of p150, p60, and PCNA molecules that had become resistant to Triton extraction by Western blotting (Fig. 6 B). In these experiments, the appearance of Triton-resistant PCNA served as a positive control to monitor the cellular response to UV irradiation (Fig. 6 B, PCNA). Consistent with previous reports (Toschi, 1988; Aboussekhra and Wood, 1995), the PCNA protein became Triton-resistant after UV irradiation of both G1 and G2 cells (Fig. 6 B, PCNA). Interestingly, unlike in G2 cells, in nonirradiated G1 cells the phosphorylated forms of the p60 subunit of CAF-1 were not selectively extracted by Triton (Fig. 6 B, p60 and pAb1). This is in striking contrast with nonirradiated G2 cells in which phosphorylated p60 was almost completely absent from the detergent-resistant structures (Fig. 5 B and Fig. 6 B, G2). However, after UV irradiation of both G1 and G2 cells, an increase in the Triton-resistant phosphorylated forms of p60 was observed (Fig. 6 B, p60 and pAb1). For both normal G1 and UV-irradiated G2 cells, we analyzed the nuclear retention of the p60 subunit of CAF-1 using DNase I as described in Fig. 3. As a control, the same blots were reprobed to detect the presence of the NuMA protein (Lydersen and Pettijohn, 1980), a protein whose association with the nuclear matrix is insensitive to DNase I treatment (Fig. 6 C). In the Triton-resistant fraction, we found that both phosphorylated forms of the p60 subunit, present in normal G1 cells and UV-irradiated G2 cells, behaved similarly and were sensitive to DNase I treatment (Fig. 6 C). We conclude that, in both cases, the phosphorylated form of p60 is associated with chromatin.
Rapid Dose-dependent Response of CAF-1 to UV Irradiation
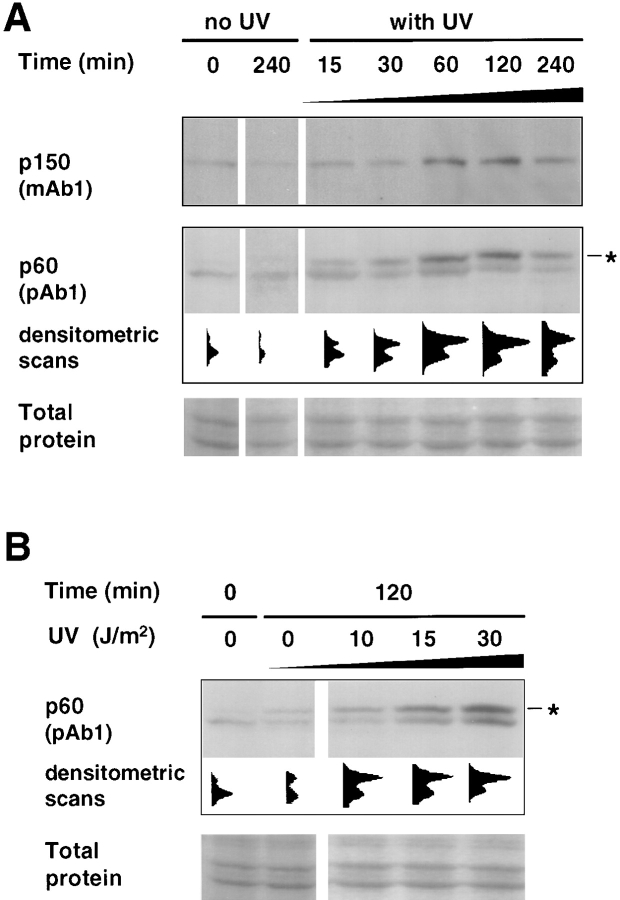
When cells are subjected to UV irradiation, a stress response is induced (Herrlich et al., 1994), cell cycle progression is arrested (Fig. 4 A), and lesions in the DNA are repaired. To gain some insight into the role of CAF-1 in this cellular response to UV irradiation, we first examined the kinetics of appearance of the phosphorylated forms of the p60 subunit of CAF-1 in the Triton-resistant fraction of G2 cells. As shown in Fig. 7 A, a UV dose of 30 J/m2 induced a significant accumulation of phosphorylated CAF-1 p60 as early as 30 min after UV irradiation, which is also clear from the densitometric scans corresponding to the individual lanes of this Western blot. This increase in phosphorylation occurred in parallel to an increase in the Triton-resistant fraction of p150 detected by Western blotting the same samples (Fig. 7 A, top). Thus, the appearance of Triton-resistant and phosphorylated CAF-1 subunits occurred as an early event after UV irradiation, consistent with the timing of early repair events (Pfeifer, 1997). We also noticed that these changes persisted for several hours postirradiation, a period during which the cells were still arrested in G2 phase of the cell cycle (data not shown). The accumulation of Triton-resistant phosphorylated forms of the p60 subunit of CAF-1 was also detected for lower UV doses (Fig. 7 B). A dose of 10 J/m2 already induced a significant increase in the amount of p60 protein retained after Triton extraction. The appearance of Triton-resistant CAF-1 p60 proportional to the UV dose (Fig. 7 B) was consistent with a possible stoichiometric targeting of CAF-1 to sites of DNA lesion/repair.
Figure 7.
Time course and UV dose response in the Triton-resistant fraction of p150 and p60 in G2 cells. (A) Time course analysis: G2 cells, either UV-irradiated at 30 J/m2 (with UV) or not irradiated (no UV) were harvested after various recovery time periods (min) and extracted with Triton X-100. Lysates (equivalent to 2.5 × 105 cells) were analyzed by Western blotting using both a monoclonal anti-p150 (p150, mAb1) and a polyclonal anti-p60 (p60, pAb1) on the same filter. (B) UV dose response: G2 cells irradiated with increasing doses of UV-C (0, 10, 15, or 30 J/m 2) were harvested after a 2-h recovery period and processed as in A. Asterisk, phosphorylated form of the p60 subunit of CAF-1. Densitometric scans of the p60 signal are presented underneath. Protein recovery (Total protein) and loading for each sample was assessed by ponceau S staining of the filters used to detect p60.
Phosphorylation Stimulates the Stoichiometric Action of CAF-1 In Vitro
We used an in vitro assay for chromatin assembly coupled to UV-induced DNA synthesis (Gaillard et al., 1996) to test whether CAF-1 functions either catalytically or stoichiometrically with respect to the number of lesions present in the DNA substrate. A cytosolic extract derived from nonirradiated HeLa cells grown in tissue culture dishes was prepared as described in Materials and Methods.
When added to our extract in the presence of a radiolabeled nucleotide, DNA synthesis preferentially occurred in the UV-irradiated plasmid when compared with a control nonirradiated plasmid (Fig. 8 A, compare − with + UV). Since DNA recovery at the end of these reactions was comparable (Fig. 8 A, Total DNA), these data demonstrated that our extract was competent to process UV photoducts. Under these conditions, we did not detect in the labeled/repaired plasmid DNA a significant proportion of supercoiled molecules. This could reflect either a defect in chromatin assembly (the accumulation of supercoiled molecules is proportional to the number of nucleosomes formed on a circular DNA molecule) (Germond et al., 1979), or a defect in ligation of repaired molecules. Western blotting analysis of an aliquot removed from these reaction mixtures showed that only traces of the p60 subunit of CAF-1 were provided by the extract (Fig. 8 C, p60, lanes 1 and 2), whereas the p150 was at an undetectable level (data not shown). Since the p150 and p60 subunits of CAF-1 are both essential for chromatin assembly in vitro (Kaufman et al., 1995), these data suggested that the limiting factor for supercoiling in this system was CAF-1. Nucleosome assembly and supercoiling of the repaired plasmid could indeed be complemented by the addition of purified recombinant CAF-1 (Fig. 8 A, CAF-1). Importantly, this supercoiling was preferentially observed on labeled/repaired material compared with the bulk population of plasmid DNA (Fig. 8 A, compare Labeled with Total DNA). A careful titration of the amount of CAF-1 added per reaction allowed us to define the amount of CAF-1 required to achieve maximal supercoiling (Fig. 8 A, lane 7).
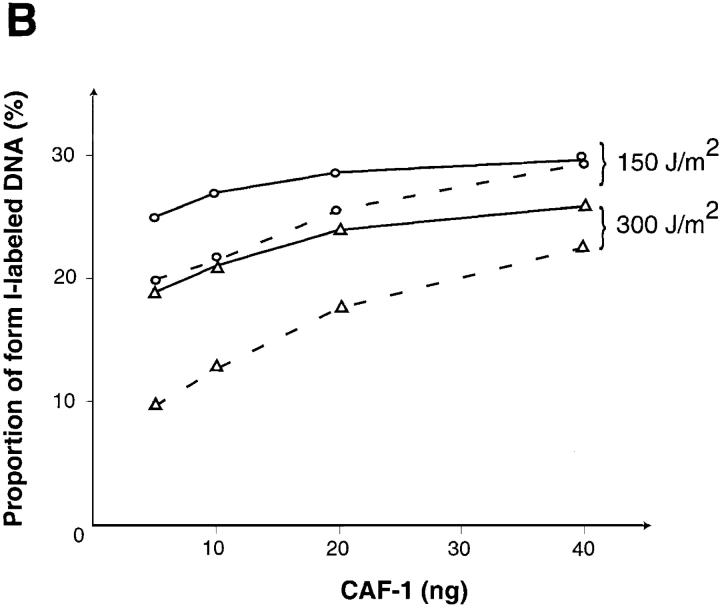
To proceed further and analyze the competence of various phosphorylated forms of CAF-1, the same amounts of the recombinant protein were treated (a) with Lambda phosphatase to completely dephosphorylate p60 (Fig. 8 B, lane 1); (b) with the phosphatase in the presence of β-glycerophosphate to produce phosphorylation intermediates of p60 (Fig. 8 B, lane 2); or (c) mock-treated (Fig. 8 B, lane 3). We also compared the migration properties of the p60 subunit in our recombinant complex with respect to the p60 subunit from mitotic extracts (Fig. 8 B, lane 4) or interphasic nuclear extracts (Fig. 8 B, lane 6). These data showed that the migration properties of the recombinant form were very distinct from those of the mitotic p60 and rather could be compared with the forms with the lowest migration properties in the interphasic nuclear extract. We thus used in the in vitro assay the three different forms of recombinant CAF-1 as produced in Fig. 8 B, lanes 1, 2, and 3. Under the limiting condition defined in Fig. 8 A, we found that when CAF-1 was treated with phosphatase before its addition to the extract, it was somewhat less active in promoting nucleosome assembly on labeled/repaired DNA (Fig. 8 C, lane 4). The fact that CAF-1 remained dephosphorylated was verified by Western blotting of an aliquot removed at the end of each reaction (Fig. 8 C, p60). Interestingly, when β-glycerophosphate was initially present during the phosphatase treatment to produce phosphorylation intermediates of p60, supercoiling was comparable to the control (Fig. 8 C, compare lane 3 with 5). We can thus discard possible effects due to contaminants in the phosphatase preparation, and further conclude that even intermediate phosphorylation forms can be competent in the assay. Since the phosphorylation state of the recombinant CAF-1 protein purified from baculovirus- infected Sf9 cells may not necessarily reflect the precise modification of the protein present in UV-irradiated HeLa cells, we verified that similar observations could be made when using equivalent amounts of CAF-1 from HeLa cell nuclear extracts (data not shown). A close examination of the supercoiling obtained using plasmids irradiated at 150 and 300 J/m2 for various amounts and forms of CAF-1 (Fig. 9 A as indicated) suggested that the number of labeled/repaired sites could be a critical parameter for CAF-1 requirement (Fig. 9 A, compare 150 with 300 J/m2). Quantification of the data further evidenced the correlation between the number of labeled repaired sites and the requirement of CAF-1 as well as the importance of the form of CAF-1 provided in the reaction (Fig. 9 B).
Figure 9.
The capacity of CAF-1 to support nucleosome assembly during UV- induced synthesis depends on both the number of lesion/repair sites and the phosphorylation form of CAF-1. (A) Plasmid DNA, either treated with UV-C (150 and 300 J/m2) or not treated (0) were incubated in a cytosolic extract in the presence of [α-32P] dCTP. Complementation in the supercoiling assay was achieved with increasing amounts of recombinant CAF-1, which was either not treated (solid triangle) or treated before its use with Lambda phosphatase (open triangle): 5 ng (lanes 3, 7, 11, and 15), 10 ng (lanes 4, 8, 12, and 16), 20 ng (lanes 5, 9, 13, and 17) or 40 ng (lanes 6, 10, 14, and 18). Deproteinized DNA was purified and analysed by agarose gel electrophoresis. The incorporation of radiolabel due to the UV-induced DNA synthesis was visualized by autoradiography (Labeled DNA) and the total population of DNA molecules by ethidium bromide staining of the gel (Total DNA). Plasmids were processed as indicated in Materials and Methods. The positions of the supercoiled (form I), nicked (form Ir), and closed circular (form II) forms of plasmid DNA are indicated. Control supercoiled DNA (C) was run in parallel. The presence and amount of the phosphorylated p60 subunit of CAF-1 (exogenous and endogenous) at the end of each reaction was assessed by Western blotting using the polyclonal anti-p60 antibody (p60). Asterisk, slowest migrating phosphorylated form of p60. (B) Data from experiments as described above using plasmid DNA treated with UV-C (150, 300, and 500 J/m2) and increasing amounts of recombinant CAF-1 either not treated (___) or treated before its use with Lambda phosphatase (- - -) were quantified using a PhosphorImager and ImageQuant software. The percentage of form I in the labeled plasmid (%) is represented as a function of the amount of CAF-1 provided in various phosphorylation status (ng).
Discussion
Dynamic reorganization of chromatin structure is a recurrent feature associated with many changes in cellular physiology. During NER, changes in nucleosome structure are likely to occur (Smerdon, 1991) both to make the DNA lesion accessible to the repair machinery and to facilitate reformation of a regular chromatin structure following the repair process (Gaillard et al., 1996, 1997). Although CAF-1 was originally identified as a protein that promotes nucleosome assembly during DNA replication in vitro (Smith and Stillman, 1989), CAF-1 is thought to play a similar role during the repair of DNA damage (Gaillard et al., 1996; Kaufman et al., 1997). This paper provides the first evidence that the activity of CAF-1 is regulated in response to DNA damage in mammalian cells.
Comparison of the Behavior of p60 and p150 Subunits of CAF-1 to PCNA
The behavior of the p60 and p150 subunits of CAF-1 shares a number of similarities with that of PCNA, an essential protein of proliferating cells which plays crucial roles in both DNA replication and repair (Cox, 1997). Both the p150 and p60 subunits of CAF-1 rapidly become resistant to detergent extraction during DNA replication or repair of nuclei (Figs. 2, 6, and 7). Furthermore, the characteristics of the UV response (timing and dose dependence), for the binding of p150 and phosphorylated p60 to chromatin (Fig. 7) is strikingly similar to those observed for PCNA during NER (Miura et al., 1992; Prosperi et al., 1993; Aboussekhra and Wood, 1995). Interestingly, both the timing and UV dose response are somewhat distinct from those reported for the p34 subunit of replication protein A whose phosphorylation is only detected with higher doses and at a later time after UV irradiation (Liu and Weaver, 1993; Carty et al., 1994). Overall, our data provide in vivo support for a dual role of CAF-1 during S phase and DNA repair. It is intriguing that this parallel between PCNA and CAF-1 may extend beyond their dual function in DNA replication and DNA repair, since both proteins also play a role in the inheritance of transcriptionally silent heterochromatin.
In Drosophila, chromosomal translocations that bring euchromatic genes next to heterochromatin generally result in transcriptional silencing (Pirrotta, 1997). The silent state is rather stable through mitosis but, with a low frequency, the translocated gene can switch to the expressed state, which can also be inherited in a stable fashion. This form of chromosomal position effect, known as position effect variegation in Drosophila, is functionally analogous to the silencing of reporter genes introduced next to telomeric heterochromatin in S. cerevisiae (Aparicio et al., 1991). Interestingly, a PCNA mutation in Drosophila, mus 209 (mutagen-sensitive 209), results in suppression of position-effect variegation (Henderson et al., 1994; Yamamoto et al., 1997), i.e., heterochromatin-mediated silencing is impaired. This may relate to the unexpected function of CAF-1 in silencing recently uncovered through genetic studies in S. cerevisiae (Enomoto et al., 1997; Kaufman et al., 1997; Monson et al., 1997; Enomoto and Berman, 1998). These findings suggest that the role of CAF-1 during nucleotide excision repair may be critical in either restoring or setting de novo repressive chromatin structures. PCNA also plays a role in repair mechanisms other than NER such as double-strand break repair (Ayyagari et al., 1995; Henderson and Glover, 1998) and one of the base excision repair pathways (Frosina et al., 1996). Our previous work using Drosophila embryo extracts (Gaillard et al., 1997) would suggest that the DNA synthesis event is not strictly required for an assembly machinery to be recruited to specific repair sites. Thus, CAF-1 may be recruited by DNA structures and/or protein factors present during earlier stages of repair. The parallel behavior of CAF-1 with PCNA could thus imply some interaction between these molecules within such a complex. This raises the possibility that CAF-1 may also play some role in DNA repair pathways other than NER. The binding of CAF-1 to chromatin and the phosphorylation of the p60 subunit of CAF-1 described here may provide useful markers to monitor the CAF-1 response to various genotoxic agents in repair-proficient cells or cells with mutations in various DNA repair pathways (Friedberg et al., 1995).
Chromatin Association and the Phosphorylation of the p60 Subunit of CAF-1 in Response to UV Irradiation
The changes in CAF-1 after UV irradiation reported here provide a novel element to the cellular response to UV irradiation. We defined two features of this CAF-1 response to UV irradiation: (a) the chromatin retention of both the p150 and p60 subunits of CAF-1 and (b) the predominance of phosphorylated forms of p60 in this chromatin-associated fraction. The chromatin association most likely reflects an interaction with a protein component of chromatin, rather than with DNA itself, since the CAF-1 protein purified from human cells was reported to present no apparent affinity for either single- or double-stranded DNA (Smith and Stillman, 1989). The p60 subunit of CAF-1 has been previously shown to exist in various phosphorylated forms in asynchronous cells, (Smith and Stillman, 1991) and several consensus sites for protein kinase C, casein kinase 2, and tyrosine kinase phosphorylation can be found. However, no information is available concerning the possible physiological significance of the posttranslational modification of p60. Although the relative abundance of the nonphosphorylated and phosphorylated forms of p60 in the chromatin-associated fraction was found to vary during the cell cycle, a significant increase in phosphorylated p60 bound to chromatin occurred in response to UV irradiation in G1, S, or G2 phase (Fig. 6 and data not shown). Whether the chromatin-associated phosphorylated forms of p60 that are constitutively present in G1 and S phase serve similar functions to those found after UV irradation remains to be determined. It is not as yet clear whether the sites of p60 phosphorylation in normal and UV-irradiated cells are the same or, for that matter, whether phosphorylation of the p60 subunit of CAF-1 plays a similar function during normal progression through S phase and in the cellular response to UV irradiation.
In normal interphasic cells, it is remarkable that the phosphorylation of the Triton-resistant form of the p60 subunit of CAF-1 is high in G1, progressively decreases throughout S phase, and is essentially absent during G2. The significance of this observation is unclear but this progressive dephosphorylation of CAF-1 could potentially serve as a marking system to avoid DNA rereplication. Alternatively, since progression through S phase is accompanied by a decrease in the number of DNA replication foci, the level of p60 phosphorylation may simply be an indication of the amount of CAF-1 actively involved in nucleosome assembly at the DNA replication fork. This in turn would suggest that p60 phosphorylation may modulate the nucleosome assembly activity of CAF-1 in normal cells. In that respect, it is also interesting to note that a distinct modified form only observed in mitosis (see Fig. 8 B) was reported to be impaired in nucleosome depostion during complementary strand synthesis (Marheineke and Krude, 1998).
Our data do not preclude the possibility of a de novo phosphorylation of the p60 subunit of CAF-1 in response to UV irradiation. However, the increase in the phosphorylated form of p60 that is associated with chromatin after UV irradiation may be simply mediated through recruitment of a preexisting pool of phosphorylated molecules. This mechanism of recruitment could be advantageous for a rapid response to UV damage. Importantly, although both phosphorylated and nonphosphorylated forms of p60 are present in roughly equal amounts in G2 cells (Fig. 5 B, Total), phosphorylated p60 is almost completely absent from the chromatin-associated pool (Fig. 5 B and Fig. 6). Thus, phosphorylation of p60 per se is not sufficient to trigger association of CAF-1 with chromatin. Remarkably, the fraction of the p60 subunit of CAF-1 recruited to chromatin appeared to be proportional with the UV dose applied to the cells (Fig. 7). These data suggest that the UV signal triggering this recruitment could operate via the sensing of the UV lesion/repair sites into DNA which would be consistent with our data in vitro (Fig. 8).
Importance of the Phosphorylated Form of the p60 Subunit of CAF-1 in Interphase
Our data in vitro argue that phosphorylation of CAF-1 increases the ability of the factor to promote nucleosome assembly during nucleotide excision repair (Fig. 8). Although this effect is rather modest in vitro, it may become important in vivo under conditions of severe DNA damage, when the concentrations of CAF-1 and repair factors may be limiting. Potentially, the modification by phosphorylation of CAF-1 may be important for its role in nucleosome assembly activity during both DNA synthesis and DNA repair.
Phosphorylation of the p60 subunit may facilitate either binding or dissociation of CAF-1 from acetylated core histones H3 and H4. Alternatively, phosphorylation of p60 may promote recognition of the specific protein factors or DNA structures necessary to attract the CAF-1 protein to sites of DNA repair. Finally, in addition to its effect on the nucleosome assembly activity of CAF-1, the increase in phosphorylation of the p60 subunit of CAF-1 in response to UV irradiation may act as a signal to prevent progression through the cell cycle until DNA lesions have been repaired and/or the chromatin structure has been restored to its original state. This potential signaling function of phosphorylated CAF-1 would provide an interesting link between cell cycle regulation and DNA repair. These issues should be explored in the future.
Acknowledgments
We thank J.-P. Quivy, J.G. Moggs (both from Institut Curie, Paris, France), T. Krude (University of Cambridge Wellcome/CRC Institute, Cambridge, UK), and A. Annunziato (Boston College, Chestnut Hill, MA) for critical reading, I. Coulter (Institut Curie) for useful suggestions, and our colleagues, especially S. Nocentini (Institut Curie), for numerous stimulating discussions. We are most grateful to B. Stillman (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) and his laboratory for the generous collaboration in providing us with antibodies as well as recombinant human CAF-1. We thank D. Rouillard (Institut Curie) for FACScan™ analysis.
This work was supported by the Association pour la Recherche sur le Cancer (ARC), an Action Thématique et Incitative sur programme et Equipes (ATIPE) no. 7, “Biologie du Developpement” from the Centre National de la Recherche Scientifique, and the European Economic Community. E. Martini has been supported by la Ligue National Contre le Cancer and ARC fellowships, K. Marheineke is supported by a Marie Curie Research Fellowship of the European Union, and A. Verreault is a postdoctoral fellow of the Jane Coffin Childs Memorial Fund for Medical Research.
Abbreviations used in this paper
- CAF-1
chromatin assembly factor 1
- CSK
cytoskeleton
- DAPI
4′,6-diamidino-2-phenylindole
- NER
nucleotide excision repair
- NuMa
nuclear matrix-associated protein
- PCNA
proliferating cell nuclear antigen
- UV-C
ultraviolet C
References
- Aboussekhra A, Wood RD. Detection of nucleotide excision repair incisions in human fibroblasts by immunostaining for PCNA. Exp Cell Res. 1995;221:326–332. doi: 10.1006/excr.1995.1382. [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. . Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PMJ. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA-replication and DNA-repair. Mol Cell Biol. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo R, Macdonald-Bravo H. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: association with DNA replication sites. J Cell Biol. 1987;105:1549–1554. doi: 10.1083/jcb.105.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carty MP, Zernik-Kobak M, McGrath S, Dixon K. UV light- induced DNA synthesis arrest in HeLa cells is associated with changes in phosphorylation of human single-stranded DNA-binding protein. EMBO (Eur Mol Biol Organ) J. 1994;13:2114–2123. doi: 10.1002/j.1460-2075.1994.tb06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LS. Who binds wins: competition for PCNA rings out cell-cycle changes. Trends Cell Biol. 1997;7:493–498. doi: 10.1016/S0962-8924(97)01170-7. [DOI] [PubMed] [Google Scholar]
- Enomoto S, McCune-Zierath P, Geraminejad M, Sanders M, Berman J. Rfl2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–363. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey EG, Krochmalnic G, Penman S. The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and RNP- depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J Cell Biol. 1986;102:1654–1665. doi: 10.1083/jcb.102.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg, E.C., G.C. Walker, and W. Siede. 1995. DNA Repair and Mutagenesis. E.C. Friedberg, editor. ASM Press, Washington, D.C. 291–294.
- Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J Biol Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- Gaillard P-H, Martini EM, Kaufman PD, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- Gaillard P-H, Moggs JG, Roche DMJ, Quivy J-P, Becker PB, Wood RD, Almouzni GA. Initiation and bidirectional propagation of chromatin assembly from a target site for nucleotide excision repair. EMBO (Eur Mol Biol Organ) J. 1997;16:6281–6289. doi: 10.1093/emboj/16.20.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond JE, Rouvière-Yaniv J, Yaniv M, Brutlag D. Nicking-closing enzyme assembles nucleosome-like structures in vitro. Proc Natl Acad Sci USA. 1979;76:3779–3783. doi: 10.1073/pnas.76.8.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson DS, Banga SS, Grigliatti TA, Boyd JB. Mutagen sensitivity and suppression of position-effect variegation result from mutations in mus209, the Drosophilagene encoding PCNA. EMBO (Eur Mol Biol Organ) J. 1994;13:1450–1459. doi: 10.1002/j.1460-2075.1994.tb06399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson HS, Glover DM. Chromosome fragmentation resulting from an inability to repair transposase-induced DNA double-strand breaks in PCNA mutants of Drosophila. . Mutagenesis. 1998;13:57–60. doi: 10.1093/mutage/13.1.57. [DOI] [PubMed] [Google Scholar]
- Herrlich P, Sachsenmaier C, Radlerpohl A, Gebel S, Blattner C, Rahmsdorf HJ. The mammalian UV response-mechanism of DNA damage induced gene-expression. Adv Enzyme Regul. 1994;34:187–223. doi: 10.1016/0065-2571(94)90024-8. [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Kamakaka RT, Bulger M, Kaufman PD, Stillman B, Kadonaga JT. Postreplicative chromatin assembly by Drosophilaand human chromatin assembly factor 1. Mol Cell Biol. 1996;16:810–817. doi: 10.1128/mcb.16.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- Kaufman PD. Nucleosome assembly: the CAF and the HAT. Curr Opin Cell Biol. 1996;8:369–373. doi: 10.1016/s0955-0674(96)80012-3. [DOI] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiaecells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- Krude T. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp Cell Res. 1995a;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- Krude T. Nucleosome assembly during DNA replication. Curr Biol. 1995b;5:1232–1234. doi: 10.1016/s0960-9822(95)00245-4. [DOI] [PubMed] [Google Scholar]
- Krude T, Jackman M, Pines J, Laskey RA. Cyclin/Cdk-dependent initiation of DNA replication in a human cell-free system. Cell. 1997;88:109–119. doi: 10.1016/s0092-8674(00)81863-2. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskey RA, Earnshaw WC. Nucleosome assembly. Nature. 1980;286:763–767. doi: 10.1038/286763a0. [DOI] [PubMed] [Google Scholar]
- Li JJ, Kelly TJ. Simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1984;81:6973–6977. doi: 10.1073/pnas.81.22.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M, Goldman AS. Meiotic recombination hotspots. Annu Rev Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- Liu VF, Weaver DT. The ionizing radiation-induced replication protein A phosphorylation response differs between ataxia telangiectasia and normal human cells. Mol Cell Biol. 1993;13:7222–7231. doi: 10.1128/mcb.13.12.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2. 8 Angstrom resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Lydersen BK, Ettijohn DE. Human-specific nuclear protein that associates with the polar region of the mitotic apparatus: distribution in a human/hamster hybrid cell. Cell. 1980;22:489–499. doi: 10.1016/0092-8674(80)90359-1. [DOI] [PubMed] [Google Scholar]
- Marheineke K, Krude T. Nucleosome assembly and intracellular localization of human CAF-1 changes during the cell division cycle. J Biol Chem. 1998;24:15279–15286. doi: 10.1074/jbc.273.24.15279. [DOI] [PubMed] [Google Scholar]
- Miura M, Domon M, Sasaki T, Kondo S, Takasaki Y. Two types of proliferating cell nuclear antigen (PCNA) complex formation in quiescent normal and xeroderma pigmentosum group A fibroblasts following ultraviolet light (UV) irradiation. Exp Cell Res. 1992;201:541–544. doi: 10.1016/0014-4827(92)90308-u. [DOI] [PubMed] [Google Scholar]
- Monson EK, de Bruin D, Zakian VA. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc Natl Acad Sci USA. 1997;94:13081–13086. doi: 10.1073/pnas.94.24.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. Checkpoint pathways come of age. Cell. 1997;91:865–867. doi: 10.1016/s0092-8674(00)80476-6. [DOI] [PubMed] [Google Scholar]
- O'Keefe RT, Henderson SC, Spector DL. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP. Formation and processing of UV photoproducts: effects of DNA sequence and chromatin environment. Photochem Photobiol. 1997;65:270–283. doi: 10.1111/j.1751-1097.1997.tb08560.x. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Chromatin-silencing mechanisms in Drosophilamaintain patterns of gene expression. Trends Genet. 1997;13:314–318. doi: 10.1016/s0168-9525(97)01178-5. [DOI] [PubMed] [Google Scholar]
- Prosperi E, Stivala LA, Sala E, Scovassi AI, Bianchi L. Proliferating cell nuclear antigen complex formation induced by ultraviolet irradiation in human quiescent fibroblasts as detected by immunostaining and flow cytometry. Exp Cell Res. 1993;205:320–325. doi: 10.1006/excr.1993.1092. [DOI] [PubMed] [Google Scholar]
- Roth SY, Allis CD. Histone acetylation and chromatin assembly: a single escort, multiple dances? . Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- Sancar A. DNA repair in humans. Annu Rev Genet. 1995;29:69–105. doi: 10.1146/annurev.ge.29.120195.000441. [DOI] [PubMed] [Google Scholar]
- Smerdon MJ, Lieberman MW. Nucleosome rearrangement in human chromatin during UV-induced DNA-repair synthesis. Proc Natl Acad Sci USA. 1978;75:4238–4241. doi: 10.1073/pnas.75.9.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerdon MJ. DNA repair and the role of chromatin structure. Curr Opin Cell Biol. 1991;3:422–428. doi: 10.1016/0955-0674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- Smith S, Stillman B. Immunological characterization of chromatin assembly factor I, a human cell factor required for chromatin assembly during DNA replication in vitro. . J Biol Chem. 1991;266:12041–12047. [PubMed] [Google Scholar]
- Sogo, J.M., and R.A. Laskey. 1995. In Chromatin Replication and Assembly. S.C.R. Elgin, editor. Oxford University Press, New York. 49–71.
- Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986;45:555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- Todorov IT, Attaran A, Kearsey SE. BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995;129:1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toschi L, Bravo R. Changes in cyclin/proliferating cell nuclear antigen distribution during DNA repair synthesis. J Cell Biol. 1988;107:1623–1628. doi: 10.1083/jcb.107.5.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Chromatin remodeling and transcription. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- van Holde, K.E. 1988. Chromatin. Springer-Verlag, New York.
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodeling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- Wolffe, A. 1995. Chromatin: Structure and Function. Academic Press, Harcourt Brace & Co., San Diego, CA.
- Wood, R.D., M. Biggerstaff, and M.K.K. Shivji. 1995. Detection and measurement of nucleotide excision repair synthesis by mammalian cell extracts in vitro. Methods: Comp. Methods Enzymol. 7:163–175.
- Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem. 1997;272:23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Girard F, Bello B, Affolter M, Gehring WJ. The cramped gene of Drosophilais a member of the Polycomb-group, and interacts with mus209, the gene encoding proliferating cell nuclear antigen. Development (Camb) 1997;124:3385–3394. doi: 10.1242/dev.124.17.3385. [DOI] [PubMed] [Google Scholar]