Abstract
We examined intercadherin interactions in epithelial A-431 cells producing endogenous E-cadherin and recombinant forms of E-cadherin tagged either by myc or by flag epitopes. Three distinct E-cadherin complexes were found. The first is a conventional E-cadherin–catenin complex consisting of one E-cadherin molecule linked either to β-catenin/α-catenin or to plakoglobin/α-catenin dimers. The second is a lateral E-cadherin complex incorporating two E-cadherin– catenin conventional complexes combined in parallel fashion via dimerization of the NH2-terminal extracellular domain of E-cadherin. The third complex is likely to contain two E-cadherin–catenin conventional complexes derived from two opposing cells and arranged in an antiparallel fashion. Formation of the antiparallel but not lateral complex strictly depends on extracellular calcium and E-cadherin binding to catenins. Double amino acid substitution Trp156Ala/Val157Gly within the extracellular NH2-terminal E-cadherin domain completely abolished both lateral and antiparallel inter–E-cadherin association. These data support an idea that the antiparallel complex has the adhesion function. Furthermore, they allow us to suggest that antiparallel complexes derive from lateral dimers and this complex process requires catenins and calcium ions.
Keywords: cadherins, catenins, p120, intercellular adhesion, epithelial cells
The classic cadherins (e.g., E-, N-, and P-cadherins) are single transmembrane domain proteins involved in calcium-dependent homophilic cell–cell recognition and adhesion. In the intercellular adherens junctions, cytoplasmic portions of these proteins orchestrate assembly of the electron-dense plaque that anchors microfilaments to the plasma membrane. It is thought that the resulting junctional system plays a pivotal role in the establishment and maintenance of the unique tissue architecture (Vasiliev and Gelfand, 1981; Edelman et al., 1990; Takeichi, 1991; Hynes, 1993; Gumbiner, 1996; Hubar et al., 1996). Furthermore, it is now clear that classic cadherins are important elements of the complex signaling pathways controlling cellular motility, growth, and differentiation (Geiger and Ayalon, 1992; Klymkowsky and Parr, 1995; Peifer, 1995). Defects in cadherin-dependent intercellular adhesion accompany neoplastic transformation and tumor progression (Takeichi, 1993; Birchmeier and Behrens, 1994). Despite these critical functions, little is known about the physical interactions between cadherin molecules leading to the establishment of cell–cell contacts, intracellular signals, or the regulation of these interactions by extracellular or intracellular factors.
The extracellular region of classic cadherins consists of five repeating domains (EI–EV). Together they coordinate several calcium ions that maintain the rod-like conformation of the entire extracellular region and are also required for cadherin's adhesion activity (Ozawa et al., 1990a ; Pokutta et al., 1994; Maurer et al., 1996). Experiments with cadherin mutants and chimeric molecules showed that the NH2-terminal EI domain governs the binding specificity of cadherins (Blaschuk et al., 1990; Nose et al., 1990). Recently, X-ray crystallographic studies determined the three-dimensional structure of the EI domain of N- and E-cadherin (Overduin et al., 1995; Shapiro et al., 1995) and the EI–EII domains of E-cadherin (Nagar et al., 1996). All reports confirmed that calcium ions stabilize the interdomain organization of the extracellular cadherin region. Furthermore, analysis of cadherin crystals suggested that these proteins may form lateral homodimers. The models of organization of such dimers, however, differed significantly in two reports. According to one model (Shapiro et al., 1995) two cadherin molecules extending from the same cell surface laterally interact through hydrophobic interactions. A major feature of this interaction is the mutual incorporation of the conserved Trp residue, localized at the second position in mature classic cadherins, into the hydrophobic core of the paired molecule. This model was supported by experiments with the recombinant extracellular portion of Xenopus C-cadherin showing that homodimerization is calcium independent (Brieher et al., 1996). In the alternative model (Nagar et al., 1996), lateral cadherin dimers are stabilized by mutually coordinating calcium ions. The second model is consistent with the observation that at high concentrations the recombinantly expressed extracellular region of mouse E-cadherin dimerized in a calcium-dependent manner (Koch et al., 1997). The existence of any of such dimers on the cell surface was not confirmed by direct experiments. Dimerization of the extracellular E-cadherin segment, however, was demonstrated in pentameric structures assembled by chimeric proteins consisting of the extracellular portion of E-cadherin and the transmembrane region of cartilage oligomeric matrix protein (Tomschy et al., 1996).
It has been proposed that lateral cadherin dimers associate into antiparallel complexes that result in cell–cell adhesion (Shapiro et al., 1995). The structural basis of this interaction is not clear. Available data suggest that lateral cadherin dimers are more efficient in adhesive interactions than monomers and their formation may regulate intercellular adhesion (Brieher et al., 1996; Tomschy et al., 1996). Extracellular calcium ions were shown to be required for cadherin-dependent cell–cell adhesion (Kartenbeck et al., 1982; Volberg et al., 1986; Ozawa et al., 1990a ). Some evidence, however, suggested that disruption of cadherin adhesion by calcium removal may have a complex mechanism and may be triggered via remodeling of the microfilament cytoskeleton (Citi, 1992; Citi et al., 1994; Shapiro et al., 1995).
Another well-established feature of classic cadherins is their interaction with cytoplasmic proteins, catenins. The intracellular cadherin region directly interacts either with β-catenin or with plakoglobin (γ-catenin). Both these proteins associate with α-catenin, which is thought to link the cadherin–catenin complex to the actin cytoskeleton. Deletion of the catenin-binding domain completely abolished the adhesion activity of classic cadherins (Nagafuchi and Takeichi, 1988; Ozawa et al., 1989, 1990b ), and the attachment of cells to immobilized lateral cadherin dimers (Brieher et al., 1996). A possible explanation for these observations is that association with the cytoskeleton via catenins is an important prerequisite to the formation of E-cadherin clusters which, in turn, are required for lateral dimerization (Tomschy et al., 1996).
To further our understanding of classic cadherin-based adhesion, we developed an approach allowing us to directly observe the lateral and antiparallel (adhesive) cadherin complexes. We demonstrated that a relatively small pool of E-cadherin does form lateral homodimers. Features of these dimers are consistent with the model proposed by Shapiro et al. (1995). In addition to lateral dimers, we were able to detect adhesive complexes formed by cadherins extending from two opposing cells. E-cadherin association with catenins was required for the formation of the adhesive but not lateral dimers. We present evidence that lateral and adhesive cadherin complexes are the distinct and consecutive steps of cadherin-mediated adhesion and discuss a possible role of catenins in the establishment of the adhesion interactions.
Materials and Methods
Cloning of Human E-cadherin and Its Mutants
Fragments of the human E-cadherin gene between nucleotides 89 and 982 (fragment 1); 756 and 1,652 (fragment 2); 1,372 and 2,363 (fragment 3); 2,105 and 2,802 (fragment 4; nucleotide and amino acid sequences of E-cadherin are numbered accordingly to the sequence with GenBank/ EMBL/DDBJ accession number Z13009, Bussemakers et al., 1993) were amplified by PCR from a human salivary gland cDNA (Clontech, Palo Alto, CA) using the following pairs of primers: 5′-CCAGCCATGGGC-CCTTGGAGCCGC-3′ and 5′-GTAGGTGTTCACATCATCGTCCGC-3′; 5′-ATAGAGAACGCATTGCCACATACA-3′ and 5′-TCTGTTCCATAAATGTGTCTGGCT-3′; 5′-CACAAATCCAGTGAACAACGATGG-3′ and 5′-CATCATAGTAATAAACGTTGTCCC-3′; 5′-GATAACCAGAATAAAGACCAAGTG-3′ and 5′-GAACGCTGATTTC-TGCATTTCCCA-3′. The fragment 4 was blunt ended, cleaved by EcoRI, and then ligated with the EcoRI-SmaI–digested Bluescript II KS+ vector (Stratagene, La Jolla, CA). The resulting plasmid was cleaved by KpnI-EcoRI and ligated with the KpnI-EcoRI–digested fragment 3 that generates a plasmid BlEc3-4. In parallel experiments, the fragment 1 was blunt ended, treated with BamHI, and then ligated with the BamHI-EcoRV– digested Bluescript vector. The resulting plasmid was combined with the fragment 2 using overlapping EcoRI site that generates a plasmid BlEc1-2. Finally, to construct the BlEcwt plasmid encoding a full-length E-cadherin cDNA, the KpnI fragment of the plasmid BlEc1-2 was inserted into the KpnI-cleaved BlEc3-4 plasmid. E-cadherin cDNA of this plasmid was completely identical to the 89–2,802 nucleotide sequence of E-cadherin published by Bussemakers et al. (1993).
To facilitate detection and immunoprecipitation, E-cadherin or its mutants were tagged COOH terminally by 6× myc or by a single flag epitope. For this procedure, the BamHI site was substituted for the E-cadherin stop codon in the BlEcwt using PCR-mediated mutagenesis. This site was used for ligation of the E-cadherin gene with the unique BamHI site of 6× myc sequence in the plasmid CS26MT (Chitaev and Troyanovsky, 1997) or with the BglII site of the Flag sequence of the plasmid pFLAG-CTS (Sigma Chemical Co., St. Louis, MO). PCR-mediated mutagenesis was used to construct the plasmid BlEc1M encoding E-cadherin containing the internal deletion in the cytosolic domain between His773 and Leu791 (Ec1M). This deletion as was shown in preliminary experiments specifically eliminates the recognition site of anti–E-cadherin mAb (clone C20820, Transduction Laboratories, Lexington, KY). To introduce this deletion, two E-cadherin fragments (1,699–2,406 and 2,468–2,740 nucleotides) were amplified by PCR using primers 5′-TAATCCGGACACTGGT and 5′-AAACAATTGGCTCAGTCAAAGTC and 5′-AAACAATTGATGAGTGTCCCCCGGTAT and 5′-GGATCCGTCGTCCTCGCCGCCTCC. They were then ligated through the unique MunI site (underlined) and the resulting fragment was inserted between the BspEI and BamHI sites of the BlEc1M plasmid.
To construct the deletion mutant Ec1Δ(159–536)M, the unique NcoI/ BspEI fragment (94–1,702 nucleotides) in the BlEc1M was replaced with the fragment (94–573) resulting from PCR performed with primers 5′-CGGCCAGCCATGGGCCCT and 5′-AAAGTCCGGAGGAATAACCCAGTCTCT. Plasmids BlEc1Δ(772–882) and BlEc1Δ(748–882) encoding two different COOH terminally truncated E-cadherin mutants were also constructed using PCR. The BspEI/BamHI fragment (1,703–2,745) of the BlEc1M plasmid was replaced with the fragments amplified between 1,699 and 2,410 or between 1,699 and 2,339 nucleotides of E-cadherin cDNA, respectively. Identical sense primer 5′-TAATCCGGACACTGGT and different antisense primers 5′-AAAGGATCCCAGCTGGCTCAAGTC and 5′-AAAGGATCCGGTGTATCCTCTGGGGG were used. PCR-mediated mutagenesis was also used to introduce point mutations. In the mutant Ec1WVM, a nucleotide sequence GACTGGGTT encoding the Asp155TrpVal amino acid sequence was replaced with GACGCCGGC, resulting in a new NaeI site. In the mutant Ec1QNM, the nucleotide sequence GATCAGAAT encoding Asp254GlnAsn tripeptide was replaced with GATGCTGCA. For transfection of A-431 cells, the corresponding Bluescript clones were further subcloned into the eukaryotic expression vector pRcCMV (Invitrogen, Carlsbad, CA) containing a neomycin resistance gene and cytomegalovirus (CMV) promoter.
The integrity of the constructs was confirmed by restriction endonuclease mapping. All fragments obtained by PCR were completely sequenced using the sequencing core facility of Washington University Medical School (St. Louis, MO).
Cell Culture, DNA Transfection, and Immunofluorescence Microscopy
HaCat human keratinocytes (Boukamp et al., 1988) were provided by W.W. Franke (German Cancer Research Center, Heidelberg, Germany). Transfection of human epidermoid carcinoma A-431 cells (CRL1555; American Type Culture Collection, Rockville, MD) and selection, growth, and immunofluorescence microscopy were done as described (Chitaev et al., 1996). The following mouse monoclonal antibodies were used: anti-plakoglobin (clone 11E4; Zymed Laboratories, San Francisco, CA); anti-desmoglein (clone 3.10; provided by W.W. Franke); anti-myc (clone 9E10; provided by R. Kopan, Washington University, St. Louis, MO); rabbit anti-myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA); anti-flag (Sigma Chemical Co.), and anti–α-catenin, anti–E-cadherin (mAbs C20820 and C37020), anti-β-catenin, and anti-p120 (Transduction Laboratories). In all cases, anti–E-cadherin antibody C20820 was used except where indicated.
Immunoprecipitation and Sedimentation Analysis
For most immunoprecipitation experiments, 2 × 106 cells were cultured in 10-cm tissue culture dishes at 37°C for ∼72 h. In coculture experiments, 6 × 106 cells producing myc- and flag-tagged forms of E-cadherin mixed in a ratio 1:1 were cultured in a 10-cm dish for 24 h. The confluent monolayer (∼107 cells) was then washed with PBS and extracted in 1.5 ml of lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM DTT, 20 μM p-APMSF, 2 mM EDTA, and 1% NP-40) for 10 min. After clarification in a microfuge (model 5415C; Eppendorf Scientific, Inc., Hamburg, Germany) for 10 min at 14,000 rpm, the lysates were immunoprecipitated for 1.5 h at 4°C in the presence of specific antibody (∼1 μg per sample). The antigene–IgG complexes were precipitated with sequential incubation with protein A–Sepharose as described previously (Chitaev et al., 1996). For the E-cadherin immunodepletion experiments, cell lysates were incubated with 5 μg of anti–E-cadherin (clones C37020 or C20820) or anti-Dsg 3.10 monoclonal antibodies. Then the antibodies were collected by two rounds of anti-mouse IgG–agarose (Sigma Chemical Co.) chromatography. After repeating this step once (second round of immunodepletion), the lysates were immunoprecipitated by anti-plakoglobin antibody as described above. For sucrose gradient centrifugation, confluent monolayer cells from three 10-cm dishes, obtained as described above, were washed with PBS and lysed with 2 ml of lysis buffer. For metabolic labeling experiments, cells were cultivated in the presence of [35S]methionine, 50 μCi/ml for 16 h. Lysates (1 ml), precleaned by centrifugation (model TL-100; Beckman Instrs., Inc., Palo Alto, CA) at 100,000 g for 1 h, were loaded on top of a 12-ml linear 5–20% (wt/wt) sucrose gradient prepared in lysis buffer. In some experiments, either 10 mM EGTA was added into gradients or EDTA was replaced with 1 mM CaCl2. Gradients, centrifuged at 200,000 g for 17 h in a rotor (model SW40Ti; Beckman Instrs., Inc., Palo Alto, CA) at 4°C, were fractionated from bottom to top into 12 fractions (1 ml each), and then analyzed by coimmunoprecipitation. 50-μl aliquots of each gradient fraction before immunoprecipitation were mixed with concentrated (5×) SDS-PAGE loading buffer and analyzed by SDS-PAGE (Laemmli, 1970) and immunoblotting. 5 μl of each fraction was loaded. The following protein standards of known S values were centrifuged on replicate gradients: BSA, 4.5 S; IgG, 7.5 S; catalase, 11.35 S, apoferritin, 17 S.
Biotinylation of Cell Surface Proteins
The cells of three 10-cm dishes reaching near confluent growth were washed with ice-cold PBS containing 0.5 mM CaCl2 (PBS-C). Each plate was incubated at 4°C with 7 ml of 0.5 mg/ml of sulfo-NHS-LC-biotin (Pierce Chemical Co., Rockford, IL) in PBS-C for 1 h. The reaction was quenched by washing the cells with 1 M Tris/100 mM glycine buffer, pH 7.5. In some experiments, cells were dissociated into a single cell suspension by 5 min of treatment with 10 mM EGTA in PBS at 37°C after biotinylation. Then cells were lysed in lysis buffer and analyzed by sucrose gradient centrifugation and coimmunoprecipitation assays as described above. Biotinylated proteins were visualized with streptavidin-HRP conjugate (Pierce Chemical Co.) in conjunction with enhanced chemiluminescence (Boehringer Mannheim, Indianapolis, IN).
Results
Lateral Dimers of E-cadherin Are Exposed on the Surface of Epithelial Cells
The cytoplasmic domain of E-cadherin binds to either β-catenin or plakoglobin (Butz and Kemler, 1994; Hinck et al., 1994) that in turn, interacts directly with α-catenin (Jou et al., 1995). This gives rise to two distinct E-cadherin–catenin complexes, E-cadherin–β-catenin–α-catenin and E-cadherin–plakoglobin–α-catenin. The presence of a plakoglobin–β-catenin complex may indicate the assembly of these conventional cadherin–catenin complexes into higher order structures. To test this hypothesis, we subjected Triton X-100–soluble proteins from A-431 or HaCat cells to sucrose gradient fractionation followed by immunoprecipitation with anti-plakoglobin (Fig. 1 a) or anti-β-catenin (data not shown) mAbs. Both types of experiments showed that the conventional E-cadherin–catenin complexes sedimented at ∼9 S, in agreement with published results (Nelson et al., 1990; Ozawa and Kemler, 1992; Hinck et al., 1995). However, the plakoglobin–β-catenin complex separated into a distinct peak, sedimenting at 13 S. Both complexes were stable upon recentrifugation (Fig. 1, b and c), showing that the 9 S complex does not associate into a 13 S complex during in vitro manipulation. To determine whether E-cadherin is part of the β-catenin– plakoglobin complex, the 13 S fraction from metabolically labeled proteins of A-431 cells was immunoprecipitated with either anti–E-cadherin or anti-plakoglobin mAbs. Immunoprecipitates were found to contain an identical set of proteins, E-cadherin, plakoglobin and α- and β-catenins (Fig. 2 a). Furthermore, depletion of the 13 S fraction with any of two anti–E-cadherin mAbs used in this work before anti-plakoglobin immunoprecipitation strongly reduced β-catenin coimmunoprecipitation. Depletion with mAb against desmoglein, that is desmosome-specific cadherin of the A-431 cells, had no effect (Fig. 2 b). This suggests that the E-cadherin found in the 13 S fraction is complexed with plakoglobin and β-catenin. The plakoglobin– β-catenin complex detectable after E-cadherin immunodepletion is likely to contain P-cadherin, another classic cadherin of A-431 cells.
Figure 1.
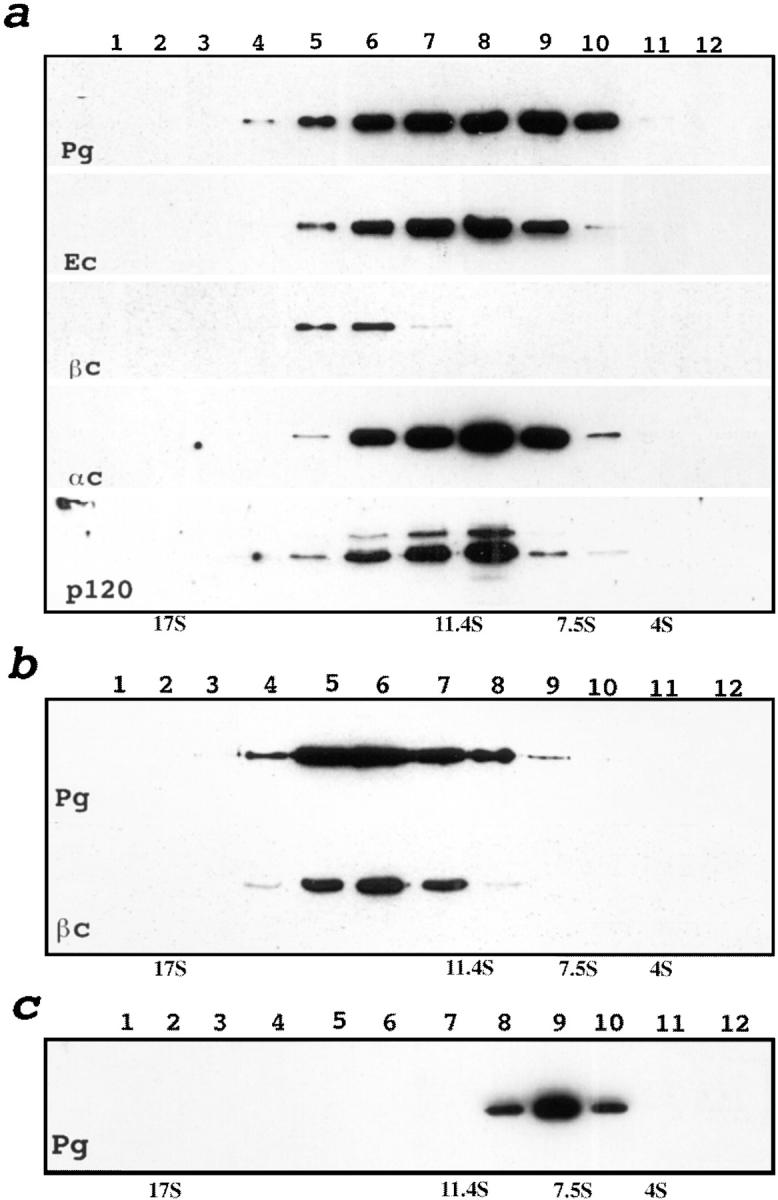
Sedimentation analysis of plakoglobin-associated complexes. (a) Total lysates of A-431 cells were subjected to sucrose gradient centrifugation (refer to Materials and Methods). Each fraction was coimmunoprecipitated with anti-plakoglobin mAb 11E4, separated by SDS-PAGE, and then analyzed by immunoblotting with plakoglobin (Pg), E-cadherin (Ec), β-catenin (βc), α-catenin (αc), and p120 (p120) antibodies. Note the plakoglobin–β-catenin complex appears in fractions 5–7. Two bands in the blot developed by anti-p120 mAb represent distinct splice forms of this protein. (b and c) Fraction 6 (b) or 9 (c) of the gradient was dialyzed for 6 h against lysis buffer and loaded onto a new gradient. After refractionation each fraction was analyzed as in a. Anti-β-catenin staining shows the stability of the plakoglobin/ β-catenin complex upon recentrifugation. The peak distribution of protein standards in a parallel gradient of known S values (bovine serum albumin, 4 S; rabbit IgG, 7.5 S; catalase, 11.4 S; apoferritin, 17 S) is shown at the bottom.
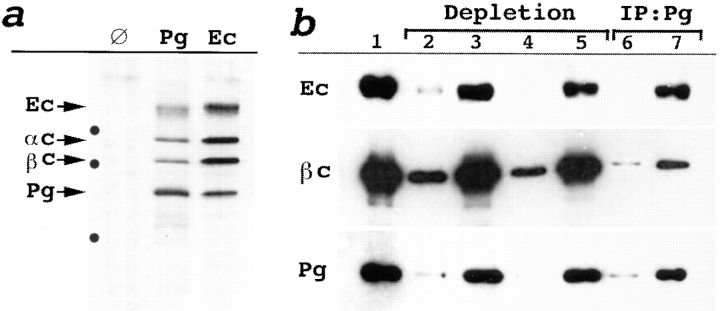
Figure 2.
Coimmunoprecipitation and immunoblot analysis of proteins collected from fraction 6 (13 S). (a) Autoradiograms of [35S]methionine proteins coimmunoprecipitated with mock (∅), anti-Pg (Pg), and anti–E-cadherin (Ec) antibodies and separated by SDS-PAGE. Identified proteins (Ec, E-cadherin; αc, α-catenin; βc, β-catenin; Pg, plakoglobin) are shown by arrows. Molecular weight markers are indicated by dots from top to bottom: 116, 97.4, and 67 kD. P-cadherin apparently comigrates with E-cadherin in the anti-plakoglobin immunoprecipitate. (b) Western blot analysis of the total proteins from fraction 6 developed with anti–E-cadherin (Ec), β-catenin (βc), and plakoglobin (Pg) antibodies before (lane 1), and after depletion with anti–E-cadherin C20820 (first round, lane 2; second round, lane 4) or anti-desmoglein (first round, lane 3; second round, lane 5) mAbs. The samples depleted with anti–E-cadherin (lane 6) or anti-desmoglein (lane 7) mAbs were coimmunoprecipitated with anti-plakoglobin mAbs (IP:Pg) and analyzed for the presence of E-cadherin, β-catenin, and plakoglobin. Depletion with E-cadherin antibody (lane 6), but not desmoglein antibody (lane 7), reduces recovery of the plakoglobin–β-catenin complex.
To show that the formation of this complex requires dimerization of E-cadherin, we expressed in A-431 cells human E-cadherin containing a 19-amino acid (aa)1 (His773–Leu791) long internal deletion in the cytoplasmic domain and COOH-terminal 6× myc epitope (Ec1M, see Fig. 3 a for details). As demonstrated by Western blotting (Fig. 3 b), this deletion completely abolished binding of E-cadherin to mAb C20820. Thus, cells stably transfected with the Ec1M DNA construct produce two forms of E-cadherin that could be specifically recognized by mAb C20820 (endogenous form) and by mAb against the myc epitope (Ec1M form). Only cell subclones producing Ec1M at the level similar to the level of endogenous E-cadherin, as estimated by immunoblotting of the total lysates of the selected clones with C37020 antibody recognizing both E-cadherin forms (data not shown), were studied. Double immunofluorescent microscopy of the Ec1M-producing cells using rabbit anti-myc serum and anti–E-cadherin mAb C20820 showed that the recombinant protein, despite the presence of this deletion, was incorporated efficiently into intercellular junctions (see Fig. 6). Western blot analysis of the immunoprecipitates obtained with anti-myc antibody showed that Ec1M, like endogenous E-cadherin, coimmunoprecipitated plakoglobin, β-catenin, α-catenin, and p120 (Fig. 4). Both endogenous and mutated E-cadherins have the same sedimentation properties (Fig. 3 c and not shown). Thus, the His773–Leu791 deletion and addition of the 6× myc tag does not influence the subcellular localization, sedimentation properties, and association with catenins of E-cadherin. These results allowed us to study dimerization of E-cadherin using the immunoprecipitation assay. Sucrose gradient sedimentation of Triton X-100–soluble proteins of the A-431 cells expressing Ec1M and subsequent immunoprecipitation of the obtained fractions with anti-myc (Fig. 3 b) or C20820 (data not shown) antibodies showed that the 13 S, but not the 9 S complex, incorporates both endogenous E-cadherin and Ec1M. Furthermore, biotinylation of the cell surface proteins from these cells before lysis and after sucrose gradient fractionation and anti-myc (data not shown) or mAb C20820 (Fig. 3 c) immunoprecipitation, clearly showed that the E-cadherin present in both 9 S and 13 S complexes is exposed on the cell surface.
Figure 3.
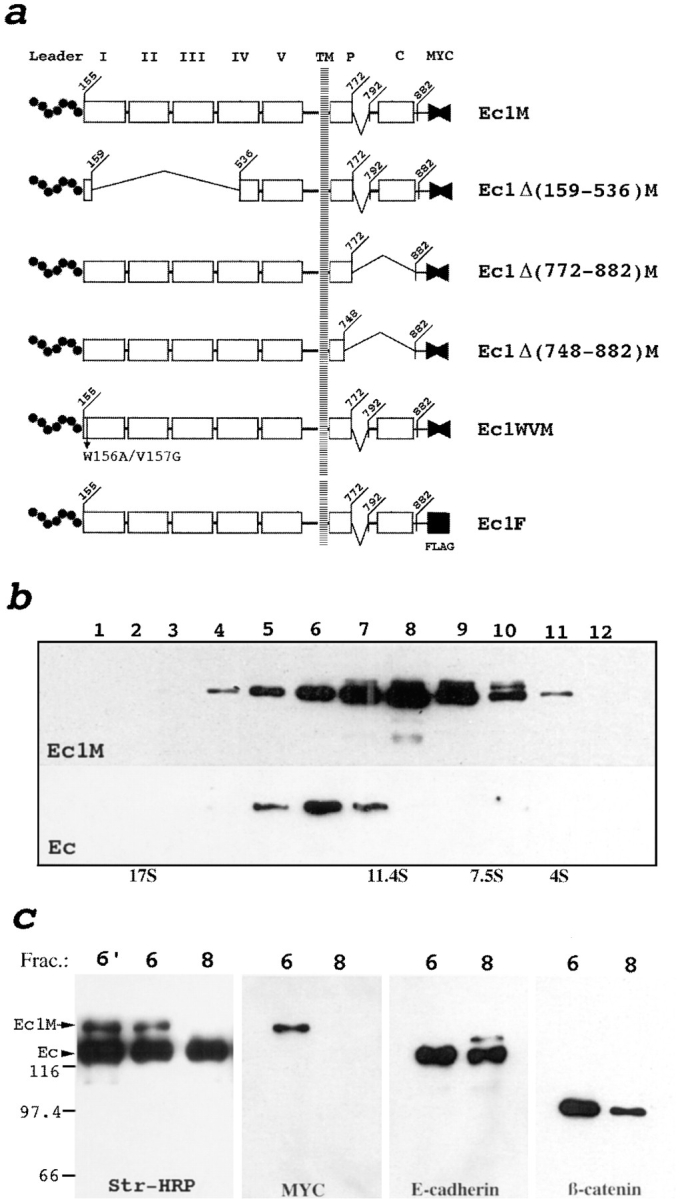
Schematic representation of human E-cadherin gene constructs (a) and sedimentation/coimmunoprecipitation analysis showing E-cadherin dimerization (b and c). (a) Five extracellular cadherin-like repeats (numbered I–V), the intracellular p120-binding site (P), the catenin-binding domain (C), and myc (fused solid triangles) or flag (solid square) epitopes are indicated. Deletions are depicted by brackets. Leader propeptide (Leader) is shown by the dotted line. Numbers show positions of the corresponding amino acids. (b) Fractions of the total lysate of the A-431 cells producing Ec1M were separated in a sucrose gradient. The fractions were then coimmunoprecipitated with anti-myc mAb and analyzed by immunoblotting with anti-myc and anti–E-cadherin mAbs for the presence of Ec1M (Ec1M) and endogenous E-cadherin (Ec), respectively. Sedimentation of the Ec1M–E-cadherin complex is the same as the plakoglobin–β-catenin complex (refer to Fig. 1 a). (c) Surface proteins of Ec1M-expressing cells were biotinylated and then the lysate was separated by sucrose gradient and fractions were immunoprecipitated by anti–E-cadherin mAb. Only the 13 S (fraction 6) and 9 S (fraction 8) fractions are shown. Immunoprecipitates were analyzed by immunoblotting using streptavidin-HRP conjugate (Str., HRP), anti-myc (myc), anti– E-cadherin, and anti–β-catenin antibodies. Note that anti–E-cadherin antibody coimmunoprecipitates biotinylated Ec1M only in fraction 6. This coimmunoprecipitation is stable after dissociation of the labeled cells into single-cell suspension (lane 6′). The absence of catenin biotinylation in E-cadherin–catenin complexes indicates specific incorporation of biotin only into surface proteins. Positions of endogenous E-cadherin (Ec) and Ec1M are indicated by arrows. Molecular weight markers are shown in kD.
Figure 6.
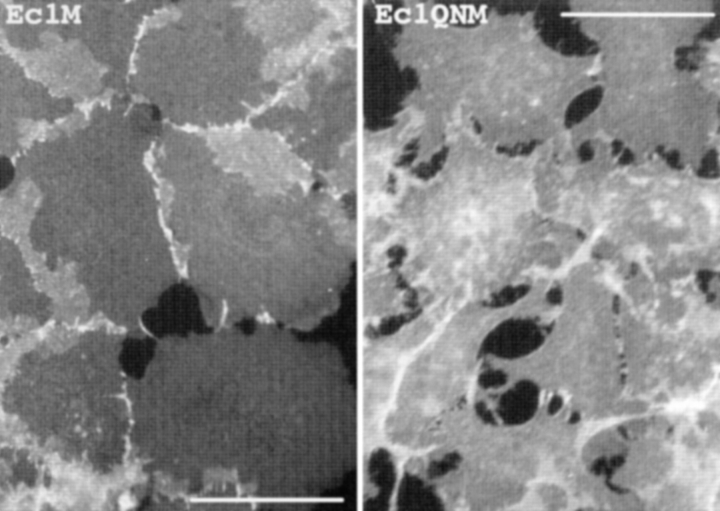
Immunofluorescent microscopy of cells expressing Ec1M (Ec1M) and Ec1QNM (Ec1QNM) stained with anti-myc antibody shows cell contact localization of Ec1M versus Ec1QNM protein. Note the strong morphological abnormalities of the Ec1QNM-expressing cells. Bar, 40 μm.
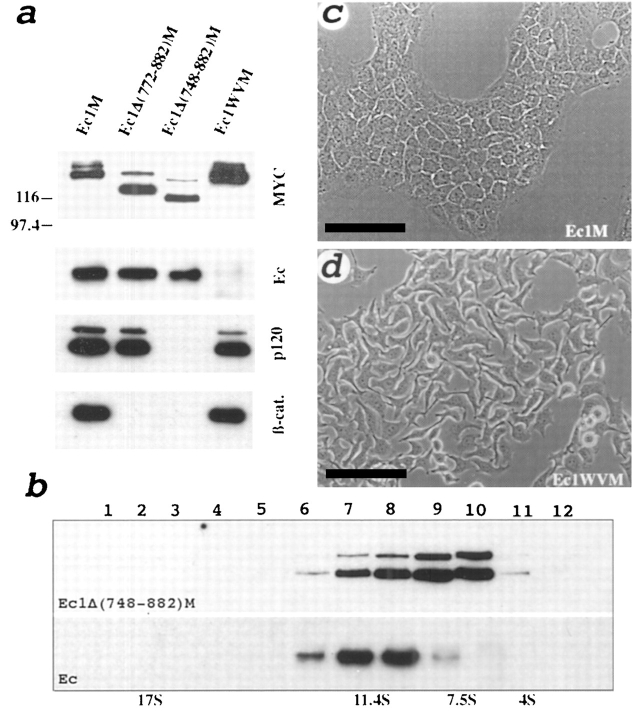
Figure 4.
Analysis of E-cadherin mutants stably expressed in A-431 cells. (a) Western blot of coimmunoprecipitates obtained using anti-myc 9E10 antibody from A-431 cells transfected to produce Ec1M, Ec1Δ(772–882)M, Ec1Δ(748–882)M, and Ec1WVM. Blots were developed with anti-myc (myc), anti– E-cadherin (Ec), anti-p120 (p120), and anti–β-catenin (β-cat) antibodies. (b) Sucrose gradient centrifugation of the lysate obtained from Ec1Δ(748–882)M-expressing cells. Each fraction of the gradient was coimmunoprecipitated with anti-myc antibody and analyzed by immunoblotting. Staining with anti-myc, Ec1Δ(748–882)M, and anti–E-cadherin (Ec) shows the distribution profile of the Ec1Δ(748–882)M protein, and its association with endogenous E-cadherin. Note that sedimentation of the hybrid complex shifted from 13 to 9 S. The upper band on Ec1Δ(748–882)M apparently represents precursor protein. (c and d) Phase-contrast photomicrographs of A-431 cells expressing Ec1M (c, Ec1M), and Ec1WVM (d, Ec1WVM) proteins showing a strong dominant-negative effect of the Ec1WVM mutant on cell morphology. Bars, 100 μm.
Experiments described above demonstrate that 13 S and 9 S E-cadherin complexes consist of the same proteins, E-cadherin, α- and β-catenins, and plakoglobin, suggesting that the 13 S complex is a heterodimer of the E-cadherin– β-catenin–α-catenin and E-cadherin–plakoglobin–α-catenin (9 S) complexes. To determine whether the E-cadherin found in the 13 S complex originates from the same or from two opposing cells, the surface proteins of Ec1M-expressing cells were biotinylated and the cells dissociated into a single cell suspension using 10 mM EGTA before lysis. The E-cadherin/Ec1M composition of the 13 S complex was not affected by complete dissociation of the cells (Fig. 3 c). This shows that at least a significant portion of the 13 S complex consists of E-cadherin molecules derived from the same cell. Taken together, these data demonstrate a presence on the cell surface of the E-cadherin complex in which both E-cadherin molecules are aligned in parallel orientation.
The Extracellular EI Domain, But Not Binding to Catenins or p120, Is Required for Lateral E-cadherin Dimerization
Next, we examined whether binding to cytosolic proteins is essential for lateral association of Ec1M with endogenous E-cadherin. The truncated version of Ec1M, the Ec1Δ(772– 882)M mutant (refer to Fig. 3 a and Fig. 4 for details), lacking the catenin-binding domain was expressed in A-431 cells. In agreement with the previous data (Nagafuchi and Takeichi, 1988; Ozawa et al., 1990b ), immunofluorescent microscopy and degradation of this mutant by trypsin treatment of the intact cells indicated that despite deletion of the catenin-binding domain, the Ec1Δ(772–882)M protein was transported to the cell surface. Results presented in Fig. 4 show that this mutant associates efficiently with endogenous E-cadherin. Besides catenins, in agreement with previous observations (Reynolds et al., 1994; Shibamoto et al., 1995), minor fractions of the 9 S and 13 S E-cadherin– catenins complexes were found to be associated with the tyrosine kinase substrate p120 (refer to Fig. 1 a and Fig. 4 a). Deletion of the catenin-binding domain in the Ec1Δ(772– 882)M mutant did not affect association with p120 (Fig. 4 a), whereas it was completely abolished by a deletion of an additional 24-aa residues from the COOH terminus of the Ec1Δ(772-882)M protein (Fig. 4 a). The resulting deletion mutant Ec1Δ(748–882)M was unable to bind p120 but formed hybrid complexes with endogenous E-cadherin (Fig. 4, a and b). As expected, the mobility of the hybrid complex in a sucrose gradient shifted from fraction 6 to 7 due to the absence of the catenin-binding site in the mutant. This complex was not affected by EGTA-induced dissociation of the cells. These experiments show that lateral dimerization of E-cadherin is mediated by its extracellular domain and does not depend on an interaction with the cytoplasmic proteins p120, plakoglobin, and catenins.
X-ray crystallographic analysis (Shapiro et al., 1995; Nagar et al., 1996) showed that the extracellular domain of cadherin forms homodimers. Shapiro et al. (1995) suggested that the side chain of Trp156 of each cadherin molecule protrudes into the hydrophobic core of the neighbor molecule. In the second model, the lateral dimers are stabilized by mutually coordinating calcium ions (Nagar et al., 1996). Both models are consistent with our observation that deletion of the extracellular cadherin-like repeats 1–3 completely abolished association of the resulting Ec1Δ(159–536)M with endogenous E-cadherin (data not shown). High stability of the 13 S complex in 10 mM EGTA does not support the role of Ca2+ in lateral dimerization. To determine whether the Trp156 residue is required for E-cadherin dimerization, we constructed an Ec1WVM combinatorial mutant in which Trp156 and a neighbor residue, Val157, were substituted for Ala and Gly, respectively (refer to Fig. 3 a). In A-431 cells, this mutant was translocated to the cell surface (data not shown) and formed a conventional 9 S trimer, but not the 13 S complex (Fig. 4 a). Additionally, the Ec1WVM mutant and the other extracellular mutant Ec1Δ(159–536)M, induced a strong dominant-negative effect on cell–cell adhesion (Fig. 4, c and d). A similar effect has been described previously for different cadherin mutants lacking the extracellular region (Kintner, 1992; Fujimori and Takeichi, 1993). Because this phenotypic effect may abolish lateral E-cadherin association indirectly, we determined whether these cells contain the 13S plakoglobin/β-catenin complex incorporating endogenous E-cadherin. This was done by analyzing the association between plakoglobin and β-catenin using anti-plakoglobin (or anti-β-catenin) coimmunoprecipitation assays as described above. The data showed that the amount and sedimentation properties of the 13 S complex were unchanged in control and in transfected cells (data not shown), suggesting that a failure of the Ec1WVM mutant to interact with endogenous E-cadherin was not caused by a dominant-negative effect. Thus, our data imply that lateral interaction between two E-cadherin molecules in the 13S complex is mediated by the mechanism proposed by Shapiro et al. (1995).
Adhesive, Head to Head, and Not Lateral E-cadherin Interaction Requires Catenins and Calcium
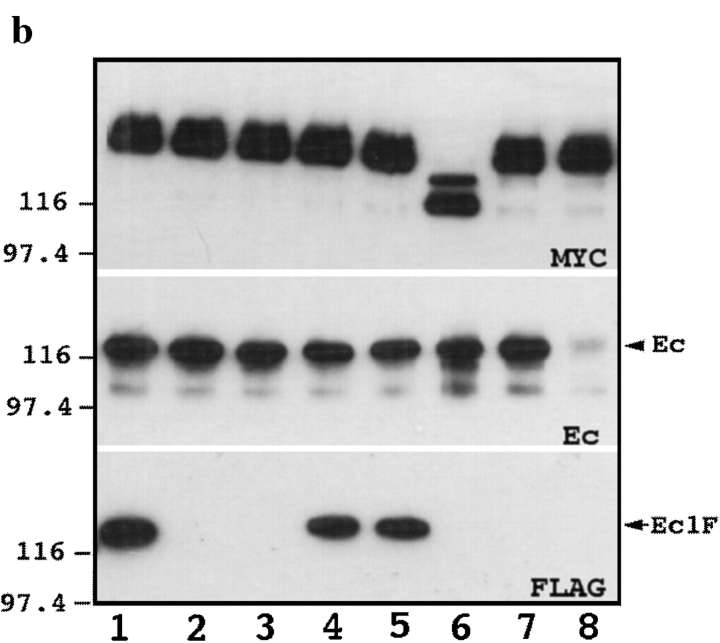
The data presented provide clear evidence that the lateral dimerization of the E-cadherin is independent of calcium and catenins. In contrast, homophilic cadherin-mediated cell–cell adhesion strictly depends on both these factors, suggesting that catenin and calcium may directly control adhesive, but not lateral association between cadherins. Our experiments with Ec1M protein, however, failed to demonstrate any multicadherin complexes other than lateral E-cadherin–catenin hexamers. The adhesive intercadherin association, however, could be hidden in the experiments described above if adhesive and lateral E-cadherin complexes have the same compositions and, hence, similar sedimentation properties. To test for the formation of adhesive complexes in which E-cadherin–catenin complexes are arranged in antiparallel orientations, we applied an approach successfully used in our recent experiments with desmosomal cadherins (Chitaev and Troyanovsky, 1997). A-431 cells expressing Ec1M were cocultured overnight with A-431 cells stably producing the Ec1F form of E-cadherin. The latter protein contained the same internal deletion as Ec1M and was selectively tagged by flag, but not the myc epitope. In this coculture system, specific coimmunoprecipitation of Ec1F with anti-myc antibody would indicate the adhesive association between two forms of E-cadherin, whereas the presence of endogenous E-cadherin in the same immunoprecipitates, assessed by mAb C20820, would show both lateral and adhesive types of E-cadherin complexes.
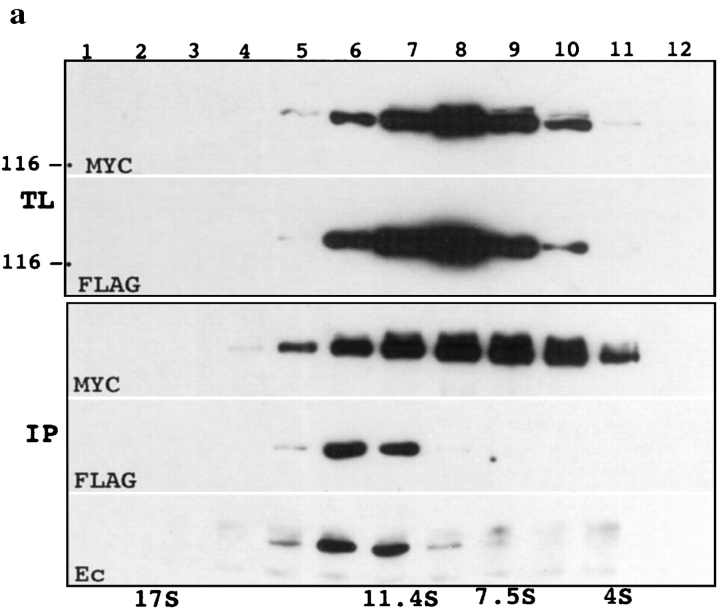
Sedimentation–immunoprecipitation experiments with the total lysates obtained from such cocultures demonstrated that in the 13 S fractions anti-myc antibody did coimmunoprecipitate both flag-tagged and endogenous E-cadherin (Fig. 5). To show that the association between flag- and myc-tagged forms of E-cadherin depends on the integrity of cell–cell contacts, the cocultured cells were incubated for 10 min in 10 mM EGTA at 37°C before lysis in the regular lysis buffer. Complete disappearance of Ec1F, but not endogenous E-cadherin, in the anti-myc immunoprecipitates after EGTA treatment (Fig. 5 b, lanes 1 and 2) showed that the 13 S fraction contained two distinct lateral and adhesive E-cadherin complexes. To study whether reestablishment of the cell–cell contacts is accompanied by restoration of adhesive E-cadherin complexes, the cocultured cells, after incubation in 10 mM EGTA at 37°C, were further cultivated for the next 30 min either in low calcium or in normal medium (Fig. 5 b, lanes 3 and 4). The data show that the normal medium completely restored the amount of adhesive dimers (Ec1M–Ec1F complexes), whereas only Ec1M–E-cadherin complexes were detected in low calcium medium. Surprisingly, depletion of the calcium ions by EGTA did not reduce the amount of the E-cadherin–Ec1M complexes, which is a sum of the lateral and adhesive complexes (Fig. 5 and refer to Fig. 3). It can be suggested that the removal of extracellular calcium, while causing dissociation of the adhesive complexes, might increase the solubility of the lateral complexes which would account for the unchanged amount of the total complexes detected.
Figure 5.
Analysis of the adhesive E-cadherin–catenin complex. (a) Two independent A-431 cell subclones producing either Ec1F or Ec1M were cocultivated overnight and subjected to sucrose gradient centrifugation. Immunoblot analysis of the total lysates (TL) of each gradient fraction (numbered at the top) with anti-myc (MYC), or anti-flag (FLAG) antibodies, shows that both proteins were similarly distributed along the gradient and were present approximately in the same amounts. Anti-myc immunoprecipitates (IP) obtained from each fraction were separated by SDS-PAGE, transferred to nitrocellulose, and then stained with anti-myc (MYC), anti-flag (FLAG) and anti-E-cadherin (Ec) antibody. Note that the same sucrose gradient fractions contain E-cadherin–Ec1M and Ec1M–Ec1F complexes. (b) Cells expressing Ec1F were cocultivated as in a with cells producing Ec1M (lanes 1–5) or with its mutants (lanes 6–8). In lane 2, cells before lysis were treated with 10 mM EGTA at 37°C; in lanes 3 and 4, cells after treatment as in lane 2 were further incubated for 30 min in low calcium or in normal medium, respectively; in lane 5, 10 mM EGTA was added into the lysis buffer; in lanes 6–8 Ec1F-expressing cells were cocultured with cells expressing Ec1Δ(768– 879)M (lane 6); Ec1QNM (lane 7); and Ec1WVM (lane 8). Total lysates of these cocultures were coimmunoprecipitated with anti-myc antibody and assayed for presence of the myc-tagged forms of E-cadherin (MYC), endogenous E-cadherin (E-cadherin), or Ec1F by epitope-specific antibody.
To further evaluate the function of calcium ions in adhesive E-cadherin interactions, two additional experiments were performed. First, the cocultured cells were incubated before lysis for 10 min with EGTA at 4°C. At this temperature, in agreement with published observations (Citi, 1992), EGTA did not affect cell–cell junctions. In the second experiment, 10 mM EGTA was added to the lysis buffer. In this case, the cells were not incubated with EGTA before lysis. Surprisingly, we found that in both these conditions EGTA did not decrease the adhesive association between flag- and myc-tagged forms of cadherins (Fig. 5 b, lane 5 and data not shown). Taken together, these data strongly suggest that calcium ions do not directly participate in the formation of the structure of the adhesive interface between two E-cadherin molecules. This is consistent with the crystallography model reported by Shapiro at al. (1995). Our experiments, however, did not exclude the possibility that calcium might be important for one of the steps leading to the formation of adhesive dimers. To test this assumption, a new Ec1M mutant, Ec1QNM, in which residues Gln255 and Asn256 were replaced with two alanines, was constructed. According to X-ray analysis, the replaced amino acids participate in coordination of two of the three Ca2+ ions filling the cleft between the ECI and ECII domains of E-cadherin (Nagar et al., 1996). Furthermore, it was shown that ablation of one of these sites blocked E-cadherin cell–cell adhesion activity (Ozawa et al., 1990a ). We found that the Ec1QNM mutant upon expression in A-431 cells, similar to the Ec1WVM mutant described above, was delivered to the cell surface and induced a severe dominant-negative effect on cell–cell adhesion (Fig. 6). This mutant normally associated with endogenous E-cadherin, but not with Ec1F (Fig. 5 b, lane 7), suggesting that defects in the Ca2+-binding sites, while unable to change lateral dimerization, affect the adhesion interaction between two E-cadherin molecules. However, we could not discard the possibility that the absence of such an interaction is caused indirectly by the dominant-negative effect of the Ec1QNM mutant on the cellular phenotype. Although additional experiments are needed to completely understand the role of calcium in cadherin-mediated adhesion, our data imply that binding of calcium ions to cadherins is important for the formation of adhesive dimers, that become independent of calcium after their formation.
Finally, we determined whether mutations constructed for the characterization of lateral E-cadherin dimerization abolish the adhesive cadherin interaction. For this, we cocultivated the cells expressing these mutants with Ec1F-producing cells as described above. Anti-myc immunoprecipitation experiments presented in Fig. 5 b clearly show that the adhesive Ec1M–Ec1F association was completely abolished either by deletion of the catenin-binding region or by Trp156Ala/Val157Gly replacement (Fig. 5 b, lanes 6 and 8, respectively). The same factors, catenin and calcium, are therefore required for E-cadherin to form adhesive dimers and to establish cell–cell adhesion. This strongly suggests that the revealed dimers are structural elements of cadherin-mediated cell–cell adhesion.
Discussion
It is well established that the transmembrane protein E-cadherin forms complexes with intracellular β-catenin (or plakoglobin) and with α-catenin (in this paper they are designated conventional E-cadherin–catenin complexes). This association is absolutely required for E-cadherin to mediate intercellular adhesion (Nagafuchi and Takeichi, 1988; Ozawa et al., 1990b ). The exact role of catenins in this process is still unknown. It was also suggested (Shapiro et al., 1995; Brieher et al., 1996; Nagar et al., 1996; Tomschy et al., 1996), although never shown directly, that E-cadherin may form lateral dimers on the cell surface. These lateral complexes, in which two E-cadherin molecules are aligned in parallel orientation, were proposed to participate, in turn, in the antiparallel interactions resulting in cell–cell adhesion. The structural basis of both adhesive and lateral interactions is not clear. In the present investigation, we analyzed intercadherin interactions using an immunoprecipitation approach. To trace such interactions, we expressed in A-431 cells producing endogenous E-cadherin recombinant forms of the same protein tagged either by myc or by flag epitopes. Our findings not only present compelling evidence for two distinct lateral and adhesive E-cadherin complexes, but also allows us to clarify their structures.
First, we obtained a strong evidence that the conventional E-cadherin–catenin complexes may self-associate on the cell surface through E-cadherin dimerization. Our data strongly support the idea that this process is driven by hydrophobic interactions between the extracellular EI domains of E-cadherin. This mechanism was previously proposed by Shapiro et al. (1995). As suggested by sedimentation analysis (refer to Figs. 1 and 3), only a relatively small pool of E-cadherin is incorporated into the lateral E-cadherin complexes. The formation of these E-cadherin complexes is independent of interactions of E-cadherin with cytoplasmic proteins, such as catenins or p120. The fact that even the longest deletion within the intracellular E-cadherin region did not abolish lateral dimerization suggests that cytoplasmic factors are unlikely to regulate assembly of these dimers. The amount of the lateral complexes was also independent of cell–cell interactions, since it was stable (a) upon EGTA treatment leading to separation of cells, (b) in cells cultivated up to 1 wk in low calcium medium preventing establishment of cell–cell contacts (data not shown), and (c) in the cells with strong dominant-negative abnormalities in cell–cell adhesion caused by Ec1QNM expression. These data imply that lateral E-cadherin dimerization is not triggered in response to head to head interaction with a protein(s) extended from the opposing cell surface. Thus, if the E-cadherin lateral complex is not formed spontaneously, its assembly is regulated either by soluble (humoral) factor(s) or by transmembrane protein(s) extended from the same cellular surface.
Interestingly, independence of E-cadherin dimerization from catenins indicates that the cell surface may contain dimers in which E-cadherin molecules are linked either to the same or to different catenins (plakoglobin and β-catenin). This possibility was supported by our experiments (Chitaev et al., 1998) with A-431 cells expressing myc-tagged versions of β-catenin or plakoglobin. In both cell types, anti-myc coimmunoprecipitation shows that the 13 S complex may contain two β-catenin or two plakoglobin molecules. Diversity of the lateral E-cadherin complexes might even be larger since some may also incorporate different isoforms of p120. It is not clear whether such variety has any biological significance. In theory, E-cadherin dimers associated with different sets of catenins may have distinct signaling or adhesive properties.
The second type of inter–E-cadherin interaction detected in our work is an adhesive interaction leading to the formation of antiparallel E-cadherin complexes. Our experiments did not allow us to evaluate the exact structure of such complexes, since these and lateral complexes were present in the same fractions of the sucrose gradient. On the other hand, several observations strongly suggest that, similar to lateral complexes, they consist of two conventional E-cadherin–catenin complexes, but arranged in an antiparallel fashion. First, both lateral and antiparallel complexes have the same sedimentation value. Second, no differences were detected in protein composition of anti– E-cadherin (or anti–β-catenin) radioimmunoprecipitates obtained from the 13 S fraction from cells exhibiting (control), and not exhibiting (EGTA-treated) antiparallel complex (data not shown). Finally, immunoblotting experiments failed to reveal association with 13 S E-cadherin complex other possible E-cadherin/catenin–associated proteins, such as EGF-receptor, α-actinin, vinculin, or actin. These observations indicate similar protein composition of the lateral and antiparallel E-cadherin complexes, although we cannot exclude that some of the peripheral proteins dissociate from these complexes during their solubilization in Triton X-100. The most important and specific feature of adhesive complexes is the requirement for their assembly of the same factors, extracellular calcium and catenins, necessary for cell–cell adhesion. Such similarities strongly suggest that the revealed antiparallel complexes are structural units of cadherin-mediated cell–cell adhesion and their clustering, via interaction with the actin cytoskeleton (Knudsen et al., 1995; Rimm et al., 1995), may be a step in adherens junction assembly.
The critical question is whether adhesive and lateral complexes are the consecutive steps of one process, or are uninterrelated, and participate in distinct processes, such as signaling and adhesion. Although additional experiments are needed to understand this question, there is evidence in favor of the first hypothesis. We show that the Trp156Ala/Val157Gly substitution which, according to Shapiro et al. (1995), leaves an intact adhesive interface, abolishing both lateral and adhesive association. The latter association, however, could be specifically blocked by removal of the catenin- or calcium-binding sites. This suggests that the adhesive complex is derived from lateral complexes. A similar scenario was suggested by Brieher et al. (1996), who found that lateral dimers of the recombinant extracellular portion of C-cadherin, participate in adhesion much more efficiently than monomers. Examination of intra–E-cadherin interactions by chimeric ECAD-COMP molecules also supports this view (Tomschy et al., 1996). The obvious problem is, that the head to head association of two lateral complexes must result in a structure containing at least four conventional E-cadherin–catenin complexes. Our attempts to reveal E-cadherin complexes greater than 13 S using different centrifugation protocols were unsuccessful. Thus, the mechanism of the assembly of the adhesive complexes remains open. One interpretation of our results is that a hypothetical antiparallel complex consisting of the four conventional E-cadherin–catenin units is very unstable. In normal cells this complex dissociates at a high rate into two equal adhesive complexes characterized in our work. Alternatively, a lateral E-cadherin complex may dissociate, giving rise to two activated E-cadherin–catenin complexes. They are subsequently used in adhesive associations. Importantly, both models would predict a special step, lateral-adhesive transition, in which two lateral E-cadherin dimers form two antiparallel dimers. Catenins and calcium ions might be functionally important for E-cadherin to proceed to this transition. In the simplest model, catenins in response to the lateral dimerization of E-cadherin would trigger a conformational change of each cadherin resulting in the activation of the E-cadherin adhesive site and concomitant dissociation of the lateral dimers. This model is consistent with our data suggesting that calcium ions, which are known to stabilize overall cadherin conformation (Pokutta et al., 1996), could be involved only in formation, but not in maintenance of the adhesive association.
In summary, our data present direct experimental evidence that E-cadherin may form lateral and adhesive complexes. They also show that interactions of E-cadherin with catenins and Ca2+ are essential for adhesive, but not for lateral intercadherin interactions. The major points for further investigation are the precise mechanisms leading to the formation of adhesive complexes and the role of catenins and other intracellular factors in this process.
Acknowledgments
This work has been supported in part by National Institutes of Health grant AR44016-03 and a research grant from the Dermatology Foundation.
Abbreviation used in this paper
- aa
amino acid(s)
Footnotes
N. Chitaev wanted to dedicate this work in memory of his mother V. Chitaeva. We thank A. Averbackh, G. Goldberg, A. Eisen, R. Troyanovsky, and J. Klingelhöfer (all from Washington University, St. Louis, MO), and A. Ljubimov (Cedars-Sinai Medical Center, Los Angeles, CA) for valuable discussion and W.W. Franke for providing us with HaCat cells and anti-desmoglein antibody.
Address all correspondence to Sergey Troyanovsky, Division of Dermatology, Washington University Medical School, Campus Box 8123, 660 South Euclid Ave., St. Louis, MO 63110. Tel.: (314) 362-8154. Fax: (314) 362-8159. E-mail: sergeyt@im.wustl.edu
References
- Birchmeier W, Behrens J. Cadherin expression in carcinomas: role in formation of cell junctions and the prevention of invasiveness. Biochim Biophys Acta. 1994;1198:11–26. doi: 10.1016/0304-419x(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Blaschuk OW, Sullivan R, David S, Pouliot Y. Identification of a cadherin cell adhesion recognition sequence. Dev Biol. 1990;139:227–229. doi: 10.1016/0012-1606(90)90290-y. [DOI] [PubMed] [Google Scholar]
- Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieher WM, Yap AS, Gumbiner BM. Lateral dimerization is required for the homophilic binding activity of C-cadherin. J Cell Biol. 1996;135:487–496. doi: 10.1083/jcb.135.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussemakers MJ, van Bokhoven A, Mees SG, Kemler R, Schalken JA. Molecular cloning and characterization of the human E-cadherin cDNA. Mol Biol Rep. 1993;17:123–128. doi: 10.1007/BF00996219. [DOI] [PubMed] [Google Scholar]
- Butz S, Kemler R. Distinct cadherin-catenin complexes in Ca2+- dependent cell-cell adhesion. FEBS (Fed Eur Biochem Soc)Lett. 1994;355:195–200. doi: 10.1016/0014-5793(94)01205-9. [DOI] [PubMed] [Google Scholar]
- Chitaev NA, Leube RE, Troyanovsky RB, Eshkind LG, Franke WW, Troyanovsky SM. The binding of plakoglobin to desmosomal cadherins: patterns of binding sites and topogenic potential. J Cell Biol. 1996;133:359–369. doi: 10.1083/jcb.133.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitaev NA, Troyanovsky SM. Direct Ca2+-dependent heterophilic interaction between desmosomal cadherins, desmoglein and desmocollin, contributes to cell–cell adhesion. J Cell Biol. 1997;138:193–201. doi: 10.1083/jcb.138.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitaev NA, Averbakh AZ, Troyanovsky R, Troyanovsky SM. Molecular organization of the desmoglein-plakoglobin complex. J Cell Sci. 1998;111:1941–1949. doi: 10.1242/jcs.111.14.1941. [DOI] [PubMed] [Google Scholar]
- Citi S. Protein kinase inhibitors prevent junction dissociation induced by low extracellular calcium in MDCK epithelial cells. J Cell Biol. 1992;117:169–178. doi: 10.1083/jcb.117.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citi S, Volberg T, Bershadsky AD, Denisenko N, Geiger B. Cytoskeletal involvement in the modulation of cell-cell junctions by the protein kinase inhibitor H-7. J Cell Sci. 1994;107:683–692. [PubMed] [Google Scholar]
- Edelman, G., B. Cunningham, and J.-P. Thiery. 1990. Morphoregulatory molecules. J. Wiley & Sons. New York. 648 pp.
- Fujimori T, Takeichi M. Disruption of epithelial cell-cell adhesion by exogenous expression of a mutated nonfunctional N-cadherin. Mol Biol Cell. 1993;4:37–47. doi: 10.1091/mbc.4.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B, Ayalon O. Cadherins. Annu Rev Cell Biol. 1992;8:307–332. doi: 10.1146/annurev.cb.08.110192.001515. [DOI] [PubMed] [Google Scholar]
- Gumbiner GM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- Hinck L, Nathke IS, Papkoff J, Nelson WJ. Dynamics of cadherin/catenin complex formation: Novel protein interactions and pathways of complex assembly. J Cell Biol. 1994;125:1327–1340. doi: 10.1083/jcb.125.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubar O, Bierkamp C, Kemler R. Cadherins and catenins in development. Curr Opin Cell Biol. 1996;8:685–691. doi: 10.1016/s0955-0674(96)80110-4. [DOI] [PubMed] [Google Scholar]
- Hynes RO. Specificity of cell adhesion in development: the cadherin superfamily. Curr Opin Gen Dev. 1992;2:621–624. doi: 10.1016/s0959-437x(05)80182-0. [DOI] [PubMed] [Google Scholar]
- Jou T-S, Stewart DB, Stappert J, Nelson WJ, Marrs JA. Genetic and biochemical dissection of protein linkages in the cadherin-catenin complex. Proc Natl Acad Sci USA. 1995;92:5067–5071. doi: 10.1073/pnas.92.11.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartenbeck J, Schmid E, Franke WW, Geiger B. Different modes of internalization of proteins associated with adherens junctions and desmosomes: experimental separation of lateral contacts induces endocytosis of desmosomal plaque material. EMBO (Eur Mol Biol Organ) J. 1982;1:725–732. doi: 10.1002/j.1460-2075.1982.tb01237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintner C. Regulation of embryonic cell adhesion by the cadherin cytoplasmic domain. Cell. 1992;69:225–236. doi: 10.1016/0092-8674(92)90404-z. [DOI] [PubMed] [Google Scholar]
- Klymkowsky M, Parr B. The body language of cells: the intimate connection between cell adhesion and behavior. Cell. 1995;83:5–8. doi: 10.1016/0092-8674(95)90226-0. [DOI] [PubMed] [Google Scholar]
- Knudsen KA, Peralta A, Soler, Johnson KR, Wheelock MJ. Interaction of α-actinin with the cadherin/catenin cell-cell adhesion complex via α-catenin. J Cell Biol. 1995;130:67–77. doi: 10.1083/jcb.130.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch AW, Pokutta S, Lustig A, Engel J. Calcium binding and homoassociation of E-cadherin domains. Biochemistry. 1997;36:7697–7705. doi: 10.1021/bi9705624. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–687. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maurer P, Hohenester E, Engel J. Extracellular calcium-binding proteins. Curr Opin Cell Biol. 1996;8:609–617. doi: 10.1016/s0955-0674(96)80101-3. [DOI] [PubMed] [Google Scholar]
- Nagafuchi A, Takeichi M. Cell binding function of E-cadherin is regulated by the cytoplasmic domain. EMBO (Eur Mol Biol Organ) J. 1988;7:3679–3684. doi: 10.1002/j.1460-2075.1988.tb03249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagar B, Overduin M, Ikura M, Rini JM. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380:360–364. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Shore EM, Wang AZ, Hammerton RW. Identification of a membrane–cytoskeletal complex containing the cell adhesion molecule uvomorulin (E-cadherin), ankyrin, and fobrin in Madin-Darby canine kidney epithelial cells. J Cell Biol. 1990;110:349–357. doi: 10.1083/jcb.110.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nose A, Tsuji K, Takeichi M. Localization of specificity determining sites in cadherin cell adhesion molecules. Cell. 1990;61:147–155. doi: 10.1016/0092-8674(90)90222-z. [DOI] [PubMed] [Google Scholar]
- Overduin M, Harvey TS, Bagby S, Tong KI, Yau P, Takeichi M, Ikura M. Solution structure of the epithelial cadherin domain responsible for selective cell adhesion. Science. 1995;267:386–389. doi: 10.1126/science.7824937. [DOI] [PubMed] [Google Scholar]
- Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO (Eur Mol Biol Organ) J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M, Engel J, Kemler R. Single amino acid substitutions in one Ca2+-binding site of uvomorulin abolish the adhesive function. Cell. 1990a;63:1033–1038. doi: 10.1016/0092-8674(90)90506-a. [DOI] [PubMed] [Google Scholar]
- Ozawa M, Ringwald M, Kemler R. Uvomorulin-catenin complex formation is regulated by a specific domain in the cytoplasmic region of the cell adhesion molecule. Proc Natl Acad Sci USA. 1990b;87:4246–4250. doi: 10.1073/pnas.87.11.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M, Kemler R. Molecular organization of the uvomorulin-catenin complex. J Cell Biol. 1992;116:989–996. doi: 10.1083/jcb.116.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M. Cell adhesion and signal transduction: the Armadillo connection. Trends Cell Biol. 1995;5:224–229. doi: 10.1016/s0962-8924(00)89015-7. [DOI] [PubMed] [Google Scholar]
- Pokutta S, Herrenknecht K, Kemler R, Engel J. Conformational changes of the recombinant extracellular domain of E-cadherin upon calcium binding. Eur J Biochem. 1994;223:1019–1026. doi: 10.1111/j.1432-1033.1994.tb19080.x. [DOI] [PubMed] [Google Scholar]
- Reynolds AB, Daniel J, McCrea PD, Wheelock MJ, Wu J, Zhang Z. Identification of a new catenin: the tyrosine kinase substrate p120casassociates with E-cadherin complexes. Mol Cell Biol. 1994;14:8333–8342. doi: 10.1128/mcb.14.12.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimm DL, Koslov EP, Kebriaei P, Cianci CD, Morrow JS. Alpfa 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci USA. 1995;92:8813–8817. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L, Fannon AM, Kwong PD, Thompson A, Lehmann MS, Grubel G, Legrand J-F, Als-Neilsen J, Colman DR, Hendrickson WA. Structural basis of cell-cell adhesion by cadherins. Nature. 1995;374:327–337. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- Shibamoto S, Hayakawa M, Takeuchi K, Hori T, Miyazawa K, Kitamura N, Johnson KR, Weelock MJ, Matsuyoshi N, Takeichi M, Ito F. Association of p120, a tyrosine kinase substrate, with E-cadherin/catenin complexes. J Cell Biol. 1995;128:949–957. doi: 10.1083/jcb.128.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulators. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol. 1993;5:806–811. doi: 10.1016/0955-0674(93)90029-p. [DOI] [PubMed] [Google Scholar]
- Tomschy A, Fauser C, Landwehr R, Engel J. Homophilic adhesion of E-cadherin occurs by a co-operative two-step interaction of N-terminal domains. EMBO (Eur Mol Biol Organ) J. 1996;15:3507–3514. [PMC free article] [PubMed] [Google Scholar]
- Vasiliev, J.M., and I.M. Gelfand. 1981. Neoplastic and Normal Cells in Culture. Cambridge University Press, Cambridge, UK. 346 pp.
- Volberg T, Geiger B, Kartenbeck J, Franke WW. Changes in membrane-microfilament interaction in intercellular adhesion junctions upon removal of extracellular Ca2+ions. J Cell Biol. 1986;102:1832–1842. doi: 10.1083/jcb.102.5.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]