Abstract
Cells of the mononuclear phagocyte lineage have the capability to adhere to and fuse with each other and to differentiate into osteoclasts and giant cells. To investigate the macrophage adhesion/fusion mechanism, we focused our attention on CD44, a surface glycoprotein known to play a role in hematopoietic cell–cell adhesion. We report that CD44 expression by macrophages is highly and transiently induced by fusogenic conditions both in vitro and in vivo. We show that CD44 ligands, hyaluronic acid, chondroitin sulfates, and osteopontin prevent macrophage multinucleation. In addition, we report that the recombinant extracellular domain of CD44 binds fusing macrophages and prevents multinucleation in vitro. These data suggest that CD44 may control the mononucleated status of macrophages in tissues by virtue of mediating cell–cell interaction.
Keywords: CD44, macrophage, adhesion, fusion, osteoclast, giant cell
Mononuclear phagocytes are cells which have the potential, in specific instances, to fuse and differentiate into either osteoclasts or multinucleated giant cells, in bone or in chronic inflammatory reactions, respectively. Multinucleation appears to endow mononuclear phagocytes with an added value, i.e., a powerful resorptive capability of extracellular components, i.e., bone or infectious agents (Vignery et al., 1989). Indeed, osteoclasts are responsible for the loss of bone that leads to osteoporosis.
Although osteoclasts and giant cells have long been recognized, the molecular mechanism by which their mononucleated precursors adhere and fuse with each other, a key step in their differentiation, remains poorly understood. Indeed, cell–cell fusion itself, whether it concerns that of sperm–oocyte or myoblast–myoblast, leading to fertilization and muscle development, respectively, has not been thoroughly investigated. It is thought that cell–cell adhesion leading to fusion involves a set of proteins similar to those used by viruses to fuse with host cells and inject their DNA or RNA (Hernandez et al., 1997). It has been hypothesized that viruses have stolen the fusion protein machinery from their target cells. It is now well accepted that virus–cell fusion requires both an attachment mechanism and a fusion peptide. One such example is HIV gp120 from the human immunodeficiency virus (HIV)1 which binds CD4 on T lymphocytes and macrophages (Dalgleish et al., 1984; Klatzmann et al., 1984), whereas the fusion molecule gp40, which arises from the same precursor molecule (gp160), is thought to trigger the actual fusion event. Although putative fusion molecules mediating sperm–oocyte and myoblast fusion have been reported (Blobel et al., 1992; Wakelam 1989), the actual protein machinery governing the attachment and fusion of these cells remains unknown.
Increasing evidence suggests that the molecular machinery mediating virus–cell and cell–cell adhesion/fusion is more complicated than anticipated and involves numerous players. Although it had been thought that HIV needed only the T lymphocyte receptor CD4 to bind and infect cells, several chemokines have now been demonstrated to slow the growth of HIV in cultures. It has been determined that the chemokine family of G protein–coupled receptors, most notably CC-CKR4 and CC-CKR5, are involved in HIV infection (Alkhatib et al., 1996; Deng et al., 1996; Dragic et al., 1996; Feng et al., 1996). There are now at least 10 chemokine receptors identified as HIV coreceptors (Dimitrov, 1997). Furthermore, the interaction between the adhesion molecules leukocyte function-associated antigen-1 (LFA-1) and intercellular adhesion molecule-1 (ICAM-1) has been described with respect to both virus–cell and cell–cell adhesion/fusion events. HIV-induced syncytium formation is blocked by a monoclonal antibody directed against the β subunit of LFA-1 (Hildreth and Orentas, 1989), whereas cytokine induced multinucleate giant cell formation from peripheral blood monocyte (Möst et al., 1990; Kazazi et al., 1994) as well as osteoclast development in vitro (Kurachi et al., 1993) are both inhibited by antibodies directed against LFA-1 and ICAM-1. The expression level of these molecules, however, does not correlate with multinucleation (Kurachi et al., 1993). Members of the cadherin family of homophilic cell adhesion molecules have also been suggested to play a role in cell–cell multinucleation. Although N-cadherin appears necessary for myoblast fusion (Mege et al., 1992), inhibition of E-cadherin function prevents the fusion of osteoclast precursors in vitro (Mbalaviele et al., 1995). A set of proteins thought to enhance or induce cell fusion, initially termed FRP-1 and FRP-2 and now known to be CD98 and integrin α3, respectively (Ohgimoto et al., 1995; Higuchi et al., 1998), have been recently identified in a number of cell lines infected with several different viruses as well as on the surface of monocytes and macrophages. Monoclonal antibodies directed against these proteins stimulate polykaryocyte formation in CD4+ U937 cells transfected with the HIV gp160 gene (Ohta et al., 1994) and in HeLa and FL cells infected with Newcastle disease virus (Ito et al., 1992). In addition, anti-FRP antibodies inhibit giant cell formation in cultures of peripheral blood monocytes (Tabata et al., 1994). Most recently, overexpression of the purigenic P2Z/P2X7 has been reported to trigger cell–cell fusion but also leads to cell death (Falzoni et al., 1995; Chiozzi et al., 1997). Although none of these proteins appear as actual fusion proteins and may not therefore mediate the actual fusion event, together they suggest that the fusion mechanism of viruses and mammalian cells may involve both regulatory proteins and adhesion molecules.
CD44 is an integral membrane glycoprotein which plays an important role in both cell–cell and cell–substrate adhesion (Belitsos et al., 1990; Stamenkovic et al., 1991; Sy et al., 1991). CD44 exists as different isoforms in a wide variety of cells and tissues due to posttranslational modifications and alternative splicing (Tolg et al., 1993; for review see Naor et al., 1997). Our interest in CD44 as a protein that might be important for macrophage multinucleation was prompted by the following criteria: first, CD44 is involved in cell migration, lymphopoiesis and lymphocyte homing, in which it mediates cell–cell and cell–substrate interactions. A substantial body of data has been published suggesting that certain molecules involved in cell–cell adhesion, such as CD4 and LFA-1, play an important role in the infectivity and cytopathicity of viruses. (Pantaleo et al., 1991). Second, CD44 recognizes and binds to extracellular matrix elements such as hyaluronate (Lesley et al., 1990; Miyake et al., 1990), collagen type I, fibronectin (Jalkanen and Jalkanen, 1992) and osteopontin (Weber et al., 1996). The interaction of CD44 with extracellular components, in addition to cell surface molecules, could provide an additional regulatory mechanism to control multinucleation in macrophages. Third, the 100-kD form of CD44, the most common so-called standard form expressed by hematopoietic cells, has been demonstrated to be involved not only in the attachment of poliovirus to HeLa cells (Shepley and Racaniello, 1994) but also in the infection of mononuclear phagocytes by HIV (Rivandeneira et al., 1995). CD44 does not, however, act as a viral receptor in either of these two instances. We therefore set out to investigate whether CD44 played a role in macrophage multinucleation. We report here that CD44 expression by macrophages is highly and transiently induced by fusogenic conditions, and that CD44 ligands prevent multinucleation. We also show that the recombinant soluble extracellular domain of CD44 binds macrophages and prevents multinucleation in vitro. This suggests that CD44 and its putative cell–surface ligand may participate in the adhesion/fusion event of macrophages.
Materials and Methods
Cells
Rat alveolar macrophages were obtained from 12-wk-old Fisher rats (Charles River, Kingston, NY) by tracheobronchial lavage and cultured in fusogenic conditions as previously described (Vignery et al., 1990). Osteoclast-like cells were elicited in vivo by implanting syngeneic bone particles intramuscularly for 10 d in 12-wk-old Sprague Dawley rats (Charles River) as previously described (Vignery et al., 1989; 1990). The National Research Council's guide for the care and use of laboratory animals were followed. COS-7 cells were a gift of M. Solimena (Yale School of Medicine, New Haven, CT).
Chemicals
Hyaluronic acid from human umbilical cord was purchased from Calbiochem-Novabiochem (La Jolla, CA) and Pharmacia Biotech (Uppsala, Sweden). Chondroitin sulfate A from bovine trachea and chondroitin sulfate B were purchased from Calbiochem-Novabiochem. Osteopontin was a kind gift of W.T. Butler (Texas Medical Center Dental Branch, Houston, TX). Unless otherwise stated, all chemicals were purchased from Sigma Chemical Co. (St. Louis, MO).
Antibodies
Mouse anti–rat macrophage CD44 (MRC OX8), CD4 (W3/25), and MHCII (RT1B) which are of the IgG1 isotype, were obtained from Serotec (Raleigh, NC). Fluorescein isothiocyanate (FITC)-conjugated F(ab)′2 goat anti–mouse IgG (H + L chains) was obtained from Boehringer Mannheim Biochemicals (Indianapolis, IN). Indocarbocyanine (Cy3)-conjugated F(ab)′2 goat anti–mouse IgG (H + L chains) and goat anti–mouse IgG horseradish peroxidase conjugate were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). Goat anti-GST and rabbit anti–goat IgG conjugated to horseradish peroxidase were obtained from Pharmacia Biotech. Mouse anti-myc was purchased from Invitrogen (Carlsbad, CA), and sheep anti–mouse horseradish peroxidase conjugate was purchased from Amersham (Arlington Heights, IL).
Immunolocalization
Tissues were prepared from both control and experimental rats implanted with bone particles (Vignery et al., 1989). The implants and the rat tissues were quick frozen and cut to 6-μm-thick frozen sections using a Reichert-Jung cryostat (2800 Frigocut; Leica, Deerfield, IL). The sections were first incubated overnight in PBS-milk (PBS supplemented with 5% nonfat dry milk [Carnation, Los Angeles, CA]), then for 2 h in PBS-milk containing anti-CD44, anti-CD4, anti-MHCII, or mouse IgG1. Sections were then incubated for 1 h in PBS-milk containing a 1:400 dilution of goat anti–mouse Cy3-F(ab)′2. After three washes of 10 min each with PBS, the sections were imaged at 550 nm using the Cy3 excitation filter block on an Olympus microscope (Melville, NY) equipped with UV light. Cells were cultured on glass coverslips for the indicated time in MEM containing 5% human serum, fixed in formaldehyde for 1 h at 4°C, and then washed for 60 min in PBS-FCS (PBS + 10% FCS). The cells were incubated overnight in PBS-FCS supplemented or not with anti-CD44, anti-CD4, anti-MHCII, or mouse IgG1. After four washes of 15 min each in PBS-FCS (PBS with 10% FCS), the cells were incubated for an additional hour with FITC-conjugated F(ab)′2 goat anti–mouse IgG (1:100 and 1:400 dilutions, respectively) in the same buffer. The cells were imaged at 488 nm using the FITC excitation filter block on an Olympus microscope equipped with UV light.
RNA Isolation and Northern Blot Analysis
Total RNA was isolated from alveolar and peritoneal macrophages cultured or not in fusogenic milieu for 72 h using a modification (Maniatis et al., 1989) of the methods described by Glisin et al. (1974) and Ullrich et al. (1977) or the RNeasy kit (QIAGEN, Santa Clarita, CA). In each case, guanidinium thiocyanate homogenization buffer was added to the freshly isolated, and the cultured cells after rapid removal of culture medium. The cell lysates were sheared using a syringe with a 23-gauge needle (Becton Dickinson, Franklin Lakes, NJ). For separation by cesium chloride, 2.5 ml-aliquots of the lysate were layered onto a 2-ml cushion of 5.7 M cesium chloride (American Bioanalytical, Natick, MA) in RNase-free 5.1-ml polyallomer centrifuge tubes which were centrifuged at 150,000 g for 20 h using a Ti 55 SW rotor. The supernatants were aspirated and the pellets dissolved in Tris-EDTA, pH 7.4, containing 0.1% SDS by freezing and thawing the samples twice and then warming to 45°C. RNA was precipitated by the addition of 0.3 M sodium acetate and 3 vol of ethanol. The pellets were resuspended in diethylpyro carbonate (DEPC)-treated water and the concentration determined using optical density measurements taken in a Perkin Elmer UV/VIS spectrophotometer (Foster City, CA).
For Northern blot analysis, 8 μg of each RNA sample was electrophoretically separated in formaldehyde-agarose gels, blotted onto a nylon membrane (GeneScreen Plus; New England Nuclear, Boston, MA) and hybridized with a 32P-labeled PCR-generated DNA probe corresponding to the full-length CD44 cDNA. The signals on the autoradiogram were quantitated using a Linotype-Hell scanner (Eschborn, Germany) with a multianalyst software for Macintosh (Bio-Rad Laboratories, Hercules, CA).
cDNA Synthesis and PCR
First-strand cDNA was synthesized as follows: 1 μg of total RNA was reverse transcribed using 200 U of MMLV reverse transcriptase (Boehringer Mannheim Biochemicals) in a 20-μl reaction primed with oligo dT15 primer (Promega, Springfield, NJ). The protocol followed was that published in the instruction manual from Clontech (Palo Alto, CA). The cDNA was aliquoted and stored at −70°C. For PCR reaction, 1 μl of cDNA was used as template in a 25-μl reaction mix containing 2.5 U of AmpliTaq DNA polymerase (Perkin Elmer), 1.25 mM MgCl2, and 0.1 mM DNTP mix (New England Biolabs, Beverly, MA). The buffer condition used for the PCR was a 1× dilution of the 10× mix provided with the polymerase. The sequences of the primer pairs (used at a concentration of 0.1 μg per reaction) were as follows: full-length CD44: primer forward (nucleotide [nt] 87–122): GATCCTTTGGTTGCTAGCTGCACATCATGGACAAGG; reverse primer (nt 1187–1225): AAGTTAGTGGCACTCGAGCTACACCCCAATCTTCATATC; CD44 extracellular domain: primer forward (nt 86–122): AGATCCTTTGGCTCGAGCCTGCACATCATCGACAAGG; reverse primer (nt 878–913): AGGTCTCCTCGCGAATTCAGAAGTTGTGGTCACTCC; CD44 intracellular domain: primer forward (nt 994–1012): TAGGAGAAGGTGTGGGCAG; reverse primer (nt 1195–1212): AGGCACTACACCCCAATC. Cycle parameters were: 3 min at 95°C, 1 min at 50°C, and 3 min at 72°C for 30 cycles. The PCR fragment was gel purified using Geneclean (Bio 101; Branford, CT) and cloned into PCR II TA cloning vector (Invitrogen, San Diego, CA). The cloned DNA insert was sequenced (W.M. Keck Biotechnology Resource Laboratory, Yale University) using AmpliTaq DNA polymerase and fluorescent dideoxy terminators (Perkin Elmer) in a cycle sequencing method. The resulting DNA fragments were gel purified and analyzed using an automated Applied Biosystems 373A Stretch or 377 DNA sequencer (Foster City, CA).
CD44, CD4, and MHCII ELISA
The levels of CD44, CD4, and MHCII cell surface expression were quantitated by ELISA as follows: 5 × 104 alveolar macrophages per well plated at 5 × 106 cells/ml in 96-well dishes were cultured for the indicated times. The minimum culture time after plating in each experiment was 1 h in order to secure the adherence of the cells to the wells. They were fixed at room temperature in 4% paraformaldehyde for 10 min, then incubated in 100 μl of PBS supplemented with 5% dry milk for 2 h. The cells were subsequently reacted overnight with IgG1, anti-rat CD44 (10–50 μg/ml), anti-rat CD4 (100 μg/ml), or anti-rat MHCII (10–50 μg/ml). After three washes of 10 min each with PBS, the cells were incubated at room temperature for 2 h with goat anti–mouse IgG horseradish peroxidase conjugate (1:5,000 dilution). The cells were washed three times for 10 min with PBS. Surface CD44 expression was quantitated by incubating the cells for 5 min in 100 μl of 3,3′5,5′-tetramethylbenzidine (HRP substrate; Moss, Pasadena, MD). Optical density measurements were made using a kinetic reader (Molecular Devices, Sunnyvale, CA).
Treatment of Cells with Glycosaminoglycans and Osteopontin
10-μl aliquots of 5 × 106 rat alveolar macrophages/ml were plated in flat bottom 96-well tissue culture dishes that had been treated overnight at 37°C with hyaluronic acid (1 mg/ml) or chondroitin sulfate A or B (1 mg/ml). Glycosaminoglycans and osteopontin were stored in lipopolysaccharide (LPS)-free H2O as 10-mg/ml and 10-μM stock solutions, respectively, which were diluted in MEM and filter-sterilized just before use. The cells were cultured for 4 d in 100 μl of MEM supplemented with different concentrations of hyaluronic acid, chondroitin sulfate A or B, and osteopontin. In experiments in which HA was obtained from Pharmacia, HA was added to the cells every day in fresh medium. Competition binding studies were performed by supplementing mAb anti-CD44 with 1 mg/ml hyaluronic acid, chondroitin sulphate A, chondroitin sulphate B, or 1 μM osteopontin. CD44 expression was determined by ELISA, as described above.
Production of CD44e: The Recombinant Soluble Extracellular Domain of CD44
The recombinant extracellular domain of CD44 (CD44e) was expressed using two different systems. CD44e was expressed as a fusion protein using the GST fusion protein system (Pharmacia Biotech). PCR amplification of this region was performed with a sense (5′TTACAGTTGAGCGAATTCCAGCAGCAGATCGATTTGAA; nt 158–195), and an antisense primer (5′AGGTCTCCTCGCCTCGAGAGAAGTTGTGGTCACTCC; nt 878–913), using rat macrophage cDNA as template (made from total RNA) and PWO as polymerase (Boehringer Mannheim, Mannheim, Germany). The primers were designed to allow digestion of the resulting PCR fragment with EcoR1 and Xho1 by means of which it was ligated in frame into an EcoR1-Xho1 cut pGEX-4T-1 vector. The resulting construct encoded a fusion protein of ∼50 kD and was used to transform the protease-deficient Escherichia coli strain BL-21. Soluble GST–CD44e was isolated from 1 liter of bacterial culture using the bulk GST purification module as described by the manufacturer. The eluted protein was extensively dialyzed against PBS and stored at −70°C until ready for use. As a control, a pGEX-calreticulin construct was obtained courtesy of A. Helenius (Yale University) and GST-calreticulin was isolated and stored as described above.
CD44e was expressed in mammalian cells with a Myc-His fusion tag using the mammalian expression vector pcDNA 3.1/Myc-His A, B, C (Invitrogen, Carlsbad, CA). PCR amplification of the extracellular region was performed with a sense primer (5′AGATCCTTTGGTGAATTCCTGCACATCATGGACAAGG; nt 86–122) an antisense primer (5′AGGTCTCCTCGCGAATTCAGAAGTTGTGGTCACTCC; nt 878–913), and rat macrophage cDNA as a template and PWO polymerase. The PCR product was digested with EcoRI and ligated into the EcoRI site of pcDNA 3.1/Myc-His B. This construct was used to transfect COS-7 cells using lipofectamine (GIBCO BRL, Gaithersburg, MD) according to the manufacturer's instructions. Transfected cells were cultured for 72 h in Opti-MEM I reduced serum medium (GIBCO BRL). The recombinant protein was purified from culture supernatant using Invitrogen's Xpress protein purification system. Recombinant GST–CD44e was quantified by running 5 μl on a 10% SDS-polyacrylamide gel and staining with Coomassie brilliant blue. The intensity of the stained band was tested against that of a serial dilution of bovine serum albumin (BSA) run on the same gel. The concentration of Myc-His–CD44e was then determined by immobilizing serial dilutions of both GST–CD44e and Myc-His–CD44e on nitrocellulose using a Bio-Dot microfiltration apparatus (Bio-Rad Laboratories). The immobilized proteins were reacted with anti–CD44 mAb followed by horseradish peroxidase (HRP)-linked sheep anti–mouse IgG (ECL kit; Amersham). The enzyme reaction was performed according to the manufacturer's instructions and the blots exposed for 30 s on X-ray films. Recombinant Myc-His-CD44e concentration was determined by comparative scanning densitometry of the dots against the concentration of GST–CD44e.
Deglycosylation of Recombinant Myc-His-CD44e
Approximately 250 μg of recombinant Myc-His-CD44e was deglycosylated in its native form by incubation with 10 U of N-glycosidase F for 48 h at 37°C in 0.1 M sodium phosphate, pH 7.4. Deglycosylation was analyzed by comparative Western blot analysis of both native and N-glycosidase F (Boehringer Mannheim, Indianapolis, IN)-treated Myc-His-CD44e using anti-myc antibody (Invitrogen).
Fusion Assay Using Recombinant Proteins
Freshly isolated rat alveolar macrophages were plated at 5 × 106 cells/ml and cultured in fusogenic milieu supplemented with either GST–CD44e, GST-Cal, or GST at the indicated concentrations. The cells were examined daily until day 4 to determine the effects of fusion proteins on macrophage multinucleation.
Binding Assay Using Recombinant Fusion Proteins
Freshly isolated rat alveolar macrophages were plated at 5 × 106 cells/ml in triplicate wells using 96-well dishes (5 × 104 cells/well). The cells were cultured overnight in fusogenic milieu. At the indicated times, duplicate sets of cells were supplemented with GST–CD44e, GST–Cal, or GST at the indicated concentrations. Binding proceeded overnight at 4°C. Media was removed from one set of cells and replaced with fresh media lacking recombinant protein to allow for dissociation. Dissociation proceeded for 3 h at 4°C. The media from all wells was then removed and the cells were fixed for 30 min in 4% paraformaldehyde. Cells were rinsed three times with PBS and blocked for 1 h in 5% milk/PBS. After three washes of 5 min in PBS, the cells were incubated with goat anti-GST for 30 min at room temperature. After three washes of 5 min each in PBS, the cells were incubated with rabbit anti–goat IgG-HRP for 30 min. After three final washes of 5 min each in PBS, 100 μl of peroxidase substrate (Moss) was added to each well and the optical density (OD) was read at 650 nm using an ELISA plate reader. Specific binding was determined as the difference in average OD values between the two sets of cells.
Statistical Analysis
Data are expressed as mean ± SD of mean. Comparative analyses of the means were performed with appropriate controls using independent Student's t test to determine the 99% confidence level (P < 0.01).
Results
High Expression of CD44 by Fusing Macrophages In Vivo and In Vitro
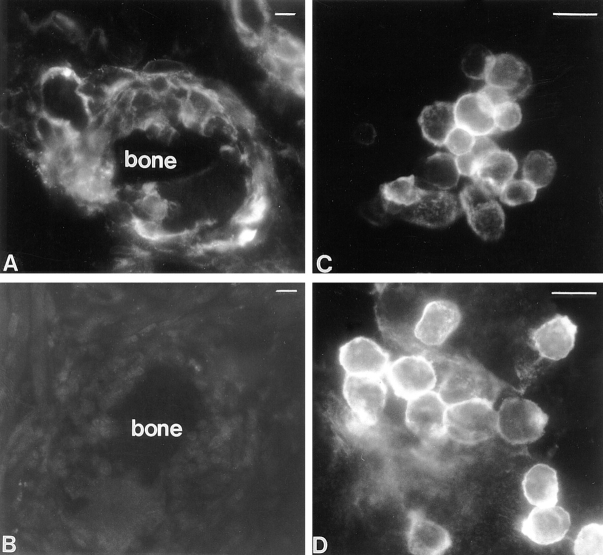
To investigate whether fusing macrophages expressed detectable levels of CD44, rat osteoclast-like cells were induced in vivo by implanting syngeneic bone particles intramuscularly as previously described (Vignery et al., 1989, 1990). Sections obtained from bone implants, as well as from long bones, brain, liver, spleen, lymph nodes, kidney, lung, skin, striated muscle, and pancreas were reacted with mAb anti-CD44, anti-CD4, anti-MHCII, or IgG1, followed by goat anti–mouse IgG conjugated to Cy3. Although each of these tissues hosts resident macrophages and many of them contain cells such as lymphocytes and epithelial cells known to express CD44, only osteoclast-like cells from the bone implants exhibited a strong fluorescent signal (Fig. 1 A) that was not detected in the presence of anti-CD4, anti-MHCII, or IgG1 (Fig. 1 B). The signal was restricted to both mono- and multinucleated cells that were closely apposed to the bone implants. In the larger cells attached to the implants, the signal appeared stronger distal to the bone, i.e., concentrated in the nonadherent domain of the plasma membrane. This is the domain that faces the incoming fusing macrophages. None of the surrounding cells, such as muscle cells, fibroblasts, or endothelial cells expressed a detectable level of fluorescence.
Figure 1.
High expression of CD44 on fusing macrophages in vivo (A) and in vitro (C and D). Multinucleation was induced in vivo by implanting rats intramuscularly with syngeneic bone particles that were recovered 10 d later and processed for immunolocalization. Frozen sections from bone implants were incubated with either mAb anti-rat CD44 (A) or mouse IgG1 (B) followed by goat anti–mouse IgG F(ab′)2 fragments conjugated to Cy3. Multinucleation was induced in vitro by plating alveolar macrophages on glass coverslips and culturing them under fusogenic conditions for either 1 (C) or 3 d (D). Cells were subjected to immunocytochemistry using anti-rat CD44 followed by goat anti–mouse IgG F(ab′)2 fragments conjugated to Cy3. Note the differences in fluorescence intensity between fusing cells. Bars, 20 μm.
To investigate whether CD44 expression by macrophages was associated with cell–cell interaction leading to fusion, alveolar macrophages were isolated and cultured in fusogenic conditions, i.e., plated at confluency, as previously described (Vignery et al., 1990). Macrophages cultured in such conditions initiate fusion a few hours after plating and reach 99% fusion within 3–4 d. Although both resident and freshly plated alveolar macrophages failed to express detectable level of CD44 expression by immunocytochemistry (data not shown), their fusion was associated with a strong fluorescent signal that was detected as soon as 24 h after plating (Fig. 1 C). By day three, the plasma membrane of the unfused mononucleated macrophages exhibited a strong signal whereas that of the multinucleated ones had become lesser (Fig. 1 D). Neither CD4 nor MHCII, which are both expressed by macrophages, were detectable by this technique suggesting that their level of expression remained low, i.e., similar to that of CD44 in nonfusing macrophages.
CD44 Expression Is Induced by Fusogenic Conditions
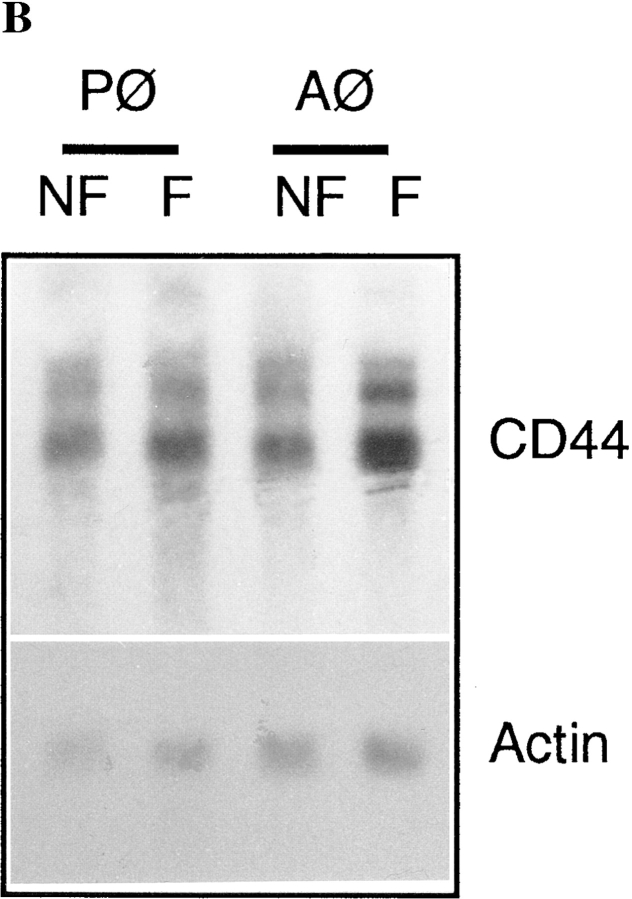
To investigate the kinetics of CD44 expression during induction of fusion, rat alveolar macrophages were plated in fusogenic conditions and CD44 expression was determined by ELISA as a function of time after plating. As shown in Fig. 2 A, CD44 expression increased to reach a peak at a time that varied between 24 and 48 h. CD44 declined thereafter. In some experiments, a dramatic increase was detected as early as 5–10 h after plating (data not shown). This confirmed our morphological observation that CD44 expression was less abundant in multinucleated than mononucleated fusing macrophages (Fig. 1 D). In contrast to CD44, the expression of CD4 and MHCII remained low, and was not altered by fusogenic conditions in macrophages (Fig. 2 A). When subjected to Northern blot analysis, CD44 transcripts were found to be abundant in freshly isolated macrophages, and increased by 36 and 13% in fusing alveolar and peritoneal cells, respectively (Fig. 2 B). This suggested that the regulation of CD44 mRNA expression in macrophages may be both transcriptional and posttranscriptional.
Figure 2.
Kinetics of CD44 expression in macrophages induced to fuse in vitro (A). Rat alveolar macrophages were isolated, plated at 5 × 106 cells/ml in quadruplicate wells, using 96-well dishes (5 × 104 cells/well), and cultured in fusogenic conditions for the indicated times. CD44 expression on paraformaldehyde fixed cells was determined by ELISA using mAb mouse anti-rat CD44 followed by goat anti–mouse IgG-conjugated to horseradish peroxidase (A and B). Mouse mAbs anti-rat CD4 and MHCII were used as controls. Northern blot analysis of CD44 (B). Each lane contained ∼8 μg of total RNA from freshly isolated as well as alveolar and peritoneal macrophages that had been cultured for 72 h under fusogenic conditions. Hybridization was performed using 32P-labeled PCR generated CD44 and β-actin probes.
CD44 Ligands Inhibit Macrophage Adhesion/Fusion
To analyze whether CD44 ligands, hyaluronic acid (HA), chondroitin sulfate A (CSA) and osteopontin (OP), all of which are extracellular matrix components, altered multinucleation, alveolar macrophages were cultured in fusogenic conditions in the presence of increasing concentrations of HA, CSA, or OP. Although alveolar macrophages are all fused after 4 d of culture under fusogenic conditions, the addition of HA, CSA, or OP prevented multinucleation in a dose-dependent manner (Fig. 3 A). In the presence of 1 mg/ml of HA, and to a lesser degree 1 mg/ml CSA and 1 μM OP, macrophages failed to form giant cells. To investigate whether CD44 played a role in macrophage attachment to their substrate, cells were plated onto surfaces coated with PBS, HA, or CSA. Although neither PBS nor CSA altered the adherence of macrophages, the coating of the dish with HA strongly prevented macrophages from adhering (Fig. 3 B). Interestingly, this inhibition was reversed by the addition of 1 mg/ml of HA to the culture medium, and to a lesser extent by the same concentration of CSA (Fig. 3 B, center panel). This suggested that the interaction of CD44 with HA was not required for macrophage attachment to their substrate. Indeed, it suggested that the possible clustering of CD44 receptors to the adherent domain of macrophages driven by HA prevented macrophage attachment. If so, soluble HA added to the culture medium may have allowed for the redistribution of CD44 to the nonadherent domain of the plasma membrane and thereby secured the adherence of the macrophages. This could even suggest that given the high expression level of CD44 molecules, their clustering to one (adherent) domain of the plasma membrane prevented (mechanistically) adhesion molecules (non-CD44) from reaching and clustering there as well, thereby preventing macrophage attachment. Altogether, our data suggested that macrophage attachment was not facilitated by HA-CD44 interaction, and that indeed other adhesion molecules might mediate their adhesion to a substrate.
Figure 3.
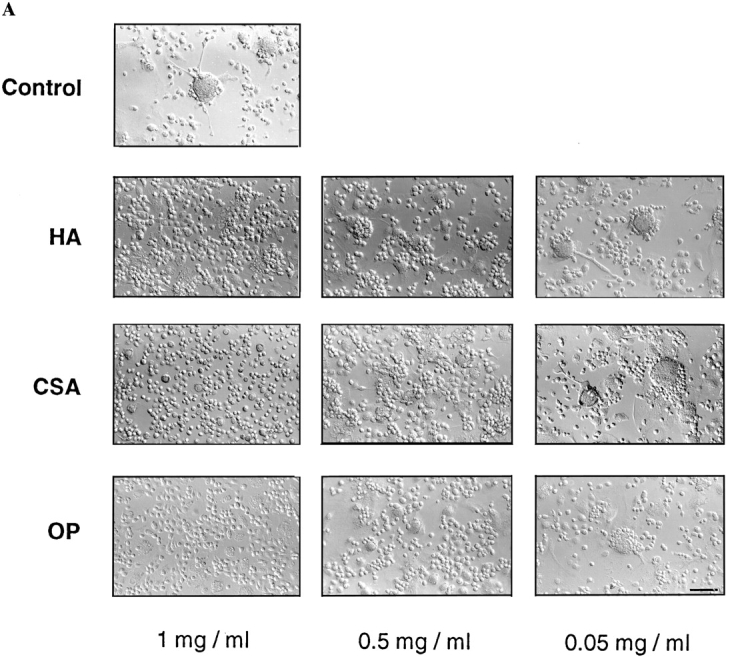

CD44 ligands inhibit macrophage adhesion/ fusion. (A) Rat alveolar macrophages were isolated, plated at 5 × 106 cells/ml in quadruplicate wells, using 96-well dishes (5 × 104 cells/ well), and cultured in fusogenic conditions for the indicated times, in the absence or presence of hyaluronic acid (HA), chondroitin sulfate A (CSA), or osteopontin (OP) at the indicated concentrations. (B) Alveolar macrophages were cultured as in A, but in dishes coated with either HA (1 mg/ml) or CSA (1 mg/ml). Bars, 100 μm.
HA Reduces CD44 Increased Expression by Fusing Macrophages
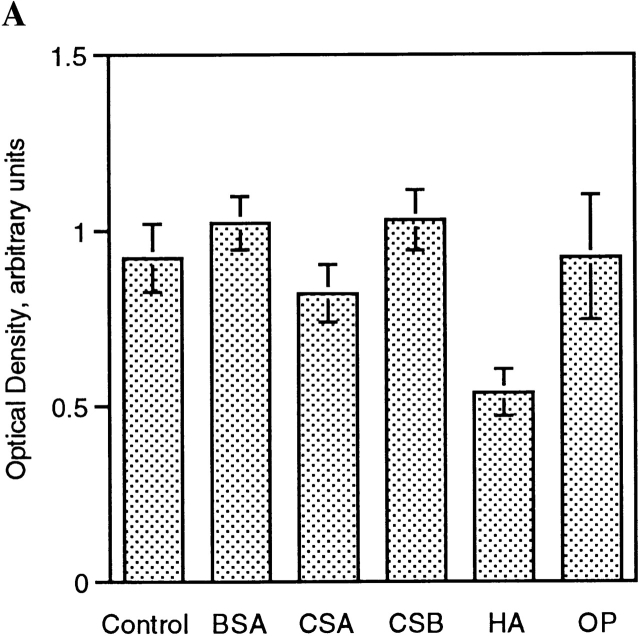
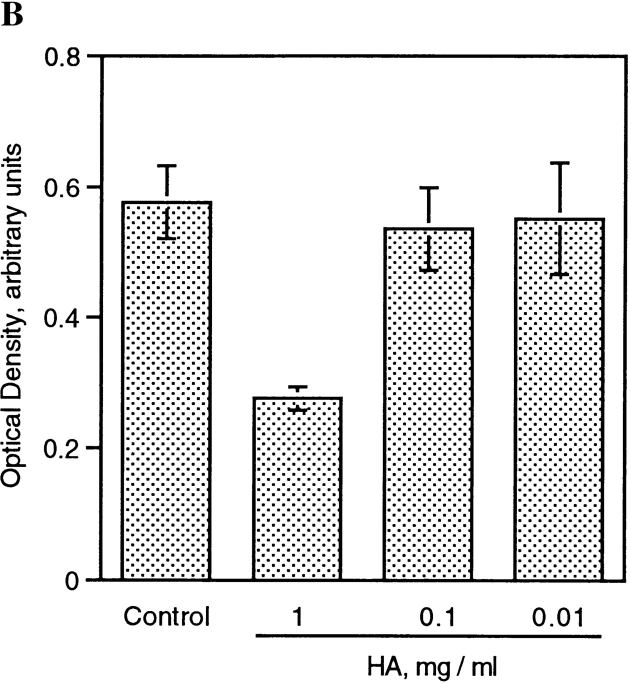
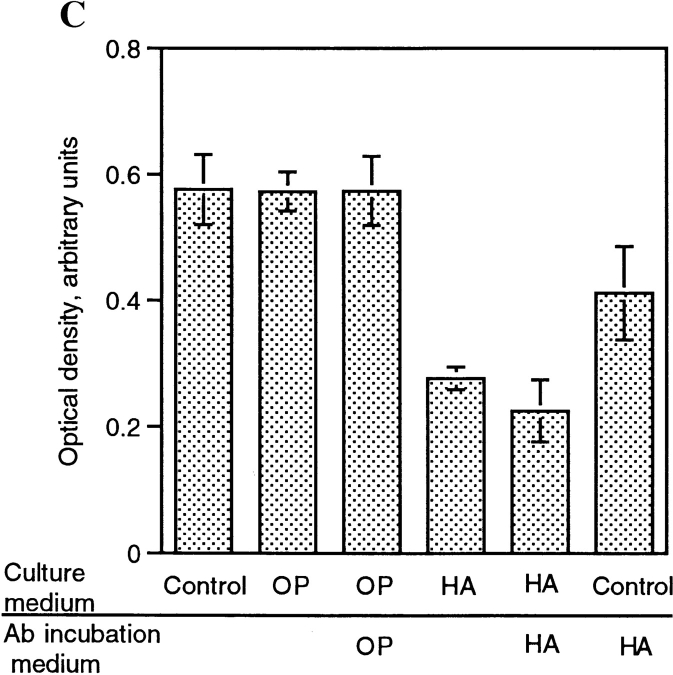
To initiate studies on the mechanism by which HA, and to a lesser extent CSA and OP inhibited macrophage multinucleation, fusing alveolar macrophages were cultured in the presence of CSA, HA, or OP, and CD44 expression was determined by ELISA. As shown in Fig. 2, the culturing of macrophages in fusogenic conditions was associated with a rapid and dramatic increase in CD44 expression. Fig. 4 shows that HA appeared to significantly (Fig. 2 A) and dose dependently (Fig. 2 B) prevent the increase in CD44 expression that accompanies the onset of fusion. This inhibition did not occur with either CSA or OP. BSA, used as a negative control, also failed to alter the binding of mAb anti-CD44 (Fig. 4 A). Because of the possibility that mAb anti-CD44 binding was prevented by HA itself which competed for the same site on CD44, fusing cells were cultured in the absence or presence of HA, and further treated or not with HA by adding HA to the mAb anti-CD44 incubation buffer. Fig. 4 C shows that mAb anti- CD44 binding was significantly reduced when mAb anti-CD44 incubation buffer was supplemented with HA. This suggested that HA and mAb anti-CD44 competed for the same epitope on CD44. But mAb anti-CD44 binding was further reduced when these cells were cultured in the presence of HA. This suggested that HA did reduce CD44 expression in fusing macrophages. Altogether, these data indicated that the interaction of CD44 with its extracellular ligands participated in the adhesion process leading to fusion of macrophages.
Figure 4.
HA decreases CD44 surface expression. (A) Rat alveolar macrophages were isolated, plated at 5 × 106 cells/ml in quadruplicate wells, using 96-well dishes (5 × 104 cells/well), and cultured in fusogenic conditions for 2 d, in the absence or presence of 1 mg/ml BSA, chondroitin sulfate A (CSA) or B (CSB), hyaluronic acid (HA), or 1 μM osteopontin (OP). CD44 expression was determined by ELISA using mAb mouse anti–rat CD44 followed by goat anti–mouse IgG-conjugated to horseradish peroxidase. (B) Cells were cultured as in A in the absence or presence of decreasing concentrations of HA and CD44 expression was determined by ELISA. (C) Cells were cultured as in A in the absence or presence of either OP (1 μM) or HA (1 mg/ml) for 3 d. CD44 expression was determined by ELISA in which mAb anti-CD44 incubation buffer was supplemented or not with either OP (1 μM) or HA (1 mg/ml).
The Extracellular Domain of CD44 Binds Fusing Macrophages and Prevents Multinucleation
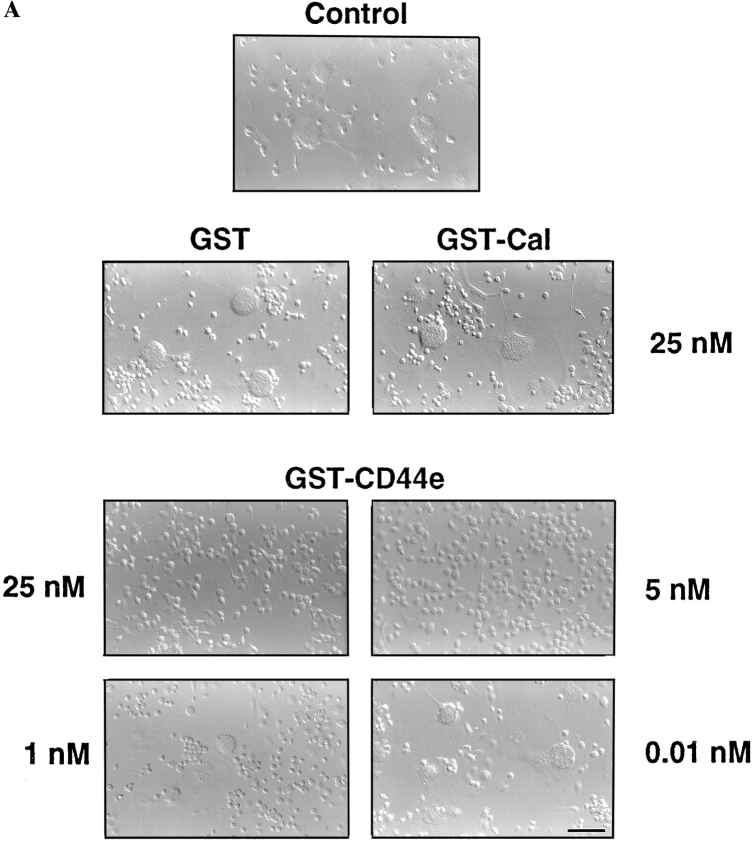
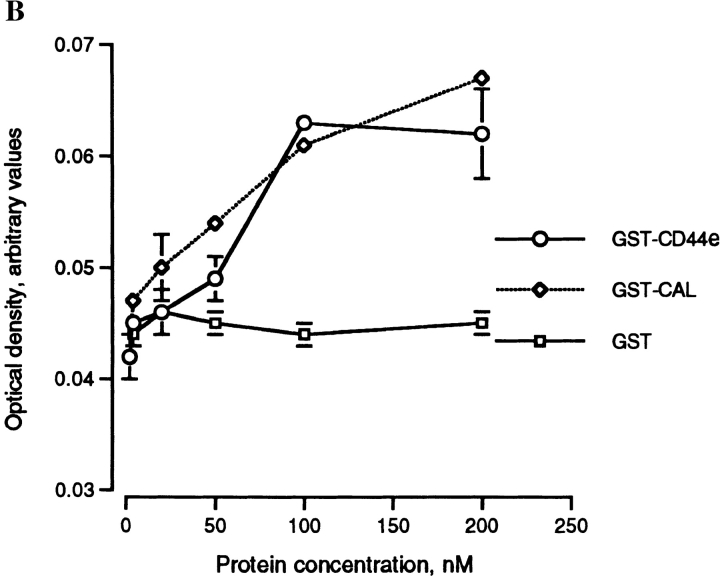
Since the fusion of macrophages requires close membrane–membrane attachment/adhesion, and since fusing macrophages express high levels of CD44 on their surface, we hypothesized that the extracellular domain of CD44 itself may bind a putative cell surface determinant on macrophages and by so doing interfere with adhesion/fusion. We therefore generated a recombinant soluble form of the extracellular domain of CD44 attached to GST, hence a GST–CD44e fusion protein. The addition of GST–CD44e to fusing macrophages strongly and dose dependently prevented multinucleation (Fig. 5 A). Neither GST–calreticulin (Fig. 5, GST-Cal) nor GST used as controls altered multinucleation. Of interest, GST–CD44e and GST–Cal bound macrophages (Fig. 5 B). Although calreticulin is known to bind many cell types in vitro, our data suggest that CD44, and possibly its cell surface ligand, participates in macrophage–macrophage interaction leading to fusion.
Figure 5.
Recombinant extracellular CD44 fusion protein (GST-CD44e) inhibits macrophage fusion. (A) Rat alveolar macrophages were isolated, plated at 5 × 106 cells/ml in quadruplicate wells, using 96-well dishes (5 × 104 cells/well), and cultured in fusogenic conditions for 3 d, in the presence of increasing concentrations of GST–CD44e, GST–Cal, and GST. (B) Cells were cultured as in A for 17 h and then incubated for 3 h at 4°C with GST–CD44e. Binding was determined by ELISA using anti-GST antibody coupled to horseradish peroxidase as described in Materials and Methods. Specific binding was determined by subtracting nonspecific binding from total binding for each concentration of GST–CD44e. Bar, 100 μM.
Fusing Macrophages Express the Standard (Hematopoietic) Form of CD44
To identify the CD44 isoform expressed by fusing macrophages, total RNA was isolated from fusing rat alveolar macrophages was subjected to reverse transcription (RT)- PCR. The use of primers located in the most 3′ and 5′ regions of rat CD44 generated a PCR product of 1,138 bp (data not shown). Using primers located at either end of the extra- and intracellular domains generated PCR products of 827 and 218 bp, respectively. This suggested that both the extra- and intracellular domains of CD44 from fusing macrophages belonged to the standard form. To ensure that these PCR products encoded CD44, the full-length RT-PCR 1,138 bp product was cloned into TA cloning vector and subjected to DNA sequence analysis. DNA sequencing confirmed that fusing rat alveolar macrophages express the standard form of CD44 (data not shown).
Discussion
We have presented evidence that surface expression of CD44 is highly and transiently induced by macrophages at the onset of cell–cell adhesion/fusion in vitro and in vivo. We have also shown that CD44 ligands, HA, CSA, and OP, and the recombinant extracellular domain of CD44 prevent multinucleation. We therefore propose that CD44, along with its putative cognate cell surface ligand, is possibly a potential member of a protein machinery that participates in macrophage–macrophage interaction leading to fusion.
Although multinucleated osteoclasts and giant cells have long been recognized, the mechanism by which their mononucleated precursors adhere to and fuse with each other remains unclear. Although a number of adhesion molecules and regulatory proteins have been suggested to participate in macrophage multinucleation, the nature of the assays reported in the literature used to identify these molecules did not allow one to investigate the direct role of these molecules in cell–cell adhesion and fusion. These assays used bone marrow cells and monocytes; thus, cells that are impure and have not yet acquired the status of the macrophage. These cells require culturing time to further differentiate and become fusion competent. In contrast, we have used a highly pure and fast assay whereby cells adhere to each other to initiate fusion within hours after plating, and reach 99% multinucleation within 3–4 d. This assay allows the investigation of molecules involved in early cell–cell interaction and possibly fusion (Vignery et al., 1989). Indeed, using this assay, we reported earlier the purification of a protein the expression of which is induced by fusogenic conditions in macrophages (Saginario et al., 1995). The subsequent cloning of the cDNA coding for that protein revealed that it belongs to the superfamily of IgG (Saginario et al., 1998). The question as to whether that protein associates with CD44 during cell–cell interaction remains open.
Although CD44 was long known to mediate cell–cell interaction/aggregation, its possible involvement in macrophage–macrophage adhesion leading to fusion had not yet been investigated. From our data, it appears that fusion-competent macrophages express, in an inducible and transient manner, a very high level of surface CD44. This suggests that constitutive CD44 may secure the mononucleated status of macrophages in tissues. It also suggests that macrophage adhesion/fusion requires an overexpression of CD44 in order to override extracellular ligands and provide unoccupied CD44. This “ligand-free” CD44 may in turn participate in cell–cell interaction by virtue of binding to a putative macrophage surface determinant. Although the nature of that putative cell surface ligand for CD44 has not been identified, it appears to bind the core peptide of CD44. The engineering of myc-his-CD44e and its expression by COS cells leads to the production of glycosylated CD44 that failed to block fusion and bind macrophages (data not shown). This might have been be due to the glycosylation pattern imposed by COS cells on the extracellular domain of CD44 that differed from that imposed by rat alveolar macrophages. It is possible that COS cell sugar moieties added to CD44 hide its binding site and prevent CD44 binding. It is also likely that COS cell-added sugar moieties bind nonspecifically to other sites on macrophages, thereby preventing CD44 from interacting with its ligand. Indeed, we observed that partially deglycosylated COS cell produced CD44e-recovered binding of macrophages and prevented their multinucleation (data not shown). This suggested that sugar moieties added to CD44 by cells other than alveolar macrophages may not be functionally relevant. Macrophages appear to process CD44 posttranslationally in a tissue-specific manner (Camp et al., 1991), and although circumstantial, alveolar macrophages do not fuse with peritoneal macrophages (our unpublished observation). The role of glycosylation of CD44 in macrophage–macrophage interaction remains an important subject to investigate.
It is interesting to note that the expression of macrophage surface molecules known to participate in cell–cell interaction such as CD4 and MHCII was not altered by fusogenic conditions. Although their lack of induced expression does not exclude their participation in adhesion/fusion, this could be further suggestive of a possible role for CD44 in macrophage–macrophage interaction leading to fusion.
The question as to how HA prevents macrophage multinucleation is interesting. One possibility is that HA activates macrophages (Nobel et al., 1993) and we have previously reported that activation of macrophages prevents fusion (Vignery et al., 1990). Another possibility is that HA competes with the putative cell surface ligand for CD44, thereby blocking CD44-ligand interaction and cell– cell adhesion.
The fact that macrophages do not attach to HA-coated surfaces suggests that CD44 may not be required for macrophages to adhere onto a substrate. Indeed, it suggests that the possible clustering of CD44 molecules to the adherent domain of the plasma membrane of macrophages may mechanistically prevent adhesion molecules from accessing that domain, thereby preventing macrophages from adhering. This possibility is further suggested by the fact that the addition of HA, and to a lesser extent CSA, to the culture medium reverses this inhibitory effect of HA-coated surfaces on macrophage attachment. Indeed, this may be the case when macrophages adhere to, and fuse onto bone that contains OP. There again, the presence of HA and CSA in the extracellular microenvironment may prevent the clustering of CD44 on the adherent domain of the macrophages.
Although cell–cell adhesion leading to fusion appears to involve a complicated protein machinery, the present findings open possibilities to investigate the role of CD44 and its putative cell surface ligand in the interaction of not only macrophages but also potentially those of sperm cells with oocytes and myoblasts with myoblasts, the other developmental cell–cell adhesion/fusion events. Indeed, the mammalian oocyte–cumulus complex contains an extracellular matrix rich in HA (Cherr et al., 1990), and HA has been suggested to interact with the sperm surface, at the time of initiation of acrosome reaction (Vandevoort et al., 1997). Our observation that CD44 is highly expressed by fusing macrophages and that both CD44 ligands and soluble CD44 prevent multinucleation, opens avenues to study the role of CD44 in osteoclast and giant cell differentiation.
Acknowledgments
We thank M. Rho for her technical help and S. Whitaker for her photographic work. We are grateful to J.A. Elias (Yale University, New Haven, CT) for helpful discussion on CD44.
This work was supported by a grant from the National Institutes of Health (NIH) to Agnès Vignery (RO1 AM-35004). H. Sterling was the recipient of a National Research Service Award (AR08395) and training support for C. Saginario was provided by the NIH (5T32 GM-07223) and the National Osteoporosis Foundation.
Abbreviations used in this paper
- CSA
chondroitin sulfate A
- Cy3
indocarbocyanine
- HA
hyaluronic acid
- HIV
human immunodeficiency virus
- LFA-1
leukocyte function-associated antigen-1
- nt
nucleotide(s)
- OP
osteopontin
- RT
reverse transcription
References
- Alkhatib G, Combadiere C, Broder C, Feng Y, Kennedy P, Murphy P, Berger E. CC CKR5: A RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- Belitsos PC, Hildreth JE, August JT. Homotypic cell aggregation induced by anti-CD44 (Pgp-1) monoclonal antibodies and related to CD44 (Pgp-1) expression. J Immunology. 1990;144:1661–1670. [PubMed] [Google Scholar]
- Blobel CP, Wolfsberg TG, Turk CW, Miles DG, Primakoff P, White JM. A potential fusion peptide and an integrin ligand domain in a protein active in sperm-egg fusion. Nature. 1992;356:248–252. doi: 10.1038/356248a0. [DOI] [PubMed] [Google Scholar]
- Cherr GN, Yudin AI, Katz DF. Organization of the hamster cumulus extracellular matrix A hyaluronate glycoprotein gel which modulates sperm access to the oocyte. Dev Growth Differ. 1990;32:353–366. doi: 10.1111/j.1440-169X.1990.00353.x. [DOI] [PubMed] [Google Scholar]
- Chiozzi P, Sanz J, Ferrari D, Falzoni S, Aleotti A, Buell G, Collo G, Di Virgilio F. Spontaneous cell fusion in macrophage cultures expressing high levels of the P2Z/P2X7receptor. J Cell Biol. 1997;138:697–706. doi: 10.1083/jcb.138.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgleish AG, Beverley P, Clapham P, Crawford D, Greaves M, Weiss R. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R, Hill C, Davis C, Peiper S, Schall T, Littman D, Landau N. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- Dimitrov D. How do viruses enter cells? The HIV coreceptors teach us a lesson in complexity. Cell. 1997;91:721–730. doi: 10.1016/s0092-8674(00)80460-2. [DOI] [PubMed] [Google Scholar]
- Dragic T, Litwin V, Allaway G, Martin S, Huang Y, Nagashima K, Cayanan C, Maddon P, Koup R, Moore J, Paxton W. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- Falzoni S, Munerati M, Ferrari D, Spisani S, Moretti S, Di Virgilio F. The purinergic P2Zreceptor of human macrophage cells. Characterization and possible physiological role. J Clin Invest. 1995;95:1207–1216. doi: 10.1172/JCI117770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Broder C, Kennedy P, Berger E. HIV-1 entry cofactor: functional cDNA cloning of a seven transmembrane, G-protein coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- Glisin V, Crkvenjakov R, Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974;13:2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Hernandez LD, Hoffman LR, Wolfsberg TG, White JM. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- Hildreth J, Orentas R. Involvement of a leukocyte adhesion receptor (LFA-1) in HIV-induced syncyium formation. Science. 1989;244:1075–1078. doi: 10.1126/science.2543075. [DOI] [PubMed] [Google Scholar]
- Higuchi S, Tabata N, Tajima M, Ito M, Tsurudome M, Sudo A, Uchida A, Ito Y. Induction of human osteoclast-like cells by treatment of blood monocytes with anti-fusion regulatory protein-1/CD98 monoclonal antibodies. J Bone Min Res. 1998;13:44–49. doi: 10.1359/jbmr.1998.13.1.44. [DOI] [PubMed] [Google Scholar]
- Ito Y, Komada H, Kusagawa S, Tsurudome M, Matsumura H, Kawano M, Ohta H, Nishio M. Fusion regulation proteins on the cell surface: Isolation and characterization of monoclonal antibodies which enhance giant polykaryocyte formation in Newcastle Disease virus-infected cell lines of human origin. J Virol. 1992;66:5999–6007. doi: 10.1128/jvi.66.10.5999-6007.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalkanen S, Jalkanen M. Lymphocyte CD44 binds the COOH terminal heparin-binding domain of fibronectin. J Cell Biol. 1992;116:817–825. doi: 10.1083/jcb.116.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazi F, Chang J, Lopez A, Vaas M, Cunningham A. Interleukin 4 and human immunodeficiency virus stimulate LFA-1-ICAM-1-mediate aggregation of monocytes and subsequent giant cell formation. J Gen Vir. 1994;75:2795–2802. doi: 10.1099/0022-1317-75-10-2795. [DOI] [PubMed] [Google Scholar]
- Klatzmann D, Champagne E, Chamaret S, Grust J, Guetard D, Hercent T, Gluckmann JC, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- Kurachi T, Morita I, Murota S. Involvement of adhesion molecules LFA-1 and ICAM-1 in osteoclast development. Biochem Biophys Acta. 1993;1178:259–266. doi: 10.1016/0167-4889(93)90202-z. [DOI] [PubMed] [Google Scholar]
- Lesley J, Schulte R, Hyman R. Binding of hyaluronic acid to lymphoid cell lines is inhibited by monoclonal antibodies against Pgp-1. Exp Cell Res. 1990;187:224–233. doi: 10.1016/0014-4827(90)90085-o. [DOI] [PubMed] [Google Scholar]
- Mbalaviele G, Chen H, Boyce B, Mundy G, Yoneda T. The role of cadherin in the generation of multinucleated osteoclasts from mononuclear precursors in murine marrow. J Clin Invest. 1995;95:2757–2765. doi: 10.1172/JCI117979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mege R, Goudou D, Diaz C, Nicolet M, Garcia L, Geraud L, Rieger F. N-cadherin and N-CAM in myoblast fusion: compared localisation and effect of blockade by peptides and antibodies. J Cell Sci. 1992;103:897–906. doi: 10.1242/jcs.103.4.897. [DOI] [PubMed] [Google Scholar]
- Miyake K, Underhill C, Lesley J, Kincade R. Hyaluronate can function as a cell adhesion molecule and CD44 participates in hyaluronate recognition. J Exp Cell Med. 1990;172:69–75. doi: 10.1084/jem.172.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möst J, Neumayer H, Dierich M. Cytokine induced generation of multinucleated giant cells in vitro requires interferon-γ and expression of LFA-1. Eur J Immunol. 1990;20:1661–1667. doi: 10.1002/eji.1830200807. [DOI] [PubMed] [Google Scholar]
- Naor D, Sionov VR, Ish-Shalom D. CD44: structure function and association with the malignant process. Adv Can Res. 1997;71:241–319. doi: 10.1016/s0065-230x(08)60101-3. [DOI] [PubMed] [Google Scholar]
- Noble P, Lake F, Henson P, Riches D. Hyaluronate activation of CD44 induces insulin-like growth factor-1 expression by a tumor necrosis factor-α-dependent mechanism in murine macrophages. J Clin Invest. 1993;91:2368–2377. doi: 10.1172/JCI116469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgimoto S, Tabata N, Suga S, Nishio M, Ohta H, Tsurudome M, Komada H, Kowano M, Watanabe N, Ito Y. Molecular characterization of fusion regulatory protein-1 (FRP-1) that induces multinucleate giant cell formation of monocytes and HIV gp-160 mediated cell fusion. J Immunol. 1995;155:3585–3592. [PubMed] [Google Scholar]
- Ohta H, Tsurudome M, Matsumura H, Koga Y, Morikawa S, Kawano M, Kusugawa S, Komada H, Nishio M, Ito Y. Molecular and biological characterization of fusion regulatory proteins (FRPs): anti-FRP mAbs induced HIV mediated cell fusion via an integrin system. EMBO (Eur Mol Biol Organ) J. 1994;13:2044–2055. doi: 10.1002/j.1460-2075.1994.tb06479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleo G, Butini L, Graziosi C, Poli G, Schnittman SM, Greenhouse J, Gallin J, Fauci AS. Human Immunodeficiency Virus (HIV) infection in CD4+T lymphocytes genetically deficient in LFA-1: LFA-1 is required for HIV-mediated cell fusion but not for viral transmission. J Exp Med. 1991;173:511–514. doi: 10.1084/jem.173.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivadeneira E, Sauls D, Yu Y, Haynes B, Weinberg J. Inhibition of HIV type I infection of mononuclear phagocytes by anti-CD44 antibodies. AIDS Res Hum Ret. 1995;11:541–546. doi: 10.1089/aid.1995.11.541. [DOI] [PubMed] [Google Scholar]
- Saginario C, Qian H-Y, Vignery A. Identification of an inducible surface molecule specific to fusing macrophages. Proc Natl Acad Sci USA. 1995;92:2210–2214. doi: 10.1073/pnas.92.26.12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saginario C, Sterling H, Beckers C, Kobayashi R-J, Solimena M, Ullu E, Vignery A. MFR, a putative receptor mediating the fusion of macrophages. Mol Cell Biol. 1998;18:6213–6223. doi: 10.1128/mcb.18.11.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 545 pp.
- Shepley M, Racaniello VR. A monoclonal antibody that blocks polovirus attachment recognizes the lymphocyte homing receptor CD44. J Virology. 1994;68:1301–1308. doi: 10.1128/jvi.68.3.1301-1308.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamenkovic I, Aruffo A, Amiot M, Seed B. The hematopoietic and epithelial forms of CD44 are distinct polypeptides with different adhesion potentials for hyaluronate bearing cells. EMBO (Eur Mol Biol Organ) J. 1991;10:343–348. doi: 10.1002/j.1460-2075.1991.tb07955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy MS, Guo YJ, Stamenkovic I. Distinct effects of two CD44 isoforms on tumor cell growth in vivo. J Exp Med. 1991;174:859–866. doi: 10.1084/jem.174.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata N, Ito M, Shimokata K, Suga S, Ohgimoto S, Tsurudome M, Kawano M, Matsumura H, Komada H, Nishio M, Ito Y. Expression of fusion regulatory proteins (FRPs) on human peripheral blood monocytes. J Immunol. 1994;153:3256–3266. [PubMed] [Google Scholar]
- Tolg C, Hofmann M, Herrlich P, Ponta H. Splicing choice from ten variant exons establishes CD44 variability. Nucl Acids Res. 1993;21:1225–1229. doi: 10.1093/nar/21.5.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich A, Shine J, Chirgwin J, Pictet R, Tischer E, Rutler WJ, Goodman H. Rat insulin genes: construction of plasmids containing the coding sequences. Science. 1977;196:1313–1319. doi: 10.1126/science.325648. [DOI] [PubMed] [Google Scholar]
- Vandevoort CA, Cherr GN, Overstreet JW. Hyaluronic acid enhances the zona pellucida-induced acrosome reaction of macaque sperm. J Andrology. 1997;18:1–5. [PubMed] [Google Scholar]
- Vignery A. Macrophage multinucleation is accompanied by the expression of new soluble and membrane antigens in mice. Am J Pathol. 1989;135:565–570. [PMC free article] [PubMed] [Google Scholar]
- Vignery A, Niven-Fairchild T, Ingbar D, Caplan M. Polarized distribution of Na+ K+-ATPase in giant cells elicited in vivo and in vitro. J Histochem Cytochem. 1989;35:1265–1271. doi: 10.1177/37.8.2546991. [DOI] [PubMed] [Google Scholar]
- Vignery A, Niven-Fairchild T, Shepard M. Recombinant murine interferon-gamma inhibits the fusion of mouse alveolar macrophages in vitro but stimulates the formation of osteoclast like cells on implanted syngeneic bone particles in mice. J Bone Miner Res. 1990;5:637–644. doi: 10.1002/jbmr.5650050613. [DOI] [PubMed] [Google Scholar]
- Wakelam MJ. Myoblast fusion. A mechanistic approach. Curr Top Membr Transp. 1989;32:87–112. [Google Scholar]
- Weber G, Ashkar S, Glimcher M, Cantor H. Receptor-ligand interaction between CD44 and osteopontin (Eta-1) Science. 1996;271:509–512. doi: 10.1126/science.271.5248.509. [DOI] [PubMed] [Google Scholar]