Abstract
Extracting isolated Drosophila centrosomes with 2 M KI generates salt-resistant scaffolds that lack the centrosomal proteins CP190, CP60, centrosomin, and γ-tubulin. To clarify the role of these proteins in microtubule nucleation by centrosomes and to identify additional centrosome components required for nucleation, we have developed an in vitro complementation assay for centrosome function. Centrosome aster formation is reconstituted when these inactive, salt-stripped centrosome scaffolds are supplemented with a soluble fraction of a Drosophila embryo extract. The CP60 and CP190 can be removed from this extract without effect, whereas removing the γ-tubulin destroys the complementing activity. Consistent with these results, we find no evidence that these three proteins form a complex together. Instead, γ-tubulin is found in two distinct protein complexes of 240,000 and ∼3,000,000 D. The larger complex, which is analogous to the Xenopus γ-tubulin ring complex (γTuRC) (Zheng, Y., M.L. Wong, B. Alberts, and T. Mitchison. 1995. Nature. 378:578–583), is necessary but not sufficient for complementation. An additional factor found in the extract is required. These results provide the first evidence that the γTuRC is required for microtubule nucleation at the centrosome.
Keywords: centrosome, γ-tubulin, microtubule nucleation, microtubule-organizing center, cytoskeleton
In animal cells, centrosome-nucleated microtubule arrays are essential for a wide variety of cellular processes including cell division and chromosome segregation, directed cell movement and interphase cytoplasmic organization (for reviews see Mazia, 1987; Vorobjev and Nadezhdina, 1987; Kellogg et al., 1994). EM studies have shown that centrosomes consist of a pair of centriolar cylinders surrounded by an electron-dense cloud of pericentriolar material (PCM),1 and that the PCM originates the many microtubules that are nucleated by the centrosome (Rieder and Borisy, 1982; Vorobjev and Chentsov, 1982; Keryer et al., 1984).
The molecular characterization of the centrosome and its ability to nucleate microtubules is still in its early stages. The centrosome may contain as many as 100 different proteins (for review see Kalt and Schliwa, 1993; Kellogg et al., 1994), but it is not known how many of these are actual components of the PCM, with direct or indirect roles in microtubule nucleation. The identification of centrosomal components is further confounded by the fact that the centrosome, as the focus of the cell's microtubule array, is also a hub for intracellular trafficking. This makes it difficult to distinguish actual components of the PCM from molecules recruited by the microtubule array. To simplify this problem, we define the “core” centrosome as the structure that remains when all of its microtubules have been depolymerized.
The discovery of one such core centrosomal protein, γ-tubulin, has led to a breakthrough in our understanding of microtubule nucleation by centrosomes. γ-Tubulin is a highly conserved member of the tubulin family shown to be involved in microtubule nucleation (Oakley et al., 1990; Stearns et al., 1991; Zheng et al., 1991; Joshi et al., 1992; Felix et al., 1994; Stearns and Kirschner, 1994). Recently, a γ-tubulin–containing ring complex (γTuRC), capable of nucleating microtubules in vitro, was purified from Xenopus eggs (Zheng et al., 1995). EM tomography on centrosomes isolated from Drosophila revealed the presence of rings containing γ-tubulin in the PCM in both the presence and absence of nucleated microtubules. The γ-tubulin rings are found at the microtubule minus ends in centrosome-nucleated microtubule asters (Moritz et al., 1995b ). Ring structures are also visible in the PCM of centrosomes of the surf clam, Spisula (Vogel et al., 1997). These results suggest that the γTuRC is a highly conserved structure responsible for the microtubule-nucleating capacity of the PCM (Moritz et al., 1995b ; Zheng et al., 1995).
Although these studies indicate that the γTuRC is likely to be essential for microtubule nucleation by centrosomes, many important questions remain. These include: What is the structural organization of the PCM and how is it assembled? How is the γTuRC anchored within the PCM? Is the attachment of the γTuRC to the centrosome matrix important for its activity? Do other centrosomal proteins contribute to microtubule nucleation?
Other core centrosomal proteins that may have direct or indirect roles in microtubule nucleation include pericentrin, CP190, CP60, and centrosomin (CNN). Pericentrin is a human autoimmune antigen that has also been identified in mouse and Xenopus. It is thought to be a structural component of the PCM that may play an essential role in its organization (Doxsey et al., 1994). CP60 and CP190 are two core centrosomal proteins of unknown function identified in Drosophila. CP190 is a novel, zinc-finger–containing protein identified by microtubule affinity chromatography (Kellogg et al., 1989; Whitfield et al., 1995). Native CP190 localizes primarily to nuclei during interphase, becoming prominent at centrosomes upon nuclear envelope breakdown at the onset of mitosis (Frasch et al., 1986; Whitfield et al., 1988; Oegema et al., 1997). CP60, a novel protein identified by immunoaffinity chromatography by virtue of its ability to interact with CP190 (Kellogg and Alberts, 1992), also localizes to both nuclei and centrosomes in a cell cycle–dependent manner, but with slightly different timing (Kellogg et al., 1995; Oegema et al., 1997). Previous work from our laboratory suggested that CP190, CP60, and γ-tubulin are components of a soluble protein complex present in embryo extracts (Raff et al., 1993), but further studies show that this is not the case (see below). In Drosophila, the centrosome core also contains at least one developmentally important component, called CNN, which is the target of a homeotic gene and is essential for proper centrosome function (Li and Kaufman, 1996).
To examine the role of the γTuRC in microtubule nucleation at centrosomes and to test the potential contributions to nucleation by other known centrosomal components, we sought to develop an in vitro complementation assay for aster formation using isolated Drosophila centrosomes (Moritz et al., 1995a ; Moritz and Alberts, 1998). Previous work has shown that the microtubule-nucleating activity of mammalian centrosomes can be destroyed by salt or urea treatments, and that the activity can be restored by injecting the treated centrosomes into Xenopus eggs, or by mixing them with egg extract. This suggests that factors in the egg cytoplasm can associate with the damaged centrosomes, restoring their ability to nucleate microtubules (Klotz et al., 1990; Buendia et al., 1992). With this information in mind, we developed an assay in which microtubule nucleation by Drosophila centrosomes is reconstituted from two components, inactive salt-stripped centrosome scaffolds and the high speed supernatant of a Drosophila embryo extract.
In this paper, we characterize both the salt-stripped scaffolds and the soluble components in the extract that are necessary for nucleation. In particular, we test for a role in nucleation for CP190, CP60 and the Drosophila γTuRC. Our assay also allows us to begin to address what components, if any, are required for attachment of the γTuRC to the salt-stripped scaffolds.
Materials and Methods
Buffers
BRB80: 80 mM K-Pipes, pH 6.8, 1 mM MgCl2, 1 mM Na3EGTA (prepare as a 5× stock, dilute to 1× for use). Hepes buffer: 50 mM K-Hepes, pH 7.6, 1 mM MgCl2, 1 mM Na3EGTA. Hepes block: Hepes buffer + 100 mM KCl, 10 mg/ml BSA (fraction V; Sigma Chemical Co., St. Louis, MO), and 1 mM β-mercaptoethanol. Embryo extract buffer for making complementing extract: Hepes buffer + 100 mM KCl, 10% glycerol, 1:100 protease inhibitor stock, and 1 mM PMSF. Protease inhibitor stock: 10 mM benzamidine-HCl, 0.1 mg/ml phenanthroline, 1 mg/ml aprotinin, 1 mg/ml leupeptin, 1 mg/ml pepstatin A in ethanol. GTP stock: 0.5 M GTP (Sigma Chemical Co.) in 1× BRB80. Tubulin dilution buffer (TDB): 1× BRB80, 10% glycerol, 1 mM GTP. TDB wash: TDB + 10 mg/ml BSA (fraction V; Sigma Chemical Co.). Extract buffer for characterizations of CP60, CP190, γ-tubulin protein complexes: 50 mM K-Hepes, pH 7.6, 75 mM KCl, 1 mM Na3EGTA, 1 mM EDTA, 0.05% NP-40, 1:50 protease inhibitor stock, 2 mM PMSF. Gradient buffer: Hepes buffer + 1 mM β-mercaptoethanol, 1:200 protease inhibitor stock. Column buffer: Hepes buffer + 2% wt/vol glycerol, 1 mM β-mercaptoethanol, 1:200 protease inhibitor stock. PBS: 5.4 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, adjusted to pH 7.2. PBST: PBS + 0.1% Tween-20. Sample buffer: 63 mM Tris-HCl, pH 6.8, 3% SDS, 5% β-mercaptoethanol, 10% glycerol. TBS: 10 mM Tris-Cl, pH 8, 150 mM NaCl, 0.05% Tween-20.
Centrosome Isolation
Drosophila centrosomes were isolated on sucrose gradients from 0–3.5-h-old embryos and tested for activity as previously described (Moritz et al., 1995a ; Moritz and Alberts, 1998).
Tubulin
Tubulin was purified from bovine brain (Mitchison and Kirschner, 1984) and labeled with N-hydroxy-succininimidyl-rhodamine (Hyman et al., 1991) as described previously.
Acid-washed, Poly-lysine–coated Coverslips
Acid-washed, 12-mm round glass coverslips were prepared in large batches by incubating the coverslips in a large glass beaker with 1 N HCl at 65°C for 4 h to overnight with occasional swirling. The coverslips were rinsed extensively in ddH2O, until the pH was neutral, and then incubated in 0.1% wt/vol poly-l-lysine for 20 min. The coverslips were dried by laying them out on a large piece of filter paper, or in a drying oven.
On-Glass Complementation Assay
To destroy the microtubule-nucleating activity of centrosomes, an equal volume of 4 M KI in 1× BRB80 was mixed with the centrosomes, and the mixture was incubated on ice for 10 min. 20 μl of this mixture was then applied to a 12-mm round acid-washed, poly-lysine–coated glass coverslip, which was placed on a piece of Parafilm inside a humidified Petri dish kept in a 30°C water bath. The centrosomes were allowed to bind to the coverslip for 5 min, and then washed briefly by pipetting on and aspirating off three times with 60 μl Hepes block. (For controls in which the centrosomes were omitted, the coverslips were washed in the same way with Hepes block, and then the sample was applied and treated in the same way as coverslips with centrosomes.) The final wash was allowed to incubate on the coverslip for 5 min. Depending on the experiment, the dish containing the coverslips was either kept at 30°C, or transferred to a 0°C ice bath. The final wash was then replaced with 10–60 μl of the sample to be tested. The centrosomes were incubated with the sample for 10 min (at 0° or 30°C), and then the sample was washed away briefly three times with 60 μl TDB wash. A 25-μl mixture of unlabeled and rhodamine-labeled tubulin (usually in a 7:1 ratio) diluted to 2 mg/ml in TDB was then incubated on the coverslips for 10 min at 30°C. Any resulting microtubules or asters were fixed by a 3-min incubation with 60 μl 1% glutaraldehyde in BRB80 (EM grade; Ted Pella, Inc., Redding, CA), followed by a 3-min incubation with −20°C methanol. The coverslips were then inverted and mounted on slides on drops of mounting medium (80% glycerol in PBS + 1 mg/ml para-phenylenediamine). The slides were viewed on a Nikon Microphot-FXA, 100× objective (1.4 NA), and either photographed using Kodak Ektachrome 400 Elite or Ektachrome P1600 film, or on a Nikon Optiphot-2, 60× or 100× objective (1.4 NA) using a cooled CCD camera (Princeton Scientific Instruments, Inc., Monmouth Junction, NJ). Micrographs were processed using Photoshop (Adobe Systems, Inc., Mountain View, CA). For quantitation, asters were counted in 50 randomly selected, 100× microscope fields.
For immunofluorescence, unlabeled tubulin was used during the microtubule regrowth step, and the samples on coverslips were rehydrated after methanol fixation by washing in TBS. Residual glutaraldehyde from the fixation step was reduced by incubation with 0.1% sodium borohydride in TBS for 7 min. The samples were washed and blocked in TBS + 3% BSA for 5 min, and incubated simultaneously for 1 h with rabbit anti–γ-tubulin and DM1α (mouse anti–α-tubulin, T2096; Sigma Chemical Co.), each diluted 1:1,000. After washing, the coverslips were incubated for 1 h with a mixture of fluorescein-labeled goat anti–rabbit (1:500) and Texas red– labeled donkey anti–mouse (1:50), washed, and then mounted for viewing under the fluorescence microscope. Images were obtained on a Nikon Optiphot-2 (60× objective, 1.4 NA) using a cooled CCD camera (Princeton Scientific Instruments, Inc.). WinView software (Princeton Scientific Instruments, Inc.) was used to quantitate fluorescence intensity (see Fig. 3 h).
Figure 3.
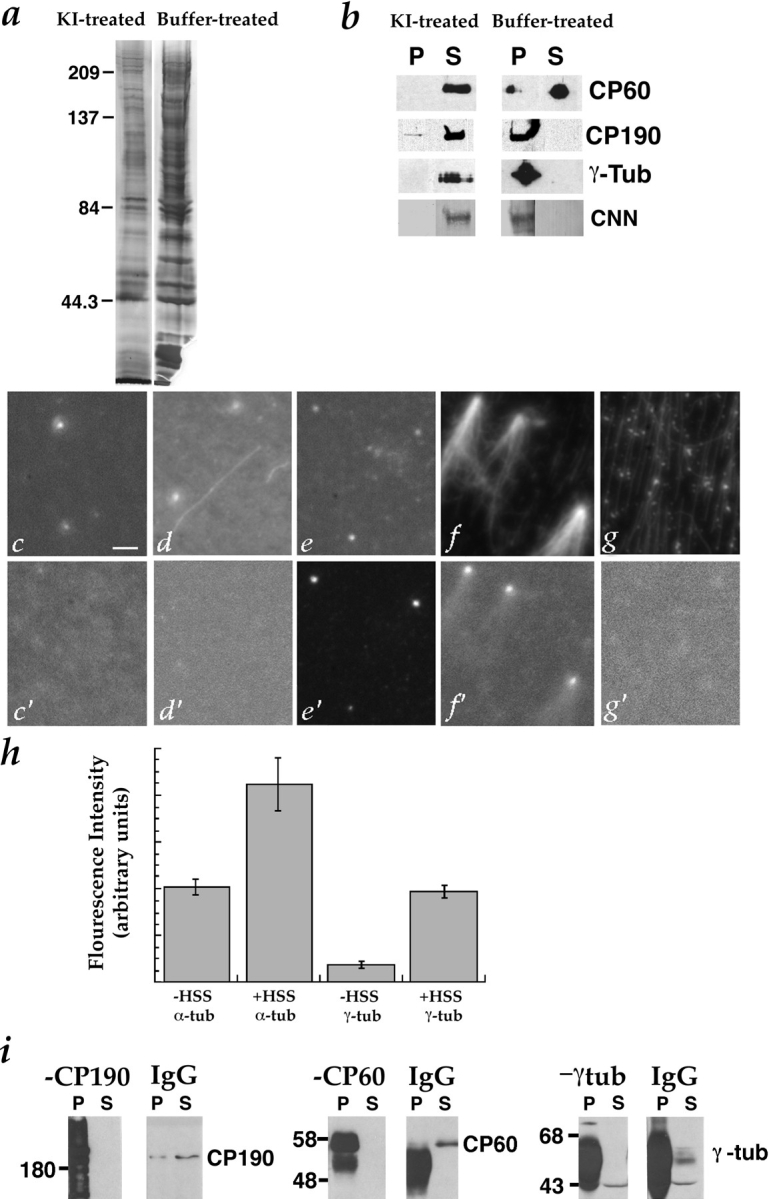
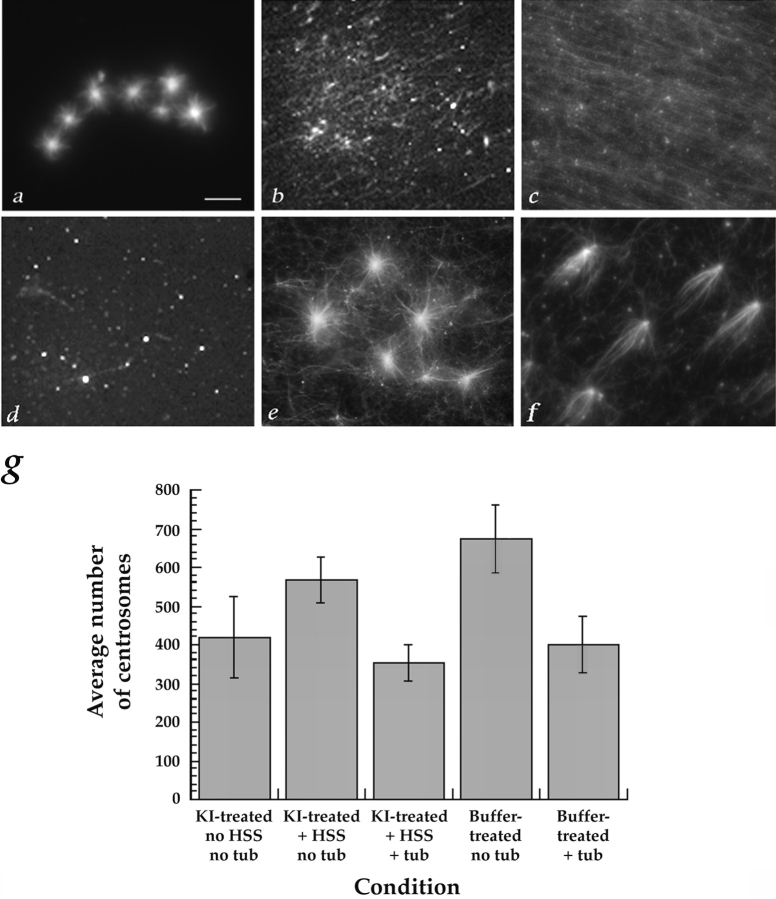
Effects of 2 M KI treatment on centrosomes. (a) The profile of centrosomal proteins is simplified by treatment with 2 M KI. Isolated centrosomes (∼3 × 107) were mixed with an equal volume of 1× BRB80 + 4 M KI (left) or 1× BRB80 (right), incubated on ice for 10 min, and then pelleted by centrifugation at 30,000 g for 1 h. The pellets were washed three times with 1× BRB80 and resuspended in sample buffer, boiled, and then separated by SDS-PAGE on a 10% gel. The gel was silver stained. (b) Removal of CP60, CP190, CNN, and γ-tubulin from centrosomes by 2 M KI. ∼5 × 106 centrosomes were mixed with an equal volume of either 1× BRB80 + 4 M KI (left-hand panels) or 1× BRB80 (right-hand panels), and incubated on ice for 10 min. Centrosomes were pelleted by centrifugation at 30,000 g for 15 min, washed with 1× BRB80, and then resuspended in sample buffer for SDS-PAGE. Proteins released from centrosomes into the supernatants by the KI or buffer treatments were precipitated with 10% TCA and resuspended in sample buffer for SDS-PAGE. The presence of the centrosomal proteins CP60, CP190, CNN, and γ-tubulin in the pellets (P) and supernatants (S) was determined by immunoblotting. Top, CP60 was completely solubilized by KI, and partly solubilized by buffer. Middle, Most CP190 was solubilized by KI, but not by buffer. Bottom, γ-tubulin and CNN were completely solubilized by KI, but not by buffer. (c–g)Immunofluorescence (Texas red, donkey anti– mouse) localization of α-tubulin (DM1α, to label centrioles and microtubules) on KI-treated centrosomes before and after complementation and with and without regrown microtubules, under the conditions indicated below. (c′–g′) Immunofluorescence (fluorescein, goat anti–rabbit) localization of γ-tubulin (rabbit anti–γ-tubulin) in the same fields shown in (c–g). The samples were stained after the following complementation assays were performed: (c, c′)KI-treated centrosomes incubated with buffer alone (no extract, no tubulin). Note that α- (c) but not γ-tubulin (c′) is present, indicating that KI has removed γ-tubulin from the centrosomes. (d, d′) KI-treated centrosomes incubated with buffer followed by tubulin. (e, e′)KI-treated centrosomes incubated with complementing extract followed by buffer (no additional tubulin). Note that γ-tubulin has re-associated with the centrosomes. (f, f′)KI-treated centrosomes incubated with complementing extract followed by tubulin. (g, g′) Coverslip without centrosomes, incubated with complementing extract followed by tubulin. Note the lack of γ-tubulin–staining foci. (h) 2 M KI removes some of the α-tubulin and most of the γ-tubulin from centrosomes, but these proteins reassociate with centrosomes during incubation with complementing extract (HSS). Fluorescence intensity was quantitated on KI-treated centrosomes costained with antibodies against α- and γ-tubulin. 50 centrosomes were measured for each condition. (i)Immunoblots showing quantitative immunodepletion of CP190, CP60, or γ-tubulin from the 228,000 g supernatant of a 0–2 h embryo extract that complements KI-treated centrosomes (see Materials and Methods for details). Left, CP190 was completely immunodepleted from the extract by affinity-purified anti-CP190 antibody, but not by non-immune IgG. Multiple forms of CP190 are visible in the −CP190 pellet. Middle, CP60 was immunodepleted by CP60 antibody, but not by non-immune IgG. (Bands in pellet lanes are antibody and/or CP60, which are of similar size). Right, γ-tubulin was immunodepleted by γ-tubulin antibody, but not by non-immune IgG. (γ-Tubulin comigrates with antibody, so bands in pellet lanes are antibody and/or γ-tubulin. In addition, the γ-tubulin antibody used for immunoblotting cross-reacts with a second ∼43-kD band). Bar, 10 μm.
Drosophila Embryo Extracts
Drosophila embryos between 0- and 2-h old (for preparation of complementing extract), or 0- and 4.5-h old (for characterization of protein complexes) were harvested, dechorionated, and then washed as described previously (Moritz and Alberts, 1998). The embryos were dried by blotting with paper towels, weighed, and then resuspended in 1 vol of extract buffer. The embryos were immediately homogenized by five passes of a motor-driven Teflon pestle in a glass Dounce homogenizer. The extract could be frozen in liquid nitrogen at this point and stored at −80°C. To prepare high speed supernatant for complementation tests and their associated immunodepletions, the crude extract was centrifuged for 20 min at ∼228,000 g (TL100; Beckman Instruments, Inc., Fullerton, CA), the supernatant was transferred to a new tube, and then centrifuged again in the same way. For complementation tests, nocodazole (100 μM final concentration) was added to the extract before centrifugation. To prepare the supernatant for characterizations of protein complexes, the crude extract was centrifuged for 10 min at 30,000 rpm in a Beckman TLA 100.3 rotor, transferred to a new tube and centrifuged again at 100,000 rpm for 8 minutes in the same rotor.
Antibodies
The rabbit antibodies to CP60 and to amino acids 385–508 of CP190 have been previously described (Kellogg et al., 1995; Oegema et al., 1995). The rabbit antibody to amino acids 705–789 of CP190 was prepared according to Oegema et al. (1995). One of the rabbit anti–γ-tubulin antibodies used was raised against the full-length maternal form of Drosophila γ-tubulin (these sequence data are available from GenBank/EMBL/DDBJ under accession number P42271) expressed in baculovirus. The second antibody recognizing γ-tubulin was raised against the COOH-terminal peptide QIDYPQWSPAVEASKAG of the maternal form of Drosophila γ-tubulin. The production and purification of these antibodies will be described elsewhere.
Immunoprecipitations
To prepare the antibodies used for immunoprecipitation, 20–30 μg of antibody was coupled to 50 μl of packed Affiprep protein A beads (Bio-Rad Laboratories, Hercules, CA). The beads were first mixed by gentle rotation with antibody in PBST for 0.5–1 h at room temperature, and then washed three times with PBST, followed by three washes and resuspension in 0.2 M sodium borate, pH 9.0. To covalently attach the antibodies to the beads, dimethyl pimelimidate was added to 20 mM and the beads were incubated while rotating the tube gently for 0.5–1 h at room temperature. To inactivate residual cross-linker, the beads were washed into 0.2 M ethanolamine, pH 8.0, and rotated at room temperature for 2 h to overnight before use. The beads were then pre-eluted three times with 100 mM glycine, pH 2.3, before washing into extract buffer. To begin the immunoprecipitation, 50 μl of packed beads were rotated with 300 μl of concentrated embryo extract for 1 h at 4°C. The beads were pelleted and the supernatants sampled. The beads were washed four times with column buffer (or with extract buffer) plus 75 mM KCl, 0.05% NP-40 or 0.05% Triton X-100, and 1:200 protease inhibitor stock, and then once with the same buffer without detergent. In some cases (for immunoprecipitations to characterize protein complexes), proteins were eluted three times sequentially with 150 μl of 100 mM glycine, pH 2.3. The elutions were pooled and neutralized by addition of 200 μl 0.5 M K-Hepes, pH 7.6. For gel analysis, 20 μg of porcine insulin was sometimes added as carrier and the samples were precipitated with TCA.
In experiments to test the ability of immunodepleted extracts to complement salt-stripped centrosomes, the beads were pelleted, washed as above, and then boiled in sample buffer for SDS-PAGE and Western analysis. Samples of the supernatants were kept for this purpose as well, and the remainder of the supernatant was used in the on-glass complementation assay.
Sucrose Gradient Sedimentation and Gel-filtration Chromatography
Sucrose gradients were poured as step gradients (five 950-μl steps) that were allowed to diffuse into continuous gradients overnight at 4°C before use. The gradients were formed from 5–20% or 5–40% sucrose (Ultra-pure; ICN Biomedicals, Costa Mesa, CA) in gradient buffer plus 75 mM, 100 mM, or 500 mM KCl, as indicated for each experiment. A 50–75-μl aliquot of sample was loaded onto each gradient, and the gradients were centrifuged at 4°C at 50,000 rpm in a Beckman SW55 rotor for 4 to 8 h, as indicated. The gradients were fractionated from the top by hand into 16 300-μl fractions. Protein standards (0.5 mg/ml each) were loaded in an equivalent volume and were run in parallel over identical gradients for each experiment.
Gel-filtration chromatography was carried out on a Superose-6 column by FPLC (Pharmacia Biotech Sevrage, Uppsala, Sweden) in column buffer plus 75 mM, 100 mM, or 500 mM KCl, as indicated. The column was calibrated with standards of known Stokes radii as indicated in the legend to Fig. 5. The size and shape (Stokes radii) of protein complexes were estimated according to (Siegel and Monty, 1966). The axial ratios of the equivalent prolate ellipsoids of revolution, {a/b}p, were estimated according to (Laue et al., 1992), using the method of Kuntz (1971) to estimate the degree of hydration from amino acid sequence.
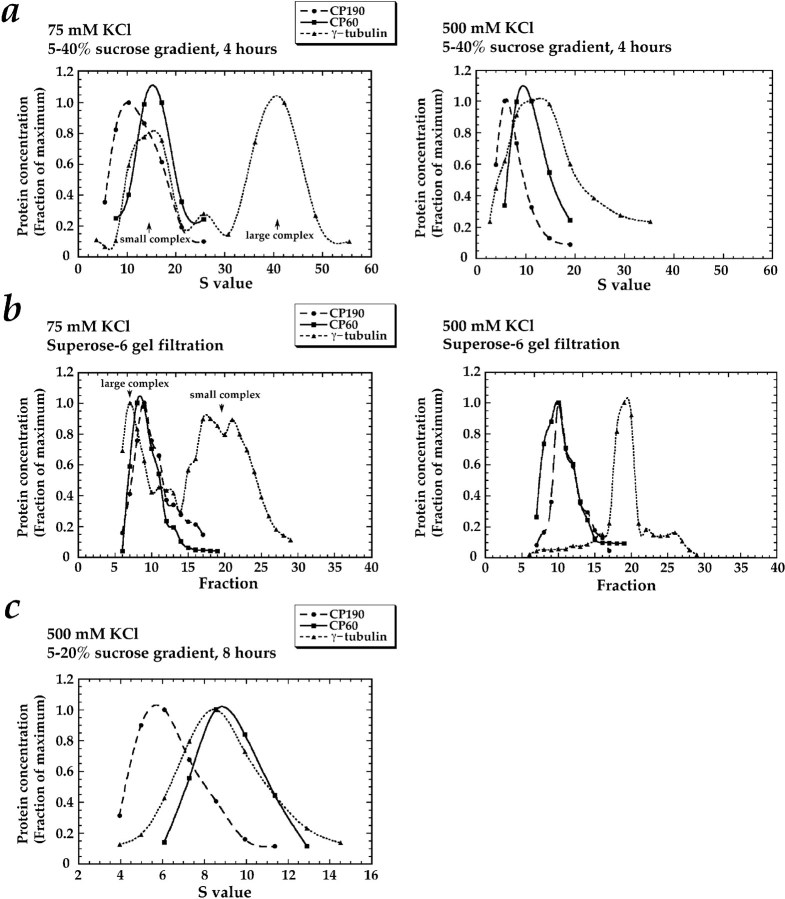
Figure 5.
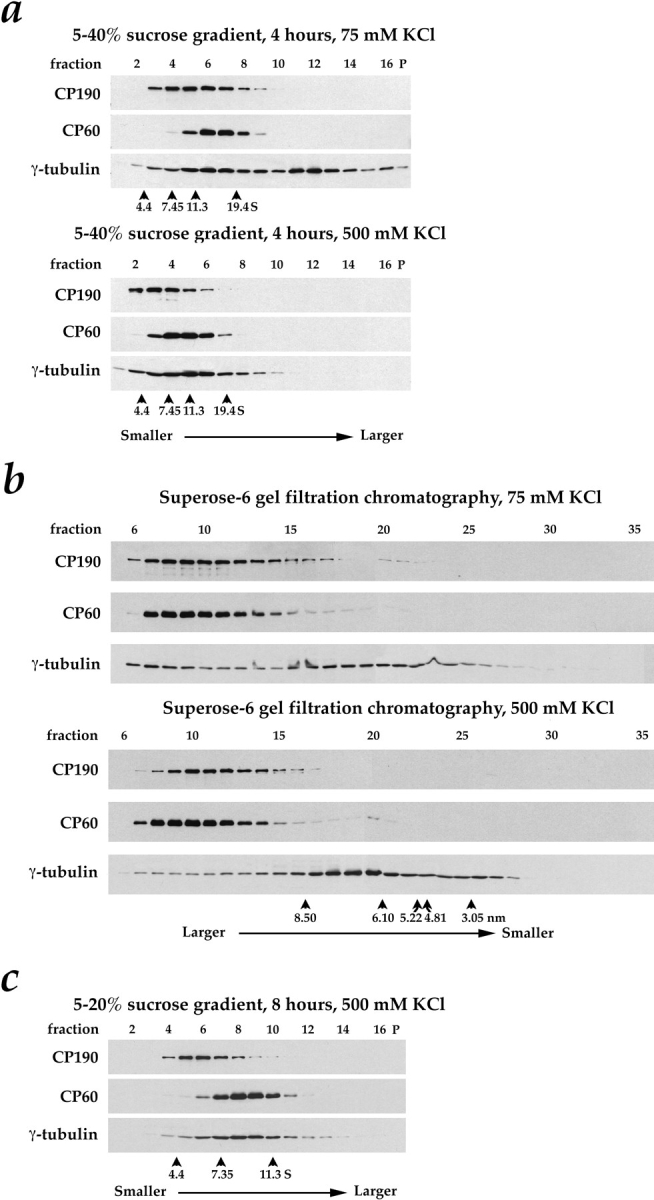
Behavior of CP60, CP190, and γ-tubulin during sucrose gradient sedimentation and Superose-6 gel filtration of concentrated Drosophila embryo extracts. Each fraction was immunoblotted for CP190, CP60, and γ-tubulin after separation by SDS-PAGE on an 11% gel. A single extract was first buffer exchanged using spin columns into column buffer containing either 75 mM or 500 mM KCl. An aliquot was then sedimented through 5–40% sucrose gradients for 4 h (a)or fractionated by Superose-6 gel filtration chromatography (b)in buffers containing 75 mM or 500 mM KCl. In c, a separate extract was sedimented on a 5–20% sucrose gradient for 8 h in 500 mM KCl to increase the separation between the smaller complexes present in the high salt. Sucrose gradient fractions were collected from the top of the gradient; gradient pellets are also shown (P). Standards were run in parallel with the extract over identical sucrose gradients. The location of the peak for each standard is indicated with an arrowhead above its S value (see Materials and Methods for determination of peak fractions). The sucrose gradient standards used were BSA (4.4 S), rabbit muscle aldolase (7.35 S), bovine liver catalase (11.3 S), and porcine thyroglobulin (19.4 S). The Superose-6 column was calibrated with bovine thyroglobulin (Stokes radius = 8.5 nm), horse spleen ferritin (6.1 nm), bovine liver catalase (5.22 nm), rabbit muscle aldolase (4.81 nm), and hen egg ovalbumin (30.5 nm). The location of the peak for each standard is indicated with an arrowhead above its Stokes radius in b.
Sucrose Gradient Quantitation
Standards were precipitated by the addition of TCA to 10%, resuspended in sample buffer, separated by 8.5 or 11% PAGE, and then stained with Coomassie blue. Gels were scanned into the computer using a UMAX scanner (Fremont, CA), and the program NIH Image was used to quantitate band intensities. The peak fraction was assigned for each standard using Kaleidagraph (Synergy Software, Reading, PA). Standard curves of peak fraction versus sedimentation coefficient were then used to convert fraction number to S value (essentially S20,w) for each sucrose gradient to allow direct comparison of protein complexes sedimented in 75 mM and 500 mM KCl. This use of standards to correct to S20,w from different buffers is valid as long as the partial specific volumes are the same for the standard proteins and the protein complexes being studied (Martin and Ames, 1961).
Quantitative Immunoblotting
For immunoblots, samples were precipitated by the addition of TCA to 10% and resuspended in Sample buffer before separation by SDS-PAGE on 10 or 11% gels. Proteins were then transferred to nitrocellulose (pore size 0.1 μm) in the presence of 25% methanol, 0.15 M glycine, 0.02% SDS. The blots were incubated for 20 min in block (TBS + 0.1% Tween-20, 3% nonfat dry milk, 10% glycerol). A chemiluminescent substrate system (SuperSignal CL-HRP; Pierce Chemical Co., Rockford, IL) was used to detect the HRP-conjugated secondary antibodies. The developed film was scanned into the computer using a UMAX scanner and NIH Image was used to quantitate band intensities. Serial dilutions of CP190, CP60, and γ-tubulin were blotted simultaneously with all experimental fractions, allowing us to determine the relative concentrations of CP190, CP60, and γ-tubulin in each fraction, as shown in Fig. 6.
Figure 6.
A graphical representation of the sucrose gradient (a)and gel filtration data (b and c) in Fig. 5. The γ-tubulin is present in two distinct complexes; CP190 and CP60 are not components of either complex. For each fraction, standard curves were used to determine the corresponding S value and the relative concentrations of CP190, CP60, and γ-tubulin (see Materials and Methods).
Results
In Vitro Complementation of Salt-stripped Centrosomes
To study the role of the γTuRC in microtubule nucleation at centrosomes and to test for potential contributions to nucleation by other centrosomal components, we developed an in vitro assay in which the nucleating activity of salt-stripped Drosophila centrosomes is restored by incubation with embryo extract (Fig. 1). Initially, several different salts, including NaCl, KCl, and KI, as well as urea at various concentrations were tested for their ability to inactivate Drosophila centrosomes. Whereas all of the salts and urea were destructive to some extent (data not shown), we found that treatment with 2 M KI consistently destroyed the microtubule-nucleating activity of the centrosomes.
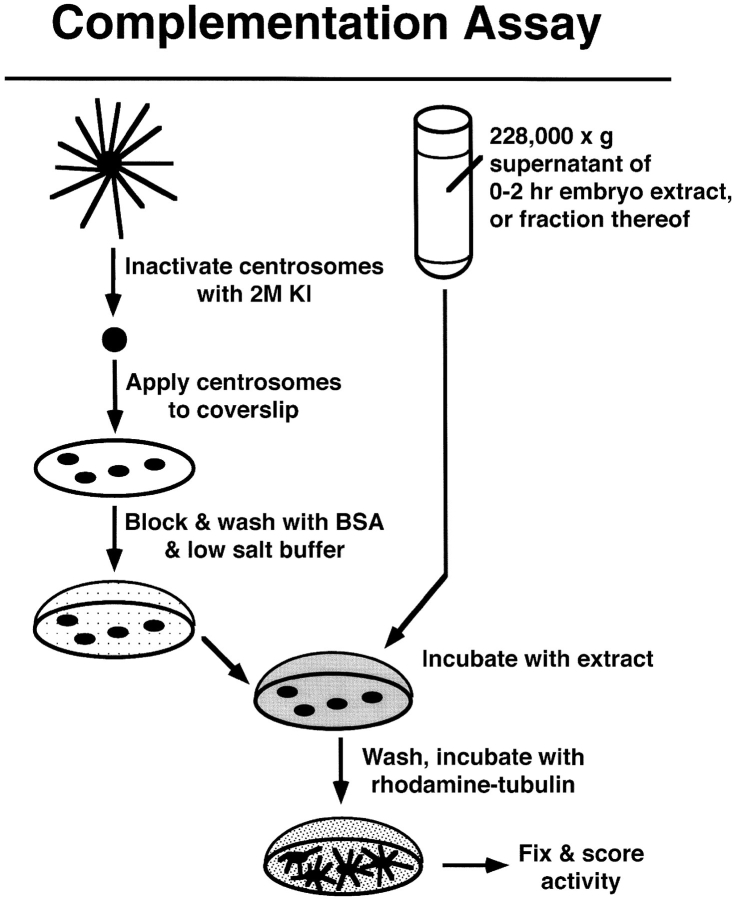
Figure 1.
Complementation assay. Centrosomes isolated from Drosophila embryos are inactivated by incubation with 2 M KI. The KI-treated centrosomes are allowed to bind to a glass coverslip. The coverslip is washed and blocked with a low-salt, BSA-containing buffer, and then incubated with the extract or fraction to be tested. The extract/fraction is washed away and the coverslip is incubated with rhodamine-labeled tubulin. Any resulting asters are fixed sequentially with glutaraldehyde and methanol. The number of asters per 50 microscope fields (100× objective) is determined by counting samples while viewing through a fluorescence microscope. See Materials and Methods for details.
Using this complementation assay, we determined that a 228,000 g supernatant of an extract made from 0–2-h-old embryos was able to complement the KI-stripped centrosomes (Fig. 2). When extract was incubated on the coverslip in the absence of centrosomes, many microtubules, but virtually no asters, formed (Fig. 2, b and c). When KI-stripped centrosomes were incubated with buffer instead of extract, very few microtubules and no asters formed (Fig. 2 d). When KI-stripped centrosomes were incubated with the complementing extract before the tubulin incubation, asters that look very similar to those that formed on buffer-treated centrosomes were produced (compare Fig. 2, a and e). Complementation occurred when the KI-treated centrosomes were incubated with extract containing nocodazole at either 0° or 30°C, indicating that aster regrowth was not merely due to the elongation of microtubules initiated during incubation with the extract (Fig 2, e and f).
Figure 2.
Examples of complementation of KI-treated centrosomes. The complementation assay was carried out as outlined in Fig. 1 and Materials and Methods. (a) Microtubule asters regrew on buffer-treated centrosomes that were incubated with rhodamine-labeled tubulin at 30°C. (b and c) Microtubules, but no asters, formed when a 228,000 g supernatant from a 0–2 h embryo extract was incubated at 30°C (b) or 0°C (c) on coverslips in the absence of centrosomes, followed by a 30°C incubation with rhodamine-tubulin. (d) When KI-treated centrosomes were incubated with buffer instead of extract, followed by rhodamine-tubulin, few microtubules and no asters formed. (e and f) Asters formed when KI-treated centrosomes were first incubated with extract at 30°C (e) or 0°C (f) and then with rhodamine-tubulin at 30°C. (g) KI- and buffer-treated centrosomes bind consistently to coverslips. The number of centrosomes bound to coverslips under the typical experimental conditions used throughout this study was determined by counting structures that were stained with antibodies against α- and/or γ-tubulin. The average number counted in 50 and 100× microscope fields is shown. Three to five separate experiments were counted for each condition. Bar, 10 μm.
To be confident that centrosomes were indeed present even when few or no asters could be found, the number of centrosomes on the coverslips was independently verified by immunofluorescent staining of α-tubulin in the centrioles (and γ-tubulin where possible). The number of centrosomes (whether intact or salt-stripped) bound to individual coverslips was found to be quite consistent (Fig. 2 g).
An initial characterization of the complementing extract showed that its activity could be destroyed by heating to 60°C or boiling, and that ATP was not required for complementation. In addition, when the extract was prepared under conditions that promote microtubule polymerization and the microtubules were removed by centrifugation, the extract did not complement. This suggests that at least one component required for the complementation binds to microtubules (data not shown).
The Centrosomal Proteins CP60, CP190, CNN, and γ-Tubulin Are Removed from Centrosomes by 2 M KI
Since it was possible to complement the salt-stripped centrosomes with soluble factors present in embryo extract, yet these same factors were not capable of inducing aster formation in the absence of the KI-treated centrosomes (Fig. 2), it appeared that a remnant structure must persist after treatment with 2 M KI. This remnant might be a scaffolding to which the soluble factors necessary for restoring microtubule nucleation attach. Therefore, we characterized what remains at the centrosomes after salt treatment, and how the known centrosomal proteins behave under such conditions. We began by determining whether the salt treatment removed the known centrosomal proteins CP60, CP190, CNN, and γ-tubulin (Kellogg et al., 1989; Oakley and Oakley, 1989; Kellogg and Alberts, 1992; Whitfield et al., 1995; Li and Kaufman, 1996). Centrosomes were incubated with 2 M KI, as described above for the complementation assay, and then pelleted. The centrosomal protein profile becomes simpler after KI treatment (Fig. 3 a). Immunoblotting of the pellets and supernatants of buffer- and KI-treated centrosomes shows that virtually all detectable CP60, CP190, CNN, and γ-tubulin are removed by the salt (Fig. 3 b). Electron microscopic examination of the KI-stripped centrosomes shows that the centrioles are destroyed to varying degrees by the salt, but that there is little obvious structural abnormality in the PCM (data not shown).
γ-Tubulin, but Not CP60 or CP190, Are Required for Restoring Microtubule-nucleating Activity to KI-treated Centrosomes
The fact that treatment of centrosomes with KI both led to a loss of microtubule-nucleating activity and extracted CP60, CP190, CNN, and γ-tubulin, suggested that one or more of these proteins might be needed for this activity. Therefore, we tested the effect of immunodepleting CP60, CP190, and γ-tubulin on the ability of the extract to complement salt-stripped centrosomes (we were not able to deplete CNN to a sufficient extent for this assay). Each protein was quantitatively depleted (Fig. 3 i), and the resulting extracts were tested in the in vitro assay (Fig. 1). Only the depletion of γ-tubulin had an effect on the ability of the extract to complement KI-inactivated centrosomes, and this activity was consistently destroyed by the removal of γ-tubulin (Table I; see Fig. 8 c).
Table I.
Ability of Immunodepleted Extracts to Restore Microtubule-nucleating Activity to KI-treated Centrosomes
| KI-treated centrosomes incubated with: | No. of asters per 50 fields* | |
|---|---|---|
| Buffer | 0 | |
| Extract + random IgG | 94 | |
| Extract + protein A beads | 93 | |
| Extract − γ-tubulin | 5 | |
| Extract − CP60 | 88 | |
| Extract − CP190 | 100 |
The microtubule-nucleating activity of centrosomes was destroyed by incubation with 2 M KI. Inactivated centrosomes were incubated with the indicated buffer or extract followed by rhodamine-labeled tubulin. Samples were then fixed and examined under a fluorescence microscope and the No. of asters per 50 microscope fields (100× objective) was counted. (As described in Fig. 1.)
Averages of 3 experiments.
Figure 8.
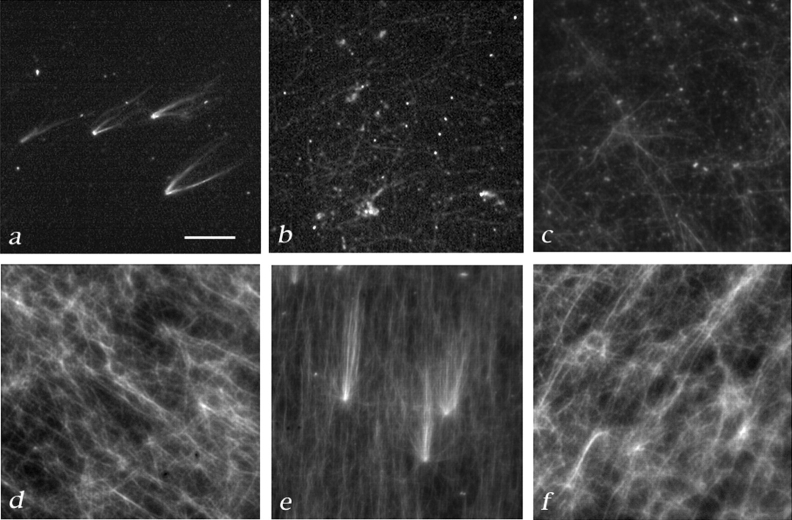
The γTuRC is necessary, but not sufficient for complementation of salt-stripped centrosomes. Complementation tests were carried out as described in Fig. 1 and Materials and Methods. (a) Asters formed when KI-treated centrosomes were incubated with the partially purified large γ-tubulin complex from a Superose-6 column followed by rhodamine-tubulin. (b) Some free microtubules, but no asters formed when the same fraction was incubated without KI-treated centrosomes. (c) KI-treated centrosomes were not complemented by γ-tubulin–depleted extract. (4 asters were counted in 50 100× microscope fields). (d)KI-treated centrosomes were not complemented by immunoaffinity-purified γTuRC, although microtubules could form (6 asters/50 microscope fields). (e) Asters formed after incubation of KI-treated centrosomes with a 1:1 mixture of immunoaffinity-purified γTuRC and γ-tubulin–depleted extract (151 asters/50 fields). (Other ratios of extract to γTuRC were tested; although some activity is still detectable at a 20:1 ratio of extract to γTuRC (data not shown). (f) Microtubules, but no asters formed when a 1:1 mixture of immunoaffinity-purified γTuRC and γ-tubulin–depleted extract were incubated in the absence of centrosomes. Bar, 10 μm.
We used immunofluorescence to further examine the effect of the KI treatment and extract complementation on the presence of γ-tubulin at centrosomes. In these experiments, staining centrosomes with antibodies against γ-tubulin and α-tubulin (which recognize PCM and centrioles, respectively) confirmed that γ-tubulin is removed from centrosomes by treatment with KI (Fig. 3, c and d; c′ and d′). We also found that γ-tubulin reassociates with centrosomes during incubation with extract (Fig. 3, e and f; e′ and f′). To rule out the possibility that spurious asters form in the absence of centrosomes resulting from clustering of γ-tubulin into foci, we also stained coverslips that were incubated with extract followed by tubulin in the absence of centrosomes, and found no foci of γ-tubulin staining (Fig. 3, g and g′). These data were quantitated by measuring the fluorescence intensity of α- and γ-tubulin staining at intact (buffer-treated) and KI-treated centrosomes (Fig. 3 h).
Cumulatively, these results suggest that γ-tubulin is required for aster formation, whereas CP60 and CP190 are not. We had initially expected that CP60 and CP190 might also be at least indirectly required since our laboratory had previously found evidence for the existence of a protein complex containing these three proteins (Raff et al., 1993). These results led us to re-evaluate the interactions of CP60, CP190, and γ-tubulin. In addition, we were interested in characterizing further the γ-tubulin component required for complementation.
Drosophila γ-Tubulin Is in a Protein Complex That Is Similar to the Xenopus γTuRC
We used immunoprecipitations, gel filtration, and sucrose gradient sedimentation to investigate centrosomal protein complexes containing γ-tubulin, CP60, and/or CP190, which might restore microtubule-nucleating activity to salt-stripped centrosomes. CP60, CP190, or γ-tubulin were each immunoprecipitated from concentrated embryo extracts and analyzed by SDS-PAGE (Fig. 4 a) and immunoblotting (Fig. 4, b and c). Antibodies recognizing γ-tubulin (Zheng, Y., unpublished observations) immunoprecipitated γ-tubulin and a group of associated proteins that are components of the Drosophila γTuRC. The protein profile of this complex is very similar to the that of the Xenopus γTuRC (Fig. 4 a, right two lanes; Oegema, K., and Y. Zheng, manuscript in preparation) (Zheng et al., 1995). CP190 antibodies brought down CP190 and a large fraction of CP60 (Fig. 4 a, middle two lanes). Antibodies to CP60 immunoprecipitated CP60 and a small fraction of the CP190 found in extracts (Fig. 4 a, second lane from left).
Figure 4.
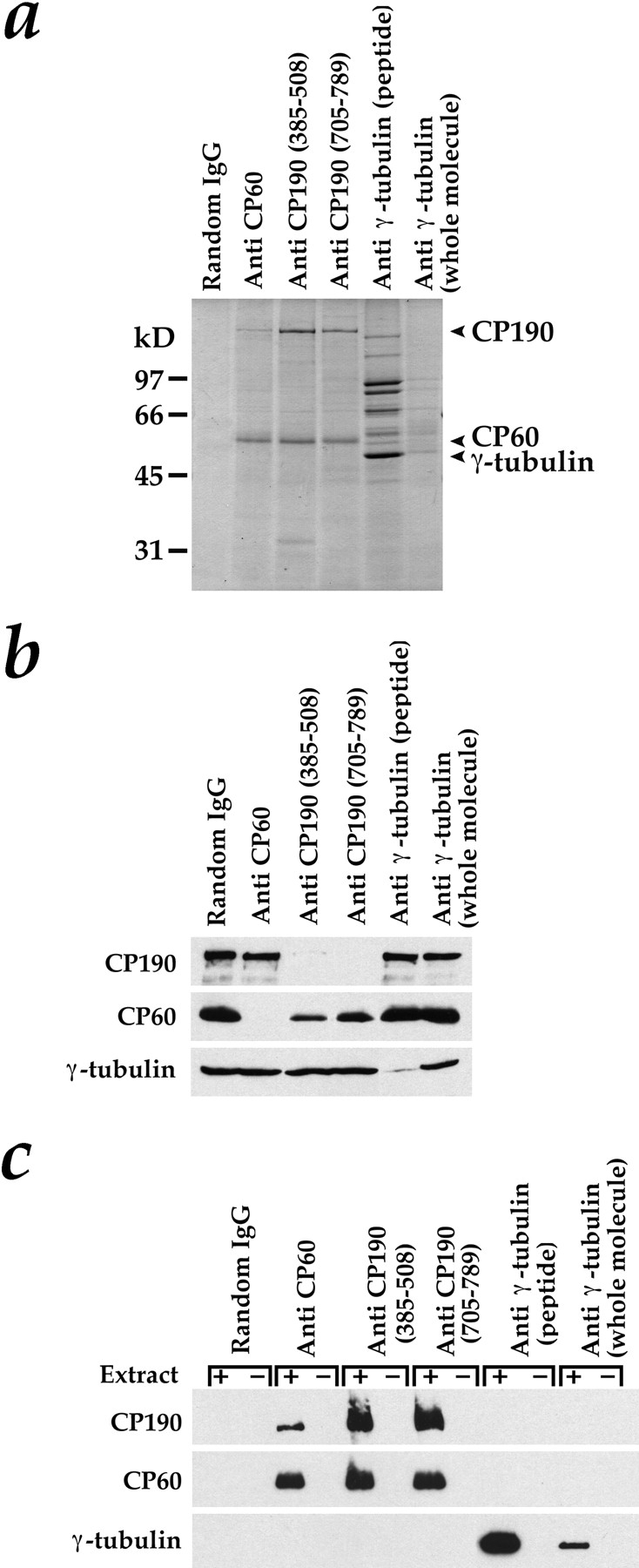
Tests for complex formation by immunoprecipitation. Since Drosophila γ-tubulin is the same size as IgG heavy chain, the immunoprecipitations in a–c were carried out with special care to avoid any IgG contamination in the pellets (see Materials and Methods). (a) Immunoprecipitation pellets after separation by SDS-PAGE on an 11% gel and staining with Coomassie blue. Anti-CP60 immunoprecipitated CP60 and a small fraction of the CP190. Both anti-CP190 antibodies immunoprecipitated CP190 and a large percentage of the CP60. Antibodies to γ-tubulin immunoprecipitated γ-tubulin and a group of γ-tubulin–associated proteins that are components of the Drosophila γTuRC. The anti-γ–tubulin COOH-terminal peptide antibody was much more effective in immunoprecipitations than the antibody made to the whole γ-tubulin molecule. (b and c) Western blots to detect CP190, CP60, and γ-tubulin in immunoprecipitation supernatants (b) and pellets (c). In c, beads were incubated in the presence (+) or absence (−) of extract to control for antibody contamination in the pellets.
Immunoblots of the supernatants and pellets from the immunoprecipitations confirmed the identity of the Coomassie-stained bands. Analysis of the immunoprecipitation supernatants showed that each protein was depleted by its corresponding antibody (Fig. 4 b). In addition, CP60 is largely depleted in the supernatants of extracts treated with antibodies against CP190 (Fig. 4 b).
Immunoprecipitation pellets from concentrated extract (+) or from buffer controls (−) are shown in Fig. 4 c. Neither CP190 nor CP60 was detected in the pellets from immunoprecipitations performed with the antibodies to γ-tubulin, nor was any γ-tubulin present in the pellets of immunoprecipitations performed with antibodies against CP60 or CP190. The majority of the CP60 in extracts coimmunoprecipitated with CP190, however, and only a small fraction of the CP190 coimmunoprecipitated with CP60. These results suggest that, although CP60 and CP190 associate in these extracts, neither CP190 nor CP60 are in a cytoplasmic complex with γ-tubulin.
In Embryo Extracts, γ-Tubulin Is Found in Two Distinct Complexes, Neither of Which Contain CP190 or CP60
To characterize potential protein complexes involving γ-tubulin, CP60, and CP190 further, we analyzed the behavior of these proteins by gel-filtration chromatography and by sucrose gradient sedimentation. The same concentrated embryo extract was simultaneously fractionated by both techniques in identical buffers containing either 75 or 500 mM KCl (Fig. 5). To facilitate the comparison between complexes containing γ-tubulin and those containing CP190 and CP60, we used a quantitative blotting technique that allowed us to determine the relative concentrations of CP190, CP60 and γ-tubulin in each fraction (Fig. 6). Sucrose gradients run in different salt concentrations cannot be directly compared because of differences in buffer density and viscosity that affect sedimentation rates. Therefore, standard curves of peak fraction versus S value, generated by loading proteins of known S value on identical sucrose gradients run in parallel with each experimental gradient, were used to convert fraction number to S value for each gradient.
In buffer containing 75 mM KCl, most of the γ-tubulin is found in two complexes that can be separated by both sucrose gradient sedimentation and by gel filtration (Figs. 5, a and b; and 6, a and b, left panels). The large γ-tubulin complex can be converted to the small γ-tubulin complex by raising the KCl concentration to 500 mM (compare γ-tubulin migration in Fig. 5 a, top and bottom; b, top and bottom; Fig. 6, a and b, compare left and right). In addition, the gel-filtration peak corresponding to the small γ-tubulin complex, which appears heterogeneous in 75 mM KCl, becomes much more homogeneous in 500 mM KCl. These results suggest that the small γ-tubulin complex is a subunit of the large γ-tubulin complex.
The sedimentation coefficients of the large and small γ-tubulin complexes are 36.9 S and 8.5 S, respectively. (The sedimentation coefficient of the large γ-tubulin complex was determined on a separate sucrose gradient using the 30 S ribosome particle as a standard, in addition to the standards mentioned in the legend to Fig. 5 [data not shown]). The Stokes radii of the small and large γ-tubulin complexes, estimated from our gel-filtration results, are 6.9 nm and ∼20 nm, respectively (the large γ-tubulin complex fractionates close to the void volume of the Superose-6 column, preventing a more precise determination). From these sedimentation coefficients and Stokes radii, the masses of the small and large γ-tubulin complexes were estimated to be 240,000 and ∼3,000,000 D, respectively (Siegel and Monty, 1966).
On sucrose gradients, CP60 comigrates with the small γ-tubulin complex in both high and low salt (compare CP60 with γ-tubulin in Figs. 5 a, and 6 a, left panel). However, under the same buffer conditions, CP60 can be separated easily from the small γ-tubulin complex by gel filtration (compare CP60 and γ-tubulin peaks in Figs. 5 b and 6 b). By gel filtration in 75 mM KCl (Figs. 5 b, and 6 b, left panel), CP60 elutes with the large γ-tubulin complex, but under identical conditions, CP60 and the γ-tubulin large complex are easily separated on sucrose gradients (Figs. 5 a, and 6 a, left panel). Similarly, CP190 can be separated from the small γ-tubulin complex by gel filtration (Figs. 5 b, and 6 b), and from the large γ-tubulin complex on sucrose gradients (Figs. 5 a, and 6 a, left panel). These experiments indicate that neither CP60 nor CP190 are among the components of either the large or small γ-tubulin complexes. Interestingly, even in high salt conditions where CP190 and CP60 do not associate (data not shown), both proteins are found in large complexes. The properties of these centrosomal protein complexes are summarized in Table II.
Table II.
Properties of Centrosomal Protein Complexes in Extracts
| S20,w | Rs | Mol wt (estimated) | {a/b}p | |||
|---|---|---|---|---|---|---|
| Å | ||||||
| γ-Tubulin large complex (75 mM KCl) | ||||||
| 36.9 S | ∼200 | ∼3,000,000 | ||||
| γ-Tubulin small complex (500 mM KCl) | ||||||
| 8.5 S | 68.7 | 240,000 | ||||
| CP190 (500 mM KCl) | ||||||
| 5.7 S | 146 | 343,000 | 19 | |||
| CP60 (500 mM KCl) | ||||||
| 8.9 S | 149 | 546,000 | 23* | |||
Suggests that the CP60 oligomer is highly asymmetric.
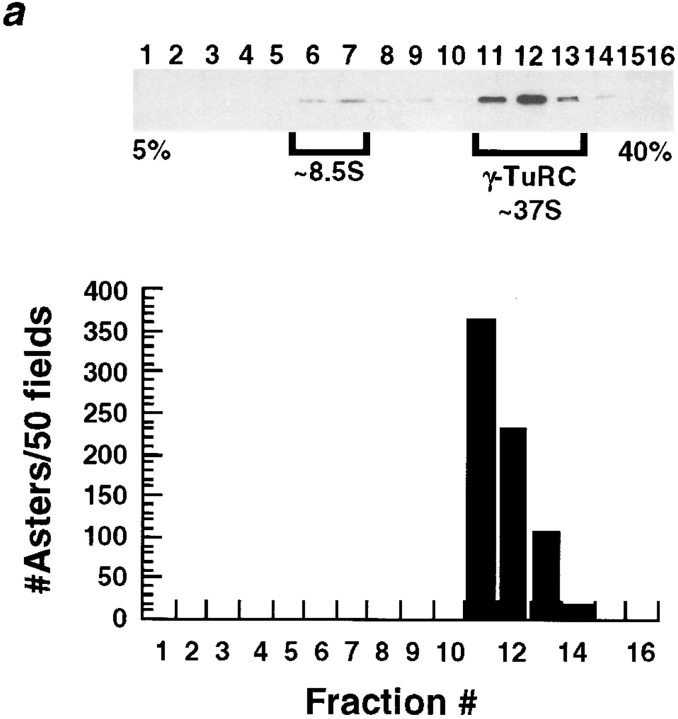
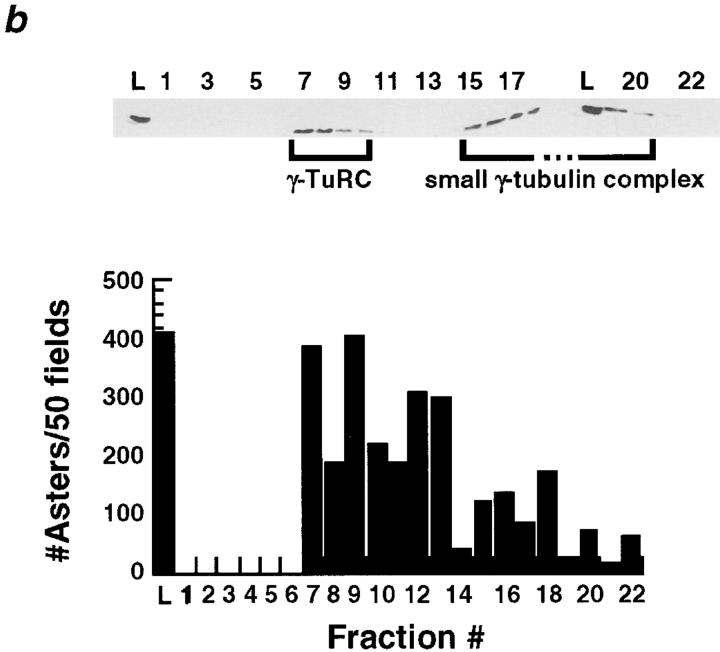
The γ-Tubulin Ring Complex Is Necessary, but Not Sufficient for Complementation of Salt-stripped Centrosomes
Since our immunodepletion studies demonstrated a requirement for soluble γ-tubulin in the complementation assay, we used the assay to determine if either the small or large γ-tubulin–containing complexes could complement the centrosome scaffolds. For this test we fractionated embryo extract on sucrose gradients or by gel filtration and then assayed the fractions for complementing activity. The sucrose gradient fractions containing the large complex complemented the salt-stripped centrosomes, whereas fractions containing the small complex did not (Fig. 7 a). Gel filtration fractions containing the large complex also gave good complementation; in addition, we observed some less robust complementation by the gel filtration fractions containing the small complex (Fig. 7 b).
Figure 7.
(a) The γTuRC partially purified on a 5–40% sucrose gradient complements KI-treated centrosomes. The small γ-tubulin complex does not. 100 μl of the extract that complements KI-treated centrosomes was loaded on a 5-ml 5–40% gradient made in column buffer + 100 mM KCl. See Materials and Methods for details on running and fractionating gradients. Top, immunoblot showing that two γ-tubulin–containing complexes can be separated on a 5–40% sucrose gradient. Bottom, sucrose gradient fractions were tested (Fig. 1) for their ability to complement KI-treated centrosomes and the number of asters that formed in 50 microscope fields was counted. Fractions 11–14, which contain the γTuRC were able to complement KI-treated centrosomes. (b) The γTuRC partially purified by FPLC on a Superose-6 gel-filtration column run in Column buffer + 100 mM KCl complements KI-treated centrosomes. Top, immunoblot showing that two γ-tubulin–containing complexes can be separated by gel filtration. The γTuRC elutes just after the void volume. 50 μl of the extract that complements KI-treated centrosomes was loaded on a 24 ml Superose-6 column. 0.5-ml fractions were collected. The column load (L) and fractions 1–22 are shown. Bottom, column fractions were tested for their ability to complement KI-treated centrosomes (Fig. 1). Fractions containing the γTuRC were able to complement.
These results suggested that the γ-tubulin large complex is not only essential, but possibly sufficient for complementation. To determine if this was the case, we used an antibody that recognizes the COOH-terminal 17 amino acids of the Drosophila maternal form of γ-tubulin to purify the large complex (we will refer to this subsequently as the Drosophila γTuRC) in a manner similar to that described for the purification of the Xenopus γTuRC (Zheng, Y., manuscript in preparation; Zheng et al., 1995), see Fig. 4 a for an immunoprecipitation showing the protein composition of this complex). The purified Drosophila γTuRC has a ring structure very similar to that of the Xenopus complex (Zheng, Y., manuscript in preparation). To our surprise, we found that the immunoaffinity-purified γTuRC was not able to complement.
We reasoned that an additional factor in the extract that was removed upon immunoaffinity purification of the γTuRC was required to allow the γTuRC to complement. To test this idea, equal volumes of pure γTuRC and extract that had been immunodepleted using an anti–γ-tubulin antibody were mixed, and then tested for complementation of salt-stripped centrosomes in the on-glass assay (Fig. 8). Asters formed when KI-treated centrosomes were incubated with the partially purified large γ-tubulin complex from a Superose-6 column (Fig. 8 a). Some free microtubules, but no asters formed when the centrosomes were omitted (Fig. 8 b). If KI-stripped centrosomes were incubated with a mixture of pure γTuRC and γ-tubulin– depleted extract, asters formed (Fig. 8 e). However, centrosomes incubated with pure γTuRC or γ-tubulin–depleted extract alone were not able to produce asters (Fig. 8, c and d). In the absence of centrosomes, the mixture of pure γTuRC and γ-tubulin–depleted extract formed microtubules, but no asters (Fig. 8 f).
We concluded that the immunoisolated γTuRC requires an additional factor found in extracts to complement salt-stripped centrosomes. To determine whether the order of addition of these two components is important, we incubated salt-stripped centrosomes sequentially with γ-tubulin–depleted extract followed by a washing step, and then by pure γTuRC, or vice versa. The centrosomes only regained their ability to nucleate microtubule asters when they were first incubated with the γ-tubulin–depleted extract, followed by pure γTuRC, or when they were incubated with a mixture of the two. The complementation worked best when the two components were added simultaneously (data not shown). Initial characterization by sucrose gradient sedimentation and gel filtration of this additional required factor indicates that it has an estimated molecular weight of 220,000 D (data not shown).
Discussion
To begin to understand microtubule nucleation in the context of the centrosome, we developed an in vitro complementation assay in which centrosome aster formation is reconstituted from salt-stripped centrosome scaffolds and a soluble fraction provided by a Drosophila embryo extract. This assay opens several new avenues for studying the structure and composition of the centrosome matrix. Extraction of centrosomes with the strongly chaotropic salt, KI, removes all of the known Drosophila components of core centrosomes, reducing the centrosomes to a simpler structure that appears to contain a scaffolding on which microtubule-nucleating sites may reassemble. The proteins left in this salt-resistant structure are unknown, but it should be possible to identify them by peptide sequencing.
Electron microscopy of the salt-stripped centrosome scaffolds did not reveal any striking modifications of the PCM, suggesting that CP60, CP190, CNN, and γ-tubulin are distributed throughout the PCM in intact centrosomes rather than being confined to particular regions. This is consistent with our previous EM observation that γ-tubulin is found at all levels of the PCM (Moritz et al., 1995b ).
We have also characterized the soluble components contributed by the embryo extract that are required for centrosome aster reconstitution. By immunodepleting the Drosophila centrosomal proteins CP60, CP190, and γ-tubulin from the complementing extract, we found that γ-tubulin is absolutely required for aster formation in our assay. This requirement is similar to the requirement for γ-tubulin in the aster assembly assay in Xenopus extracts (Felix et al., 1994; Stearns and Kirschner, 1994). Previous work from our laboratory suggested that CP60, CP190, and γ-tubulin are in a protein complex together (Raff et al., 1993), yet we found that CP60 and CP190 were not necessary for complementation.
We therefore characterized the γ-tubulin–containing complexes found in Drosophila embryo extracts to determine if we could identify a specific complex required for complementation and to re-evaluate the association of γ-tubulin with CP190 and CP60. Protein complexes were immunoprecipitated or fractionated by sucrose-gradient sedimentation and gel-filtration chromatography at high and low salt concentrations. We found that in low salt, most of the γ-tubulin is found in two distinct complexes of 240,000 and ∼3,000,000 D, which can be separated by either fractionation technique. The larger γ-tubulin complex can be converted into the smaller complex by 500 mM KCl, suggesting that the small γ-tubulin complex is a subunit of the larger one. The large γ-tubulin complex seen in these extracts has been purified to near homogeneity and is the Drosophila analogue of the γ-tubulin ring complex (γTuRC) isolated from Xenopus egg extracts (Zheng, Y., and K. Oegema, manuscript in preparation; Zheng et al., 1995).
These experiments also demonstrated that although CP60, CP190, and γ-tubulin are all found in large protein complexes neither CP60 nor CP190 are among the components of the γ-tubulin complexes. How can we reconcile these data with previous results favoring a cytoplasmic complex containing CP190, CP60, and γ-tubulin? In the previous study (Raff et al., 1993) the complexes were not examined by gel-filtration chromatography, experiments that turned out to be crucial in the current work to distinguish the CP60 complex from the γ-tubulin small complex. However, in the previous study, γ-tubulin was detected by Western blotting in the elutions from immunoaffinity columns constructed from antibodies to CP60 or CP190, although it was a very minor component of these elutions, since a Coomassie blue–staining γ-tubulin band was never detected (Raff et al., 1993). In contrast, in this study we were not able to immunoprecipitate γ-tubulin with antibodies to CP60 or CP190, nor could we immunoprecipitate CP60 or CP190 with antibodies to γ-tubulin. One possible explanation for these disparate results is that although γ-tubulin is not found in a soluble complex with CP60 or CP190 in extracts, these proteins may assemble with each other to form a higher order complex on immunoaffinity columns and in vivo at the centrosome under conditions where the concentrations of these centrosomal proteins are higher than they are in extracts.
To explore the role of the large and small γ-tubulin– containing complexes in aster formation, we tested embryo extracts fractionated on sucrose gradients or by gel filtration for complementing activity in our assay. Although a direct role for the small complex is unclear, both sucrose gradient and gel filtration fractions containing the large complex (the Drosophila γTuRC) can complement salt-stripped centrosomes (Figs. 7 and 8). This result was surprising because treatment with KI extracts all of the known components of the pericentriolar material in Drosophila (CP190, CP60, CNN, and γ-tubulin) and appears to substantially simplify the protein composition of the isolated centrosomes. The fact that both sucrose gradient and gel filtration fractions containing the γTuRC can complement suggests that the connection between the γTuRC present in the extract and the salt-resistant scaffold is likely to be direct.
In contrast to the γTuRC in the sucrose gradient and gel filtration fractions, purified Drosophila γTuRC is unable to complement, suggesting that a component or modification of the γTuRC that is required for complementation is lost during immunoaffinity purification. The possibilities for what this component or modification is doing include: (a) providing a physical link between the centrosome and the γTuRC, or (b) modifying the scaffolds or the γTuRC in some way so that the γTuRC can bind to, or become activated at the centrosome. The fact that the purified Drosophila γTuRC can nucleate microtubules in solution (Zheng, Y., and C. Wiese, unpublished observation) and when bound to the coverslips in our assay (Fig. 8, d and f), suggests that this component or modification probably is not necessary for nucleating activity but may instead be required for attachment of the γTuRC to the salt-stripped centrosome scaffolds. The order of addition experiment further supports the idea that γ-tubulin–depleted extract may supply an attachment factor required to link the purified γTuRC to the salt-stripped scaffolds.
We have found that this putative attachment factor has an estimated molecular weight of ∼220,000 D. Pericentrin is one possible candidate for the factor, since it is a large coiled-coil, core centrosome structural protein that may interact with γ-tubulin (Doxsey et al., 1994; Dictenberg et al., 1998). However, testing this possibility awaits the development of pericentrin reagents that work well in Drosophila.
In summary, we have developed an assay in which microtubule nucleation by Drosophila centrosomes is reconstituted from inactive salt-stripped centrosome scaffolds and soluble components derived from a Drosophila embryo extract. We conclude that the γTuRC is required for microtubule nucleation at centrosomes and that the connection between the γTuRC and the simplified scaffolds is likely to be directly mediated by a factor found in extracts that is normally loosely associated with the γTuRC. We believe that our approach of attempting to simplify centrosome structure and function by focusing on smaller centrosomal protein complexes in conjunction with an in vitro complementation assay will be valuable in understanding this important but complicated organelle.
Acknowledgments
We thank M. Welch, C. Walczak (University of California, San Francisco, CA), and C. Wiese (Carnegie Institute of Washington, Baltimore, MD) for helpful suggestions on the manuscript, and all members of the Mitchison lab (East and West) for a lively environment in which to study centrosomes and microtubules. Many thanks to Dr. T. Kaufman (Howard Hughes Medical Institute, Indiana University, Bloomington, IN) for antibodies against centrosomin, and to A. Desai (Harvard Medical School, Boston, MA) for help with centriole immunofluorescence.
This work was supported by the Herbert W. Boyer Fund (to M. Moritz) and the National Institutes of Health (No. GM23928 to B.M. Alberts).
Abbreviations used in this paper
- CNN
centrosomin
- PCM
pericentriolar material
- TDB
tubulin dilution buffer
- γTuRC
γ-tubulin ring complex
Footnotes
Address all correspondence to Michelle Moritz, Department of Biochemistry and Biophysics, University of California, San Francisco, CA 94143-0448. Tel.: (415) 476-4581. Fax: (415) 476-0806.
References
- Buendia B, Draetta G, Karsenti E. Regulation of the microtubule nucleating activity of centrosomes in Xenopusegg extracts: Role of cyclin A–associated protein kinase. J Cell Biol. 1992;116:1431–1442. doi: 10.1083/jcb.116.6.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, Doxsey SJ. Pericentrin and γ-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- Felix M-A, Antony C, Wright M, Maro B. Centrosome assembly in vitro: Role of γ-tubulin recruitment in Xenopussperm aster formation. J Cell Biol. 1994;124:19–31. doi: 10.1083/jcb.124.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M, Glover D, Saumweber H. Nuclear antigens follow different pathways into daughter nuclei during mitosis in early Drosophila embryos. J Cell Sci. 1986;82:155–172. doi: 10.1242/jcs.82.1.155. [DOI] [PubMed] [Google Scholar]
- Hyman A, Drechsel D, Kellogg D, Salser S, Sawin K, Steffen P, Wordeman L, Mitchison T. Preparation of modified tubulins. Methods Enzymol. 1991;196:478–487. doi: 10.1016/0076-6879(91)96041-o. [DOI] [PubMed] [Google Scholar]
- Joshi HC, Palacios MJ, McNamara L, Cleveland DW. γ-Tubulin is a centrosomal protein required for cell cycle-dependent microtubule nucleation. Nature. 1992;356:80–83. doi: 10.1038/356080a0. [DOI] [PubMed] [Google Scholar]
- Kalt A, Schliwa M. Molecular Components of the Centrosome. Trends Cell Biol. 1993;3:119–128. doi: 10.1016/0962-8924(93)90174-y. [DOI] [PubMed] [Google Scholar]
- Kellogg DR, Alberts BM. Purification of a multiprotein complex containing centrosomal proteins from the Drosophila embryo by chromatography with low-affinity polyclonal antibodies. Mol Biol Cell. 1992;3:1–11. doi: 10.1091/mbc.3.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR, Field CM, Alberts BM. Identification of microtubule-associated proteins in the centrosome, spindle, and kinetochore of the early Drosophilaembryo. J Cell Biol. 1989;109:2977–2991. doi: 10.1083/jcb.109.6.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR, Moritz M, Alberts BM. The centrosome and cellular organization. Annu Rev Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- Kellogg DR, Oegema K, Raff J, Schneider K, Alberts BM. CP60: a Drosophilamicrotubule associated protein that is localized to the centrosome in a cell cycle-specific manner. Mol Biol Cell. 1995;12:1673–1684. doi: 10.1091/mbc.6.12.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keryer G, Ris H, Borisy GG. Centriole distribution during tripolar mitosis in Chinese hamster ovary cells. J Cell Biol. 1984;98:2222–2229. doi: 10.1083/jcb.98.6.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz C, Dabauvalle M-C, Paintrand M, Weber T, Bornens M, Karsenti E. Parthenogenesis in Xenopuseggs requires centrosomal integrity. J Cell Biol. 1990;110:405–415. doi: 10.1083/jcb.110.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz ID. Hydration of macromolecules. IV. Polypeptide conformation in frozen solutions. J Am Chem Soc. 1971;93:516–518. doi: 10.1021/ja00731a037. [DOI] [PubMed] [Google Scholar]
- Laue, T., B. Shah, T. Ridgeway, and S. Pelletier. 1992. Computer-aided interpretation of analytical sedimentation data for proteins. In Analytical Ultracentrifugation in Biochemistry and Polymer Science. S. Harding, A. Rowe, and J. Horton, editors. The Royal Society of Chemistry, Cambridge. 90–125.
- Li K, Kaufman TC. The homeotic target gene centrosominencodes an essential centrosomal component. Cell. 1996;85:585–596. doi: 10.1016/s0092-8674(00)81258-1. [DOI] [PubMed] [Google Scholar]
- Martin R, Ames B. A method for determining the sedimentation behavior of enzymes: Application to protein mixtures. J Biol Chem. 1961;236:1372–1379. [PubMed] [Google Scholar]
- Mazia D. The chromosome cycle and the centrosome cycle in the mitotic cycle. Int Rev Cytology. 1987;100:49–92. doi: 10.1016/s0074-7696(08)61698-8. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner MW. Microtubule assembly nucleated by isolated centrosomes. Nature. 1984;312:232–237. doi: 10.1038/312232a0. [DOI] [PubMed] [Google Scholar]
- Moritz, M., and B.M. Alberts. 1998. Isolation of centrosomes from Drosophila embryos. Methods Cell Biol. In press. [DOI] [PubMed]
- Moritz M, Braunfeld MB, Fung JC, Sedat JW, Alberts BM, Agard DA. Three-dimensional structural characterization of centrosomes from early Drosophilaembryos. J Cell Biol. 1995a;130:1149–1159. doi: 10.1083/jcb.130.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Sedat JW, Alberts BM, Agard DA. Microtubule nucleation by γ-tubulin–containing rings in the centrosome. Nature. 1995b;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Yoon Y, Jung MK. Gamma-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- Oakley CE, Oakley BR. Identification of gamma-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature. 1989;338:662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- Oegema K, Whitfield WGF, Alberts BM. The cell cycle–dependent localization of the CP190 centrosomal protein is determined by the coordinate action of two separable domains. J Cell Biol. 1995;131:1261–1273. doi: 10.1083/jcb.131.5.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K, Marshall WF, Sedat JW, Alberts BM. Two proteins that cycle asynchronously between centrosomes and nuclear structures: DrosophilaCP60 and CP190. J Cell Sci. 1997;110:1573–1583. doi: 10.1242/jcs.110.14.1573. [DOI] [PubMed] [Google Scholar]
- Raff JW, Kellogg DR, Alberts BM. Drosophila gamma tubulin is part of a complex containing two previously identified centrosomal MAPs. J Cell Biol. 1993;121:823–835. doi: 10.1083/jcb.121.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Borisy GG. The centrosome cycle in PTK2 cells: asymmetric distribution and structural changes in pericentriolar material. Biol Cell. 1982;44:117–32. [Google Scholar]
- Siegel LM, Monty KJ. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Stearns T, Kirschner M. In vitro reconstitution of centrosome assembly and function: The role of γ-tubulin. Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- Stearns T, Evans L, Kirschner M. Gamma-tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- Vogel JM, Stearns T, Rieder CL, Palazzo RE. Centrosomes isolated from Spisula solidissimaoocytes contain rings and an unusual stoichiometric ratio of α/β tubulin. J Cell Biol. 1997;137:193–202. doi: 10.1083/jcb.137.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov YS. Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol. 1982;93:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev IA, Nadezhdina ES. The centrosome and its role in the organization of microtubules. Int Rev Cytol. 1987;106:227–293. doi: 10.1016/s0074-7696(08)61714-3. [DOI] [PubMed] [Google Scholar]
- Whitfield WG, Chaplin MA, Oegema K, Parry H, Glover DM. The 190 kDa centrosome-associated protein of Drosophila melanogastercontains four zinc-finger motifs and binds to specific sites on polytene chromosomes. J Cell Sci. 1995;108:3377–3387. doi: 10.1242/jcs.108.11.3377. [DOI] [PubMed] [Google Scholar]
- Whitfield WG, Millar SE, Saumweber H, Frasch M, Glover DM. Cloning of a gene encoding an antigen associated with the centrosome in Drosophila. J Cell Sci. 1988;89:467–480. doi: 10.1242/jcs.89.4.467. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Jung MK, Oakley BR. Gamma-tubulin is present in Drosophila melanogaster and Homo sapiensand is associated with the centrosome. Cell. 1991;65:817–823. doi: 10.1016/0092-8674(91)90389-g. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. A γ tubulin ring complex purified from the unfertilized egg of Xenopus laeviscan nucleate microtubule assembly in vitro. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]