Abstract
In Saccharomyces cerevisiae, the unconventional myosin Myo2p is of fundamental importance in polarized growth. We explore the role of the neck region and its associated light chains in regulating Myo2p function. Surprisingly, we find that precise deletion of the six IQ sites in the neck region results in a myosin, Myo2-Δ6IQp, that can support the growth of a yeast strain at 90% the rate of a wild-type isogenic strain. We exploit this mutant in a characterization of the light chains of Myo2p. First, we demonstrate that the localization of calmodulin to sites of polarized growth largely depends on the IQ sites in the neck of Myo2p. Second, we demonstrate that a previously uncharacterized protein, Mlc1p, is a myosin light chain of Myo2p. MLC1 (YGL106w) is an essential gene that exhibits haploinsufficiency. Reduced levels of MYO2 overcome the haploinsufficiency of MLC1. The mutant MYO2-Δ6IQ is able to suppress haploinsufficiency but not deletion of MLC1. We used a modified gel overlay assay to demonstrate a direct interaction between Mlc1p and the neck of Myo2p. Overexpression of MYO2 is toxic, causing a severe decrease in growth rate. When MYO2 is overexpressed, Myo2p is fourfold less stable than in a wild-type strain. High copies of MLC1 completely overcome the growth defects and increase the stability of Myo2p. Our results suggest that Mlc1p is responsible for stabilizing this myosin by binding to the neck region.
Keywords: myosin, polarized, stability, Myo4, cytokinesis
The Saccharomyces cerevisiae unconventional myosin Myo2p is a member of the class V myosins and has been implicated in vesicle movement and polarized growth (Johnston et al., 1991; Govindan et al., 1995). Homologues of Myo2p include mouse dilute, which plays a role in melanosome transport (Provance et al., 1996; Nascimento et al., 1997), and chicken myosin V, which has a role in neuron filopodial extension (Wang et al., 1996). Myo2p is essential for growth of S. cerevisiae, and it localizes to the bud tip during bud formation and to the bud neck during cytokinesis (Brockerhoff et al., 1994; Lillie and Brown, 1994). A temperature-sensitive mutation in MYO2 confers defects in polarized growth and in vacuole inheritance but not in general secretion (Johnston et al., 1991; Govindan et al., 1995; Hill et al., 1996).
All myosins have at least one light chain that binds to the myosin heavy chain via a light chain binding motif called an IQ site (Cheney and Mooseker, 1992; Xie et al., 1994). IQ sites are often found in tandem repeats between the head-motor domain and the tail domain (Cheney and Mooseker, 1992; Rayment et al., 1993). IQ sites are ∼25– amino acid residue motifs that bind calmodulin or myosin light chains. In many cases, the binding of these small EF-hand proteins activates the Mg2+ATPase activity of myosins. For example, calmodulin is required for the Mg2+ATPase activity of chicken myosin V (Espindola et al., 1992). Furthermore, light chains perform structural roles by affecting myosin head orientation as well as orientation of light chains to each other. As an example, the light chains of scallop myosin II are required to stabilize Ca2+ binding by the myosin head domain and alter the myosin head orientation (Fromherz and Szent-Gyorgyi, 1995). Myo2p has six tandem IQ sites (Johnston et al., 1991).
We have previously shown that calmodulin binds to Myo2p to perform an essential function in polarized growth (Brockerhoff et al., 1994). Not only is calmodulin present at sites of polarized growth (Brockerhoff and Davis, 1992), but calmodulin and Myo2p have direct physical contact through the IQ sites in the neck of Myo2p (Brockerhoff et al., 1994). CMD1 mutants show allele-specific synthetic lethality with the mutant myo2-66, thus suggesting that calmodulin and Myo2p share an essential function.
Interestingly, both class V myosins in S. cerevisiae, Myo2p and Myo4p, confer deleterious effects on cell growth when overexpressed (Haarer et al., 1994). Myo4p localizes to the bud and is essential for the polarized distribution of the asymmetry determinant, Ash1p (Bobola et al., 1996; Jansen et al., 1996). MYO4 is not essential for growth (Haarer et al., 1994). The cause of growth defects when either MYO2 or MYO4 is overexpressed is unknown.
Here we investigate the role of light chains in the function of Myo2p. Localization of calmodulin to sites of cell growth depends on the neck region of Myo2p. The S. cerevisiae genome sequencing project revealed a small protein with similarities to calmodulin and myosin light chains. We show that this protein, which we name Mlc1p (myosin light chain), binds to the myosin neck and regulates the stability of Myo2p.
Materials and Methods
Plasmids
The plasmids used in this study are listed in Table I.
Table I.
Plasmids Used in This Study
| Plasmid | Parent vector | Relevant markers and construction* | Reference or source | |||
|---|---|---|---|---|---|---|
| pBluescriptII KS (+) | ampr f1 origin | Stratagene | ||||
| pGEX-2T | Ptac with gene encoding GST | Pharmacia | ||||
| pQE30 | Ptac with 6XHis polylinker | Qiagen | ||||
| pRS306 | URA3 f1 origin | (Sikorski and Hieter, 1989) | ||||
| pRS315 | CEN6 ARSH4 LEU2 f1 origin | (Sikorski and Hieter, 1989) | ||||
| pRS316 | CEN6 ARSH4 URA3 f1 origin | (Sikorski and Hieter, 1989) | ||||
| pGF27 | pRS304 | 2-μm origin (YEp24 fragment) inserted at AatII site | G. Zhu | |||
| pGF29 | pRS306 | 2-μm origin (YEp24 fragment) inserted at AatII site | G. Zhu | |||
| pJG28 | pSB5 | ampr trc promoter, CMD1 | J. Geiser | |||
| pJP10-2B | YCp50 | URA3 MYO2 CEN4 ARS1 | (Johnston et al., 1991) | |||
| pLI831 | pBluescriptII SK (+) | ADE3 | E. Muller | |||
| myo4Δ::URA3 | pBluescriptII KS (+) | myo4Δ::URA3 | S. Brown | |||
| pRS23 | pBluescriptII SK (+) | 5.6-kb ClaI fragment of MYO2 | This study | |||
| pRS28 | pRS27 | myo2Δ::TRP1 | This study | |||
| pRS29 | pRS25 | 5.6-kb ClaI fragment of MYO2 | This study | |||
| pRS31 | pGF29 | 5.6-kb ClaI fragment of MYO2 | This study | |||
| pRS43 | pRS29 | NheI site removed | This study | |||
| pRS50 | pRS37 | 2-μm origin (YEp24 fragment) inserted at NotI site | This study | |||
| pRS72 | pRS43 | NcoI site added at codon 1 of MYO2 | This study | |||
| pRS172 | pRS43 | Internal deletion of MYO2 (S787G | This study | |||
| Δaa788–927) also known as MYO2-D6IQ | ||||||
| pRS174 | pRS306 | 5.4-kb ClaI fragment of MYO2-D6IQ | This study | |||
| pRS221 | pGF29 | 5.4-kb ClaI fragment of MYO2-D6IQ | This study | |||
| pRS276 | pBluescriptII KS (+) | 3.3-kb BamHI fragment carrying MLC1 | This study | |||
| pRS286 | pRS285 | mlc1Δ::TRP1 | This study | |||
| pRS289 | pRS316 | 1.4-kb BamHI-SacI fragment of MLC1 | This study | |||
| pRS290 | pGF27 | 1.4-kb BamHI-SacI fragment of MLC1 | This study | |||
| pRS296 | pQE30 | ampr promoter 6XHis-MLC1 | This study | |||
| pRS321 | pRS315 | 1.4-kb BamHI-SacI fragment of MLC1 | This study | |||
| pSB6 | pACYC177 | kanr lysis genes of lambda | (Brockerhoff et al., 1994) | |||
| pSB20 | pGEX-3X | MYO2 (aa740–1457) fused to GST | (Brockerhoff et al., 1994) | |||
| pSB21 | pGEX-3X | MYO2 (aa740–1116) fused to GST | (Brockerhoff et al., 1994) | |||
| pSB24 | pGEX-3X | MYO2 (aa790–924) fused to GST | (Brockerhoff et al., 1994) | |||
| pSB25 | pGEX-3X | MYO2 (aa740–833) fused to GST | (Brockerhoff et al., 1994) | |||
| pSB27 | pGEX-3X | MYO2 (aa247–740) fused to GST | (Brockerhoff et al., 1994) | |||
| pTD28 | pTD17 | 2-μm origin LYS2 CMD1 | (Davis and Thorner, 1989) | |||
| pTD29 | YEp24 | 2-μm origin LYS2 | (Geiser et al., 1993) |
Unless otherwise stated, all markers from the parent plasmid are present in the new construct. MYO2-Δ6IQ encodes an internal deletion in Myo2p with the mutation S787G and missing amino acids 787–927. MLC1 is identical to the Saccharomyces cerevisiae open reading frame YGL106w.
Plasmid pRS28, used to precisely delete MYO2, was made in several steps. The 5.6-kb ClaI fragment of pJP10-2B, containing MYO2, was cloned into the ClaI site of pBluescriptII KS+ (Stratagene, La Jolla, CA) to make pRS23. In pRS24, the 4.1-kb NdeI-AflII fragment of MYO2 was removed. Plasmid pRS24 was digested with EcoRI, the ends were filled in with the Klenow fragment of DNA polymerase to destroy the EcoRI site, and the ends were ligated with T4 DNA ligase to make pRS26. The remaining coding sequence of MYO2 in pRS26 was replaced with an EcoRI site by oligonucleotide-directed mutagenesis (Kunkel et al., 1987) using the primer MYO2D (5′-CTCGCGCCATCAGTTGAATTCTATACGCTGTAATATCTGCTG-3′), creating plasmid pRS27. In the plasmid pRS28, a 0.8-kb EcoRI fragment containing TRP1 (Davis et al., 1986) was cloned into the EcoRI site of pRS27.
The plasmid shuffle used in this study required a plasmid encoding ADE3 and MYO2. A 5.6-kb ClaI fragment of MYO2 was cloned into the SmaI site of pLI831 (Geiser et al., 1993) to make pRS37. The shuttle vector pRS50 contains a 2-μm origin from a 2.1-kb EcoRI fragment of YEp24 ligated into the NotI site of pRS37. In both plasmids, the 5′ overhangs were filled in by treatment with the Klenow fragment of DNA polymerase.
Additional plasmids containing MYO2 were made carrying the URA3 selectable marker. Plasmid pRS25 is a derivative of pRS316 (Sikorski and Hieter, 1989) in which the HindIII-NotI fragment of the polylinker was removed, the 5′ overhangs were filled in with Klenow, and the plasmid was recircularized with T4 DNA ligase. The plasmid pRS29 contains the 5.6-kb ClaI fragment of MYO2 from plasmid pJP10-2B cloned into the ClaI site of pRS25. In pRS29, the coding sequence of MYO2 runs in the same direction as the f1 origin. For later studies, the NheI site of pRS29 was removed by digestion with NheI, filling in with Klenow, and reclosing with T4 DNA ligase to create pRS43. The high-copy number plasmid pRS31 was created by cloning the 5.6-kb ClaI fragment of MYO2 into the ClaI site of pGF29, a 2-μm plasmid.
Deletion of the region encoding the neck of MYO2 was made by oligonucleotide-directed mutagenesis (Kunkel et al., 1987). Because of the large size of MYO2, a cassette was created containing the middle one-third of the gene. The 2.6-kb BamHI-EcoRI fragment of MYO2 was cloned into the BamHI and EcoRI sites of pBluescriptII KS+ to make pRS78. The first step towards deleting the six IQ sites was to introduce NarI restriction endonuclease cleavage sites into the coding sequence of MYO2 at either end of the region encoding the IQ sites. The oligonucleotide NAR1 (5′-GCAATAAGATGCATAATGGCGCCATTGTTATGATCCAGAAG-3′) results in the addition of a glycine between amino acid residues 786 and 787 as well as the mutation S787A. The oligonucleotide NAR4 (5′-GAAATTGAAACAATTGAAGGGCGCCGACGCTAAATCAGTTAATC-3′) results in the addition of a glycine between amino acids 927 and 928. After mutating pRS78 using NAR1 and NAR4, the resulting plasmid was digested with NarI and reclosed using T4 DNA ligase. This mutant cassette was then ligated to BamHI- and EcoRI-cut pRS43 to create the plasmid pRS172, which encodes a Myo2p with an S787G mutation and lacks only amino acids 788–927. This mutant was named MYO2-Δ6IQ. Plasmid pRS221 has the 5.4-kb Asp718-SacI fragment containing MYO2-Δ6IQ cloned into pGF29.
The plasmid pRS174 was created to integrate MYO2-Δ6IQ into the genome of S. cerevisiae. pRS174 was made by ligating the 5.4-kb ClaI fragment of pRS172 containing MYO2-Δ6IQ into the ClaI site of pRS306.
The gene YGL106w encodes a 149–amino acid protein with similarities to calmodulin. Based on the work described in this paper, we have renamed YGL106w as MLC1. The 3.3-kb BamHI fragment of lambda PM-3353 (American Type Culture Collection, Rockville, MD) was cloned into the BamHI site of pBluescriptII KS+, creating plasmid pRS276. Plasmid pRS287 is a derivative of pRS276, in which the 2.0-kb NdeI-NotI fragment was removed, 5′ overhangs were filled in with Klenow, and plasmid was recircularized with T4 DNA ligase. In plasmid pRS287, MLC1 is the only open reading frame remaining in the pBluescript polylinker.
The plasmid to delete MLC1 precisely was made in a similar way to the myo2Δ construct. Plasmid pRS284 was made by digesting pRS276 with EcoRI, filling in the 5′ overhangs with Klenow, and recircularizing the plasmid with T4 DNA ligase. The coding sequence of MLC1 was replaced with an EcoRI site by oligonucleotide-directed mutagenesis (Kunkel et al., 1987) using primer MLC1D (5′-CTAAATTTGCAGTTCCGCACTCTCAGAATTCTGTTATTCTATGTATGTGCG-3′), creating pRS285. A 0.8-kb EcoRI fragment containing TRP1 (Davis et al., 1986) was cloned into the EcoRI site of pRS285 to make pRS286. In pRS286, the TRP1 gene is in the same orientation as the original MLC1.
Plasmid pRS296, designed to express 6XHis-tagged MLC1 in Escherichia coli, was made in several steps. An NcoI site was placed at the first codon of MLC1 by oligonucleotide-directed mutagenesis using primer MLC1-NCOI (5′-GGCTCTGGTGGCTGCCATGGTTATTCTATGTATGTGC-3′) (Kunkel et al., 1987) to make pRS291. This mutagenesis also created the mutation S2A. The MLC1 fragment was made by digesting pRS291 with NcoI, filling in using the Klenow fragment of DNA polymerase, digesting with SacI, and purifying the 0.8-kb fragment. Plasmid pRS296 was then made by digesting pQE30 (Qiagen, Inc., Chatsworth, CA) with BamHI, filling in using the Klenow fragment of DNA polymerase, digesting with SacI, and ligating in the 0.8-kb MLC1 fragment.
Media and Strains
Media for growth of S. cerevisiae and E. coli were described previously (Zhu et al., 1993). The yeast strains used in this study are listed in Table II. Genetic manipulations and transformations were performed essentially as described previously (Sherman et al., 1986). RSY2 carrying the deletion of MYO2 was created by one-step gene replacement (Rothstein, 1991) using the plasmid pRS28 digested with BglII and ClaI transformed into RSY1. Because strains containing ade3Δ-100 are histidine auxotrophs, ADE3 on a plasmid can be selected by histidine prototrophy (Jones and Fink, 1982). RSY2 was transformed with pRS50 carrying ADE3 and MYO2. The strain RSY2-60B has MYO2 deleted from the genome and carries pRS50.
Table II.
Yeast Strains
| Strain | Genotype* | Reference | ||
|---|---|---|---|---|
| CRY1 | MAT a ade2-1oc can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | R. Fuller (Stanford University, CA) | ||
| JGY46 | MAT a/MATα ade2-1oc/ade2-1oc can1-100/can1-100 his3- | (Geiser et al., 1991) | ||
| 11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 | ||||
| EMY55-5D | MAT a ade2-1oc ade3Δ-100 can1-100 cyh2r his3-11,15 leu2-3,112 lys2Δ::HIS3 trp1-1 ura3-1 | E. Muller (University of | ||
| Washington, Seattle, WA) | ||||
| EMY55-6B | MATα ade2-1oc ade3Δ-100 can1-100 cyh2S his3-11,15 leu2-3,112 lys2Δ::HIS3 trp1-1 ura3-1 | E. Muller (University of | ||
| Washington) | ||||
| MMY28 | MAT a ade2-1oc ade3Δ-100 can1-100 (S65T-GFP)–CMD1 his3- | (Moser et al., 1997) | ||
| 11,15 leu2-3,112 lys2Δ::HIS3 trp1-1 ura3-1 | ||||
| RSY1 | EMY55-5D X EMY55-6B | This study | ||
| RSY2 | MAT a /MATα ade2-1oc/ade2-1oc ade3Δ-100/ade3Δ-100 can1-100/can1-100 cyh2r/cyh2S | This study | ||
| his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 lys2Δ::HIS3/lys2Δ::HIS3 MYO2/myo2Δ::TRP1 | ||||
| trp1-1/-trp1-1 ura3-1/ura3-1 | ||||
| RSY2-60B | MAT a ade2-1oc ade3 Δ-100 can1-100 his3-11,15 leu2-3,112 | This study | ||
| lys2Δ::HIS3 myo2Δ::TRP1 trp1-1 ura3-1 carrying plasmid pRS50 | ||||
| RSY21 | MAT a ade2-1oc can1-100 his3-11,15 leu2-3,112 MYO2-D6IQ trp1-1 ura3-1 | This study | ||
| RSY22 | MATα ade2-1oc can1-100 his3-11,15 leu2-3,112 MYO2-D6IQ trp1-1 ura3-1 | This study | ||
| RSY38 | MAT a /MATα ade2-1oc/ade2-1oc ADE3/ade3Δ-100 can1-100/can1-100 CMD1/GFP–CMD1 | This study | ||
| his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 MYO2/MYO2-Δ6IQ MYO4/myo4Δ::URA3 | ||||
| trp1-1/trp1-1 ura3-1/ura3-1 | ||||
| RSY38-9D | MAT a ade2-1oc ade3Δ-100 can1-100 GFP–CMD1 his3-11,15 | This study | ||
| leu2-3112 lys2Δ::HIS3 MYO2-Δ6IQ trp1-1 ura3-1 | ||||
| RSY38-16A | MAT a ade2-1oc ade3Δ-100 can1-100 GFP–CMD1 his3-11,15 | This study | ||
| leu2-3,112 lys2Δ::HIS3 myo4Δ::URA3 trp1-1 ura3-1 | ||||
| RSY38-16C | MATα ade2-1oc ade3Δ-100 can1-100 his3-11,15 leu2-3,112 | This study | ||
| lys2Δ::HIS3 MYO2-Δ6IQ myo4Δ::URA3 trp1-1 ura3-1 | ||||
| RSY38-17C | MATα ade2-1oc ade3Δ-100 can1-100 GFP–CMD1 his3-11,15 | This study | ||
| leu2-3,112 lys2Δ::HIS3 trp1-1 ura3-1 | ||||
| RSY105 | MAT a/MATα ade2-1 oc/ade2-1 oc can1-100/can1-100 his3- | This study | ||
| 11,15/his3-11,15 leu2-3,112/leu2-3,112 MLC1/mcl1D::TRP1 | ||||
| trp1-1/trp1-1 ura3-1/ura3-1 carrying plasmid pRS289 | ||||
| RSY105-6A | MATα ade2-1oc can1-100 his3-11,15 leu2-3,112 mcl1Δ::TRP1 | This study | ||
| trp1-1 ura3-1 carrying plasmid pRS289 | ||||
| RSY107 | RSY21 X RSY105-6A carrying plasmid pRS289 | This study | ||
| RSY112 | MAT a/MATα ade2-1oc/ade2-1oc ade3Δ-100/ade3Δ-100 can1-100/can1-100 his3-11,15/his3-11,15 | This study | ||
| leu2-3,112/leu2-3,112 lys2Δ::HIS3/lys2Δ::HIS3 MLC1/mcl1Δ::TRP1 | ||||
| MYO2/myo2Δ::TRP1 trp1-1/trp1-1 ura3-1/ura3-1 carrying plasmid pRS289 |
GFP–CMD1 encodes a fusion protein of green fluorescent protein and calmodulin.
The mutant MYO2-Δ6IQ was integrated into yeast by two-step gene replacement (Rothstein, 1991) using plasmid pRS174 digested with BamHI. Strains RSY21 and RSY22 were shown to contain MYO2-Δ6IQ by PCR and by Southern blot analysis. RSY22 was mated to strain MMY28 carrying an integrated form of GFP–CMD1 (Moser et al., 1997) to make the diploid strain RSY33. A myo4Δ::URA3 construct (a gift from Susan Brown, University of Michigan, Ann Arbor, MI) was transformed into RSY33 to create strain RSY38, heterozygous for GFP–CMD1, MYO2-Δ6IQ, and myo4Δ.
Strain RSY105 carrying a deletion of MLC1 was difficult to create because MLC1 displays haploinsufficiency. To create the disruption, we first transformed JGY46 with plasmid pRS289, carrying the MLC1 gene. The resulting strain had three functioning copies of MLC1. If one copy of MLC1 were removed by one-step gene replacement, then the resulting strain would still have two copies of the gene. The 1.7-kb BamHI-NdeI fragment of plasmid pRS286 encoding mlc1Δ::TRP1 was transformed into JGY46(pRS289). Of the 42 colonies obtained, four were unable to lose the URA3-marked MLC1 plasmid pRS289 when streaked onto minimal medium containing 5′-fluoro orotic acid (5′-FOA).1 These four colonies were the likely candidates for heterozygous mlc1Δ strains. To test this hypothesis, the four strains were transformed with the LEU2-marked MLC1 plasmid pRS321 and streaked onto minimal medium containing 5′-FOA. Three of the four strains were able to lose pRS289 when given an alternative source of MLC1 from pRS321. RSY105 was shown to be heterozygous MLC1/mlc1Δ::TRP1 by Southern blot analysis. RSY105 must carry a plasmid source of MLC1 to survive.
To study the null phenotype of mlc1Δ, strains RSY105(pRS289) and JGY46 were sporulated on SpoIII medium, and 50 tetrads were dissected from each strain. After 24 h, spores were examined for germination and colony growth. Images of spores and colonies were obtained as described below using a fluorescent microscope (model Axioplan; Carl Zeiss, Inc., Thornwood, NY) with 0.3-s, low-light exposures. Germinating spores were transferred to 20 μl minimal medium containing 3.7% formaldehyde. After 60 min at room temperature, 4,6′-diamidino-2-phenylindole (DAPI) was added to a final concentration of 50 μg/ml, and nuclei were examined as described below.
Immunoblot Analysis
SDS-PAGE and immunoblot analysis were performed as described (Geiser et al., 1991) with the following exceptions. Strains to be analyzed were grown to 50 Klett units in the appropriate medium. After centrifugation, cells were washed in lysis buffer (50 mM Tris, pH 8.0, 1× proteinase inhibitor cocktail [Drubin et al., 1988]). Cells were lysed by mixing equal volumes of cells, glass beads (Sigma Chemical Co., St. Louis, MO), and lysis buffer and vortexing five times for 1 min alternating with 1-min incubations on ice. After centrifugation for 15 min in a microcentrifuge at 4°C, pellets were washed once in lysis buffer and solubilized using cracking buffer (10 mM sodium phosphate, pH 7.2, 1% β-mercaptoethanol, 1% SDS, and 6 M urea). For detection of Myo2p, 30 μg of protein was loaded into each lane of a 6% SDS–polyacrylamide gel. Proteins were transferred to nitrocellulose membranes using a wet-transfer apparatus (Bio-Rad Laboratories, Hercules, CA) following the instructions provided by the manufacturer. The membranes were incubated with a 1:400 dilution of affinity-purified anti-Myo2p antibody (Lillie and Brown, 1994). Secondary antibody was blotting grade goat anti–rabbit IgG horseradish peroxidase conjugate (Bio-Rad Laboratories) at a 1:5,000 dilution. Signal was detected using Renaissance luminol reagent (DuPont/NEN, Boston, MA) and Hyperfilm-MP (Amersham Corp., Arlington Heights, IL).
Gel Overlays
Production of 35S-labeled 6XHis-Mlc1p was as follows. The E. coli strain GM1 (Coulondre and Miller, 1977) containing the plasmids pRS296 and pSB6 was grown in 30 ml of M9 medium (Miller, 1972) with 1.0 mM MgCl2 (instead of MgSO4), 0.4% glucose, 1 μg/ml thiamine, 50 μg/ml ampicillin, 15 μg/ml kanamycin, and 8 mCi carrier-free Na35SO4 (DuPont/ NEN) to 20 Klett units and then induced by adding isopropyl-β-d-thiogalactopyranoside to a final concentration of 2 mM. After 5 h at 37°C, the cells were collected by centrifugation and resuspended in 400 μl lysis buffer (50 mM Tris, pH 7.5, 1 mM MgCl2, and 1 mM phenylmethylsulfonyl fluoride). The sample was frozen and thawed several times, alternating between −20 and 20°C to lyse the cells, and then centrifuged at 10,000 g for 30 min in a 4°C microcentrifuge. The supernatant fraction was batch mixed for 10 min with 700 μl preequilibrated Ni-NTA beads (Qiagen, Inc.). The beads were poured to form a column and washed with 10 ml lysis buffer containing 25 mM imidazole. Purified 6XHis-Mlc1p was eluted from the column using lysis buffer containing 500 mM imidazole. The imidazole buffer was exchanged with 100 mM ammonium bicarbonate with a PD-10 G25 column (Pharmacia Biotech, Piscataway, NJ). Protein was stored at −80°C until needed. Protein concentration was determined using bicinchoninic acid (Sigma Chemical Co.). The purified 35S-labeled 6XHis-Mlc1p had a specific activity of 27 dpm/fmol.
Purified 35S-labeled calmodulin was made as previously described (Brockerhoff et al., 1992), except that only 8 mCi Na35SO4 was used in the labeling. The purified 35S-labeled calmodulin had a specific activity of 39 dpm/fmol.
The gel overlay assays for calmodulin and Mlc1p binding were performed as previously described (Brockerhoff et al., 1994) with slight modifications. In brief, extracts were made from the E. coli strain GM1 (Coulondre and Miller, 1977) containing either plasmid pGEX-2T, pSB20, pSB21, pSB24, pSB25, or pSB27. Approximately 30 μg total protein was loaded into each lane for SDS-PAGE. Proteins were transferred to Immobilon membranes (Millipore Corp., Bedford, MA) using a wet-transfer apparatus (Bio-Rad Laboratories) following the instructions provided by the manufacturer. After transfer, the proteins were renatured by washing the Immobilon membranes for a total of 40 min (four changes) in buffer A (20 mM Hepes, pH 7.2, 100 mM NaCl) and blocked for 6 h in buffer A containing 3% bovine serum albumin and 0.05% Tween-20. The membranes were probed for 16 h at 21°C with either 140 mM 35S-labeled 6XHis-Mlc1p or 156 mM 35S-labeled calmodulin in buffer C (buffer A with 5 mM CaCl2 and 0.05% Tween-20). The membranes were then washed for 40 min (four changes) in buffer C. Next, the membranes were air dried, dipped in 7% 2,5-diphenyloxazole in acetone, air dried, and exposed to Hyperfilm-MP (Amersham Corp.) for 5–8 d.
Northern Analysis of MYO2 Transcript
Total mRNA was purified from strains of CRY1 carrying either pRS43, pRS172, pRS31, or pRS221 using a previously described procedure (Wise, 1991). From 500-ml cultures, ∼7.0 mg RNA was obtained. Total mRNA was purified from the samples using the Promega PolyA Tract mRNA Isolation System I (Madison, WI) with MagneSphere particles. Approximately 50 μg poly A–containing mRNA was purified from each culture.
Northern blot analysis was performed using standard procedures (Sambrook et al., 1989) with the following modifications. 10 μg mRNA was loaded in each lane of a prerun formaldehyde gel. After removing the formaldehyde by washing 2× in DEPC dH2O, mRNA was transferred to a nitrocellulose membrane using capillary transfer (Sambrook et al., 1989). The membrane was baked at 65°C for 1 h. Probes were made using the 3.4-kb NcoI-EcoRI fragment of MYO2 from pRS72 and a PCR product containing a 0.5-kb fragment of TRX2. After incubating the membrane with ∼1 × 107 cpm of each probe (pooled) for 48 h, the membrane was washed and analyzed using a Molecular Dynamics PhosphorImager (model 400S; Sunnyvale, CA) using 176-micron pixel size.
Indirect Immunofluorescence and Green Fluorescent Protein Fusions
The immunolocalization of Myo2p in strains CRY1 and RSY21 was performed as previously described (Brockerhoff et al., 1994; Lillie and Brown, 1994).
The localization of GFP–Cmd1p in strains RSY38-9D, RSY38-16A, RSY38-16C, and RSY38-17C was performed using a fluorescent microscope (model Axioplan; Carl Zeiss, Inc.) (Moser et al., 1997). 200 cells from each strain were scored for the presence of polarized calmodulin. Calmodulin was judged to be polarized if the fluorescent signal was stronger in the bud than in the mother portion of each cell. Images were captured using an Imagepoint-cooled CCD video camera (Photometrics, Tucson, AZ) fitted to the microscope in conjunction with IP Lab software (Signal Analytics, Vienna, VA). Images for GFP–calmodulin are 5-s exposures. Previous studies have shown that expression of GFP alone in yeast resulted in a uniform distribution of fluorescence throughout the cell, excluding the vacuole (Niedenthal et al., 1996).
Results
Myo2p Functions without a Neck Domain
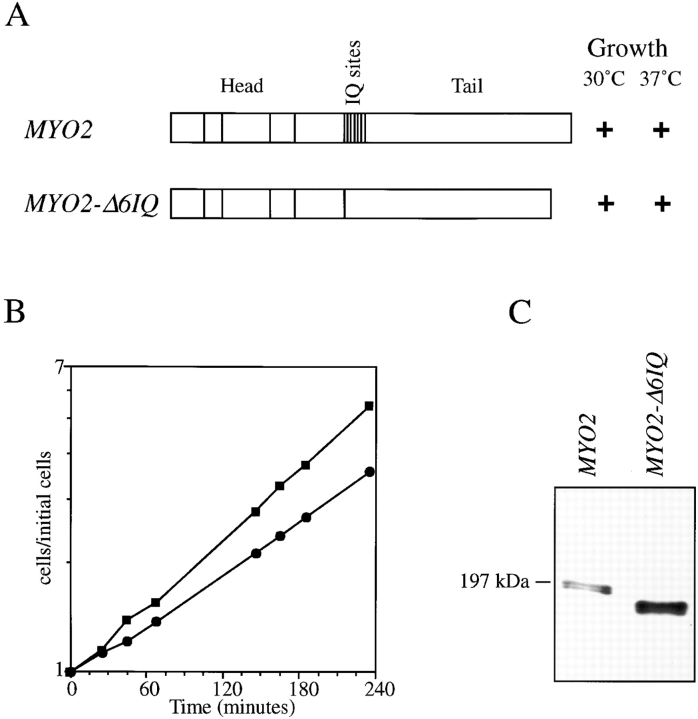
The neck of Myo2p contains six IQ sites in a tandem array spanning amino acids 790–921 (Johnston et al., 1991). Myo2p is essential for growth, but the six IQ sites are not. The mutant MYO2-Δ6IQ, lacking the sequence encoding amino acids 787–927, supports nearly normal growth as the only source of Myo2p, causing only a 10% reduction in growth rate (Fig. 1). An immunoblot confirmed that neckless Myo2-Δ6IQp is stably produced and that no wild-type Myo2p is present in the strain (Fig. 1 C). The mutant cells have a normal budding index but on average are 10% larger in size. The distribution of bud sizes was similar to that seen in wild-type cultures. The mutant cultures contained about 5% large unbudded cells not found in the wild-type cultures. A strain carrying MYO2-Δ6IQ does not grow at 38.5°C, whereas an isogenic wild-type strain grows poorly. A strain carrying MYO2-Δ6IQ grows well on medium with or without 1 M sorbitol (data not shown).
Figure 1.
The mutant MYO2-Δ6IQ allows growth. Strains carrying wild-type MYO2 (CRY1) or MYO2-Δ6IQ (RSY21) were plated on YPD medium and incubated for 3 d at 30 and 37°C (A). Log-phase cultures were diluted to 10,000 cells/μl in YPD medium, and growth was monitored over 4 h. Cell number was determined on a Coulter counter (Coulter Corporation, Hialeah, FL). (B) ▪, wild-type MYO2 strain, CRY1; •, MYO2-Δ6IQ strain, RSY21. An immunoblot of extracts made from log-phase cultures of strains CRY1 and RSY21 (C). The antibody was anti-Myo2p (Lillie and Brown, 1994).
The neckless mutant protein localizes indistinguishably from that of wild-type Myo2p (Fig. 2). In both cases, unbudded cells have either no localization or a small patch of Myo2p. In small- and medium-budded cells, the Myo2p is present in a concentrated region at the bud tip. In large-budded cells, the Myo2p is either at the bud tip or at the bud neck (Fig. 2). These results are consistent with previous analysis of Myo2p localization (Brockerhoff et al., 1994; Lillie and Brown, 1994).
Figure 2.
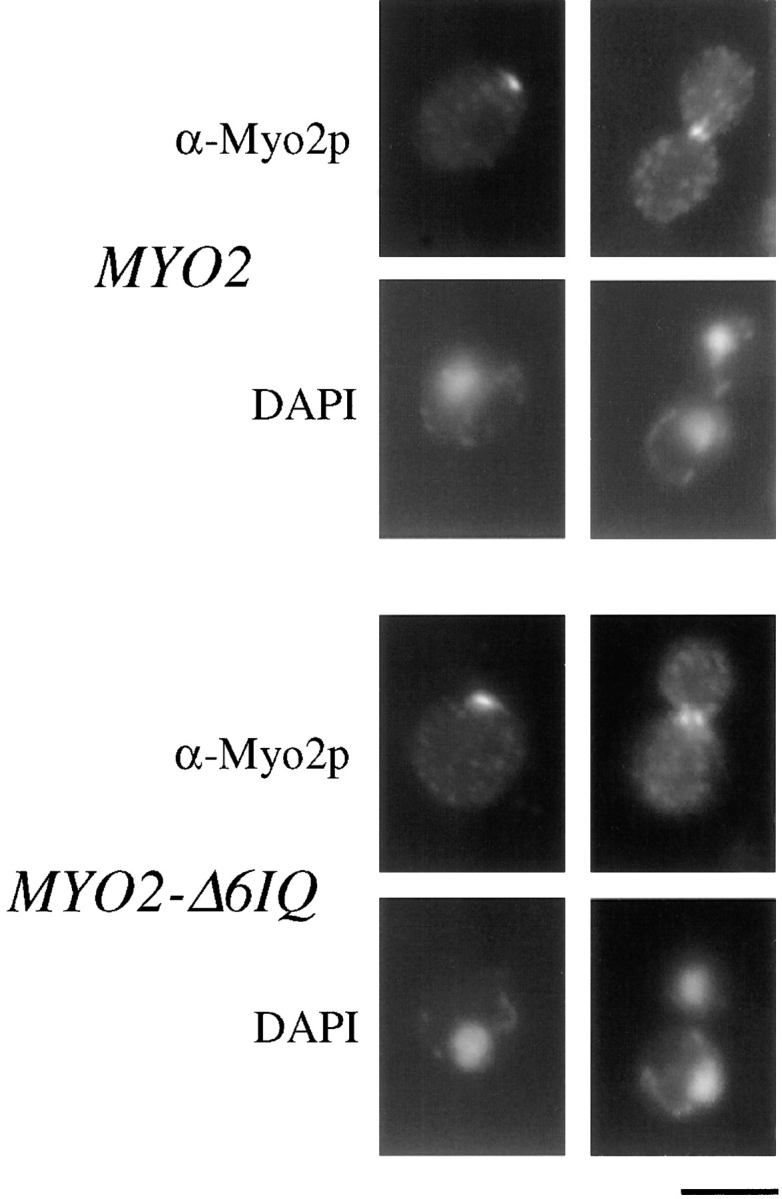
Comparison of the localization of Myo2p and Myo2-Δ6IQp. Cells were stained with affinity-purified anti-Myo2p antibody as described in Materials and Methods. Cells were simultaneously stained with DAPI to stain DNA. Bar, 5 μm.
Neckless MYO2-Δ6IQ Affects Calmodulin Localization
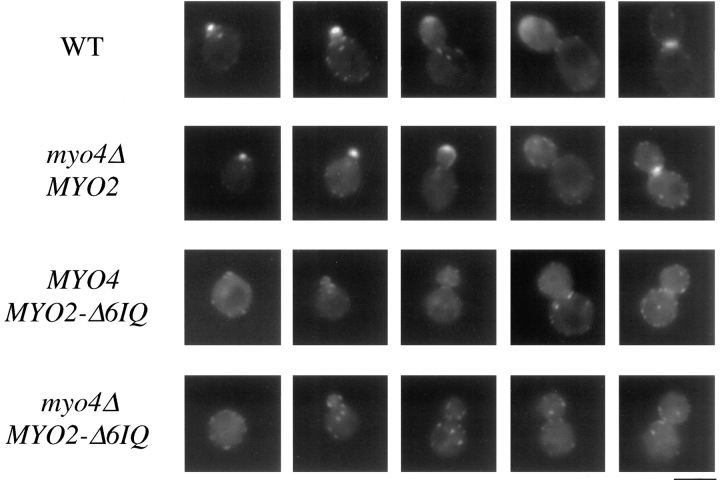
Our previous results strongly suggest calmodulin is a light chain of Myo2p. Calmodulin localizes to the sites of cell growth, and calmodulin mutants display defects in polarized growth (Brockerhoff and Davis, 1992; Davis, 1992; Ohya and Botstein, 1994). Mutations in CMD1 exacerbate defects caused by myo2-66 in an allele-specific manner. The viability of the MYO2-Δ6IQ strain allows us to test if the localization of calmodulin at sites of cell growth depends on the six IQ sites of Myo2p. Localization of a fusion of GFP to calmodulin was examined in wild-type and neckless mutant strains. GFP–calmodulin functionally replaces calmodulin and localizes properly (Moser et al., 1997). In a strain carrying wild-type MYO2, GFP–calmodulin is found at sites of cell growth in 84% of the cells (Fig. 3 and Table III). This number is equivalent to values obtained by immunolocalization (Brockerhoff and Davis, 1992). The cells without polarized calmodulin are either large-budded cells in the process of moving calmodulin from the bud tip to the bud neck or unbudded cells that have not yet started bud formation. In a strain carrying MYO2-Δ6IQ, only 15% of the cells have polarized calmodulin with greatly reduced and more diffuse signal even in these cells. Myo4p localizes diffusely in the yeast bud (Jansen et al., 1996). We tested whether the remaining GFP–calmodulin in the bud of the MYO2-Δ6IQ strain was dependent on Myo4p, which is not essential for growth. Strains deleted for MYO4 and containing wild-type MYO2 show the same localization of GFP–calmodulin as a wild-type strain containing both MYO4 and MYO2. However, in a strain carrying MYO2-Δ6IQ and myo4Δ, GFP–calmodulin is polarized in only 4% of the cells (Fig. 3 and Table III). Thus, calmodulin localization to sites of polarized growth is largely dependent on the six IQ sites of Myo2p, although Myo4p contributes to a small extent.
Figure 3.
Localization of GFP–calmodulin in myo4Δ strains. Cultures of RSY38-17c (WT), RSY38-16A (myo4Δ MYO2), RSY38-9D (MYO4 MYO2-Δ6IQ), and RSY38-16C (myo4Δ MYO2-Δ6IQ) were grown to log phase. Samples were collected, mixed with equal volumes of 1% agarose dissolved in SDC, and mounted on microscope slides for image collection as described in the Materials and Methods. Bar, 5 μm.
Table III.
Calmodulin Localization in Myosin Mutants
| Genotype | Polarized calmodulin (%) | |
|---|---|---|
| Wild type | 84 | |
| myo2-Δ6IQ | 15 | |
| myo4Δ | 80 | |
| myo4Δ myo2-Δ6IQ | 4 |
MLC1, a New Light Chain
The newly completed genome sequence for S. cerevisiae identified a potential myosin light chain. YGL106w encodes a 149–amino acid polypeptide with significant homology to calmodulin and myosin light chains. Based on the results described below, we have renamed this gene MLC1. An alignment with vertebrate calmodulin and the most similar light chain reveals homology throughout the sequences (Fig. 4). Mlc1p shares 35% identity with vertebrate calmodulin, second to only yeast calmodulin among yeast proteins (Davis and Thorner, 1986). Mlc1p is 35% identical to a chicken light chain (MLE1) (Matsuda et al., 1981). Mlc1p is 31% identical to Acanthamoeba myosin 1C light chain, another unconventional myosin light chain (Wang et al., 1997).
Figure 4.
Alignment of the predicted amino acid sequence of MLC1 with three other small EF-hand proteins. Vertebrate calmodulin (Homo sapiens) amino acids 7–149, a chicken light chain (Gallus gallus, MLE1) amino acids 45–190, and Acanthamoeba myosin IC light chain (A. castellanii, MICLC) amino acids 7–149 are compared with the complete amino acid sequence of Mlc1p. Amino acid residues sharing identity between Mlc1p and at least one of the other sequences are shaded. Amino acids within potential Ca2+-binding loops are labeled with a solid line.
MLC1 Is Essential and Displays Haploinsufficiency
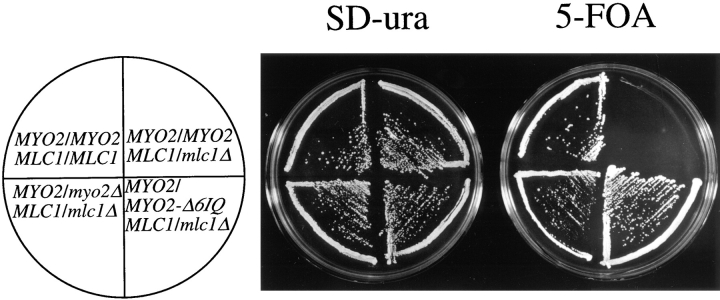
Initial attempts to make a diploid hemizygous for MLC1 were unsuccessful. Southern blot analysis revealed that one-step gene replacement (Rothstein, 1991) resulted in strains containing at least two wild-type copies of MLC1 for every one copy of the deletion construct. One possible explanation for this effect is that one copy of MLC1 per diploid genome is insufficient for viability. This phenomenon is called haploinsufficiency (Wilkie, 1994). We tested if MLC1 is haploinsufficient by repeating the gene disruption in a strain carrying two copies of MLC1 in the genome plus one copy of MLC1 on a plasmid. In this case, we were able to delete one copy of MLC1 from the genome as described in the Materials and Methods. The hemizygous diploid requires a plasmid carrying MLC1 for viability (Fig. 5). Haploid spores that contain the mlc1Δ gene disruption require MLC1 on a plasmid (data not shown). Therefore, MLC1 is haploinsufficient and essential.
Figure 5.
MLC1 displays haploinsufficiency. Strains JGY46, RSY105, RSY107, and RSY112, each carrying the MLC1 plasmid pRS289, were streaked onto both minimal medium lacking uracil and minimal medium containing 5′-FOA (which will kill any cells that require the plasmid pRS289). The plates were incubated at 30°C for 3 d.
To begin characterizing the phenotype of mlc1Δ cells, we examined spores from a hemizygous diploid strain. The diploid requires a plasmid copy of MLC1 to survive. Any spores receiving the plasmid would grow into colonies even if they carried the genomic copy of mlc1Δ. The mlc1Δ spores that did not inherit the plasmid were identified as the 20% of the spores that germinated but did not form colonies. The mlc1Δ cells grew elongated buds (Fig. 6). Cytokinesis was not complete because removal of the cell wall by treatment with zymolyase did not result in separation of the buds from the mother cells. DAPI staining revealed that the elongated cells have multiple nuclei (Fig. 6 C). Less than 1% of the spores from the control strain formed elongated cells.
Figure 6.
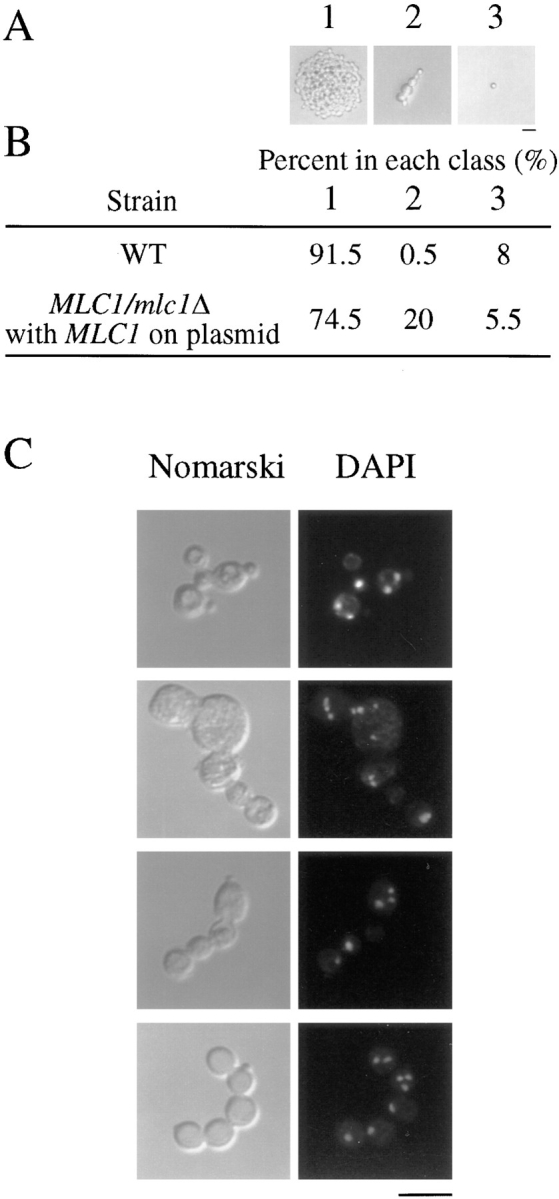
mlc1Δ spores have defects in cell separation. Strains JGY46 and RSY105(pRS289) were sporulated and tetrads were dissected. After 24 h, spores were characterized into classes: (1) germinated and formed colonies, (2) germinated and did not form colonies, or (3) did not germinate. Examples of each class are shown (A). 200 spores from each strain were classified (B). Germinated spores from RSY105(pRS289) were stained with DAPI. Elongated spores with DAPI staining are shown (C). Bars, 10 μm.
Mlc1p Interacts with the Neck of Myo2p
Two lines of evidence suggest Mlc1p is a light chain of Myo2p. First, the haploinsufficiency exhibited by MLC1 is suppressed by reduced copies of MYO2 (Fig. 5). A diploid strain hemizygous for both MYO2 and MLC1 is viable. Suppression of the haploinsufficiency by hemizygous MYO2 demonstrates that Mlc1p must ordinarily be present at the minimum level necessary for cell growth. MYO2-Δ6IQ can also suppress the haploinsufficiency of MLC1 (Fig. 5). However, MYO2-Δ6IQ cannot bypass the requirement for Mlc1p in a haploid strain (data not shown), so there must be at least one other essential function of Mlc1p in yeast.
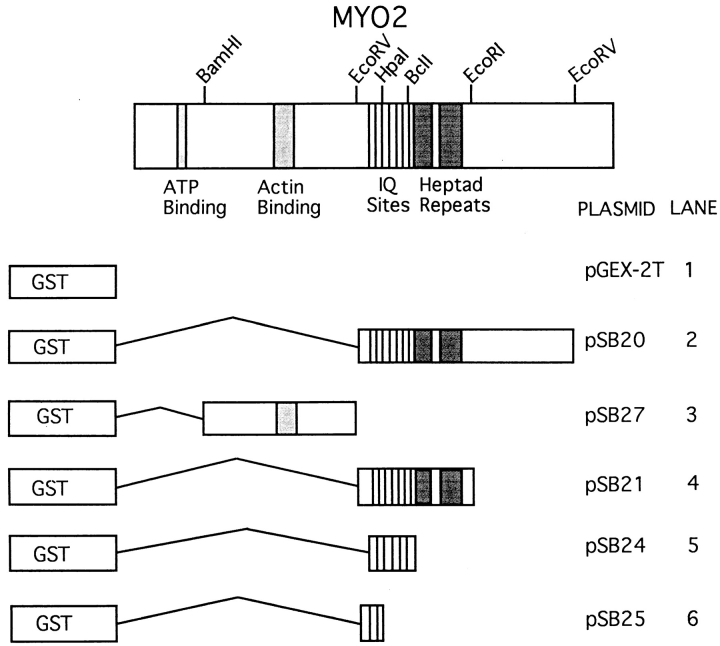
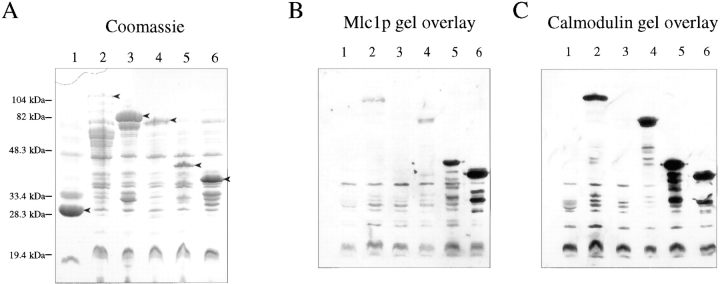
Second, Mlc1p binds directly to the IQ sites of Myo2p as assayed by a gel overlay protocol. The constructs used in this experiment are presented in Fig. 7. 6XHis-Mlc1p binds specifically to the fusion proteins that contain IQ sites (Fig. 8 B). As a control, we show calmodulin also binds specifically to fusion proteins that contain IQ sites (Fig. 8 C). Both calmodulin and 6XHis-Mlc1p bind equally well in the presence of Ca2+ or EGTA (data not shown). These data demonstrate that Mlc1p has a direct interaction with the IQ sites of Myo2p.
Figure 7.
Glutathione-S-transferase (GST)–Myo2p fusion proteins.
Figure 8.
Gel-overlay assays show in vitro binding of Mlc1p and calmodulin to the IQ sites of Myo2p. Myo2p fusion proteins were expressed and subjected to SDS-PAGE as described in Materials and Methods. Polyacrylamide gel stained with Coomassie blue. The full-length fusion protein in each extract is marked by an arrow (A). Autoradiographs of membranes incubated with 35S-labeled 6XHis-Mlc1p (B) and 35S-labeled calmodulin (C). The gel overlay assays were performed as described in Materials and Methods. Lanes 1–6 contain extracts of IPTG-induced E. coli strain GM1 containing plasmids pGEX-2T, pSB20, pSB27, pSB21, pSB24, and pSB25, respectively. See Fig. 7 and Table I for a description of the fusion proteins.
The Toxicity of Overexpressed MYO2 Is Suppressed by MLC1 but Not CMD1
We looked for a functional interaction between Mlc1p and Myo2p. A yeast strain carrying a high-copy number plasmid of MYO2 grows 40% slower than strains with normal levels of MYO2 (data not shown). Microscopy has revealed that 5% of cells in the MYO2 overexpression strain have abnormal bud necks or multiple buds (data not shown). A similar defect in high-copy number MYO4 strains may be caused by the neck region of Myo4p, and overexpression of calmodulin does not suppress this effect (Haarer et al., 1994). One possibility is that high copies of myosin light chains will overcome defects associated with MYO2 overexpression.
A strain overexpressing MYO2 shows a noticeable reduction in growth rate (Fig. 9 A). The same strain overexpressing MYO2-Δ6IQ grows normally. Northern blot analysis reveals that both MYO2 and MYO2-Δ6IQ are transcribed to similar steady-state levels in these two strains and at least fivefold over normal levels (Fig. 9 B). Surprisingly, the immunoblot for a strain overexpressing MYO2 does not show higher levels of Myo2p (Fig. 9, C and D). In fact, Myo2p seems to actually decrease in the strain overexpressing MYO2. Overexpressing MYO2-Δ6IQ results in high levels of Myo2-Δ6IQp (Fig. 9, C and D).
Figure 9.
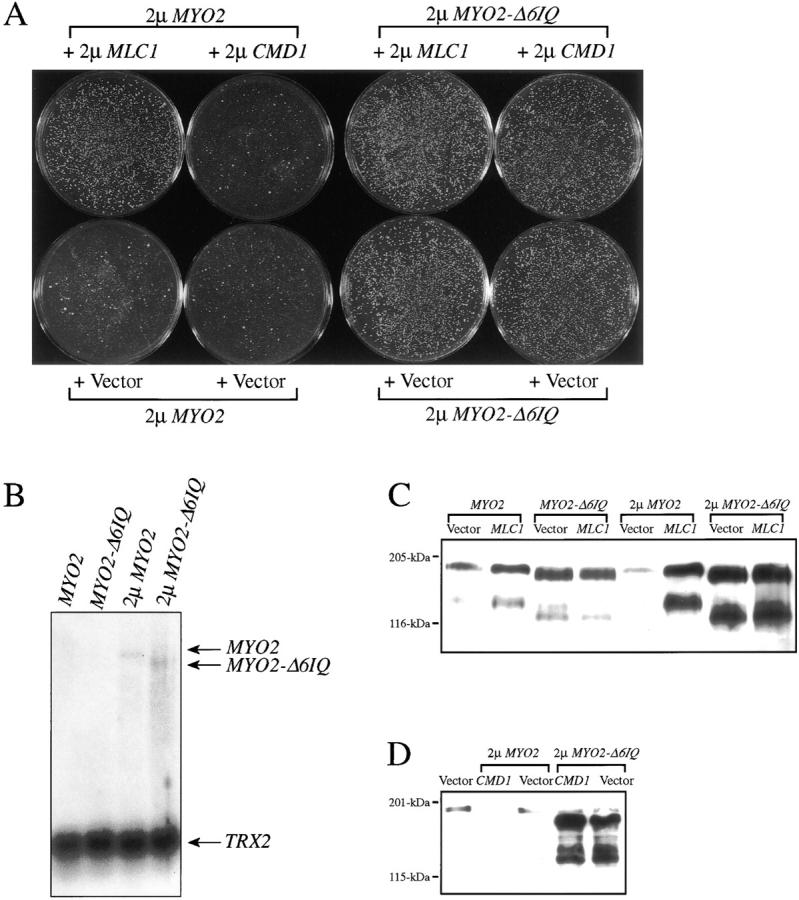
High copies of MLC1, but not high copies of CMD1, overcome the toxic effects of overexpressed MYO2. Plasmids used in these experiments are listed in Table I. EMY55-5D was transformed with plasmids encoding high copies of MYO2, MYO2-Δ6IQ, CMD1, or MLC1. Plates were incubated for 3 d at 30°C (A). CRY1 was transformed with pRS43, pRS172, pRS31, or pRS221, and mRNA was collected from log-phase cultures. 10 μg of polyA-containing RNA samples were analyzed as described in Materials and Methods. Blots were simultaneously hybridized with MYO2 and TRX2 32P-labeled probes (B). CRY1 was transformed with the indicated constructs to study the effects of high copies of MLC1 (pRS290) on strains with one extra copy of MYO2 (pRS43) or one extra copy of MYO2-Δ6IQ (pRS172), or overexpressing MYO2 (pRS31) or MYO2-Δ6IQ (pRS221) (C). CRY1 was transformed with the indicated constructs to study the effects of high copies of CMD1 (pTD28) on strains overexpressing MYO2 (pRS31) or MYO2-Δ6IQ (pRS221) (D). Log-phase extracts were made as described in Materials and Methods. Samples were assayed by immunoblot analysis using anti-Myo2p antibody (Lillie and Brown, 1994) to detect steady-state levels of Myo2p.
We hypothesized that high levels of calmodulin or Mlc1p would counteract the toxicity of overexpressed MYO2. Overexpressing CMD1 is unable to overcome the decreased growth rate (Fig. 9 A) or the low levels of Myo2p (Fig. 9 D) caused by overexpressed MYO2. In contrast, a high-copy number plasmid carrying MLC1 is able to fully suppress the overexpression MYO2 defects and results in a strain that has high levels of Myo2p (Fig. 9, A and C). These results indicate that the neck region of MYO2 is indeed responsible for an overexpression growth defect and that this toxicity is overcome by high levels of Mlc1p.
Mlc1p Stabilizes Myo2p
Because high copies of MLC1 are able to overcome defects caused by overexpression of MYO2, we tested the hypothesis that Mlc1p stabilizes Myo2p. Yeast cultures were analyzed for the stability of Myo2p by following Myo2p breakdown by immunoblot analysis (Fig. 10). In a wild-type strain, Myo2p has a half-life of greater than 8 h. In a strain carrying MYO2-Δ6IQ, the Myo2-Δ6IQp has a half-life of greater than 8 h. The neckless Myo2-Δ6IQp is at least as stable as wild-type Myo2p. When overexpressed, the half-life of Myo2p is only 2–3 h. These data explain why the Myo2p levels in the overexpression strain are not increased. In a strain overexpressing MYO2 and carrying high copies of MLC1, the half-life of Myo2p is 4 h. High copies of MLC1 are able to stabilize Myo2p, thus suggesting Mlc1p is a light chain responsible for stabilizing the neck region of Myo2p.
Figure 10.
Degradation of Myo2p. Cycloheximide was added to a final concentration of 100 μg/ml to log-phase cultures. Cultures were allowed to continue shaking, and samples were collected for immunoblot analysis as described in Materials and Methods. CRY1 containing either no plasmids (▴), plasmids pRS31 (high copies of MYO2) and pRS290 (high copies of MLC1) (▪), or pRS31 (high copies of MYO2) and pGF27 (vector alone) (•) was examined. Bands from immunoblots probed with anti-Myo2p antibody were quantified, and values were normalized for loading using a Coomassie blue–stained band that demonstrated high stability (A). The error bars represent standard deviation between two (wild type), two (with high copies of MYO2 and high copies of MLC1), or three (with high copies of MYO2) independent experiments. The immunoblots probed with anti-Myo2p antibody demonstrate Myo2p stability in CRY1 carrying either pRS31 and pRS290 (B) or pRS31 and pGF27 (C), and in CRY1 (MYO2 strain) alone (D) or in RSY21 (MYO2-Δ6IQ strain) alone (E). Numbers represent hours after cycloheximide addition.
Discussion
Electron micrographs of the chicken myosin V molecule show two heads connected to a 30-nm stalk. At the proximal end of each head is a 20-nm neck that likely corresponds to the six IQ sites (Cheney et al., 1993). This region is predicted to be an α-helix as is the neck of class II myosins. In class II myosins, the neck region is stabilized by the binding of the myosin light chains (Rayment et al., 1993; Houdusse and Cohen, 1996). Calmodulin is the previously identified light chain for both the chicken and the yeast myosin V (Cheney et al., 1993; Brockerhoff et al., 1994). However, calmodulin does not stabilize the yeast myosin. Here, we present evidence that MLC1 (YGL106w) encodes a new light chain for the yeast class V myosin Myo2p. Our results argue that Mlc1p binds to the neck region and stabilizes Myo2p.
When overexpressed, Myo2p has a decreased half-life and is toxic to yeast cells. Overproduction of calmodulin does not affect the stability of Myo2p, but overproduction of Mlc1p stabilizes Myo2p and ameliorates the toxicity. The requirement for Mlc1p to stabilize Myo2p is bypassed by deleting the six IQ sites in Myo2p. Thus, the presence of the IQ sites destabilizes Myo2p unless sufficient Mlc1p is present.
Removal of the IQ sites in Myo2p causes a slightly slower growth rate and slightly larger cells. A mutant Dictyostelium myosin II heavy chain ΔBLCBS, which lacks the two IQ sites, maintains 20% wild-type motor activity when measured in the sliding filament assay (Uyeda et al., 1996). The ΔBLCBS myosin allows cell division in vivo but demonstrates slightly slowed furrow formation during cytokinesis (Zang et al., 1997). Thus, both Myo2-Δ6IQp and the ΔBLCBS myosin confer relatively minor defects. These results indicate that the full efficiency of the myosin is not essential for myosin function in vivo.
MLC1 is essential and haploinsufficient. Haploinsufficiency is rarely observed in yeast genes. There are several reasons reduction in the number of gene copies may have deleterious effects. In some cases, reduced gene copy number affects regulatory genes working at a threshold level (Wilkie, 1994). An example of this form of regulation is dosage-dependent sex determination in Drosophila. Alternately, some proteins may be produced at the minimum level to give proper function. The ACT1 gene displays temperature-sensitive growth defects and increased osmosensitivity when present at low levels (Shortle et al., 1984). Finally, the stoichiometry of various protein components may be important. Several lines of evidence suggest that a fixed ratio between Mlc1p and Myo2p is required to confer normal growth. First, a diploid yeast strain carrying one copy of MLC1 can only grow if there is no more than one copy of wild-type MYO2. Second, the growth defects associated with a yeast strain overexpressing MYO2 are eliminated by adding extra copies of MLC1. Finally, removal of the IQ binding sites from Myo2p rescues the haploinsufficiency exhibited by MLC1.
Myo2-Δ6IQp does not overcome a requirement of Mlc1p for cell viability. This observation does not address the essential nature of the interaction between Mlc1p and Myo2p. It merely demonstrates that Mlc1p must have at least one essential function that does not involve binding to the neck of Myo2p. None of the other four myosins in yeast are essential for growth (Brown, 1997), suggesting that Mlc1p has an essential function unrelated to myosins. Inviable mlc1Δ cells show a striking defect in cytokinesis, resulting in enlarged, multinucleate cells. This phenotype is identical to defects detected in cells deleted for IQG1, which encodes a newly discovered IQGAP family member in yeast (Epp and Chant, 1997; Lippincott and Li, 1998). Iqg1p contains five IQ sites that could act as binding sites for Mlc1p. We propose that one essential function of Mlc1p is to act as a light chain for IQGAP.
Calmodulin is an additional light chain for Myo2p. We have previously shown that calmodulin and Myo2p coimmunoprecipitate from yeast cell extracts and interact in vitro. A mutation that is in the actin-binding site of Myo2p and interferes with Myo2p function dramatically worsens the phenotype conferred by mutations in calmodulin (Brockerhoff et al., 1994). Here we show that the localization of calmodulin to sites of cell growth is predominately dependent on the IQ sites of Myo2p with only a small contribution by the other yeast class V myosin Myo4p. In a MYO2-Δ6IQ strain lacking MYO4, calmodulin localizes to the spindle pole body and to fast moving patches on the cell surface. The protein target that binds calmodulin in these fast moving patches has yet to be identified. The IQGAP protein in yeast localizes to the actin ring at the bud neck during cytokinesis (Epp and Chant, 1997; Lippincott and Li, 1998). Because calmodulin localization to the bud neck during cytokinesis depends on Myo2p, either calmodulin does not bind to yeast IQGAP or calmodulin binds IQGAP at levels below detection.
Only one other noncalmodulin unconventional myosin light chain has been characterized. Biochemical studies identified MICLC as a light chain for Acanthamoeba myosin IC (Wang et al., 1997). Mlc1p and MICLC share 31% sequence identity, and both proteins share significant identity with other calmodulin/EF-hand superfamily members. The chicken class V myosin has at least three light chains, calmodulin and two additional uncharacterized small proteins that copurify with the myosin (Cheney et al., 1993). These two small proteins have yet to be identified.
In conclusion, we presented several lines of evidence that Myo2p has at least two light chains, calmodulin and a newly characterized protein, Mlc1p. Mlc1p regulates the stability of Myo2p by binding to the neck region. Our characterization of Mlc1p may aid in the identification of the additional light chains of the chicken class V myosins.
Acknowledgments
We thank Susan Brown for plasmid myo4Δ::URA3.
This work was supported by National Institutes of Health grant GM40506 (T.N. Davis). R.C. Stevens was supported by Public Health Services National Research Service Award T32 GM07270.
Abbreviations used in this paper
- 5′-FOA
5′-fluoro orotic acid
- DAPI
4′,6-diamidino-2-phenylindole
- GFP
green fluorescent protein
- GST
glutathione-S-transferase
Footnotes
Address all correspondence to Trisha N. Davis, Department of Biochemistry, Box 357350, University of Washington, Seattle, WA 98195-7350. Tel.: (206) 543-5345. Fax: (206) 685-1792. E-mail: tdavis@u.washington.edu
References
- Bobola N, Jansen RP, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- Brockerhoff SE, Davis TN. Calmodulin concentrates at regions of cell growth in Saccharomyces cerevisiae. . J Cell Biol. 1992;118:619–629. doi: 10.1083/jcb.118.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockerhoff SE, Edmonds CG, Davis TN. Structural analysis of wild-type and mutant yeast calmodulins by limited proteolysis and electrospray ionization mass spectrometry. Protein Sci. 1992;1:504–516. doi: 10.1002/pro.5560010405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockerhoff SE, Stevens RC, Davis TN. The unconventional myosin, Myo2p, is a calmodulin target at sites of cell growth in Saccharomyces cerevisiae. . J Cell Biol. 1994;124:315–323. doi: 10.1083/jcb.124.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SS. Myosins in yeast. Curr Opin Cell Biol. 1997;9:44–48. doi: 10.1016/s0955-0674(97)80150-0. [DOI] [PubMed] [Google Scholar]
- Cheney RE, Mooseker MS. Unconventional myosins. Curr Opin Cell Biol. 1992;4:27–35. doi: 10.1016/0955-0674(92)90055-h. [DOI] [PubMed] [Google Scholar]
- Cheney RE, O'Shea MK, Heuser JE, Coelho MV, Wolenski JS, Espreafico EM, Forscher P, Larson RE, Mooseker MS. Brain myosin-V is a two-headed unconventional myosin with motor activity. Cell. 1993;75:13–23. doi: 10.1016/S0092-8674(05)80080-7. [DOI] [PubMed] [Google Scholar]
- Coulondre C, Miller JH. Genetic studies of the lac repressor: IV. Mutagenic specificity in the lacI gene of Escherichia coli. . J Mol Biol. 1977;117:577–606. doi: 10.1016/0022-2836(77)90059-6. [DOI] [PubMed] [Google Scholar]
- Davis TN. A temperature-sensitive calmodulin mutant loses viability during mitosis. J Cell Biol. 1992;118:607–617. doi: 10.1083/jcb.118.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, T.N., and J. Thorner. 1986. Calmodulin and other calcium-binding proteins in yeast. In Yeast Cell Biology. Vol. 33. J.B. Hicks, editor. Alan R. Liss, Inc., New York. 477–503.
- Davis TN, Thorner J. Vertebrate and yeast calmodulin, despite significant sequence divergence, are functionally interchangeable. Proc Natl Acad Sci USA. 1989;86:7909–7913. doi: 10.1073/pnas.86.20.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TN, Urdea MS, Masiarz FR, Thorner J. Isolation of the yeast calmodulin gene: calmodulin is an essential protein. Cell. 1986;47:423–431. doi: 10.1016/0092-8674(86)90599-4. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Miller KG, Botstein D. Yeast actin-binding proteins: evidence for a role in morphogenesis. J Cell Biol. 1988;107:2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp JA, Chant J. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr Biol. 1997;7:921–929. doi: 10.1016/s0960-9822(06)00411-8. [DOI] [PubMed] [Google Scholar]
- Espindola FS, Espreafico EM, Coelho MV, Martins AR, Costa FRC, Mooseker MS, Larson RE. Biochemical and immunological characterization of p190-calmodulin complex from vertebrate brain: a novel calmodulin-binding myosin. J Cell Biol. 1992;118:359–368. doi: 10.1083/jcb.118.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromherz S, Szent-Gyorgyi AG. Role of essential light chain EF hand domains in calcium binding and regulation of scallop myosin. Proc Natl Acad Sci USA. 1995;92:7652–7656. doi: 10.1073/pnas.92.17.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, van-Tuinen D, Brockerhoff SE, Neff MM, Davis TN. Can calmodulin function without binding calcium? . Cell. 1991;65:949–959. doi: 10.1016/0092-8674(91)90547-c. [DOI] [PubMed] [Google Scholar]
- Geiser JR, Sundberg HA, Chang BH, Muller EGD, Davis TN. The essential mitotic target of calmodulin is the 110-kilodalton component of the spindle pole body in Saccharomyces cerevisiae. . Mol Cell Biol. 1993;13:7913–7924. doi: 10.1128/mcb.13.12.7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan B, Bowser R, Novick P. The role of Myo2, a yeast class V myosin, in vesicular transport. J Cell Biol. 1995;128:1055–1068. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer BK, Petzold A, Lillie SH, Brown SS. Identification of MYO4, a second class V myosin gene in yeast. J Cell Sci. 1994;107:1055–1064. doi: 10.1242/jcs.107.4.1055. [DOI] [PubMed] [Google Scholar]
- Hill KL, Catlett NL, Weisman LS. Actin and myosin function in directed vacuole movement during cell division in Saccharomyces cerevisiae. . J Cell Biol. 1996;135:1535–1549. doi: 10.1083/jcb.135.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houdusse A, Cohen C. Structure of the regulatory domain of scallop myosin at 2Å resolution: implications for regulation. Structure. 1996;4:21–32. doi: 10.1016/s0969-2126(96)00006-8. [DOI] [PubMed] [Google Scholar]
- Jansen RP, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin Myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- Johnston GC, Prendergast JA, Singer RA. The Saccharomyces cerevisiae MYO2gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, E.W., and G.R. Fink. 1982. Regulation of amino acid and nucleotide biosynthesis in yeast. In The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 181–299.
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lillie SH, Brown SS. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. . J Cell Biol. 1994;125:825–842. doi: 10.1083/jcb.125.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda G, Maita T, Kato Y, Chen JIU, Umegane T. Amino acid sequences of the cardiac L-2A, L-2B and gizzard 17000-Mr light chains of chicken muscle myosin. FEBS Lett. 1981;135:232–236. doi: 10.1016/0014-5793(81)80789-2. [DOI] [PubMed] [Google Scholar]
- Miller, J.H. 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor Laboratory Press, NY. 466 pp.
- Moser MJ, Flory MR, Davis TN. Calmodulin localizes to the spindle pole body of Schizosaccharomyces pombeand performs an essential function in chromosome segregation. J Cell Sci. 1997;110:1805–1812. doi: 10.1242/jcs.110.15.1805. [DOI] [PubMed] [Google Scholar]
- Nascimento AAC, Amaral RG, Bizario JCS, Larson RE, Espreafico EM. Subcellular localization of myosin-V in the B16 melanoma cells, a wild-type cell line for the dilutegene. Mol Biol Cell. 1997;8:1971–1988. doi: 10.1091/mbc.8.10.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenthal RK, Riles L, Johnston M, Hegemann JH. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Ohya Y, Botstein D. Diverse essential functions revealed by complementing yeast calmodulin mutants. Science. 1994;263:963–966. doi: 10.1126/science.8310294. [DOI] [PubMed] [Google Scholar]
- Provance DW, Wei M, Ipe V, Mercer JA. Cultured melanocytes from dilute mutant mice exhibit dendritic morphology and altered melanosome distribution. Proc Natl Acad Sci USA. 1996;93:14554–14558. doi: 10.1073/pnas.93.25.14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I, Rypniewshi WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 1650 pp.
- Sherman, F., G.R. Fink, and J.B. Hicks. 1986. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 186 pp.
- Shortle D, Novick P, Botstein D. Construction and genetic characterization of temperature-sensitive mutant alleles of the yeast actin gene. Proc Natl Acad Sci USA. 1984;81:4889–4893. doi: 10.1073/pnas.81.15.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda TQP, Abramson PD, Spudich JA. The neck region of the myosin motor acts as a lever arm to generate movement. Proc Natl Acad Sci USA. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FS, Wolenski JS, Cheney RE, Mooseker MS, Jay DG. Function of myosin-V in filopodial extension of neuronal growth cones. Science. 1996;273:660–663. doi: 10.1126/science.273.5275.660. [DOI] [PubMed] [Google Scholar]
- Wang Z-Y, Sakai J, Matsudaira PT, Baines IC, Sellers JR, Hammer JA, III, Korn ED. The amino acid sequence of the light chain of Acanthamoebamyosin 1C. J Muscle Res Cell Motil. 1997;18:395–398. doi: 10.1023/a:1018686428955. [DOI] [PubMed] [Google Scholar]
- Wilkie AO. The molecular basis of genetic dominance. J Med Genet. 1994;31:89–98. doi: 10.1136/jmg.31.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise JA. Preparation and analysis of low molecular weight RNAs and small ribonucleoproteins. Methods Enzymol. 1991;194:405–415. doi: 10.1016/0076-6879(91)94031-7. [DOI] [PubMed] [Google Scholar]
- Xie X, Harrison DH, Schlichting I, Sweet RM, Kalabokis VN, Szent-Gyorgyl AG, Cohen C. Structure of the regulatory domain of scallop myosin at 2.8 Å resolution. Nature. 1994;368:306–312. doi: 10.1038/368306a0. [DOI] [PubMed] [Google Scholar]
- Zang JH, Cavet G, Sabry JH, Wagner P, Moores SL, Spudich JA. On the role of myosin-II in cytokinesis division of Dictyosteliumcells under adhesive and nonadhesive conditions. Mol Biol Cell. 1997;8:2617–2629. doi: 10.1091/mbc.8.12.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Muller EGD, Amacher SL, Northrop JL, Davis TN. A dosage-dependent suppressor of a temperature-sensitive calmodulin mutant encodes a protein related to the fork headfamily of DNA-binding proteins. Mol Cell Biol. 1993;13:1779–1787. doi: 10.1128/mcb.13.3.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]