Abstract
Melanocyte differentiation characterized by an increased melanogenesis, is stimulated by α-melanocyte–stimulating hormone through activation of the cAMP pathway. During this process, the expression of tyrosinase, the enzyme that controls melanin synthesis is upregulated. We previously showed that cAMP regulates transcription of the tyrosinase gene through a CATGTG motif that binds microphthalmia a transcription factor involved in melanocyte survival. Further, microphthalmia stimulates the transcriptional activity of the tyrosinase promoter and cAMP increases the binding of microphthalmia to the CATGTG motif. These observations led us to hypothesize that microphthalmia mediates the effect of cAMP on the expression of tyrosinase. The present study was designed to elucidate the mechanism by which cAMP regulates microphthalmia function and to prove our former hypothesis, suggesting that microphthalmia is a key component in cAMP-induced melanogenesis. First, we showed that cAMP upregulates the transcription of microphthalmia gene through a classical cAMP response element that is functional only in melanocytes. Then, using a dominant-negative mutant of microphthalmia, we demonstrated that microphthalmia is required for the cAMP effect on tyrosinase promoter. These findings disclose the mechanism by which cAMP stimulates tyrosinase expression and melanogenesis and emphasize the critical role of microphthalmia as signal transducer in cAMP-induced melanogenesis and pigment cell differentiation.
Keywords: melanocytes, cAMP, microphthalmia, tyrosinase, differentiation
In mammals, melanin synthesis or melanogenesis takes place in melanocytes after differentiation of the non-pigmented precursor, the melanoblast (Le Douarin, 1982). Three melanocyte-specific enzymes, tyrosinase (Körner and Pawelek, 1982; Hearing, 1987; Prota, 1988), tyrosinase-related protein 1 (TRP1)1 (Jiménez-Cervantes et al., 1994; Kobayashi et al., 1994), and tyrosinase-related protein 2 (TRP2) (Barber et al., 1984; Jackson et al., 1992; Yokoyama et al., 1994a ) are involved in this enzymatic process that converts tyrosine to melanin pigments. The expression of these enzymes is restricted to melanocytes, suggesting that their promoters contain regulatory elements responsible for tissue-specific expression. Sequencing of tyrosinase, TRP1, and TRP2 promoters has revealed a highly conserved 10-bp motif (GTCATGTGCT) termed the M-box that plays a key role in the tissue-specific expression of tyrosinase and TRP1 genes (Lowings et al., 1992; Ganss et al., 1994; Yokoyama et al., 1994b ). The M-box core motif, CATGTG, is reminiscent of the CANNTG sequence (E-box), which binds basic-helix-loop-helix (b-HLH) transcription factor. Recently, a transcription factor belonging to the b-HLH family, named microphthalmia (Mi) and expressed in a limited number of tissues such as heart, mast cells, osteoclast precursors, and melanocytes has been involved in melanocyte survival, development, and differentiation (Hodgkinson et al., 1993; Steingrímsson et al., 1994). Indeed, mutations at the mouse mi locus lead to coat color dilution, white spotting, or complete loss of pigmentation (Hodgkinson et al., 1993; Hughes et al., 1993). Similarly, mutations in the human homologue of the mouse microphthalmia gene, microphthalmia-associated transcription factor (MITF), has been linked to abnormal pigmentation observed in Waardenburg Syndrome type II (Hughes et al., 1994; Tassabehji et al., 1994). Furthermore, studies have shown that microphthalmia through the binding to M-box strongly stimulates tyrosinase and TRP1 promoter activities, suggesting that microphthalmia is involved in the tissue-specific expression of the melanogenic genes (Bentley et al., 1994; Ganss et al., 1994; Hemesath et al., 1994; Yasumoto et al., 1994; Yavuzer et al., 1995).
In vivo, melanogenesis is stimulated by ultraviolet radiation of solar light (Ortonne, 1990) and α-melanocyte–stimulating hormone (α-MSH) (Levine et al., 1991) that increase expression of tyrosinase, the rate-limiting enzyme in melanin synthesis. In vitro, the melanogenic effects of α-MSH can be mimicked by pharmacological agents such as forskolin, cholera toxin, and IBMX, indicating that the cAMP pathway plays an important role in the regulation of melanogenesis (Wong and Pawelek, 1975; Abdel-Malek et al., 1987; Jiménez et al., 1988; Hunt et al., 1993; Hunt et al., 1994; Englaro et al., 1995). Besides its role in melanocyte survival and tissue-specific expression of the melanogenic genes, microphthalmia is also involved in the acute regulation of the tyrosinase gene during cAMP-induced melanogenesis. Indeed, using different construction of the tyrosinase promoter in a luciferase reporter system, we showed that the M-box plays a key role in the cAMP responsiveness of the tyrosinase promoter. Moreover, gel shift experiments showed that forskolin increases microphthalmia binding to the M-box (Bertolotto et al., 1996) resulting from an increased microphthalmia expression (Bertolotto et al., 1998). Taken together, these data led us to propose that microphthalmia could mediate the effect of cAMP on tyrosinase gene expression. However, it remained necessary to elucidate the mechanism by which cAMP upregulates microphthalmia expression, and to prove that microphthalmia mediates the effect of cAMP on the tyrosinase gene.
In this report, we clearly demonstrate that cAMP elevating agents increase the level of microphthalmia mRNA and protein. Furthermore, using a reporter plasmid containing a 2.1-kb fragment 5′ of the transcriptional start site of the microphthalmia promoter, we showed that cAMP-elevating agents and protein kinase A (PKA) stimulate the transcriptional activity of the promoter. Our analysis of the promoter region revealed that responsiveness to cAMP is mediated through a CRE consensus motif located between −140 and −147 bp from the initiation site. Finally, we showed that a dominant-negative form of microphthalmia, lacking the NH2-terminal activation domain, impaired the stimulation of the tyrosinase promoter activity induced by cAMP-elevating agents, providing evidence that microphthalmia activity is required for cAMP-induced stimulation of the tyrosinase promoter. Therefore, we conclude that cAMP increases microphthalmia expression, which in turn binds to the tyrosinase promoter M-box, thereby leading to the stimulation of tyrosinase expression and melanin synthesis.
Materials and Methods
Materials
Forskolin, α-melanocyte–stimulating hormone ([Nle4, D-Phe7]-α-MSH), hydrocortisone, insulin, phorbol 12-myristate 13-acetate (PMA), p-nitrophenyl phosphate (pNPP), NP-40, sodium fluoride, sodium orthovanadate, β-glycerophosphate, 4-(2-aminoethyl)-benzene-sulfonyl fluoride (AEBSF), aprotinin, and leupeptin were purchased from Sigma Chemical Co. (St. Louis, MO). Klenow fragment and T4 DNA ligase were from Biolabs (Beverly, MA). The PGL2-basic vector and the basic (b)-FGF were from Promega. DME, trypsin, and lipofectamine reagent were from GIBCO BRL (Gaithersburg, MD), and FCS was from Hyclone Laboratories Inc. (Logan, UT). CREB-1 bZIP (254–327) peptide was from Santa Cruz Biotechnology (Santa Cruz, CA). The C5 mAb was raised against a histidine fusion protein expressed from the NH2-terminal Taq-Sac fragment of human MITF cDNA and produces a specific gel mobility supershift with microphthalmia, but not with the related proteins TFE3, TFEB, and TFEC (Weilbaecher et al., 1998). Peroxidase- and FITC-conjugated anti–mouse antibodies were from Dakopatts (Glostrup, Denmark). Synthetic oligonucleotides were from Oligo (Paris, France) express.
Cell Cultures
B16/F10 murine melanoma cells and NIH3T3 fibroblast cells were grown at 37°C under 5% CO2 in DME supplemented with 7% FCS and penicillin/streptomycin (100 U/ml:50 μg/ml). Epidermal cell suspensions were obtained from foreskins of Caucasoid children by overnight digestion in PBS containing 0.5% dispase grade II at 4°C, followed by a 1-h digestion with trypsin/EDTA solution (0.05%:0.02% in PBS) at 37°C. Cells were grown in MCDB 153 medium supplemented with FCS 2%, 0.4 μg/ml hydrocortisone, 5 μg/ml insulin, 16 nM PMA, 1 ng/ml b-FGF, and penicillin/ streptomycin (100 U/ml:50 μg/ml) in a humidified atmosphere containing 5% CO2 at 37°C.
Western Blot Assays
B16 mouse melanoma cells were grown in six-well dishes with 20 μM forskolin or 1 μM α-MSH for the times indicated on the figure legends. Normal human melanocytes were grown in six-well dishes for 5 d in MCDB 153 supplemented with 5% FCS, 0.4 μg/ml hydrocortisone, and 5 μg/ml insulin before stimulation. Then, cells were treated with 20 μM forskolin or 1 μM α-MSH for the indicated times. Cells were then lysed in radioimmunoprecitpitation assay (RIPA) buffer, pH 7.5, containing 10 mM Tris-HCl, 1% sodium deoxycholate, 1% NP-40, 150 mM NaCl, 0.1% SDS, 5 μg/ml leupeptin, 1 mM AEBSF, 100 IU/ml aprotinin, 1 mM NaVO4, 5 mM NaF, 20 mM β-glycerophosphate, and 10 mM pNPP. Proteins (20 μg) were separated on 7.5% SDS–polyacrylamide gels and transferred to a nitrocellulose membrane. Microphthalmia protein was detected with the C5 mAb at a 1/10 dilution in saturation buffer and with a secondary peroxidase-conjugated anti–mouse antibody at a 1/3,000 dilution. Protein was visualized with the enhanced chemiluminescence (ECL) system from Amersham Life Sciences, Inc. (Arlington Heights, IL).
Northern Blot Analysis
poly(A)+ RNA were isolated from control and forskolin-treated B16 melanoma cells using the mRNA purification kit from Qiagen Inc. (Valencia, CA) (Oligotex™, mRNA). RNA was denatured 5 min at 65°C in a formamide/formaldehyde mixture, separated by electrophoresis in a 1% agarose-2.2 M formaldehyde gel, transferred to a nylon membrane (Hybond N.; Amersham Life Sciences, Inc.) in 20× SSPE, pH 7.7 (3.5 M NaCl, 0.2 M sodium phosphate, 0.02 M Na2EDTA), and then hybridized to microphthalmia and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe labeling by random priming with α-[32P]dCTP (Amersham Life Sciences, Inc.). The microphthalmia probe was the 1.3-kb HindIII–NotI fragment containing the full open reading frame of microphthalmia (Bertolotto et al., 1996).
Construction of the Reporter Plasmids
A 2.1-kb fragment 5′ of the transcriptional start site of the MITF gene was isolated from SpMITF 1, kindly provided by Dr. Shibahara (Tohoku University School, Sendaï, Japan; Fuse et al., 1996). The 2.1-kb BamHI–BanI (converted to blunt end) fragment was isolated from pBluescript IISK+ SpMITF 1 and subcloned into the unique BglII (compatible with BamHI)/ HindIII (converted to blunt end) restriction site of the pGL2 basic vector (pGL2B), upstream of the luciferase coding sequence (pMI: −2135/+136). Cohesive ends were filled in with Klenow fragment. The transcription initiation site was numbered +1 according to the report of Fuse et al. (1996). For the deletion constructs, pMI was first linearized with AvrII, MscI, AflII, or PstI located in the MITF promoter and filled in with Klenow fragment. The plasmids were then digested by the unique SmaI site of the pGL2B and self-ligated with T4 DNA ligase, giving, respectively, the following plasmids: pMIΔ1 (−1,812/+136), pMIΔ2 (−1,581/+136), pMIΔ3 (−680/+136), pMIΔ4 (−387/+136). pMIΔ5 and pMImCRE were constructed with the Transformer™ site-directed mutagenesis kit (CLONTECH Laboratories, Inc., Palo Alto, CA). Initially, we introduced a NheI site (5′-GATATCAACATTTAAGACC-GCTAGCAAACTCGTAGGGCTTCC-3′) just below the CRE sequence located between −147 and −140 bp upstream from the initiation start site of the microphthalmia gene in pMI. pMI was then linearized with NheI, filled in with Klenow fragment, digested by SmaI, and then the plasmid was self-ligated with T4 DNA ligase to give pMIΔ5 (−103/+136). In pMImCRE, the CRE site was mutated with an oligonucleotide that changes the TGACGTCA sequence in TGGGGTCA (5′-GAAAAAAAGCATGGGGTCAAGCCAGGGG-3′). The minimal thymidine kinase promoter (−80/+60) was cloned upstream the luciferase coding sequence in the NheI–BglII sites of pGL2B (pTK). Double-stranded oligonucleotides corresponding to the microphthalmia sequence −136/−153 and containing the CRE motif were then cloned into the SmaI–SacI sites of the pTK plasmid (pTKCRE). pTKmCRE corresponds to the pTKCRE in which the CRE motif TGACGTCA was converted to TGGGGTCA.
The reporter plasmid containing the 2.2-kb fragment of the mouse tyrosinase promoter (pTyro; −2,236/+59) as well as the cloning of the microphthalmia coding sequence (Mi) in pCDNA3 expression vector were described in our previous report (Bertolotto et al., 1996). Mi-ΔNT containing an inframe deletion extending between amino acids 11 and 704 that delete the NH2-terminal domain was obtained from pCDNA3Mi by mutagenesis with the following oligonucleotide (5′-GCTGGAAATGCTAGAATACGCAAGAGCATTGGCTAA-3′).
Transfections and Luciferase Assays
B16 melanoma cells and NIH3T3 fibroblast cells were seeded in 24-well dishes and transient transfections were performed the following day using 2 μl of lipofectamine and 0.5 μg of total plasmid DNA in a 200 μl final volume as indicated in figure legends. pCMVβGAL was transfected with the test plasmids to control the variability in transfection efficiency. 24 h after transfection, soluble extracts were harvested in 50 μl of lysis buffer and assayed for luciferase and β-galactosidase activities. In our conditions, α-MSH, forskolin (Fsk), and PKA increase slightly pCMVβGAL expression (1.5-, 1.5- and 1.8-fold, respectively). All transfections were repeated at least five times using different plasmid preparations.
In Vitro Transcription/Translation
In vitro translation of microphthalmia, Mi-ΔNT and microphthalmia mixed with Mi-ΔNT were carried out using the linked T7 transcription/ translation system from Amersham Life Science, Inc. Microphthalmia and Mi-ΔNT translation were monitored using [35S]methionine (Amersham life Science, Inc.) and SDS-PAGE analysis.
Gel Mobility Shift Assay
Double-stranded synthetic M-box (5′-GAAAAAGTTAGTCATGTGCTTTGCAGAAGA-3′), CRE (5′-AAAGCATGACGTCAAGCCAG-3′) or mutated CRE (mCRE) (5′-AAAGCATGGGGTCAAGCCAG-3′) were end labeled with γ-[32P]ATP (Amersham Life Sciences, Inc.) and T4 polynucleotide kinase. In vitro–translated proteins (microphthalmia, Mi-ΔNT, microphthalmia, plus Mi-ΔNT), CREB-1 bZIP (254–327), or nuclear extracts prepared as previously described (Bertolotto et al., 1998), were preincubated in binding buffer containing 10 mM Tris, pH 7.5, 100 mM NaCl, 1 mM DTT, 1 mM EDTA, 4% glycerol, 80 μg/ml of salmon sperm DNA, 0.1 μg poly (dIdC), 10% FCS, 2 mM MgCl2, and 2 mM spermidine for 15 min on ice. Then, 30,000–50,000 cpm of 32P-labeled probe were added to the binding reaction for 10 min at room temperature. DNA–protein complexes were resolved by electrophoresis on 5.5% polyacrylamide gel (37.5:1; acrylamide/bisacrylamide) in TBE buffer (22.5 mM Tris-borate, 0.5 mM EDTA, pH 8) for 2 h at 150 V.
Results
Stimulation of Microphthalmia Expression by cAMP-elevating Agents in B16 Melanoma Cells and in Normal Human Melanocytes
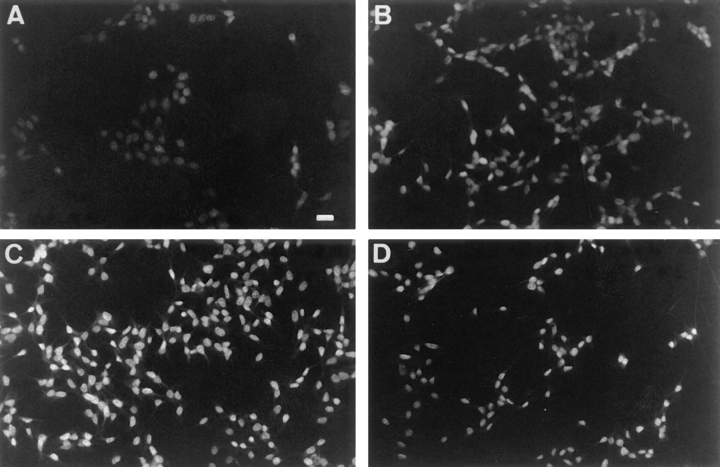
First, we wished to assess short-term effects of Fsk on microphthalmia expression. In basal conditions, immunofluorescence studies showed, a weak nuclear labeling with an anti-microphthalmia mAb (Weilbaecher et al., 1998) (Fig. 1). Interestingly, when B16 melanoma cells were incubated 2 h with Fsk, the nuclear labeling was increased and the maximal effect was obtained after 4 h. Then, after 24 h with Fsk, the nuclear labeling decreased but remained clearly above the basal level.
Figure 1.
Forskolin treatment stimulates microphthalmia gene expression in B16 mouse melanoma cells. Immunofluorescence labeling was performed with the anti-microphthalmia mAb. Unstimulated B16 mouse melanoma cells (A), B16 mouse melanoma cells stimulated with 20 μM forskolin for 3 h (B), 5 h (C), and 24 h (D). Bar, 10 μm.
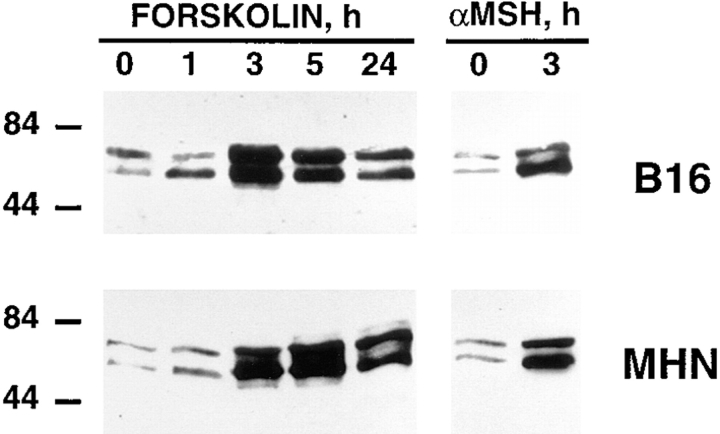
Next, we carried out Western blot experiments with the anti-microphthalmia mAb in both B16 melanoma cells (Fig. 2, top panel) and normal human melanocytes (Fig. 2, bottom panel). Similar results were obtained for both cell types. In basal conditions, microphthalmia appeared as a doublet of 55 and 60 kD that have been shown to represent a MAP kinase–specific phosphorylation of microphthalmia, based on two-dimensional tryptic mapping (Hemesath et al., 1998). After 3 h, Fsk markedly increased the level of microphthalmia protein. Maximal expression of microphthalmia with Fsk treatment was observed between 3 and 5 h, and then decreased after 24 h consistently with our findings from the immunofluorescence study. Additionally, the physiological melanogenic agent α-MSH also induced a strong stimulation of microphthalmia expression after 3 h in both B16 melanoma cells (Fig. 2, top panel) and normal melanocytes (bottom panel) thereby demonstrating that cAMP-elevating agents lead to a rapid induction of microphthalmia expression in both cell types.
Figure 2.
cAMP-elevating agents increases microphthalmia expression in B16 mouse melanoma cells and in normal human melanocytes. 20 μg of proteins from B16 mouse melanoma cells (top panel) or from normal human melanocytes (bottom panel), non-stimulated or treated for the indicated times with 20 μM forskolin or 1 μM α-MSH, were subjected to Western blot analysis using the anti-microphthalmia mAb. Molecular masses, indicated on the left, are expressed in kD.
Regulation of Microphthalmia Expression in B16 Melanoma Cells Involves a Transcriptional Mechanism
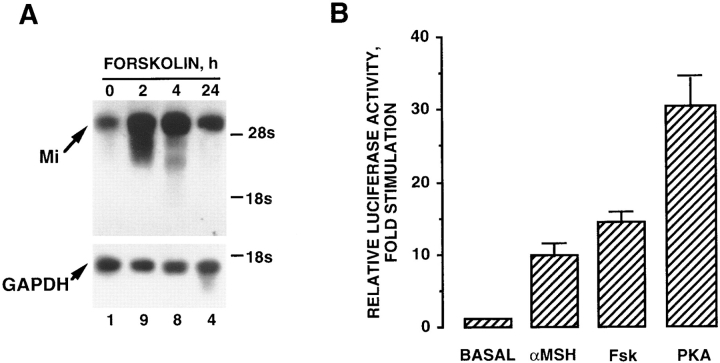
To investigate the mechanism by which Fsk increases microphthalmia expression, we first performed a Northern blot experiment. As shown in Fig. 3 A, top panel, incubation for 2 or 4 h with Fsk induced a strong increase in microphthalmia mRNA level (ninefold and eightfold, respectively). Then, the amount of microphthalmia transcripts decreased after 24 h with Fsk. The loading of each lane was controlled with a GAPDH probe (Fig. 3 A, bottom panel). Next, we wished to investigate the effect of cAMP on the transcriptional activity of the microphthalmia gene promoter. To accomplish this aim, we studied the effect of Fsk on a reporter plasmid containing a 2.1-kb fragment of the microphthalmia promoter upstream of the luciferase coding sequence (pMI). pMI was transiently transfected in B16 melanoma cells and exposed to cAMP-elevating agents for 12 h (Fig. 3 B). α-MSH and Fsk treatment caused a 7-fold and a 15-fold stimulation, respectively, of the luciferase activity of pMI. A large stimulation of the luciferase activity was obtained when pMI was cotransfected with an expression vector encoding the catalytic subunit of PKA (30-fold). Taken together, these results demonstrate that cAMP elevating agents regulate microphthalmia expression through a transcriptional mechanism and show the presence in this part of the promoter of cis-acting elements accountable for the cAMP sensitivity of the microphthalmia promoter.
Figure 3.
cAMP-elevating agents regulate microphthalmia expression through a transcriptional mechanism. (A) Northern blot analysis for microphthalmia and GAPDH mRNA transcripts from B16 cells exposed for the indicated times to 20 μM forskolin. (B) B16 cells were transfected with 0.35 μg of pMI, 0.1 μg of empty pCDNA3, and 0.05 μg of pCMVβ′-GAL. Then, cells were treated for 12 h with 20 μM forskolin or 1 μM α-MSH. To study the effects of PKA expression, B16 cells were transfected with 0.35 μg of pMI, 0.1 μg of pCDNA3 encoding PKA, and 0.05 μg of pCMVβGAL. Luciferase activity was normalized by the β-galactosidase activity and the results were expressed as fold stimulation of the basal luciferase activity from unstimulated cells. Data are means ± SE of five experiments performed in triplicate.
Localization of cis-Regulatory Elements Involved in the cAMP Response of the Microphthalmia Promoter
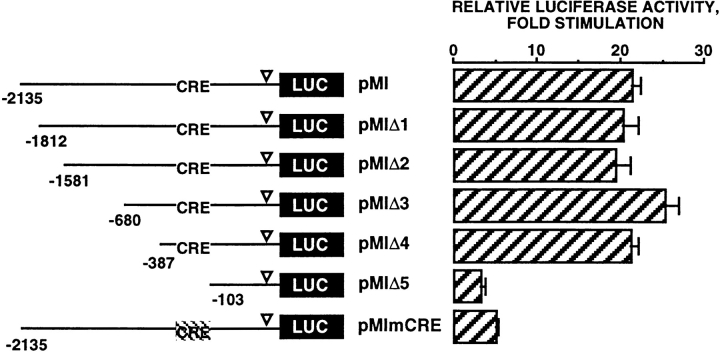
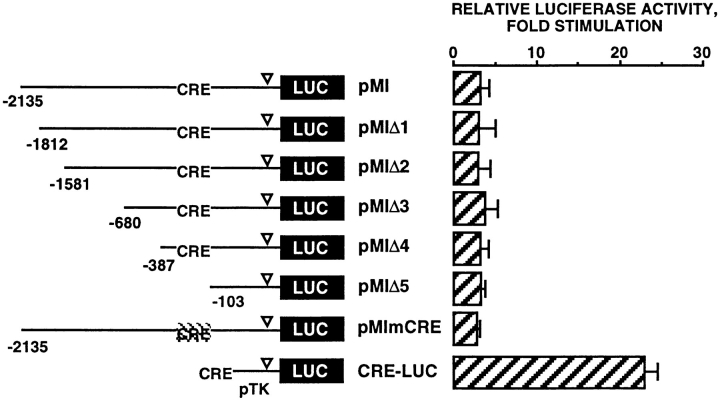
The next series of experiments were undertaken to identify the elements responsible for the cAMP sensitivity of the microphthalmia promoter. Hence, we studied the effects of Fsk on the transcriptional activity of reporter plasmids containing various deletions in the 5′ flanking region of the promoter. Luciferase activity of pMI, pMIΔ1 (−1,812/+136), pMIΔ2 (−1,581/+136), pMIΔ3 (−680/ +136), and pMIΔ4 (−387/+136) was stimulated ∼20-fold by Fsk while in the same condition, luciferase activity of pMIΔ5 (−103/+136) was only stimulated threefold (Fig. 4). Interestingly, the 2.1-kb fragment of the microphthalmia promoter contained a CRE consensus sequence (TGACGTCA) located just upstream of the pMIΔ5 deletion. Mutation of the CRE motif in pMI (pMImCRE) markedly reduced the stimulation of luciferase activity (fivefold) in response to Fsk, thus demonstrating the key role of this CRE motif in the cAMP response of the pMI.
Figure 4.
The effect of forskolin on microphthalmia promoter activity is mediated by a consensus CRE motif (−147/−140). B16 cells were transfected with 0.45 μg of pMI, pMIΔ1, pMIΔ2, pMIΔ3, pMIΔ4, pMIΔ5, or pMImCRE and 0.05 μg of pCMVβGAL. After 12 h with forskolin, luciferase activity was normalized by the β-galactosidase activity and the results were expressed as fold stimulation of the basal luciferase activity from unstimulated cells. Data are means ± SE of five experiments performed in triplicate. ▿ indicates the TATA box position.
Gel shift assay, using labeled probe corresponding to the CRE motif of the microphthalmia promoter showed that purified CREB-1 bZIP bound specifically to this CRE motif (Fig. 5 A). Furthermore, we also observed a complex between B16 nuclear extracts and the CRE probe (Fig. 5 B). This complex was totally displaced by an excess of unlabeled homologous CRE oligonucleotide and was shifted by an anti-CREB antibody. On the other hand, no binding was observed between B16 nuclear extracts and the mCRE probe in which the CRE was mutated. Additionally, transfection experiments showed that the presence of the microphthalmia promoter CRE motif strongly increased the cAMP sensitivity of the minimal thymidine kinase promoter (5-fold for pTK vs. 27-fold for pTKCRE) while no increase was observed when the CRE was mutated (4-fold for pTKmCRE) (Fig. 5 C).
Figure 5.
The CRE motif (−147/−140) binds CREB and confers cAMP sensitivity to the minimal thymidine kinase promoter. (A) Gel shift experiments were performed with CREB-1 bZIP (254–327) using a labeled oligonucleotide containing the CRE site of the microphthalmia promoter in control conditions or in presence of an excess (50-fold) of unlabeled homologous oligonucleotide. (B) B16 nuclear extracts from control (−) or forskolin-treated cells (+) were incubated with the same labeled CRE probe or with a labeled oligonucleotide in which the CRE site was mutated (mCRE). When indicated, reactions were carried out in the presence of an excess (50-fold) of unlabeled homologous CRE probe or with 0.6 μl of an anti-CREB antibody. (C) B16 cells were transfected with 0.35 μg of pTK, pTKCRE, or pTKmCRE and 0.05 μg of pCMVβGAL. After 12 h with forskolin, luciferase activity was normalized by the β-galactosidase activity and the results were expressed as fold stimulation of the basal luciferase activity from unstimulated cells. Data are means ± SE of three experiments performed in triplicate.
These results clearly show that the CRE consensus motif, located between −147 and −140 bp upstream of the initiation start site, plays a pivotal role in the cAMP responsiveness of the microphthalmia promoter. This CRE motif binds transcription factors of CREB family and is sufficient to confer cAMP responsiveness to a minimal promoter.
Stimulation of the Microphthalmia Promoter Activity by cAMP-elevating Agents Is Restricted to B16 Melanoma Cells
The 2.1-kb fragment 5′ of the transcriptional start site of the microphthalmia gene promoter was recently shown to be preferentially expressed in pigment cells (Fuse et al., 1996). However, identification of a classical CRE consensus motif as the major cis-regulatory element in the cAMP sensitivity of the microphthalmia promoter in B16 melanoma cells prompted us to assay the cAMP response of pMI in NIH3T3 fibroblast cells. In NIH3T3 cells, luciferase activity of pMI as well as the reporter plasmids containing deletions or mutation was not stimulated by Fsk treatment (Fig. 6). In contrast, in the same condition, a CRE-reporter plasmid used as control was stimulated 24-fold by Fsk. Thus, the CRE (−140/−147) of the microphthalmia promoter is not functional in NIH3T3 cells, suggesting the existence of a cell-specific mechanism that makes the microphthalmia promoter responsive to cAMP in B16 melanoma cells.
Figure 6.
The CRE motif do not confer cAMP sensitivity to microphthalmia promoter in NIH3T3 fibroblast cells. NIH3T3 fibroblast cells were transfected with 0.45 μg of pMI, pMIΔ1, pMIΔ2, pMIΔ3, pMIΔ4, pMIΔ5, pMImCRE, or CRE-LUC and 0.05 μg of pCMVβGAL. After 12 h with forskolin, luciferase activity was normalized by the β-galactosidase activity and the results were expressed as fold stimulation of the basal luciferase activity from unstimulated cells. Data are means ± SE of five experiments performed in triplicate. ▿ indicates the TATA box position.
Stimulation of the Tyrosinase Promoter Activity by cAMP-elevating Agents Is Mediated by Microphthalmia
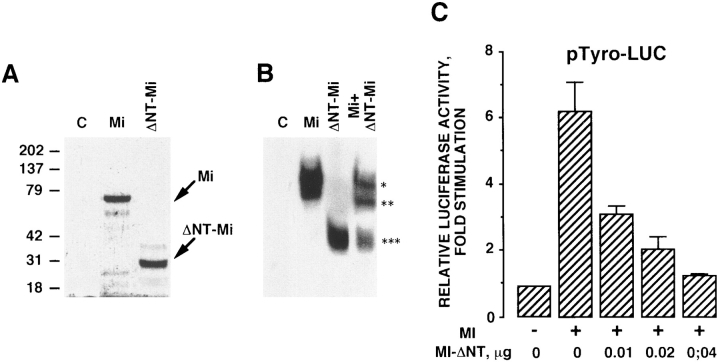
To demonstrate that microphthalmia is involved in the stimulation of the tyrosinase promoter activity by cAMP, we constructed a dominant-negative form of microphthalmia, lacking the NH2-terminal transactivation domain (Mi-ΔNT). First, we carried out in vitro transcription/ translation of Mi-ΔNT to verify the expression of the truncated protein using the intact microphthalmia as control. In vitro–translated microphthalmia (Mi) gave rise to a single band ∼60 kD and Mi-ΔNT product was at 30 kD (Fig. 7 A). Next, we performed band shift assay using M-box as probe. Microphthalmia and Mi-ΔNT formed a complex with the M-box. According to their respective molecular weight, Mi complexes migrated slower compared with the Mi-ΔNT complexes. When microphthalmia and Mi-ΔNT were mixed in transcription/translation reaction, we observed an intermediate complex corresponding to an heterodimer Mi/Mi-ΔNT bound to the M-box (Fig. 7 B). Then, transfection experiments were undertaken to study whether Mi-ΔNT exerted a dominant-negative effect on wild-type microphthalmia. The luciferase activity of the tyrosinase promoter, pMT2.2, was stimulated sixfold by transfection with 0.01 μg of an expression vector encoding microphthalmia. Cotransfection with Mi-ΔNT dose-dependently decreased the activation of the tyrosinase promoter induced by microphthalmia (Fig. 7 C), demonstrating that this mutant inhibits the activity of microphthalmia.
Figure 7.
Mi-ΔNT, a form of microphthalmia lacking the NH2-terminal transactivation domain, exerts dominant-negative effects on the wild-type microphthalmia. (A) Microphthalmia and Mi-ΔNT were in vitro translated in the presence of [35S]methionine, and then analyzed by gel electrophoresis. Molecular masses, indicated on the left, are expressed in kD. (B) For gel shift assay, 2 μl of in vitro–translated microphthalmia, Mi-ΔNT, or microphthalmia plus Mi-ΔNT were incubated with labeled tyrosinase M-box. *, Wild-type microphthalmia homodimer; ***, Mi-ΔNT homodimer; and **, microphthalmia plus Mi-ΔNT heterodimer. Autoradiogram was exposed overnight at −80°C. (C) B16 cells were transfected with 0.4 μg of pTyro, 0.01 μg of pCDNA3 encoding microphthalmia, 0.05 μg of pCMVβGAL, and different amounts of pCDNA3 encoding Mi-ΔNT. After 24 h, luciferase activity was normalized by the β-galactosidase activity and the results were expressed as fold stimulation of the basal luciferase activity from unstimulated cells. Data are means ± SE of five experiments performed in triplicate.
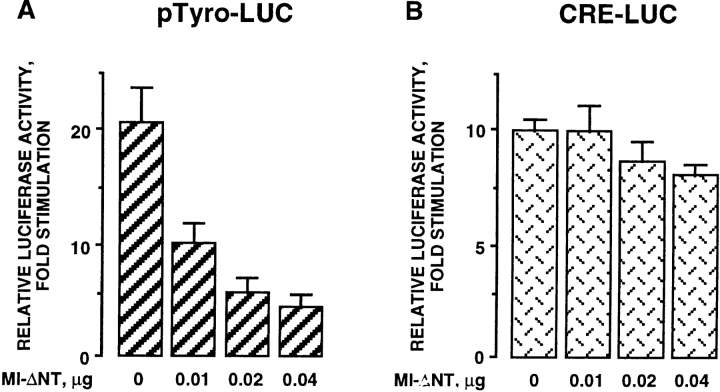
Finally, to study the role of microphthalmia in the cAMP responsiveness of the tyrosinase promoter (pMT2.2), we cotransfected pMT2.2 with increasing amounts of Mi-ΔNT. Mi-ΔNT dose-dependently inhibited the stimulation of the tyrosinase promoter activity induced by a Fsk treatment (Fig. 8 A) while Mi-ΔNT did not significantly impair the cAMP response of a CRE reporter plasmid (Fig. 8 B). The same doses of empty plasmid did not affect the stimulation of the tyrosinase promoter activity by Fsk. These results indicate that microphthalmia activity is required for the induction of tyrosinase promoter activity by cAMP.
Figure 8.
Regulation of tyrosinase promoter activity by forskolin is mediated by microphthalmia. B16 cells were transfected with (A) 0.4 μg of pTyro or (B) 0.4 μg of a CRE-LUC reporter plasmid, 0.05 μg of pCMVβGAL, and different amounts of pCDNA3 encoding Mi-ΔNT. After 24 h with forskolin, luciferase activity was normalized by the β-galactosidase activity and the results were expressed as fold stimulation of the basal luciferase activity from unstimulated cells. Data are means ± SE of five experiments performed in triplicate.
Discussion
To thoroughly dissect the mechanism of cAMP-induced melanogenesis and melanocyte differentiation, we first assessed the early effect of cAMP on the expression of microphthalmia, which has been proposed to play a pivotal role in these processes. Immunofluorescence and Western blot experiments with anti-microphthalmia antibody show that cAMP elevating agents lead to a rapid, robust, but transient increase in microphthalmia protein levels in B16 melanoma cells. Using Western blot analysis, the maximal effect of Fsk was observed between 3 and 5 h. After 24 h, we observed a marked decrease in microphthalmia expression indicating that cAMP has a transient effect on microphthalmia expression. We previously reported that Fsk did not increase microphthalmia expression (Bertolotto et al., 1996), but this observation was made after 48 h of Fsk treatment. At that time, the effect of Fsk is markedly reduced compared with shorter exposure times. Furthermore, it should be noted that in the present report, we used an mAb, whereas our previous observations were made with a polyclonal antibody that showed a stronger non-specific labeling. Thus, the time of stimulation with Fsk and the use of different antibodies easily explain the results obtained in the present report. Fsk treatment also increases microphthalmia expression in normal human melanocytes and the same effect can be obtained with a physiological cAMP-elevating agent, α-MSH, that induces pigmentation in humans when subcutaneously injected (Levine et al., 1991). These data suggest that upregulation of microphthalmia expression is a common regulatory step in response to cAMP-elevating agents in both B16 melanoma cells and normal human melanocytes, thereby demonstrating the physiological relevance of our observations. Fsk increases the amount of microphthalmia messengers and the activity of microphthalmia promoter is stimulated by α-MSH, Fsk, and co-expression of the catalytic subunit of PKA. Furthermore, deletions and mutations in the microphthalmia promoter demonstrated that cAMP effects are mediated through a CRE consensus motif located between −147 and −140 bp from the transcription start site. This CRE motif binds CREB family proteins and confers cAMP sensitivity to the minimal thymidine kinase promoter. However, in NIH3T3 fibroblast cells, Fsk does not stimulate the microphthalmia promoter activity. The lack of cAMP responsiveness of the microphthalmia promoter in NIH3T3 cells cannot be explained by a dysfunctional cAMP pathway component such as PKA, CREB or CREB binding protein (CBP), since a construct containing a somatostatin CRE upstream of the minimal thymidine kinase promoter is activated by Fsk in these cells. Thus, it appears that the CRE in the microphthalmia promoter is turned off in fibroblasts while it is made functional in B16 melanoma cells resulting from a cell-specific mechanism that remains to be elucidated. Taken together, our results demonstrate that cAMP, through a classical CRE, stimulates microphthalmia gene transcription thereby leading to an increase in microphthalmia expression. Since microphthalmia has been shown to strongly transactivate the tyrosinase promoter, it was tempting to propose that upregulation of tyrosinase amount by cAMP is the direct consequence of the stimulation of microphthalmia expression. To verify this hypothesis we constructed a mutated microphthalmia (Mi-ΔNT) containing a large deletion in its NH2 terminus part that removes the transcriptional activation domain. A similar naturally occurring mutation, classified as a semidominant mutation, has been found in the microphthalmia gene of the white spot mice (miws), as the consequence of an intragenic deletion of exon 2, 3, and 4 (Hodgkinson et al., 1993; Moore, 1995). Mi-ΔNT inhibited the transactivation of the tyrosinase promoter induced by the wild-type microphthalmia, demonstrating that Mi-ΔNT functions as a dominant-negative mutant. The effect of Mi-ΔNT can be explained both by the binding of Mi-ΔNT/Mi-ΔNT homodimer to the M-box that would prevent the binding of wild-type microphthalmia and by the dimerization of Mi-ΔNT with the wild-type microphthalmia resulting in a non-functional heterodimer.
Interestingly, Mi-ΔNT inhibits the effect of cAMP on the tyrosinase promoter activity, but affects neither the cAMP responsiveness of a construct containing one somatostatin CRE nor that of the microphthalmia promoter (data not shown). In agreement with our former hypothesis, we demonstrate that microphthalmia activity is required for the induction of the tyrosinase gene expression by cAMP. Noteworthy, upregulation of tyrosinase messengers cannot be observed before 6 h of treatment with cAMP elevating agents (Bertolotto et al., 1996; Rungta et al., 1996) while generally, the effect of cAMP on gene transcription is much more rapid (1–2 h). Hence, it has been thought for a long time that the effect of cAMP on tyrosinase expression was due to tyrosinase messenger stabilization (Ganss et al., 1994) rather than an increase in tyrosinase gene transcription. However, the delayed effect of cAMP on tyrosinase messenger can be explained by a two-step mechanism that requires first, the accumulation of microphthalmia then, an increase in tyrosinase gene transcription.
The cAMP responsiveness of the microphthalmia promoter implies a classical CRE that binds transcription factors of CREB family. Another noteworthy observation is that CREB-mediated regulation of transcription requires an interaction of phosphorylated CREB with the transcriptional coactivator CBP (Chrivias et al., 1993; Arias et al., 1994; Kwok et al., 1994). Interestingly, expression of adenovirus E1A oncoprotein in melanocytes abrogates pigmentation and expression of melanocyte-specific genes including tyrosinase, TRP1, TRP2, and microphthalmia (Yavuzer et al., 1995; Halaban et al., 1996). This observation could be explained by the binding of E1A to CBP thereby leading to the inhibition of CREB function as previously described (Arany et al., 1995). Further, microphthalmia has been recently reported to interact with CBP through the CBP2 domain that also binds E1A (Bannister and Kouzarides, 1995; Sato et al., 1997). Thus, E1A could prevent microphthalmia and CBP interaction and reduce the transactivation of the tyrosinase promoter by microphthalmia. These two mechanisms could participate in the extinction of the pigmented phenotype of melanocytes by E1A protein or by another oncoprotein such as SV40 large T antigen (Dooley et al., 1988; Larue et al., 1993; Orlow et al., 1995). However, extinction of the pigmented phenotype could involve other pathways, since the interaction of E1A with CBP seems to be dispensable for the effect of E1A (Halaban et al., 1996).
Microphthalmia is also known to play a key role in the development of the melanocyte lineage and recent observations showed that microphthalmia may convert fibroblasts to cells with melanocyte features (Tachibana et al., 1996). This protein is detected very early in the roof of the neural crest and precedes the expression of the melanocyte markers, such as TRP2, TRP1, and tyrosinase (Goding and Fisher, 1997; Opdecamp et al., 1997). The mechanism that triggers microphthalmia upregulation during embryogenesis has not been elucidated. It thought that microenvironmental factors could induce microphthalmia expression in stem cells of the melanocytes lineage (Lahav et al., 1996). That these microenvironmental factors act through the cAMP pathway to upregulate microphthalmia expression is a fascinating hypothesis that remains to be explored.
In the present report we have gathered compelling evidence demonstrating that cAMP-elevating agents, through PKA activation, increase microphthalmia expression, which in turn stimulates tyrosinase gene expression, thereby leading to an increase in melanin pigment production. These findings, demonstrating that microphthalmia acts as a signal transducer in cAMP-induced melanocyte differentiation, bring information of paramount importance on the molecular signaling pathway that controls melanogenesis and pigment cell differentiation.
Acknowledgments
We thank Dr. S. Shibahara for providing the plasmid SpMITF1 containing a fragment 5′ of the transcriptional start site of the microphthalmia-associated transcription factor gene. We also thank Dr. P. Sassone-Corsi and Dr. E. Lalli (Insititut de Genétique et de Biologie Moléculaire et Cellulaire, Illkirch, France) for providing the expression vector encoding the catalytic subunit of the PKA. We are grateful to A. Grima and C. Minghelli for illustration work, and to C. Sable (all from INSERM, Nice, France) for critical reading of the manuscript.
This work was supported by Association pour la Recherche sur le Cancer (grant 9402) and Ligue Nationale Contre le Cancer. D.E. Fisher is supported by National Institutes of Health grant AR43369 and is a Pew Foundation Scholar and a James S. McDonnall Fellow.
Abbreviations used in this paper
- α-MSH
α-melanocyte–stimulating hormone
- b-HLH
basic-helix-loop-helix
- CREB
cAMP response element binding protein
- CBP
CREB binding protein
- Fsk
forskolin
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- Mi
microphthalmia
- MITF
microphthalmia-associated transcription factor
- PKA
protein kinase A
- pNPP
paranitro-phenylphosphate
- TRP1 and TRP2
tyrosinase-related proteins 1 and 2
Footnotes
Address all correspondence to R. Ballotti, Institut National de la Sante et de la Recherche Medicale U385, Biologie et Physiopathologie de la Peau, Faculté de Médecine, Avenue de Valombrose, Paris, France. Tel.: (33) 4 93 37 77 90. Fax: (33) 4 93 81 14 04. E-mail: ballotti@unice.fr
References
- Abdel-Malek A, Swope BV, Amornsiripanitch N, Nordlund JJ. In vitro modulation of proliferation and melanization of S91 melanoma cells by prostaglandins. Cancer Res. 1987;47:3141–3146. [PubMed] [Google Scholar]
- Arany Z, Newsome D, Olread D, Livingston DM, Eckner R. A family of transcriptional adaptator proteins targeted by E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- Arias J, Alberts AS, Brindle P, Claret FX, Smeal T, Karim M, Ferasmisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a commun nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. CBP-induces stimulation of c-fos activity is abrogates by E1A. EMBO (Eur Mol Biol Organ) J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber JI, Townsend D, Olds DP, King RA. Dopachrome oxydoreductase: a new enzyme in the pigment pathway. J Invest Dermatol. 1984;83:145–149. doi: 10.1111/1523-1747.ep12263381. [DOI] [PubMed] [Google Scholar]
- Bentley NJ, Eisen T, Goding CR. Melanocyte-specific expression of the human tyrosinase promoter: Activation by the Microphthalmia gene product and role of the initiator. Mol Cell Biol. 1994;14:7996–8006. doi: 10.1128/mcb.14.12.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotto C, Bille K, Ortonne JP, Ballotti R. Regulation of tyrosinase gene expression by cAMP in B16 melanoma cells involves two CATGTG motifs surrounding the TATA box: Implication of the microphthalmia gene product. J Cell Biol. 1996;134:747–755. doi: 10.1083/jcb.134.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotto C, Buscà R, Abbe P, Bille K, Aberdam E, Ortonne JP, Ballotti R. Different cis-acting elements are involved in the regulation of TRP1 and TRP2 promoter activities by cyclic AMP: pivotal role of M boxes(GTCATGTGCT) and of microphthalmia. Mol Cell Biol. 1998;18:694–702. doi: 10.1128/mcb.18.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrivias JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Dooley TP, Wilson RE, Jones NC, Hart IR. Polyoma middle T abrogates TPA requirements of murine melanocytes and induces malignant melanoma. Oncogene. 1988;3:531–535. [PubMed] [Google Scholar]
- Englaro W, Rezzonico R, Durand-Clément M, Lallemand D, Ortonne JP, Ballotti R. Mitogen-activated protein kinase pathway and AP–1 are activated during cAMP-induced melanogenesis in B-16 melanoma cells. J Biol Chem. 1995;270:24315–24320. doi: 10.1074/jbc.270.41.24315. [DOI] [PubMed] [Google Scholar]
- Fuse N, Yasumoto KI, Suzuki H, Takahashi K, Shibahara S. Identification of a melanocyte-type promoter of the microphthalmia-associated transcription factor gene. Biochem Biophys Res Commun. 1996;219:702–707. doi: 10.1006/bbrc.1996.0298. [DOI] [PubMed] [Google Scholar]
- Ganss R, Schutz G, Beermann F. The mouse tyrosinase gene. J Biol Chem. 1994;269:29808–29816. [PubMed] [Google Scholar]
- Goding CR, Fisher DE. Regulation of melanocyte differentiation and growth. Cell Growth Differ. 1997;8:935–940. [PubMed] [Google Scholar]
- Halaban R, Böhm M, Dotto P, Moellmann G, Cheng E, Zhang YH. Growth regulatory proteins that repress differenciation markers in melanocytes also downregulate the transcription factor microphthalmia. J Invest Dermatol. 1996;106:1266–1272. doi: 10.1111/1523-1747.ep12348972. [DOI] [PubMed] [Google Scholar]
- Hearing VJ. Mammalian monophenol monooxygenase (tyrosinase): purification, properties and reactions catalyzed. Methods Enzymol. 1987;142:154–165. doi: 10.1016/s0076-6879(87)42024-7. [DOI] [PubMed] [Google Scholar]
- Hemesath TJ, Steingrímsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. Microphthalmia, a critical factor in melanocyte development, definers a discrete transcription factor family. Genes Dev. 1994;8:2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- Hemesath TJ, Price ER, Takemoto C, Badalian T, Fisher DE. MAPK links microphthalmia to c-kit signaling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrímsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Hughes AE, Newton VE, Liu XZ, Read AO. A gene for Waardenburg syndrome type II maps close to the human homologue of the microphthalmia gene at chromosome 3p12-p14.1. Nat Genet. 1994;7:509–513. doi: 10.1038/ng0894-509. [DOI] [PubMed] [Google Scholar]
- Hughes MJ, Lingrel JB, Krakowski JM, Anderson KP. A helix-loop-helix transcription factor-like gene is located at the milocus. J Biol Chem. 1993;268:20687–20690. [PubMed] [Google Scholar]
- Hunt G, Todd C, Kyne S, Thody AJ. Human melanocytes can respond to MSH and ACTH peptides in vitro. J Endocrinol. 1993;139:29–39. [Google Scholar]
- Hunt G, Todd C, Cresswell JE, Thody AJ. α-MSH and its analog Nle4DPhe7α-MSH affect morphology, tyrosinase activity and melanogenesis in cultured human melanocytes. J Cell Sci. 1994;107:205–211. doi: 10.1242/jcs.107.1.205. [DOI] [PubMed] [Google Scholar]
- Jackson JI, Cambers DM, Tsukamoto K, Copeland N, Gilbert DJ, Jenkins NA, Hearing VJ. A second tyrosinase-related protein, TRP-2, maps to and mutated at the mouse slaty locus. EMBO (Eur Mol Biol Organ) J. 1992;11:527–535. doi: 10.1002/j.1460-2075.1992.tb05083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez M, Kameyama K, Maloy WL, Tomita Y, Hearing VJ. Mammalian tyrosinase: biosynthesis, processing, and modulation by melanocyte-stimulating hormone. Proc Natl Acad Sci USA. 1988;85:3830–3834. doi: 10.1073/pnas.85.11.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Cervantes C, Solano F, Kobayashi T, Urabe K, Hearing VJ, Lozano JA, García-Borrón JC. A new enzymatic function in the melanogenic pathway: the 5,6-dihydroxyindole-2-carboxylic acid oxidase activity of tyrosinase-related protein-1 (TRP1) J Biol Chem. 1994;269:29198–29205. [PubMed] [Google Scholar]
- Kobayashi T, Urabe K, Winder A, Jiménez-Cervantes C, Imokawa G, Brewington T, Solano F, García-Borrón JC, Hearing VJ. Tyrosinase related protein 1 (TRP1) functions as a DHICA oxidase in melanin biosynthesis. EMBO (Eur Mol Biol Organ) J. 1994;13:5818–5825. doi: 10.1002/j.1460-2075.1994.tb06925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner A, Pawelek J. Mammalian tyrosinase catalyzes three reactions in the biosynthesis of melanin. Science. 1982;217:1163–1165. doi: 10.1126/science.6810464. [DOI] [PubMed] [Google Scholar]
- Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SGE, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- Lahav R, Ziller C, Dupin E, Le Douarin NM. Endothelin 3 promotes neural crest cell proliferation and mediates a vast increase in melanocyte number in culture. Proc Natl Acad Sci USA. 1996;93:3892–3897. doi: 10.1073/pnas.93.9.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue L, Dougherty N, Bradl M, Mintz B. Melanocyte culture from Tyr-SV40E transgenic mice: models for the molecular genetic evolution of malignant melanocyte. Oncogene. 1993;8:523–531. [PubMed] [Google Scholar]
- Le Douarin, N. 1982. The Neural Crest. Cambridge University Press, Cambridge, UK.
- Levine N, Sheftel SN, Eytan T, Dorr RT, Hadley ME, Weinrach JC, Ertl GA, Toth K, McGee DL, Hurby VJ. Induction of skin tanning by subcutaneous administration of a potent synthetic melanotropin. J Am Med Assoc. 1991;226:2730–2736. [PubMed] [Google Scholar]
- Lowings P, Yavuzer U, Goding R. Positive and negative elements regulate a melanocyte-specific promoter. Mol Cell Biol. 1992;12:3653–3662. doi: 10.1128/mcb.12.8.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KJ. Insight into the microphthalmia gene. Trends Genet. 1995;11:442–448. doi: 10.1016/s0168-9525(00)89143-x. [DOI] [PubMed] [Google Scholar]
- Opdecamp K, Nakayama A, Nguyen MT, Hodgkinson CA, Pavan WJ, Arnheiter H. Melanocyte development in vivo and in neural crest cell cultures: crucial dependence on the Mitf basic-helix-loop-helix-zipper transcription factor. Development (Camb) 1997;124:2377–2386. doi: 10.1242/dev.124.12.2377. [DOI] [PubMed] [Google Scholar]
- Orlow SJ, Hearing VJ, Sakai C, Urabe K, Zhou BK, Silvers WK. Changes in expression of putative antigens encoded by pigment genes in mouse melanomas at different stages of malignant progression. Proc Natl Acad Sci USA. 1995;92:10152–10156. doi: 10.1073/pnas.92.22.10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortonne JP. The effects of ultraviolet exposure on skin melanin pigmentation. J Int Med Res. 1990;18:8–17. [PubMed] [Google Scholar]
- Prota, G. 1988. Some new aspects of eumelanin chemistry. In Progress in Clinical and Biological Research. Advance in Pigment Cells Research. J.T. Bagnaras, editor. A.R. Liss, Inc., New York. 101–124. [PubMed]
- Rungta D, Corn TD, Fuller DD. Regulation of tyrosinase mRNA in mouse melanoma cells by a-melanocyte-stimulating hormone. J Invest Dermatol. 1996;107:689–693. doi: 10.1111/1523-1747.ep12365578. [DOI] [PubMed] [Google Scholar]
- Sato C, Roberts K, Gambino G, Cook A, Kouzarides T, Goding CR. CBP/p300 as a co-factor for the microphthalmia transcription factor. Oncogene. 1997;14:3083–3092. doi: 10.1038/sj.onc.1201298. [DOI] [PubMed] [Google Scholar]
- Steingrímsson E, Moore KJ, Lamoreux ML, Ferré-D'Amaré A, Burley SK, Zimring DCS, Skow LC, Hodgkinson CA, Arnheiter H, Nakayama A, et al. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat Genet. 1994;8:256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Takeda K, Nobukini Y, Urabe K, Long JE, Meyers KA, Aaranson SA, Miki T. Ectopic expression of MITF, a gene for Waardenburg syndrome type 2 converts fibroblasts to cells with melanocyte characteristics. Nat Genet. 1996;14:50–54. doi: 10.1038/ng0996-50. [DOI] [PubMed] [Google Scholar]
- Tassabehji M, Newton VE, Read AP. MITF gene mutations in patient with type 2 Waardenburg syndrome. Nat Genet. 1994;8:251–255. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]
- Weilbaecher KN, Hershey CL, Takemoto CM, Horstmann MA, Hemesath TJ, Tashjian AH, Jr, Fisher DE. Age-resolving osteopetrosis: a rat model implicating microphthalmia and the related transcription factor TFE3. J Exp Med. 1998;187:775–785. doi: 10.1084/jem.187.5.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong G, Pawelek J. Melanocyte-stimulating hormone promotes activation of pre-existing tyrosinase molecules in Cloudman S91 melanoma cells. Nature. 1975;255:644–645. doi: 10.1038/255644a0. [DOI] [PubMed] [Google Scholar]
- Yasumoto KI, Yokoyama K, Shibata K, Tomita Y, Shibahara S. Microphtalmia-associated transcriptionn factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol Cell Biol. 1994;14:8058–8070. doi: 10.1128/mcb.14.12.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavuzer U, Keenan E, Lowings P, Vachtenheim J, Currie G, Goding CR. The microphthalmia gene product interacts with the retinoblastoma protein in vitro and is a target for deregulation of melanocyte-specific transcription. Oncogene. 1995;10:123–134. [PubMed] [Google Scholar]
- Yokoyama K, Suzuki H, Tomita Y, Shibahara S. Molecular cloning of and functional analysis of a cDNA coding for human DOPAchrome tautomerase/tyrosinaserelated protein-2. Biochim Biophys Acta. 1994a;1217:317–321. doi: 10.1016/0167-4781(94)90292-5. [DOI] [PubMed] [Google Scholar]
- Yokoyama K, Yasumoto KI, Suzuki H, Shibahara S. Cloning of the human DOPAchrome tautomerase/tyrosinase-related protein 2 gene and identification of two regulatory regions required for its pigment cell-specific expression. J Biol Chem. 1994b;269:27080–27087. [PubMed] [Google Scholar]