Abstract
The nuclear pore complex (NPC), a supramolecular assembly of ∼100 different proteins (nucleoporins), mediates bidirectional transport of molecules between the cytoplasm and the cell nucleus. Extensive structural studies have revealed the three- dimensional (3D) architecture of Xenopus NPCs, and eight of the ∼12 cloned and characterized vertebrate nucleoporins have been localized within the NPC. Thanks to the power of yeast genetics, 30 yeast nucleoporins have recently been cloned and characterized at the molecular level. However, the localization of these nucleoporins within the 3D structure of the NPC has remain elusive, mainly due to limitations of preparing yeast cells for electron microscopy (EM). We have developed a new protocol for preparing yeast cells for EM that yielded structurally well-preserved yeast NPCs. A direct comparison of yeast and Xenopus NPCs revealed that the NPC structure is evolutionarily conserved, although yeast NPCs are 15% smaller in their linear dimensions. With this preparation protocol and yeast strains expressing nucleoporins tagged with protein A, we have localized Nsp1p and its interacting partners Nup49p, Nup57p, Nup82p, and Nic96p by immuno-EM. Accordingly, Nsp1p resides in three distinct subcomplexes which are located at the entry and exit of the central gated channel and at the terminal ring of the nuclear basket.
Keywords: colloidal gold, Nsp1p, nuclear pore complex, nucleoporin, yeast
The nuclear pore complex (NPC),1 a supramolecular assembly of proteins embedded in the double membrane of the nuclear envelope (NE), mediates the bidirectional transport of molecules between the cytoplasm and the nucleus of eukaryotic cells (for review see Görlich and Mattaj, 1996; Panté and Aebi, 1996a ; Nigg, 1997; Ohno et al., 1998). Extensive electron microscopy (EM) studies using amphibian NEs have revealed a consensus model of the three-dimensional (3D) structure of the NPC (for review see Davis, 1995; Panté and Aebi, 1996b ). Accordingly, the NPC consists of an eightfold symmetric basic framework (also called the spoke complex), sandwiched between a cytoplasmic and a nuclear ring. The cytoplasmic ring is decorated with eight ∼50-nm- long fibrils, whereas the nuclear ring is capped with a cage-like assembly, called the nuclear basket. The ring-like basic framework embraces the central gated channel which is implicated in signal- and energy-mediated bidirectional transport of macromolecules between the cytoplasm and the nucleus. This central gated channel is often plugged with a particle (called central plug or transporter) of highly variable appearance whose ultimate structure and functional role remain to be established.
Based on its molecular mass of ∼120 megadaltons (MDa) (Reichelt et al., 1990) and on the high degree of symmetry of the basic framework (8, 2, 2), it is assumed that the NPC is composed of multiple copies (i.e., 8 or 16) of ∼100 different proteins, called nucleoporins (Nups). Whereas in vertebrates only 11 nucleoporins have been identified and molecularly characterized to date, in yeast the number of known Nups has now reached 30 (for review see Panté and Aebi, 1996b ; Doye and Hurt, 1997; Fabre and Hurt, 1997). This rapid progress in identifying and characterizing yeast Nups is due to (a) the development of a biochemical procedure to bulk isolate yeast NPCs (Rout and Blobel, 1993), (b) the use of genetic screens designed both to identify proteins interacting with a known Nup (i.e., synthetic lethal screens) and to isolate transport defected mutants (for review see Fabre and Hurt, 1997), and (c) the recent completion of the yeast genome project. It is expected that in the near future all yeast Nups will be identified.
Biochemical and genetic interactions among nucleoporins have indicated that within the NPC different sets of nucleoporins are organized in subcomplexes which may be isolated as such. The first identified and molecularly characterized NPC subcomplex was the vertebrate p62 complex, consisting of the nucleoporins p62, p58, p54, and p45 (Dabauvalle et al., 1990; Finlay et al., 1991; Kita et al., 1993; Guan et al., 1995; Hu et al., 1996). In Xenopus egg lysates, p62 was also found engaged in a second subcomplex together with CAN/Nup214 (Macaulay et al., 1995). In addition, three other vertebrate NPC subcomplexes have also been identified but not yet well characterized. These are the Nup153 homo-oligomer, and the CAN/Nup214– Nup84 complex with molecular masses of 1 MDa and 1.5– 2.0 MDa, respectively (Panté et al., 1994), and the Nup93– p205 complex (Grandi et al., 1997). These results indicate that Nups can be organized in multiple subcomplexes. In yeast, Nsp1p (the putative homologue of p62; Carmo-Fonseca et al., 1991) has also been isolated in two distinct subcomplexes. One subcomplex consists of Nsp1p interacting with Nup49p, Nup57p, and Nic96p (Grandi et al., 1993), and the second complex contains Nsp1p and Nup82p (Grandi et al., 1995b ). The first subcomplex may represent the yeast homologue of the vertebrate p62 complex, and in the following we will refer to it as the Nsp1p complex. Another identified yeast subcomplex is the so-called Nup84p complex, consisting of the nucleoporins Nup84p, Nup85p, Nup120p, and Nup145p, and the proteins Sec13p (a protein involved in ER to Golgi transport) and Seh1p (a Sec13p homologue; Siniossoglou et al., 1996).
From the three yeast NPC subcomplexes described above, only the Nsp1p complex has been characterized at the molecular level (Schlaich et al., 1997). Accordingly, as shown by in vitro reconstitution experiments, purified recombinant Nsp1p, Nup49p, and Nup57p form a 150-kD complex containing one copy each of these three nucleoporins. Nup57p is the organizing center of this complex to which Nsp1p and Nup49p individually can bind. Although Nic96p does not bind to individual Nsp1p, Nup49p, and Nup57p, it associates to the fully assembled Nsp1p– Nup49p–Nup57p core complex (Schlaich et al., 1997). A common feature of the primary structure of Nsp1p and its interacting nucleoporins is the presence of heptad repeats that are predicted to form α-helical coiled-coil segments. These heptad repeat segments reside in the COOH-terminal domain of Nsp1p, Nup49p, and Nup57p, and are involved in the interactions among these proteins. Moreover, full-length Nic96p, but not a mutant form lacking the NH2-terminal coiled-coil domain of Nic96p, associates with the Nsp1p core complex (Grandi et al., 1995b ; Schlaich et al., 1997). Similarly, the Nsp1p–Nup82p complex is held together by α-helical coiled-coil interactions between these two proteins (Grandi et al., 1995a ).
In addition to the α-helical coiled-coil domains, the Nups of the Nsp1p core complex also contain repetitive FXFG and GLFG sequence motifs whose specific role in nuclear transport is still speculative (e.g., Radu et al., 1995). Similarly, other yeast nucleoporins and a number of vertebrate nucleoporins contain FXFG sequence motifs. However, the relationship between these vertebrate FXFG repeat nucleoporins and the yeast FXFG repeat nucleoporins is unclear. Except for the Nsp1p core complex which is considered to be the yeast homologue of the vertebrate p62 complex, p62 and Nsp1p reveal 31% identity and 50% homology in their COOH-terminal regions (Carmo-Fonseca et al., 1991), and Nup49p and Nup57p share homology with rat p58/45 and rat p54 respectively (Doye and Hurt, 1997; Fabre and Hurt, 1997). Recently, vertebrate Nic96p homologues, called Nup93, have been identified which share 27% identity and 50% similarity with yeast Nic96p (Allende et al., 1996; Grandi et al., 1997). As Nic96p is interacting with Nsp1p, Nup93 interacts with a small fraction of the p62 moiety. Similar to the p62 complex, the two Nsp1p complexes are directly implicated in nuclear transport: whereas mutation or deletion of the NSP1 or the NIC96 gene led to import defects (Mutvei et al., 1992; Nehrbass et al., 1993; Grandi et al., 1993), NUP57, NUP49, and NUP82 mutant strains are defective in both protein import and mRNA export (Sharma et al., 1996; Grandi et al., 1995b ; Hurwitz and Blobel, 1995). Moreover, synthetic lethal screens indicated genetic interactions of Nsp1p with several nucleoporins, nearly all of them being involved in mRNA export (for review see Doye and Hurt, 1997; Fabre and Hurt, 1997).
Little is known about how Nups interact with each other to build the NPC, and about the participation of different Nups in nuclear import and export. To obtain a more detailed insight into the molecular architecture of the NPC that may allow to deduce Nup function, it is necessary to map nucleoporins within the 3D structure of the NPC. In vertebrates these localization studies are limited because only few nucleoporins have been identified and molecularly characterized. Nevertheless, eight vertebrate nucleoporins have now been localized within the 3D architecture of the NPC by immuno-EM (Panté et al., 1994; Guan et al., 1995; Grandi et al., 1997). Yeast will be an excellent system for mapping Nups and studying the functional roles of nucleoporins, because 30 Nups have already been identified and molecularly characterized, and several mutant strains that affect both import of nuclear proteins and export of RNA have been created. Although these Nups have been localized to the NE rim by immunofluorescence microscopy, none of them have been localized within the 3D NPC architecture. This is in part due to limitations in EM sample preparation of yeast cells, and to the high cross-reactivity of anti-Nup antibodies caused by the presence of highly antigenic repetitive sequence motifs within the amino acid sequence of nucleoporins (e.g., the FXFG sequence motifs). The yeast system also offers the advantage of introducing genetic manipulations. Thus, knowing the molecular architecture of yeast NPCs will open the possibility to combine genetic with structural and functional studies. These type of studies could yield a more detailed picture of how NPCs transport macromolecules into and out of the nucleus.
Here we have developed a new EM sample preparation protocol that yields structurally well-preserved yeast NPCs. Using this new protocol in combination with yeast strains expressing nucleoporins tagged with protein A, we have mapped five yeast nucleoporins within the 3D architecture of the NPC. Specifically, we report the localization of Nsp1p and its interacting proteins Nup49p, Nup57p, Nup82p, and Nic96p within the 3D architecture of yeast NPCs by immuno-EM. Interestingly, our localization studies indicate that Nsp1p resides in three distinct subcomplexes within the NPC which are located at NPC sites where cargo or transport ligands can be arrested during nuclear protein import (Panté and Aebi, 1995, 1996c ; Görlich et al., 1996). Thus, we proposed that these subcomplexes are directly involved in protein import.
Materials and Methods
Yeast Strains and Plasmids
The yeast strains and plasmids used in this study are listed in Table III and shown in Fig. 2, respectively. Yeast cells were grown in complete medium (YPAD; 1% yeast extract, 2% bacto-peptone, 2% glucose, 0.003% adenine sulfate) at 30°C.
Table III.
Yeast Strains
| Strain | Genotype | References | ||
|---|---|---|---|---|
| RS453 | a /α, ade2/ade2, his3/his3, leu2/leu2, trp1/trp1, ura3/ura3 | Wimmer et al., 1992 | ||
| Wt | α, ade2, his3, leu2, trp1, ura3 (haploid derivative from RS453) | Grandi et al., 1993 | ||
| TF2 | a /α, ade2/ade2, ade8/ade8, can1-100/can1-100, his4/HIS4, his3/HIS3, leu2/leu2, lys1/lys1, | Hurt, 1988 | ||
| ura3/URA3::nsp1/NSP1 (derived from JU4-2×JR26-19B) | ||||
| JU4-2×JR26-19B | a /α, ade2/ade2, ade8/ADE8, can1-100/can1-100, his4/HIS4, his3/HIS3, leu2/leu2, lys1/lys1, ura3/URA3 | Hurt, 1988 | ||
| VD1 | a /α, ade2/ade2, his3/his3, leu2/leu2, ura3/ura3, trp1/TRP::nsp49/NSP49 (derived from RS453) | Wimmer et al., 1992 | ||
| PG2 | a /α, ade2/ade2, trp1/trp1, leu2/leu2, ura3/ura3, his3/HIS3::nup82/NUP82 (derived from RS453) | Grandi et al., 1995 | ||
| ProtA-NSP1 | α, ade2, ade8, can1-100, leu2, lys1, URA3::nsp1, his −, pSB32-ProtA-NSP1 (derived from TF2) | Grandi et al., 1993 | ||
| ProtA-NUP49 | a , ade2, his3, trp::nup49, leu2, ura3, pUN100-LEU2-ProtA-NUP49 (derived from VD1) | Wimmer et al., 1992 | ||
| NUP57-ProtA | α, ade2, trp1, leu2, ura3, HIS3::nup57, pRS316-LEU2-NUP57-ProtA | Grandi et al., 1993 | ||
| ProtA-NIC96 | α, ade2, trp1, leu2, ura3, URA3::nic96, pUN100-LEU2-ProtA-NIC96 | Grandi et al., 1993 | ||
| ProtA-NUP82 | a , ade2, trp1, leu2, ura3, his3, HIS3::nup82, pUN100-LEU2-ProtA-NUP82 (derived from PG2) | Grandi et al., 1995b | ||
| ProtA-PUS1 | a , ade2, trp1, leu2, ura3, his3, HIS3::pus1, pUN100-LEU2-ProtA-PUS1 (derived from RS453) | Simos et al., 1996 | ||
| ProtA-DHFR | a /α, ade2/ade2, ade8/ADE8, can1-100/can1-100, his4/HIS4, his3/HIS3, leu2/leu2, | Grandi et al., 1993 | ||
| lys1/lys1, ura3/URA3, YEp13-ProtA-DHFR (derived from JU4-2×JR26-19B) |
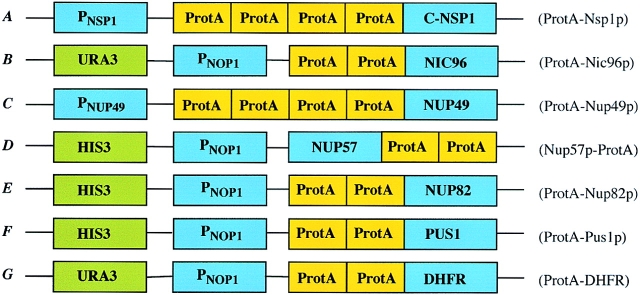
Figure 2.
Schematic diagram of the fusion constructs used in this study: (A) Four IgG-binding domains derived from S. aureus protein A (ProtA) and the essential NSP1 COOH-terminal domain. (B) Two IgG-binding domains of ProtA and the NIC96 gene under control of the NOP1 promoter and URA3 selection. (C) Four IgG-binding domains of ProtA and the NUP49 gene. (D) Two IgG-binding domains of ProtA and the NUP57 gene under control of the NOP1 promotor and HIS3 selection. (E) Two IgG-binding domains of ProtA and the NUP82 gene under control of the NOP1 promotor and HIS3 selection. (F) Two IgG-binding domains of ProtA and the PUS1 gene under control of the NOP1 promotor and HIS3 selection. (g) Two IgG-binding domains of ProtA and the DHFR gene under control of the NOP1 promotor and URA3 selection. Right, abbreviations for the yeast strains carrying protein A–tagged nucleoporins used in this paper.
Epitope Tagging Yeast Nups with Protein A
The tagging of Nups with the IgG binding domain from Staphylococcus aureus protein A was described elsewhere (NSP1, NIC96, and DHFR: Grandi et al., 1993; NUP49: Wimmer et al., 1992; NUP57: Grandi et al., 1993; NUP82: Grandi et al., 1995b ; PUS1: Simos et al., 1996).
Thin Section Electron Microscopy of Yeast Cells
Yeast cells were grown to an OD600 from 1 to 2, and all procedures were performed at room temperature unless otherwise stated. The cells were fixed by adding paraformaldehyde, pH 6.5, to a final concentration of 2% followed by incubation for 1 h. After fixation, the cells were centrifuged for 2 min at 1,000 g, washed once in water, and then incubated for 3 min in 0.1 M Tris, pH 9.5, 10 mM DTT. After washing in SP buffer (1.2 M sorbitol, 20 mM potassium phosphate buffer, pH 7.4), the cells were resuspended in this buffer to 5 OD600/ml. Spheroplasts were obtained by adding 5 U/ml zymolyase 20T (Seikagaku Corporation, Tokyo, Japan) and a 1:100 dilution of solution P (87 mg PMSF and 1.5 mg pepstatin A in 5 ml dry absolute ethanol) to the cells, followed by a 20-min incubation at 30°C while shaking with 200 rpm. The spheroplasts were then washed twice by adding 5 ml of KPi buffer (0.1 M potassium phosphate buffer, pH 6.5), followed by centrifugation at 4,000 rpm for 2 min. Samples were prefixed with 2% glutaraldehyde in KPi for 1 h, washed two times with KPi, embedded in low melting agarose, and postfixed with 1% OsO4 in KPi for 1 h. Next the fixed spheroplasts were washed two times in KPi, dehydrated with 50% ethanol for 10 min, and bloc stained in 70% ethanol/2% uranyl acetate for 1 h. The samples were further dehydrated with 90% ethanol for 10 min, followed by three times 100% ethanol (each for 10 min), and finally with 100% acetone for 10 min. Next the samples were infiltrated with mixtures of Epon (Fluka, Buchs, Switzerland) and acetone 1:1 for 1 h, 2:1 for 1 h, and finally in pure Epon for 3–4 h. Samples were placed into gelatin capsules filled with fresh pure Epon resin and polymerized over night at 60°C.
Thin sections were cut on a Reichert Ultracut microtome (Reichert-Jung Optische Werke, Vienna, Austria) using a diamond knife. The sections were collected on pallodion-coated copper EM grids and stained with 6% uranyl acetate for 1 h and 2% lead citrate for 2 min. Electron micrographs were recorded with a Hitachi H-7000 transmission electron microscope (Hitachi Ltd., Tokyo, Japan) operated at an acceleration voltage of 100 kV.
Immunogold Localization of Yeast Nups
Colloidal gold particles, ∼8 nm in diameter, were prepared by reduction of tetrachloroauric acid with sodium citrate in the presence of tannic acid (Slot and Geuze, 1985). The polyclonal anti–protein A antibody (Sigma Chemical Co., St. Louis, MO) was conjugated to colloidal gold particles as described by Baschong and Wrigley (1990). After conjugation, the complexes were centrifuged at 32,000 g for 15 min. The soft pellet was resuspended in KPi buffer containing 0.1% BSA (Merck, Darmstadt, Germany) and used for labeling spheroplasted yeast cells.
Yeast cells were grown and spheroplasted as described above. 1.5 ml of spheroplasted yeast cells were transferred in an Eppendorf tube, washed two times in 1 ml KPi, resuspended in 1 ml KPi containing 0.05% Triton X-100, and spun down immediately. The Triton X-100–extracted yeast cells were washed four times in KPi, resuspended in 100 μl anti–protein A antibody labeled with 8-nm colloidal gold (75 μg/ml) and incubated for 3 h at 30°C while shaking with 200 rpm. Next, the cells were washed twice in KPi containing 0.1% BSA, fixed, dehydrated, Epon-embedded, and then prepared for EM as described above.
Quantitation of Gold Labeling at the NPC
The position of gold particles associated with NPCs were measured from electron micrographs of cross sections along the NE. Distances of gold particles perpendicular to the central plane of the NPC and from the eightfold symmetry axis of the NPC were determined.
Stereology
Quantitation of the antibody labeling in the wild-type strain and the control strains ProtA-DHFR and ProtA-Pus1p were done by a stereological counting method described by Lucocq (1992). In brief, ultrathin sections were selected at random and mounted on an EM grid. Electron micrographs of ultrathin sections were recorded and printed on photographic paper with a final magnification of 50,000. The area of the cytoplasm and the nucleus and the gold particle per cell were estimated with a square lattice grid of 20-mm spacing. 22, 18, and 15 cells for the wild-type, ProtA-Pus1p and, ProtA-DHFR were analyzed, respectively.
Isolation of Xenopus Oocyte Nuclei and Preparation of NEs
Nuclei were isolated from mature (stage six) Xenopus oocytes, and embedded and thin-sectioned exactly as described by Panté et al. (1994).
Fluorescence Microscopy
The in vivo location of GFP-C-Nsp1p was analyzed according to Segref et al. (1997) in the thermosensitive mutant nup49-313 (Doye et al., 1994) and NUP49+ cells which were transformed with plasmid SM666 (provided by S. Bailer, BZH, Heidelberg, Germany). SM666 is an ADE2-containing ARS/CEN plasmid containing a fusion construct between the genes encoding GFP and the Nsp1p COOH-terminal domain. Cells were examined in the fluorescein channel of a Zeiss Axioskop fluorescence microscope (Carl Zeiss, Inc., Thornwood, NY). Pictures were taken with a Xillix Microimager charge-coupled device camera (Xillix Technology Corp., British Columbia, Canada). Digital pictures were processed by modules of the software Openlab (Improvision, Coventry, UK).
Results
Structural Analysis of Yeast NPCs
To establish the molecular architecture of the yeast NPC, we have first developed an EM sample preparation protocol that yields structurally well-preserved yeast NPCs (Fig. 1 A). Our strategy was to systematically test preparation conditions to remove the yeast cell wall. For this purpose, we used different combinations of chemical prefixation and enzyme-digestion conditions. The best results were found with 2% paraformaldehyde prefixation and digestion with zymolyase 20T (see Materials and Methods for details). Good results were also obtained when the yeast cells were prefixed with 0.125% glutaraldehyde for 5 min at room temperature instead (Fig. 1 A, top). However, in combination with Triton X-100 detergent extraction (a step that is necessary for pre-embedding immunolabeling, see below), the glutaraldehyde prefixation was not strong enough so that the yeast cells were completely destroyed. On the other hand, increasing the glutaraldehyde concentration (i.e., 0.25 or 0.5%) interfered with the zymolyase activity so that the cell wall was not removed. Hence, we invariably used 2% paraformaldehyde or 0.125% glutaraldehyde prefixation for structural studies, but always 2% paraformaldehyde for immunolabeling.
Figure 1.

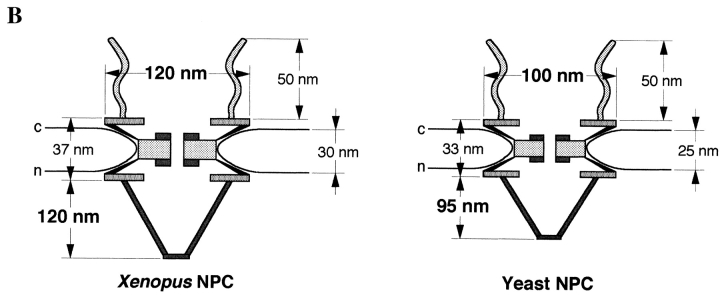
Yeast NPCs. (A) Cross section through the NE and a gallery of selected electron micrographs of NPC cross sections. Large arrowheads, nuclear baskets; small arrowheads, cytoplasmic fibrils; c, cytoplasm; n, nucleus. (B) Schematic comparison of Xenopus and yeast NPCs. Bar, 100 nm.
As shown in Fig. 1 A, similar to amphibian NPCs, our preparation protocol reveals the existence of peripheral NPC structures such as the cytoplasmic fibrils (small arrowheads) and the nuclear baskets (large arrowheads). Direct comparison of yeast and Xenopus NPCs (the best structurally characterized higher eukaryotic NPC) after using the same Epon embedding protocol, revealed that although the overall architecture of the NPC is rather conserved, yeast NPCs appear typically 15% smaller in their linear dimensions than Xenopus NPCs. As documented in Fig. 1 B and Table I, the vertical distance between the cytoplasmic and nuclear ring, the lumenal space of the NE, and the length of the cytoplasmic fibrils appear very similar in the two species, whereas the total diameter of the NPC and the height of the nuclear baskets appear significantly smaller in yeast as compared with Xenopus.
Table I.
Size of Xenopus NPC in Comparison with Yeast NPC
| Diameter of the NPC | Ring distance | Height of the nuclear baskets | Length of the cytoplasmic fibrils | Lumenal space of the NE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| nm | nm | nm | nm | nm | ||||||
| Xenopus | 119.9 ± 7.3 | 36.6 ± 5.5 | 118.6 ± 9.6 | 52.3 ± 10.0 | 29.6 ± 5.1 | |||||
| Yeast | 102.7 ± 7.9 | 32.8 ± 3.5 | 95.0 ± 11.3 | 51.3 ± 11.1 | 25.4 ± 4.7 |
Measurements were from cross sectioned yeast and Xenopus NPCs prepared for EM as described in Materials and Methods. The mean ± SD are given from all values. 21 NPCs were evaluated for each species.
Nsp1p Is Found at Three Distinct NPC Sites
After having succeeded to prepare structurally well-preserved yeast NPCs, we next wanted to examine in detail the location of yeast Nups within the 3D architecture of the NPC. Since anti-Nup antibodies frequently cross-react on the EM level, for our localization studies, we used yeast strains expressing Nups tagged with protein A in combination with a colloidal gold-conjugated anti–protein A antibody (see Materials and Methods). These location studies were started with Nsp1p, the first yeast Nup that has been identified and molecularly characterized (Hurt, 1988; Nehrbass et al., 1990). For this purpose we used a yeast strain disrupted for the chromosomal NSP1 gene, and complemented by plasmid-borne (single copy ARS/CEN plasmid) Nsp1p tagged with protein A (ProtA–Nsp1p; see Fig. 2). We have not integrated ProtA–NSP1 into the genome to guarantee chromosomal expression levels. However, we found that the amount of Nsp1p expressed from a centromeric plasmid is similar to the expression level when the NSP1 gene expression is driven by the chromosomal NSP1 gene copy (data not shown).
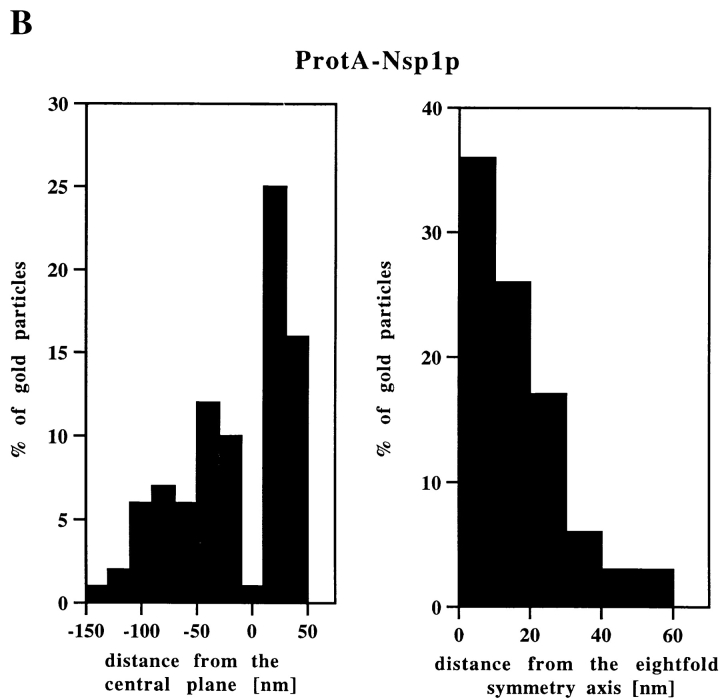
By immunogold EM we found that the anti–protein A antibody significantly labeled the NPC on both the cytoplasmic and the nuclear face of ProtA–Nsp1p cells (Fig. 3 A). To identify the NPC sites labeled by the antibody we performed quantitative analysis of the gold particle distribution along the two symmetry axis of the NPC (i.e., the central plane of the NPC and the eightfold symmetry axis). As indicated in Fig. 3 B, the distribution of the gold particles relative to the central plane of the NPC revealed three major peaks at vertical distances of ∼20, −20, and −80 nm from the central plane of the NPC which correspond to the cytoplasmic and the nuclear periphery of the central gated channel, and to the terminal ring of the nuclear basket. As these NPC components are located about the center of the eightfold symmetry axis, 93% of the gold particles were found within a radius of 30 nm from this symmetry axis (see Fig. 3 B). The quantitative analysis also revealed that the labeling of the cytoplasmic site was more abundant than the labeling of the two distinct nuclear sites (see Fig. 3 B). This is probably due to the fact that with the pre-embedding labeling technique that we used the antibody first reaches the cytoplasmic epitope, where it is bound before it can diffuse through the permeabilized NE to the nuclear epitopes.
Figure 3.
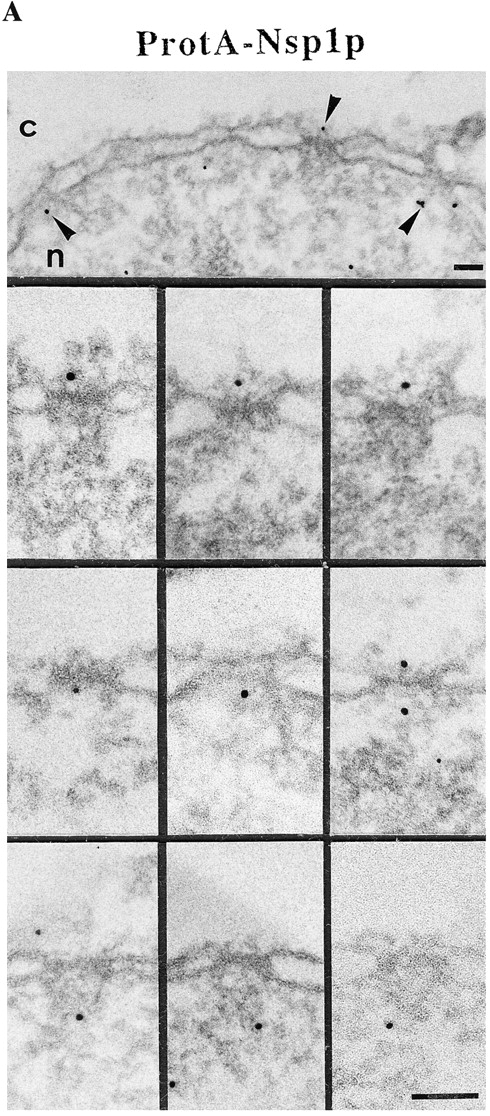
Immunogold localization of Nsp1p in ProtA–Nsp1p cells. (A) Triton X-100– extracted spheroplasts from ProtA–Nsp1p cells were pre-immunolabeled with a polyclonal anti–protein A antibody conjugated to 8-nm colloidal gold, and prepared for EM by Epon embedding and thin sectioning. Shown is a view along a cross sectioned NE stretch with labeled NPCs (arrowheads), together with a gallery of selected examples of gold-labeled NPC cross sections. The antibody labeled the cytoplasmic periphery and the nuclear periphery of the central gated channel, and the terminal ring of the nuclear baskets. c, cytoplasm; n, nucleus. (B) Quantitative analysis of the gold particles associated with the NPCs in the ProtA–Nsp1p strain after labeling with gold-conjugated anti–protein A antibody. 88 gold particles were scored. Bars, 100 nm.
To demonstrate that the gold-conjugated anti–protein A antibody specifically labeled the NPCs of ProtA–Nsp1p cells, we carried out similar labeling experiments with control strains expressing either a cytosolic or nuclear reporter protein tagged with protein A. For a cytosolic protein, a strain expressing the cytosolic mouse dihydrofolate reductase (DHFR) tagged with protein A (ProtA–DHFR, see Fig. 2, Grandi et al., 1993), whereas for a nuclear protein a strain expressing Pus1p, a protein involved in tRNA biogenesis, tagged with protein A (ProtA–Pus1p, see Fig. 2; Simos et al., 1996) were used. As expected, for ProtA– DHFR we obtained gold labeling predominantly in the cytoplasm with a very small nuclear background, and for ProtA–Pus1p the gold particles accumulated in the nucleus with some cytoplasmic background (Table II). As a control, we also used a wild-type strain expressing no protein A–tagged proteins. When these wild-type cells were prepared for immunolabeling by exactly the same protocol as the mutant strains, no gold labeling of these cells was found except for a small cytoplasmic background (Table II). In neither wild-type control nor ProtA-DHFR and ProtA-Pus1p strains we could detect any significant gold labeling of the NPCs. In addition, an antibody against Nsp1p (Hunter et al., 1998) labeled exactly the same NPC sites of wild-type cells as the anti–protein A antibody in ProtA– Nsp1p cells (Fahrenkrog, B., N. Panté, and J. Aris, unpublished results). Therefore, our immuno-EM protocol using yeast strains carrying protein A–tagged Nups is able to reveal the location of yeast Nups at the ultrastructural level.
Table II.
Quantitation of Antibody Labeling in Yeast Control Strains
| Strain | Nuclear gold particles/μm2 | Cytoplasmic gold particles/μm2 | Gold particles at the NPC | |||
|---|---|---|---|---|---|---|
| Wt | 0 | 1.9 ± 1.6 | 0 | |||
| ProtA-DHFR | 0.5 ± 1.1 | 5.7 ± 2.5 | 0 | |||
| ProtA-Pus1p | 6.7 ± 3.8 | 3.0 ± 2.0 | 0 |
Yeast cells were pre-embedding labeled with anti–protein A antibody conjugated to 8-nm colloidal gold (see Materials and Methods). The mean ± SD are given from all values. 22, 15, and 19 cells were analyzed for the wild-type (Wt), ProtA-DHFR, and ProtA-Pus1p strains, respectively. The quantitation was performed by a stereological counting method (Lucocq, 1992).
Nic96p Colocalizes with Nsp1p
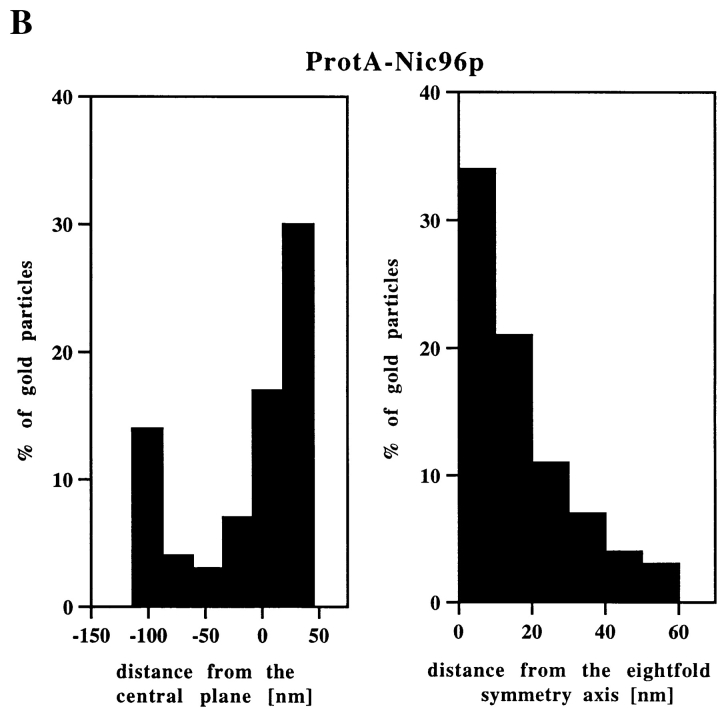
As described in the Introduction, biochemical studies have demonstrated the existence of two Nsp1p complexes. To identify these Nsp1p complexes within the architecture of the yeast NPC, we next localized the Nsp1p-interacting proteins. Using our immuno-EM protocol and a yeast strain expressing Nic96p tagged with protein A (ProtA-Nic96p, see Fig. 2), we found that the location of this Nup coincides with that of Nsp1p (i.e., at the cytoplasmic and nuclear periphery of the central gated channel and the terminal ring of the nuclear baskets; Fig. 4 A) with a preference of the cytoplasmic epitope (see Fig. 4 B). However, unlike for ProtA–Nsp1p, there was more abundant labeling at the nuclear baskets than at the nuclear periphery of the central channel (compare Fig. 3 B with Fig. 4 B). As with ProtA–Nsp1p, ∼90% of the gold particles were found within a radius of 30 nm from the eightfold symmetry axis of the NPC (Fig. 4 B).
Figure 4.
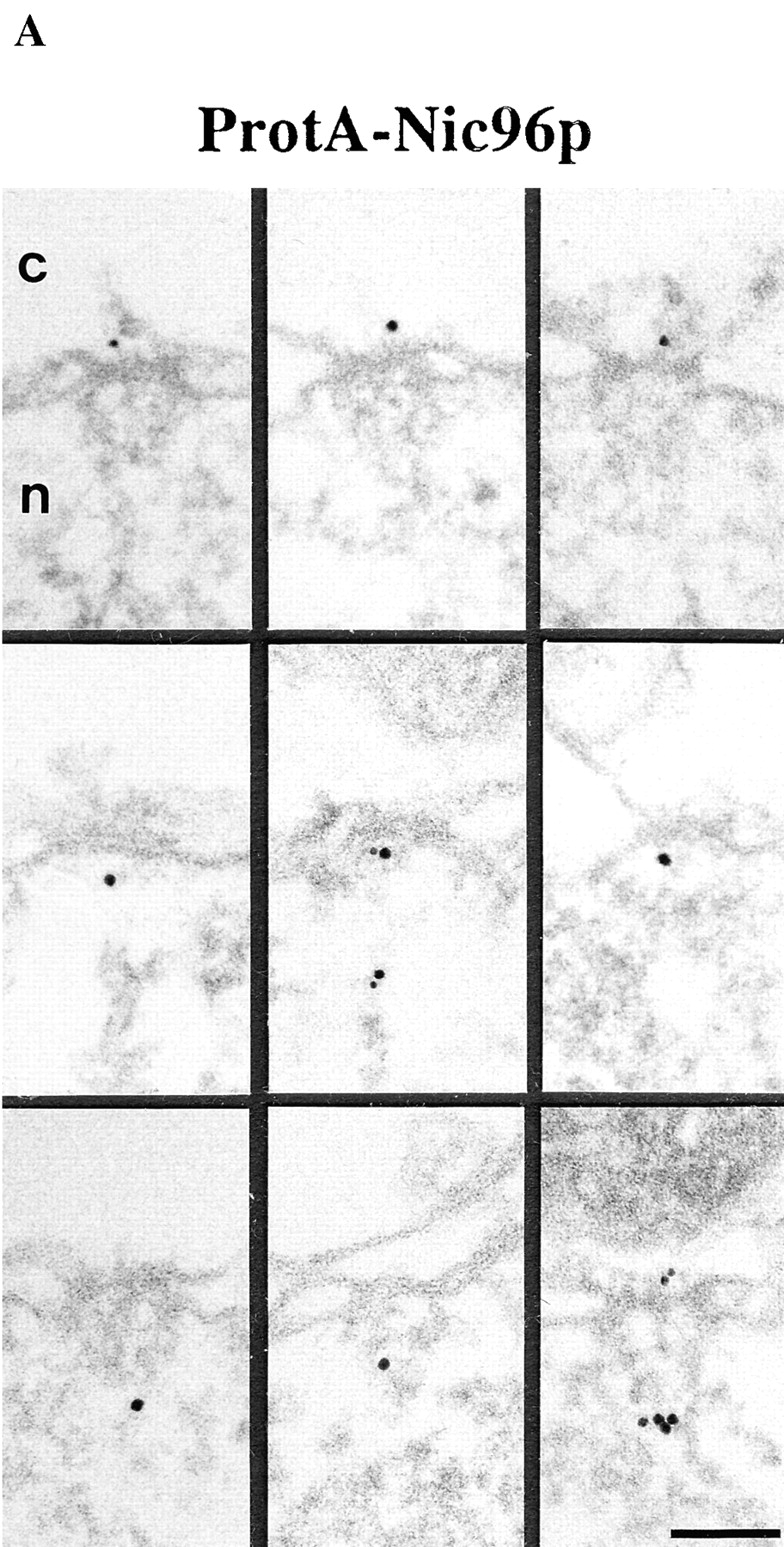
Localization of Nic96p in the ProtA–Nic96p strain. (A) Gallery of selected NPC cross sections showing labeling with the anti–protein A antibody conjugated to 8-nm colloidal gold in Triton X-100–extracted spheroplasts form ProtA–Nic96p cells. Similar to Nsp1p (see Fig. 3), Nic96p is localized at the cytoplasmic (top) and the nuclear periphery (middle) of the central gated channel, and at the terminal ring of the nuclear baskets (bottom). c, cytoplasm; n, nucleus. (B) Quantitation of the gold particle distribution associated with NPCs in the ProtA–Nic96p strain. 80 gold particles were scored. Bar, 100 nm.
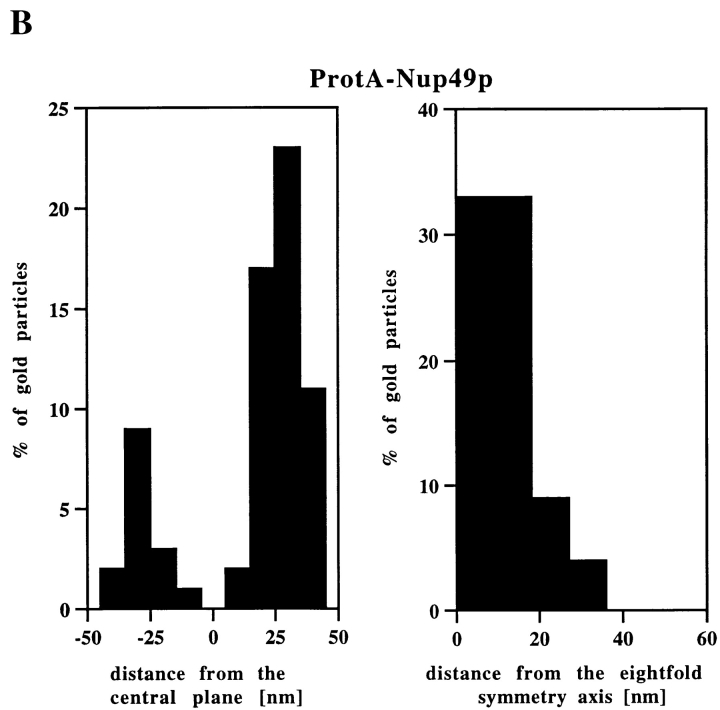
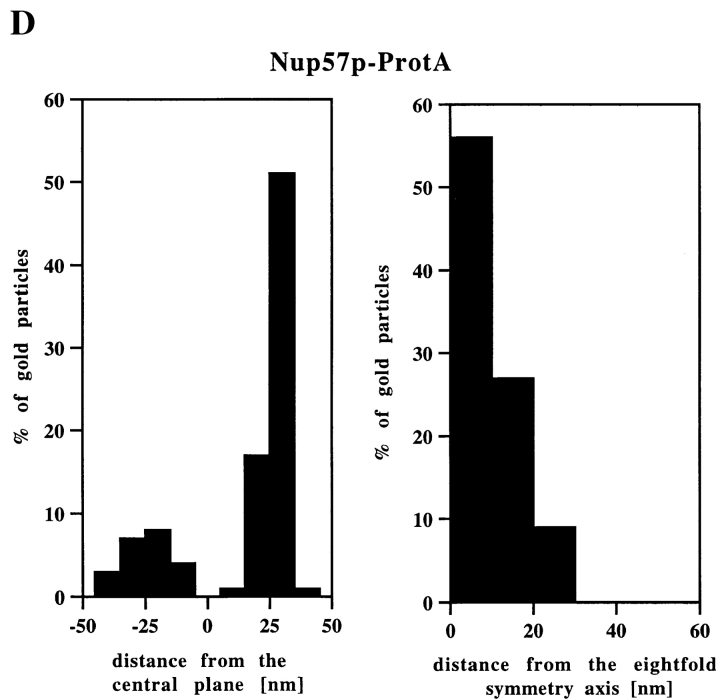
Nup49p and Nup57p Reside Both at the Cytoplasmic and the Nuclear Periphery of the Central Gated Channel
Our locations of ProtA–Nsp1p and ProtA–Nic96p (Figs. 3 and 4) suggest that the Nsp1p–Nup49p–Nup57p–Nic96p subcomplex may reside in three distinct NPC sites. To confirm this further, we localized Nup49p and Nup57p, again using yeast strains expressing these Nups tagged with protein A (see Fig. 2). As illustrated in Fig. 5, A and C, we localized ProtA–Nup49p and Nup57p–ProtA at both the cytoplasmic and the nuclear periphery of the central gated channel. However, based on our labeling data, neither Nup49p nor Nup57p were located at the terminal ring of the nuclear baskets. As already found for ProtA–Nsp1p and ProtA–Nic96p (see Fig. 3 B and Fig. 4 B), quantitative analysis of the gold particle distribution with regard to the central plane of the NPC (Fig. 5, B and D) indicates the same preference for the cytoplasmic epitope (see Fig. 5 A, top). Quantitation of the gold labeling with respect to the central plane of the NPC revealed that for Nup57p–ProtA, almost 100% of the cytoplasmic gold particles were detected in a range of 5–35 nm from this plane (see Fig. 5 D), whereas for ProtA–Nup49p, ProtA–Nsp1p, and ProtA– Nic96p, most of the gold particles were found in a range of 20–45 nm from the central pane (see Fig. 3 B, Fig. 4 B, and Fig. 5 B). Hence, Nsp1p, Nic96p, and Nup49p are located more peripherally than Nup57p. In fact, our immunolocalization data are in agreement with data obtained by in vitro reconstitution experiments, indicating that Nup57p is the organizing center of the Nsp1p–Nup57p–Nup49p core complex (Schlaich et al., 1997).
Figure 5.
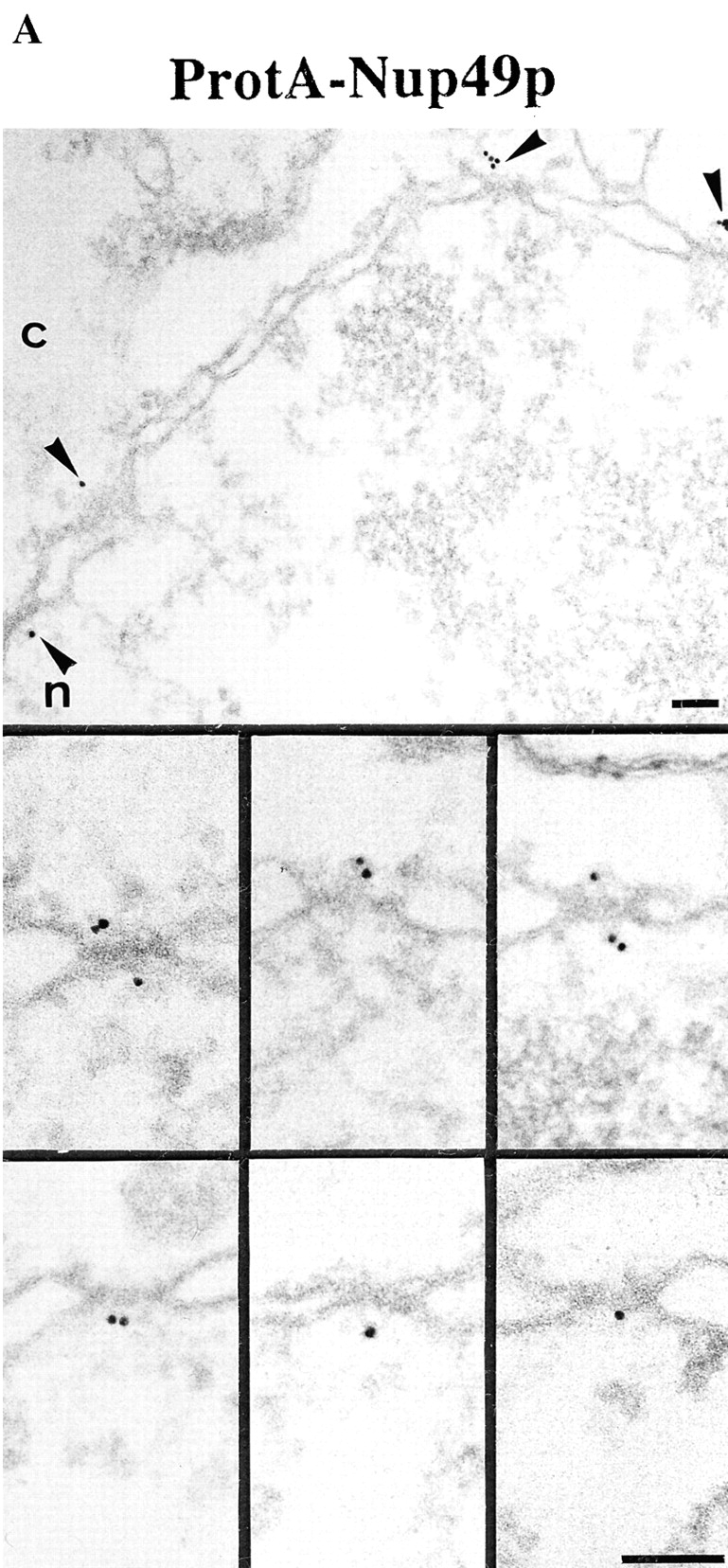
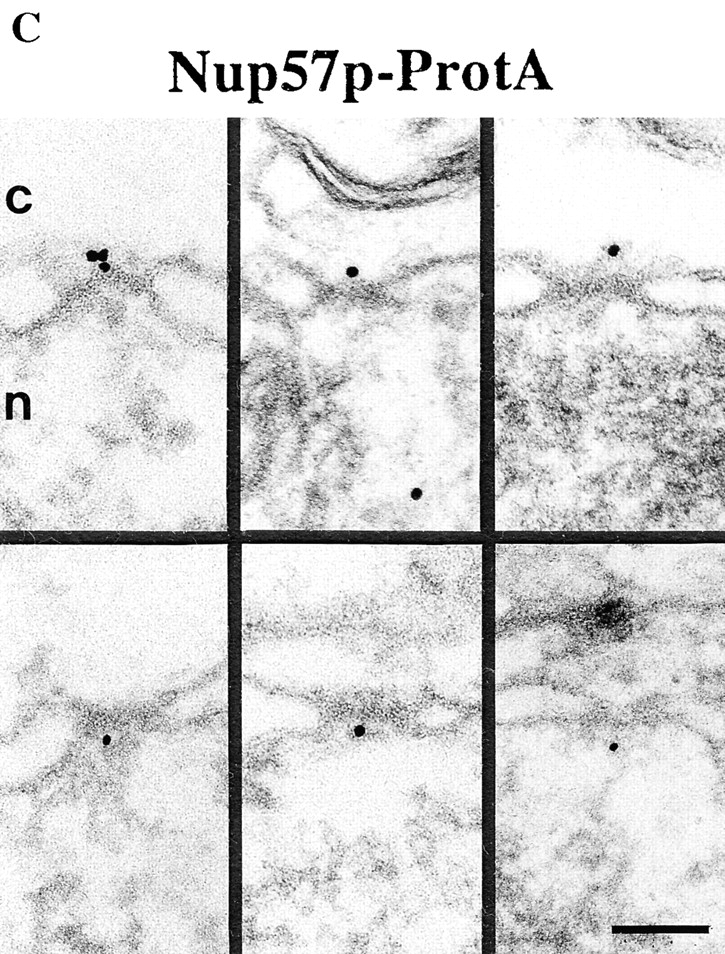
Nup49p (A) and Nup57p (C) are located at the cytoplasmic and the nuclear periphery of the central gated channel. Triton X-100–extracted spheroplasts from the ProtA–Nup49p and Nup57p–ProtA strains were incubated with an anti– protein A antibody conjugated to 8-nm colloidal gold (B and D). c, cytoplasm; n, nucleus. Distribution of the gold particles associated with NPCs in the ProtA–Nup49p and Nup57p–ProtA strains, respectively. 79 and 93 gold particles were scored, respectively. Bars, 100 nm.
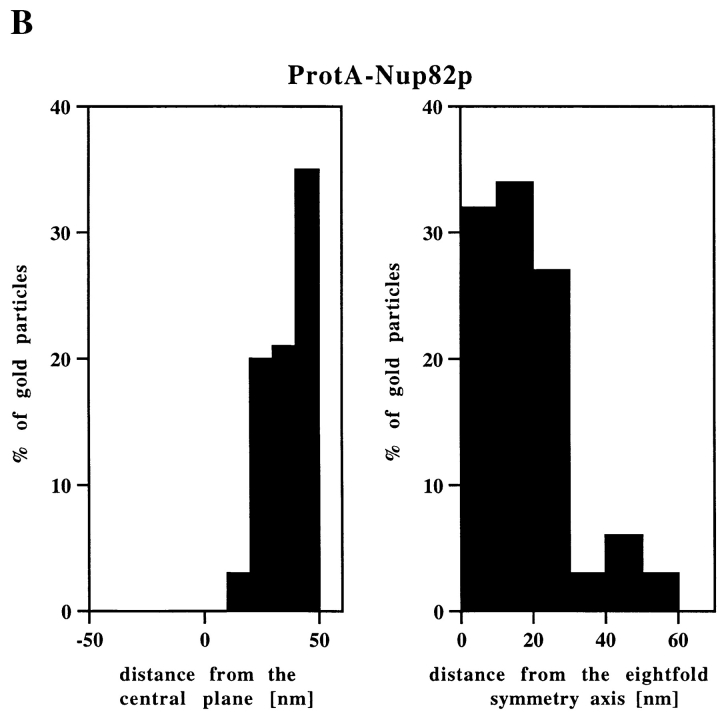
Nup82p Resides Exclusively at the Cytoplasmic Periphery of the Central Gated Channel
Nup82p, a fourth Nsp1p-interacting protein, occurs in an independent subcomplex with Nsp1p lacking Nup49p, Nup57p, and Nic96p (see Introduction). To localize Nup82p, pre-embedding labeling immuno-EM using a yeast strain expressing Nup82p tagged with protein A (ProtA–Nup82p, see Fig. 2) was carried out. As documented in Fig. 6, we found that Nup82p has only cytoplasmic epitopes with 95% of the gold particles at 38.3 ± 9.9 nm from the central plane of the NPC (Fig. 6 B). This finding might suggest that Nup82p is a constituent of the cytoplasmic fibrils. However, the analysis of the gold particle distribution along the eightfold symmetry axis revealed that 90% of the gold particles were located within a radius of 0–30 nm NPC (Fig. 6 B). As the cytoplasmic fibrils are located up to 50 nm from the eightfold symmetry axis, we concluded that Nup82p is located at the cytoplasmic periphery of the central gated channel. Since Nsp1p, Nic96p, Nup49p, and Nup57p were predominately detected within a distance of 5–30 nm from the central plane of the NPC, Nup82p is located more peripheral than these nucleoporins (compare Fig. 3 B, Fig. 4 B, and Fig. 5, B and D with Fig. 6 B).
Figure 6.
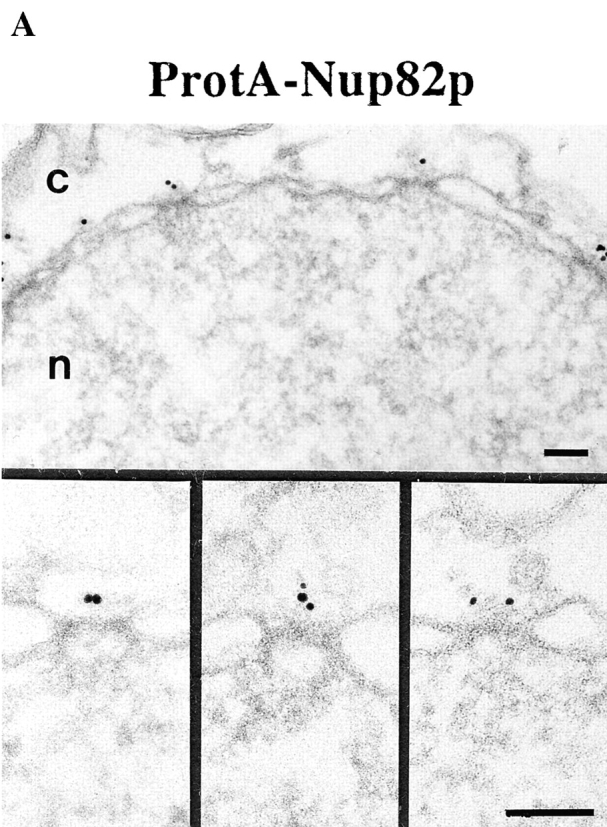
Nup82p is found only at the cytoplasmic periphery of the central gated channel. (A) Triton X-100–extracted spheroplasts from ProtA–Nup82p cells were pre-immunolabeled with an anti–protein A antibody directly conjugated to 8-nm gold. Shown are a stretch along the NE and a gallery of selected examples of gold-labeled NPC cross sections. c, cytoplasm; n, nucleus. Bars, 100 nm. (B) The quantitative analysis of the gold particle distribution revealed that Nup82p resides in the center of the NPC in a distance of ∼40 nm from the central plane of the NPC. 106 gold particles were scored for the quantitative analysis.
In Vivo GFP–Nsp1p Location in the Nup49-313 Thermosensitive Mutant
To find out whether the association of Nsp1p with the NPCs is altered if one of the interacting Nup is mutated, the in vivo location of green fluorescent protein (GFP)- tagged Nsp1p was analyzed in thermosensitive nup49-313 cells (Doye et al., 1994). Therefore, NUP49+ and nup49-313 cells were transformed with a plasmid harboring a fusion construct between GFP and the Nsp1p COOH-terminal domain. In NUP49+ cells, GFP–Nsp1p exhibits a typical NPC labeling (i.e., punctate staining of the NE) at both permissive and restrictive temperatures (Fig. 7). In contrast, the in vivo location of Nsp1p drastically changed when nup49-313 cells were shifted for 6 h to 37°C. Cells largely lost the characteristic ring- and dot-like staining of the nuclear periphery and GFP–Nsp1p accumulated in the cytoplasm (Fig. 7). However, a residual ring-like staining of the NE remained in nup49-313 cells under restrictive conditions which could be the pool of Nsp1p associated with Nup82p (Grandi et al., 1995b ). When nup49-313 cells were shifted back from 37° to 20°C, a distinct and prominent ring-like and punctate NE staining of GFP–Nsp1p reappeared. This shows that the in vivo targeting of a significant pool of Nsp1p to the NPCs requires the intactness of the other members (e.g., Nup49p) of the Nsp1p–Nup49p– Nup57p complex.
Figure 7.
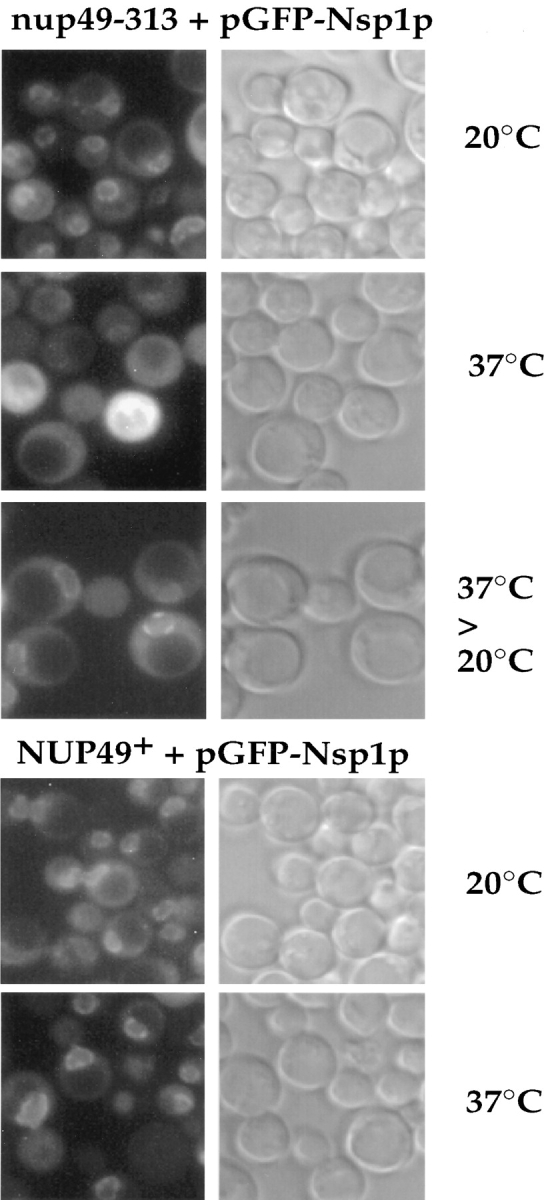
In vivo location of GFP-tagged Nsp1p in nup49-313 cells. NUP49+ and nup49-313 cells transformed with a single copy ARS/CEN/ADE2 plasmid harboring a fusion construct between GFP and the Nsp1p COOH-terminal domain were grown at permissive temperature (20°C) or shifted for 6 h to restrictive temperature (37°C). In the case of the nup49-313 strain, cells were regrown after a 6-h shift to 37°C for a further 2 h at the permissive temperature (20°C). Cells were inspected in the fluorescence microscope to detect the GFP fusion protein and also viewed by Nomarski optics.
Discussion
Although significant progress has been made to elucidate the 3D architecture of the NPC and to clone and molecularly characterize Nups, the 3D location of many of the known Nups has remained elusive. This is in part due to the high cross-reactivity of the available anti-Nup antibodies. Because yeast is an excellent system to combine biochemical and genetic manipulations with functional assays, and because most of the nucleoporins identified to date are from yeast NPCs, we have started to dissect the molecular architecture of the yeast NPC. In the present investigation, we have structurally analyzed yeast NPCs by developing an EM sample preparation protocol that for the first time reproducibly yields structurally well-preserved yeast NPCs. We then adapted this sample preparation protocol to localize Nsp1p and its interacting nucleoporins Nup49p, Nup57p, Nup82p, and Nic96p within the 3D architecture of the yeast NPC.
Our method revealed that yeast NPCs are built according to a very similar architecture as vertebrate NPCs (e.g., Xenopus, Chironomus, rat). Specifically, we have documented the existence of cytoplasmic fibrils and a nuclear basket within the yeast NPC (see Fig. 1 A). Strikingly, our analysis revealed that although yeast and Xenopus NPCs exhibit very much the same structural organization, yeast NPCs are about 15% smaller in their linear dimensions than Xenopus NPCs (see Fig. 1 B, and Table I). Hence, it appears that the overall 3D architecture of the NPC is evolutionarily conserved from yeast to higher eukaryotes (e.g., Xenopus, Chironomus, rat). The peripheral components of the yeast NPC have not been completely and reproducibly visualized in previous structural studies (Rout and Blobel, 1993; Yang et al., 1998). This is probably because these studies were performed with highly enriched, detergent-extracted yeast NPCs, a procedure that might damage or dissociate peripheral components such as the cytoplasmic fibrils and the nuclear baskets. In fact, it has been well documented that in Xenopus oocytes detergent extraction of their NE releases (nonquantitatively) the cytoplasmic and nuclear ring with attached cytoplasmic fibrils and nuclear basket from the basic framework or spoke complex (Reichelt et al., 1990; Jarnik and Aebi, 1991). Hence, it is most likely that the isolated yeast NPCs reported previously (Rout and Blobel, 1993; Yang et al., 1998) only represent their basic framework rather than the intact yeast NPC.
To map nucleoporins within the 3D structure of the yeast NPC, we have used our new sample preparation protocol of yeast cells in combination with immuno-EM. However, to avoid the high degree of cross-reactivity commonly encountered with anti-Nup antibodies (see Introduction), we used yeast strains disrupted for a given Nup gene and complemented by this Nup tagged with protein A, in combination with a high-affinity anti–protein A antibody directly conjugated to 8-nm-diam colloidal gold. With this approach, we have localized Nsp1p together with its interacting Nups: Nup49p, Nup57p, Nup82p, and Nic96p. As illustrated in Fig. 8, these location studies revealed that Nsp1p resides in three subcomplexes located at distinct NPC sites: the Nsp1p–Nup49p–Nup57p–Nic96p complex (i.e., the Nsp1p complex) located at both the cytoplasmic and the nuclear periphery of the central channel, the Nsp1p–Nup82p complex exclusively located at the cytoplasmic periphery of the central gated channel, and the Nsp1p–Nic96p complex located near or at the terminal ring of the nuclear basket. Biochemical evidence for the latter complex has not been achieved so far.
Figure 8.
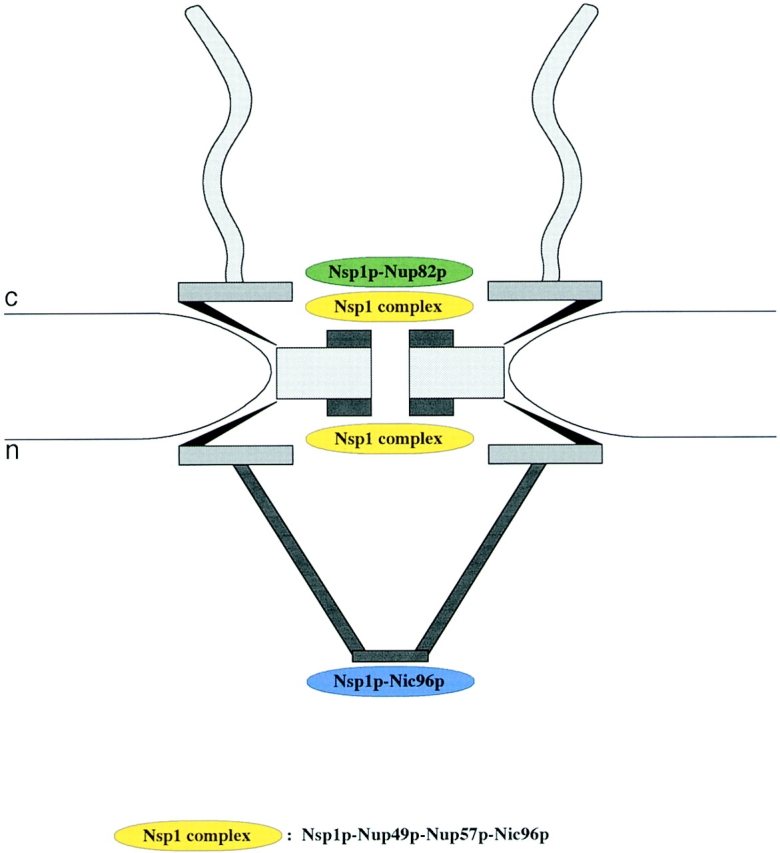
Schematic representation of the yeast NPC with the 3D localization of three Nsp1p subcomplexes. The Nsp1p–Nup49p–Nup57p– Nic96p complex (i.e., the Nsp1p complex) is localized at both the cytoplasmic and nuclear periphery of the central gated channel, and the Nsp1p–Nup82p complex resides only at the cytoplasmic periphery of the central gated channel. In addition, a third Nsp1p complex consisting of Nsp1p interacting with Nic96p is located at the terminal ring of the nuclear baskets. c, cytoplasmic, and n, nuclear side of the NE.
The location of the Nsp1p complex within the 3D architecture of the yeast NPC is similar to that of the p62 complex (Guan et al., 1995), its putative homologue within the vertebrate NPC. Moreover, similar to Nsp1p, p62 was also found near or at the terminal ring of the nuclear baskets of Xenopus oocyte NPCs (Panté and Aebi, 1993). The location of Nic96p is somewhat similar to that of its vertebrate homologue Nup93, which resides at the nuclear periphery of the central gated channel and the terminal ring of the nuclear baskets, but, unlike Nic96p, Nup93 is absent from the cytoplasmic periphery of the central gated channel (Grandi et al., 1997). This difference in the location between Nic96p and Nup93 may be a reflection of the limited sequence homology between these two Nups. These limited homologies, in turn, may result in similar but not identical function(s) of these Nups. For Nic96p and Nup93, at least, this appears to be the case: both proteins are required for nuclear pore biogenesis, but only Nic96p was shown to be involved in protein import (Zabel et al., 1996; Grandi et al., 1997).
We found Nup82p to be the most peripheral Nsp1p-interacting protein on the cytoplasmic side of the NPC. Very recently, Nup159p, a third member of the Nsp1p– Nup82p subcomplex, was identified (Belgareh et al., 1998). This Nup was previously localized to the cytoplasmic face of the yeast NPC (Kraemer et al., 1995), where it preferentially resided at a distance of ∼40 nm from the central plane of the NPC, in good agreement with our localization of Nup82p.
Since the multiple locations of Nsp1p and Nic96p represent NPC sites at which cargo or transport ligands can be arrested during nuclear protein import (Panté and Aebi, 1995, 1996c ; Görlich et al., 1996), our data provide further evidence that Nsp1p and Nic96p play central roles in nucleocytoplasmic transport. For example, Nsp1p may interact with the cargo at these three NPC sites while it crosses the NPC. Alternatively, Nsp1p may be directly involved in nuclear transport by binding transport ligands or transport factors on one side of the NPC and accompanying them to the other side of the NPC, for example, by interacting with nucleoporins which, contrary to Nsp1p, have fixed positions. Thus, for import of a protein containing a nuclear localization signal (NLS) into the nucleus, the NLS protein first binds to the import receptor complex (importin α/β in vertebrate cells, Srp1p/Kap95p in yeast) in the cytoplasm. Subsequently this NLS protein–receptor complex (i.e., the cargo) docks to a cytoplasmic fibril, before it is delivered to the cytoplasmic periphery of the central gated channel. This interaction may involve interaction of importin β with FXFG repeat sequences of Nsp1p, Nup49p, and Nup57p (for review see Fabre and Hurt, 1997), or a direct association of the NLS protein with Nsp1p (Barth and Stochaj, 1996). Alternatively, Nsp1p may interact with the cargo via NTF2, a Ran-binding protein that has been implicated in the translocation of nuclear proteins through the NPC (Clarkson et al., 1996).
In summary, we have visualized the cytoplasmic fibrils and the nuclear baskets of yeast NPCs, and we have for the first time localized five yeast nucleoporins to distinct NPC components. Our immuno-EM data suggest that Nsp1p resides in three distinct complexes within the 3D architecture of the yeast NPC. The first complex consists of Nsp1p, Nup82p, and possibly other nucleoporins (Belgareh et al., 1998) (Bailer, S., and E.C. Hurt, unpublished data), and it is located exclusively at the cytoplasmic periphery of the central gated channel. The second complex consists of Nsp1p, Nup49p, Nup57p, and Nic96p, and it resides near or at the cytoplasmic/nuclear entry/exit of the central gated channel. Finally, the third complex containing at least Nsp1p and Nic96p is located at the terminal ring of the nuclear basket. Whether Nsp1p resides permanently at this distinct sites or, in association with cargo, can sequentially move from one to the next site, remains to be established.
Acknowledgments
We thank A. Mandinova and C.-A. Schoenenberger (Muller Institute, Basel, Switzerland) for helpful suggestions concerning the experimental procedures, S. Bailer (BZH, Heidelberg, Germany) for providing the plasmid SM666, J. Aris (University of Florida, Gainesville, FL) for providing the antibody against Nsp1p, R. Wyss (Muller Institute) for help with Figs. 1 B and 8, and H. Frefel and M. Zoller (Blozentrum, Basel, Switzerland) for their expert photographic work.
This work was supported by grants from the Swiss National Science Foundation (4036-044061 and 3100-053034), by the Kanton Basel-Stadt, and by the M.E. Müller Foundation of Switzerland.
Abbreviations used in this paper
- 3D
three-dimensional
- DHFR
dihydrofolate reductase
- GFP
green fluorescent protein
- MDa
megadaltons (106 daltons)
- NE
nuclear envelope
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- Nups
nucleoporins
- ProtA
protein A
Footnotes
Address all correspondence to N. Panté, Institute of Biochemistry, Swiss Federal Institute of Technology, Universitätsstrasse 16, CH-8092 Zürich, Switzerland. Tel.: (41) 1-6323134. Fax: (41) 1-6321269. E-mail: nelly.pante@bc.biol.ethz.ch
References
- Allende ML, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development. Genes Dev. 1996;10:3141–3155. doi: 10.1101/gad.10.24.3141. [DOI] [PubMed] [Google Scholar]
- Barth W, Stochaj U. The yeast nucleoporin Nsp1 binds nuclear localization sequences in vitro. . Biochem Cell Biol. 1996;74:363–372. doi: 10.1139/o96-039. [DOI] [PubMed] [Google Scholar]
- Baschong W, Wrigley NG. Small colloidal gold conjugated to Fab fragments or to immunoglobulin G as high-resolution labels for electron microscopy: a technical overview. J Electron Microsc Technol. 1990;14:313–323. doi: 10.1002/jemt.1060140405. [DOI] [PubMed] [Google Scholar]
- Belgareh, N., C. Snay-Hodge, F. Pasteau, S. Dagher, C.N. Cole, and V. Doye. 1998. Functional characterization of a Nup159p-containing nucleus pore subcomplex. Mol. Biol. Cell. In press. [DOI] [PMC free article] [PubMed]
- Carmo-Fonseca M, Kern H, Hurt EC. Human nucleoporin p62 and the essential yeast nuclear pore protein NSP1 show sequence homology and a similar domain organization. Eur J Cell Biol. 1991;55:17–30. [PubMed] [Google Scholar]
- Clarkson WD, Kent HM, Stewart M. Separate binding sites on nuclear transport factor 2 (NTF2) for GDP-Ran and the phenylalanine-rich repeat regions of nucleoporins p62 and Nsp1p. J Mol Biol. 1996;263:517–524. doi: 10.1006/jmbi.1996.0594. [DOI] [PubMed] [Google Scholar]
- Dabauvalle M-C, Loos K, Scheer U. Identification of a soluble precursor complex essential for nuclear pore assemble in vitro. . Chromosoma. 1990;100:257–263. doi: 10.1007/BF00337603. [DOI] [PubMed] [Google Scholar]
- Davis L. The nuclear pore complex. Annu Rev Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- Doye V, Hurt EC. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- Doye V, Wepf R, Hurt EC. A novel nuclear pore protein Nup133p with distinct roles in poly (A)+RNA transport and nuclear pore distribution. EMBO (Eur Mol Biol Organ) J. 1994;13:6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E, Hurt EC. Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu Rev Genet. 1997;31:277–313. doi: 10.1146/annurev.genet.31.1.277. [DOI] [PubMed] [Google Scholar]
- Görlich D, Panté N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO (Eur Mol Biol Organ) J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Finlay DR, Meier E, Bradley P, Horecka J, Forbes DJ. A complex of nuclear pore proteins required for pore function. J Cell Biol. 1991;114:169–183. doi: 10.1083/jcb.114.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Grandi P, Doye V, Hurt EC. Purification of NSP1 reveals complex formation with ‘GLGF' nucleoporins and a novel nuclear pore protein NIC96. EMBO (Eur Mol Biol Organ) J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Schlaich N, Tekotte H, Hurt EC. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO (Eur Mol Biol Organ) J. 1995a;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Emig S, Weise C, Hucho F, Pohl T, Hurt EC. A novel nuclear pore protein Nup82p which specifically binds to a fraction of Nsp1p. J Cell Biol. 1995b;130:1263–1273. doi: 10.1083/jcb.130.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Dang T, Panté N, Shevchenko A, Mann M, Forbes D, Hurt EC. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol Biol Cell. 1997;8:2017–2038. doi: 10.1091/mbc.8.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan T, Müller S, Klier G, Panté N, Blevitt JM, Häner M, Paschal B, Aebi U, Gerace L. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol Biol Cell. 1995;6:1591–1603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, Guan T, Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, L.A., A.L. Benko, J.P. Avis, D.R. Stanford, N.C. Martin, and A.K. Hopper. 1998. S. cerevisiae Mod5p-II contains sequences antagonistic for nuclear and cytosolic locations. Genetics. In press. [DOI] [PMC free article] [PubMed]
- Hurt EC. A novel nucleoskeletal-like protein located at the nuclear periphery is required for the life cycle of Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1988;7:4324–4334. doi: 10.1002/j.1460-2075.1988.tb03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz ME, Blobel G. NUP82 is an essential yeast nucleoporin required for poly(A)+RNA export. J Cell Biol. 1995;130:1275–1281. doi: 10.1083/jcb.130.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarnik M, Aebi U. Toward a more complete 3-D structure of the nuclear pore complex. J Cell Biol. 1991;107:291–308. doi: 10.1016/1047-8477(91)90054-z. [DOI] [PubMed] [Google Scholar]
- Kita K, Omata S, Horigome T. Purification and characterization of a nuclear pore glycoprotein complex containing p62. J Biochem (Tokyo) 1993;113:377–382. doi: 10.1093/oxfordjournals.jbchem.a124054. [DOI] [PubMed] [Google Scholar]
- Kraemer DM, Strambio-de-Castillia C, Blobel G, Rout MP. The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J Biol Chem. 1995;270:19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- Lucocq J. Quantitation of gold labeling and estimation of labeling efficiency with a stereological counting method. J Histochem Cytochem. 1992;40:1929–1936. doi: 10.1177/40.12.1453009. [DOI] [PubMed] [Google Scholar]
- Macaulay C, Meier E, Forbes DJ. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J Biol Chem. 1995;270:254–262. doi: 10.1074/jbc.270.1.254. [DOI] [PubMed] [Google Scholar]
- Mutvei A, Dihlmann S, Herth W, Hurt EC. NSP1 depletion in yeast affects nuclear pore formation and nuclear accumulation. Eur J Cell Biol. 1992;59:280–295. [PubMed] [Google Scholar]
- Nehrbass U, Fabre E, Dihlmann S, Herth W, Hurt EC. Analysis of nucleo-cytoplasmic transport in a thermosensitive mutant of nuclear pore protein NSP1. Eur J Cell Biol. 1993;62:1–12. [PubMed] [Google Scholar]
- Nehrbass U, Kern H, Mutvei A, Horstmann H, Marshallsay B, Hurt EC. NSP1: a yeast nuclear envelope protein localized at the nuclear pores exerts its essential function by its carboxy-terminal domain. Cell. 1990;61:979–989. doi: 10.1016/0092-8674(90)90063-k. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Panté N, Aebi U. The nuclear pore complex. J Cell Biol. 1993;122:977–984. doi: 10.1083/jcb.122.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panté N, Aebi U. Exploring nuclear pore complex structure and function in molecular detail. J Cell Sci Suppl. 1995;19:1–11. doi: 10.1242/jcs.1995.supplement_19.1. [DOI] [PubMed] [Google Scholar]
- Panté N, Aebi U. Toward the molecular dissection of protein import into nuclei. Curr Opin Cell Biol. 1996a;8:397–406. doi: 10.1016/s0955-0674(96)80016-0. [DOI] [PubMed] [Google Scholar]
- Panté N, Aebi U. Molecular dissection of the nuclear pore complex. Crit Rev Biochem Mol Biol. 1996b;31:153–199. doi: 10.3109/10409239609106583. [DOI] [PubMed] [Google Scholar]
- Panté N, Aebi U. Sequential binding of import ligands to distinct nucleopore regions during their nuclear import. Science. 1996c;273:1729–1732. doi: 10.1126/science.273.5282.1729. [DOI] [PubMed] [Google Scholar]
- Panté N, Bastos R, McMorrow I, Burke B, Aebi U. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J Cell Biol. 1994;126:603–617. doi: 10.1083/jcb.126.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G. The peptide repeat domain of the nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Reichelt R, Holzenburg A, Buhle EL, Jarnik A, Engel A, Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J Cell Biol. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Blobel G. Isolation of the yeast nuclear pore complex. J Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaich NL, Häner M, Lustig A, Aebi U, Hurt EC. In vitroreconstitution of a heterotrimeric nucleoporin complex consisting of recombinant Nsp1p, Nup49p, and Nup57p. Mol Biol Cell. 1997;8:33–46. doi: 10.1091/mbc.8.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Hurt EC. Mex67p which is an essential factor for nuclear mRNA export binds to both poly(A)+RNA and nuclear pores. EMBO (Eur Mol Biol Organ) J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Fabre E, Tekotte H, Hurt EC, Tollerey D. Yeast nucleoporin mutants are defective in pre-tRNA splicing. Mol Cell Biol. 1996;16:294–301. doi: 10.1128/mcb.16.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G, Tekotte H, Grosjean H, Segref A, Sharma K, Tollerey D, Hurt EC. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO (Eur Mol Biol Organ) J. 1996;15:2270–2284. [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Slot JW, Geuze HJ. A new method of preparing gold probes for multiple-labeling cytochemistry. Eur J Cell Biol. 1985;38:87–93. [PubMed] [Google Scholar]
- Wimmer C, Doye V, Grandi P, Nehrbass U, Hurt EC. A new subclass of nucleoporins that functionally interact with the nuclear pore protein NSP1. EMBO (Eur Mol Biol Organ) J. 1992;11:5051–5061. doi: 10.1002/j.1460-2075.1992.tb05612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Biol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- Zabel U, Doye V, Tekotte H, Grandi P, Hurt EC. Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J Cell Biol. 1996;133:1141–1152. doi: 10.1083/jcb.133.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]