Abstract
We recently isolated a novel actin filament (F-actin)–binding protein, afadin, that has two isoforms, l- and s-afadins. l-Afadin is ubiquitously expressed and specifically localized at zonula adherens (ZA) in epithelial cells and at cell–cell adherens junction (AJ) in nonepithelial cells, whereas s-afadin is abundantly expressed in neural tissue. l-Afadin has one PDZ domain, three proline-rich regions, and one F-actin–binding domain, whereas s-afadin lacks the third proline-rich region and the F-actin–binding domain. To understand the molecular mechanism of the specific localization of l-afadin at ZA in epithelial cells and at cell–cell AJ in nonepithelial cells, we attempted here to identify an l-afadin–binding protein(s) and isolated a protein, named ponsin. Ponsin had many splicing variants and the primary structures of two of them were determined. Both the two variants had three Src homology 3 (SH3) domains and turned out to be splicing variants of SH3P12. The third proline-rich region of l-afadin bound to the region of ponsin containing the second and third SH3 domains. Ponsin was ubiquitously expressed and localized at ZA in epithelial cells, at cell–cell AJ in nonepithelial cells, and at cell–matrix AJ in both types of cells. Ponsin furthermore directly bound vinculin, an F-actin–binding protein localized at ZA in epithelial cells, at cell–cell AJ in nonepithelial cells, and at cell–matrix AJ in both types of cells. Vinculin has one proline-rich region where two proline-rich sequences are located. The proline-rich region bound to the region of ponsin containing the first and second SH3 domains. l-Afadin and vinculin bound to ponsin in a competitive manner and these three proteins hardly formed a ternary complex. These results indicate that ponsin is an l-afadin– and vinculin-binding protein localized at ZA in epithelial cells, at cell–cell AJ in nonepithelial cells, and at cell–matrix AJ in both types of cells.
Keywords: ponsin, l-afadin, vinculin, zonula adherens, cell–matrix adherens junction
The cell junction plays essential roles in various cell functions, including cell adhesion, growth, polarization, and motility (Takeichi, 1988, 1991; Luna and Hitt, 1992; Jockusch et al., 1995; Gumbiner, 1996). The cell junction is classified into two types: cell–cell and cell–matrix junctions. In polarized epithelial cells, the cell–cell junction shows a specialized membrane structure, comprised of zonula occludens (ZO1, tight junction), zonula adherens (ZA), and desmosome, which is known as the junctional complex. These three junctional structures are typically aligned from the apical to the basal side of the lateral membrane, although desmosome is independently distributed in other areas.
At ZA, cadherins interact with each other at the extracellular surface and serve as a critical adhesion molecule (Takeichi, 1988, 1991; Geiger and Ginsberg, 1991; Tsukita et al., 1992). The cytoplasmic region of cadherin interacts with β- and γ-catenins, and β-catenin binds α-catenin (Ozawa et al., 1989; Nagafuchi et al., 1991; Takeichi, 1991; Tsukita et al., 1992; Aberle et al., 1994). α-Catenin interacts directly with actin filament (F-actin) or indirectly with it through α-actinin and vinculin, other F-actin–binding proteins (Knudsen et al., 1995; Rimm et al., 1995; Weiss et al., 1998; Watabe-Uchida et al., 1998). ZA is distinguished from cadherin-based cell–cell adherens junction (AJ) in nonepithelial cells by having a thick F-actin belt, called the circumferential filament band. Furthermore, while ZA is defined as a junctional structure observed by electron microscopy, the cadherin-catenin system is not confined to ZA, but distributed along the entire lateral membrane (Tsukita et al., 1992; Gumbiner, 1996). α-Actinin is also broadly distributed (Geiger et al., 1981). In this study, therefore, we use “cell–cell AJ” as cell–cell adhesion site where cadherin is present, and distinguish “cell–cell AJ” from “ZA.” In contrast, vinculin is more highly concentrated at ZA than the cadherin-catenin system (Geiger et al., 1981; Yonemura et al., 1995).
The cell–matrix junction is comprised of cell–matrix AJ and hemidesmosome. At cell–matrix AJ where integrin interacts with matrix proteins at the extracellular surface, the cytoplasmic region of integrin β subunit directly binds talin (Horwitz et al., 1986) and α-actinin (Otey et al., 1990), both of which directly interact with F-actin. Furthermore, talin directly binds vinculin (Burridge and Mangeat, 1984). We have recently shown that a novel F-actin– binding protein, named nexilin, is localized at cell–matrix AJ (Ohtsuka et al., 1998).
Thus, many F-actin–binding proteins serve as linkers of the actin cytoskeleton to the adhesion molecules, such as cadherin and integrin. However, little is known about the mechanism of how these F-actin–binding proteins are involved in the establishment of the polarized junctional alignment in epithelial cells. It has neither fully been understood how these F-actin–binding proteins regulate the function of the adhesion molecules nor how the adhesion molecule-initiated signals are transduced to the F-actin– binding proteins.
We have recently isolated a novel cell–cell AJ component, named l-afadin, as an F-actin–binding protein (Mandai et al., 1997). l-Afadin has a small splicing variant, named s-afadin. l-Afadin is ubiquitously expressed, whereas s-afadin is abundantly expressed in neural tissue. l-Afadin has one PDZ domain at the middle region, three proline-rich regions following the PDZ domain, and one F-actin–binding domain at the COOH-terminal region. s-Afadin has one PDZ domain but lacks the third proline-rich region and the F-actin–binding domain. s-Afadin is the protein encoded by the AF-6 gene, which is originally found to be fused to the ALL-1 gene (Prasad et al., 1993), known to be involved in acute leukemia (Cimino et al., 1991). Immunofluorescence and immunoelectron microscopic analyses indicate that l-afadin is localized at ZA in small intestine absorptive epithelial cells and at cell–cell AJ in nonepithelial cells, such as intercalated disc in cardiac muscle cells (Mandai et al., 1997). In contrast to our results, another group has reported that the AF-6 protein (s-afadin) is localized at ZO in Madin-Darby canine kidney (MDCK) cells (Yamamoto et al., 1997). Recently, consistent with our previous results (Mandai et al., 1997), we have found that l-afadin is a major expressed variant and is localized at the junctional complex region in MDCK cells (Sakisaka et al., 1999). However, it is difficult to conclude that l-afadin is localized at ZO or ZA in MDCK cells because ZO and ZA are not well separated in this cell line. Furthermore, l-afadin does not directly bind to α-, β-catenin, or the cytoplasmic region of E-cadherin (Sakisaka et al., 1999).
The results thus far available suggest that there is a mechanism that determines the specific localization of l-afadin at ZA in epithelial cells and at cell–cell AJ in nonepithelial cells. To understand the molecular mechanism of this specific localization of l-afadin, we attempted here to identify an l-afadin–binding protein(s) specifically localized at these sites, and isolated a protein. The isolated protein had many splicing variants and the primary structures of two of them were determined. They turned out to be splicing variants of SH3P12, which had originally been identified as a Src homology 3 (SH3) domain-containing protein (Sparks et al., 1996). The l-afadin–binding protein isolated here as well as SH3P12 had three SH3 domains at their COOH-terminal regions and two sorbin-like regions at their NH2-terminal regions. SH3 domains have first been identified as conserved, noncatalytic regions in cytoplasmic tyrosine kinases such as Src (Cohen et al., 1995; Pawson, 1995) and subsequently shown to interact with proline-rich sequences (Yu et al., 1994). Sorbin is a peptide hormone that induces the absorption of water and sodium in upper small intestine (Pansu et al., 1981). Furthermore, we have found that the l-afadin–binding protein directly bound vinculin, an F-actin–binding protein (Jockusch et al., 1995) that is localized at ZA in epithelial cells, at cell–cell AJ in nonepithelial cells (Weiss et al., 1998; Watabe-Uchida et al., 1998), and at cell–matrix AJ in both types of cells (Burridge and Mangeat, 1984). The l-afadin– binding protein was also colocalized with vinculin at these sites. We named this protein ponsin (from the Latin word pons, meaning a bridge) because it was capable of binding both l-afadin and vinculin. We describe here the purification and characterization of ponsin.
Materials and Methods
Blot Overlay
Blot overlay was done using 35S-labeled l-afadin, ponsin, or vinculin as described (Shieh and Zhu, 1996; Chevesich et al., 1997), with a slight modification. In brief, 35S-labeled l-afadin, ponsin, and vinculin were generated with pBlue vectors containing full-length cDNAs of l-afadin, ponsin, and vinculin, respectively, using the TNT T7 quick coupled transcription/ translation system (Promega Corp.). The vinculin cDNA (Watabe-Uchida et al., 1998) was kindly supplied by Drs. Y. Imamura, A. Nagafuchi, and Sh. Tsukita (Kyoto University, Kyoto, Japan). To remove free 35S-methionine, each translated protein was applied to a Bio-Spin 6 chromatography column (Bio-Rad Laboratories). Each eluate from the column was used as a probe. The sample to be tested was subjected to SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was blocked in PBS containing 5% defatted powder milk and 1% Triton X-100. The membrane was then incubated at 4°C for 16 h with the 35S-labeled l-afadin, ponsin, or vinculin probe (∼2 × 106 cpm/ml each) in PBS containing 5% defatted powder milk and 1% Triton X-100. After the incubation, the membrane was washed with PBS containing 5% defatted powder milk and 1% Triton X-100, followed by autoradiography using an image analyzer (Fujix BAS-2000II; Fuji Photo Film Co.).
Purification of Ponsin
The AJ-enriched fraction was prepared from 80 rat livers as described (Tsukita and Tsukita, 1989), and stored at −80°C until use. Half of the AJ-enriched fraction was dissolved in an extraction solution (20 mM Tris-HCl at pH 9.0, 8 M urea, 1 mM DTT, and 1% hydrogenated Triton X-100; Calbiochem Corp.). After sonication, the solution was mildly stirred at room temperature for 1 h and centrifuged at 100,000 g at 20°C for 30 min. The supernatant was diluted with a buffer (20 mM Tris-HCl at pH 9.0 and 1 mM DTT) to give final concentrations of 6 M urea and 0.75% hydrogenated Triton X-100. This sample (135 ml, 91 mg of protein) was applied to a Mono Q HR 10/10 column (Amersham-Pharmacia Biotechnology) equilibrated with buffer A (20 mM Tris-HCl at pH 9.0, 6 M urea, 0.75% CHAPS, and 1 mM DTT). After the column was washed with 70 ml of buffer A, elution was performed with a 160-ml linear gradient of sodium chloride (0–0.3 M) in buffer A. Fractions of 1.5 ml each were collected. When each fraction was subjected to 35S-labeled l-afadin blot overlay, the radioactive bands appeared in fractions 14–25. These active fractions (18 ml, 3.1 mg of protein) were collected. The other half of the AJ-enriched fraction was also subjected to the Mono Q column chromatography in the same manner as described above. The active fractions of the two Mono Q column chromatographies were combined. The combined sample was applied to a hydroxyapatite column (0.75 × 7.5 cm; Tosoh) equilibrated with buffer B (8 mM potassium phosphate at pH 7.8, 6 M urea, 0.75% CHAPS, and 1 mM DTT). After the column was washed with 15 ml of the same buffer, elution was performed with a 25-ml linear gradient of potassium phosphate (8–100 mM) at pH 7.8 in buffer B. Fractions of 0.5 ml each were collected. When each fraction was subjected to 35S-labeled l-afadin blot overlay, the radioactive bands appeared in fractions 25–33. The active fractions (4.5 ml, 1 mg of protein) were collected and applied to a Mono S PC 1.6/5 column (Amersham-Pharmacia Biotechnology) equilibrated with buffer C (50 mM sodium acetate at pH 4.0, 6 M urea, 0.75% CHAPS, and 1 mM DTT). After the column was washed with 0.7 ml of buffer C containing 0.15 M sodium chloride, elution was performed with a 2-ml linear gradient of sodium chloride (0.15–0.4 M) in buffer C. Fractions of 0.1 ml each were collected. When each fraction was subjected to 35S-labeled l-afadin blot overlay, the radioactive bands appeared in fractions 9–13 (see Fig. 1). The active fractions were stored at −80°C.
Figure 1.
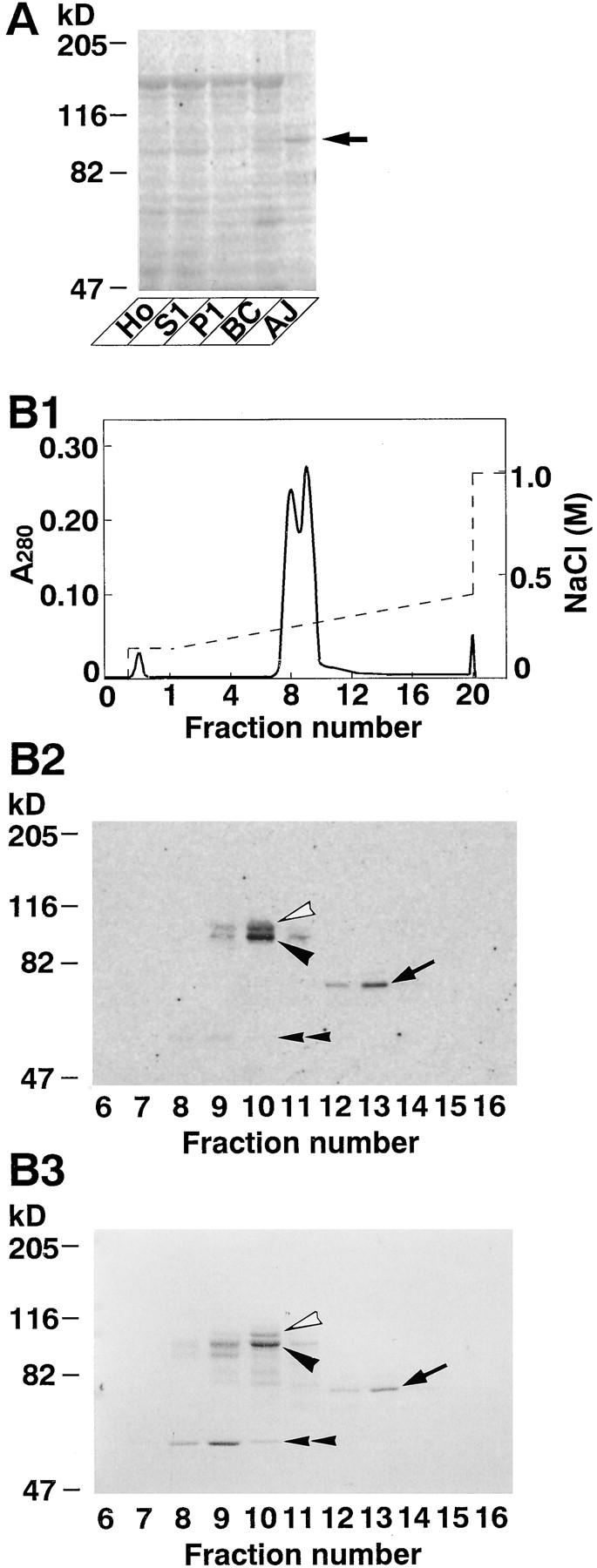
35S-Labeled l-afadin–binding proteins. (A) Subcellular distribution of 35S-labeled l-afadin–binding proteins in rat liver. An aliquot (30 μg protein) of each subcellular fraction was subjected to SDS-PAGE (8% polyacrylamide gel), followed by 35S-labeled l-afadin blot overlay. (Arrow) p93, (Ho) homogenate, (S1) soluble fraction, (P1) pellet fraction, (BC) fraction rich in bile canaliculi, and (AJ) fraction rich in AJ. (B) 35S-Labeled l-afadin–binding proteins in Mono S column chromatography. (B1) Absorbance at 280 nm (A280). (B2) 35S-Labeled l-afadin blot overlay. An aliquot (0.75 μl) of each fraction was subjected to SDS-PAGE (8% polyacrylamide gel), followed by 35S-labeled l-afadin blot overlay. (B3) Protein staining with Coomassie brilliant blue. An aliquot (2 μl) of each fraction was subjected to SDS-PAGE (8% polyacrylamide gel), followed by protein staining with Coomassie brilliant blue. (Open arrowhead) p95, (closed arrowhead) p93, (arrow) p70, and (double arrowhead) p55. The results are representative of three independent experiments.
Peptide Microsequencing and Cloning of the Ponsin cDNAs
On the Mono S column chromatography, three radioactive bands with molecular masses of ∼95 (p95), 93 (p93), and 70 (p70) kD were identified by 35S-labeled l-afadin blot overlay (see Fig. 1). The purified Mono S samples, fraction 10 for p93 and p95, and fractions 12 and 13 for p70, were subjected to SDS-PAGE (8% polyacrylamide gel). The protein bands corresponding to p95, p93, and p70 were separately cut out from the gel and digested with a lysyl endopeptidase, and the digested peptides were separated by TSKgel ODS-80Ts (4.6 × 150 mm; Tosoh) reverse phase HPLC as described (Imazumi et al., 1994). The amino acid (aa) sequences of the peptides were determined with a peptide sequencer. BLAST search of the GenBank database indicated that these sequences were almost identical to that of the mouse SH3P12 gene product (U58883; GenBank/EMBL/ DDBJ). Based on the sequence of SH3P12, oligonucleotide primers, 5′-CATTGGAAGACCTTGAGATCC-3′ and 5′-GAATGATGCTTCATCCTCCG-3′, were designed and a PCR reaction was performed using a mouse brain cDNA (Clontech) as a template. DNA sequencing was performed by the dideoxynucleotide termination method using a DNA sequencer (ABI 373).
Expression and Purification of Recombinant Proteins
Prokaryote and eukaryote expression vectors were constructed in pCMV5 (Satoh et al., 1998), pCMV-Myc (Mandai et al., 1997), pFLAG-CMV2 (Eastman Kodak Scientific Imaging Systems), pGEX (Amersham-Pharmacia Biotechnology), pMal-C2 (New England Biolabs Inc.), and pQE (QIAGEN Inc.) using standard molecular biology methods (Sambrook et al., 1989). pCMV-Myc and pFLAG-CMV2 were designed to express the proteins with the NH2-terminal Myc- and FLAG-epitopes, respectively. Various eukaryote expression constructs contained the following aa residues: pCMV5–l-afadin, 1–1829 (the full length); pCMV5–s-afadin, 1–1663 (the full length); pCMV-Myc–l-afadin, 1–1829 (the full length); pCMV-Myc–s-afadin, 1–1663 (the full length); pCMV-Myc–ponsin-1, 1–714 (the full length); pCMV-Myc–ponsin-2, 1–724 (the full length); pCMV-Myc–vinculin-N, 1–533 (the NH2-terminal half); pCMV-Myc–vinculin-C, 534–1066 (the COOH-terminal half); pCMV-Myc–vinculin-1, 1–800 (the NH2-terminal region); pCMV-Myc–vinculin-2, 801–1066 (the COOH-terminal region); pFLAG-CMV2–ponsin-1, 1–714 (the full length); and pFLAG-CMV2–ponsin-2, 1–724 (the full length). These constructs were transfected to COS7 cells with a DEAE-dextran method (Hata and Südhof, 1995). The Myc-tagged recombinant proteins were purified from the cell extracts by immunoprecipitation with an anti–Myc antibody as described below. Various glutathione S-transferase (GST)–fusion constructs contained the following aa residues: GST–ponsin-2-F, 1–724 (the full length); GST–ponsin-2-N, 1–400 (the NH2-terminal half); GST–ponsin-2-C, 401–724 (the COOH-terminal half); GST–ponsin-2-SH3(1+2), 460–631 (the first and second SH3 domains); GST–ponsin-2-SH3(2+3), 543–724 (the second and third SH3 domains); GST–ponsin-2-SH3(1), 460–558 (the first SH3 domain); GST–ponsin-2-SH3(2), 543–631 (the second SH3 domain); GST–ponsin-2-SH3(3), 648–724 (the third SH3 domain); GST– vinculin-P, 837–878 (the proline-rich region); and GST–vinculin-C-ΔP, 879–1066 (the COOH-terminal region lacking the proline-rich region). Six histidine residues (His6)–fusion constructs contained the following aa residues: His6–l-afadin-C199, 1631–1829 (the COOH-terminal 199 aa region including the third proline-rich region); His6–l-afadin-C132, 1698–1829 [the COOH-terminal 132 aa region lacking the aa residues (PPLP) of the third proline-rich region]. A maltose-binding protein (MBP)–fusion construct (MBP–vinculin-C) contained the COOH-terminal half of vinculin (aa 534–1066). The GST-, His6-, and MBP-fused proteins were purified by use of glutathione-Sepharose beads (Amersham-Pharmacia Biotechnology), TALON metal affinity beads (Clontech), and amylose resin beads (New England Biolabs Inc.), respectively.
Immunoprecipitation and Affinity Chromatography
Immunoprecipitation experiments from COS7 cells were done as follows: the eukaryote expression constructs described above were transfected to COS7 cells with the DEAE-dextran method in various combinations (Hata and Südhof, 1995). A lysis buffer (20 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, and 1 mM EGTA) containing protease inhibitors (20 μg/ml leupeptin, 1 μg/ml pepstatin A, and 1 mM PMSF) was added to the transfected COS7 cells. The cell extract (1.2 mg protein), obtained by centrifugation at 100,000 g for 1 h, was incubated with either an anti–Myc antibody (11.5 μg of protein; Takaishi et al., 1995) or an anti–FLAG M2 antibody (13.5 μg of protein; Eastman Kodak Scientific Imaging Systems) at 4°C for 3 h. 20 μl of protein G–Sepharose 4 Fast Flow beads (Amersham-Pharmacia Biotechnology) were added to this sample and incubation was performed at 4°C for 1 h. After the beads were washed with the lysis buffer, the bound proteins were eluted by boiling the beads in an SDS sample buffer for 10 min. The samples were then subjected to SDS-PAGE, followed by protein staining with Coomassie brilliant blue or by Western blot analysis.
Affinity chromatography was done as follows: various GST-fused proteins of ponsin-2 and GST alone (200 pmol each) were separately immobilized on 10 μl of glutathione-Sepharose beads (Amersham-Pharmacia Biotechnology). His6–l-Afadin-C-199 (the COOH-terminal 199 aa region including the third proline-rich region), His6–l-afadin-C132 (the COOH-terminal 132 aa region lacking the aa residues [PPLP] of the third proline-rich region), MBP–vinculin-C (the COOH-terminal half), or MBP alone (200 pmol each) was incubated at 4°C overnight with the glutathione-Sepharose beads in 100 μl of PBS containing 0.1% Triton X-100. For competition experiments, His6–l-afadin-C199 (200 pmol or 2 nmol) and MBP-vinculin-C (200 pmol or 2 nmol) were mixed and incubated with 10 μl of the GST–ponsin-2-C (the COOH-terminal half)–immobilized beads in 400 μl of PBS containing 0.1% Triton X-100. The beads were extensively washed with the same buffer. Elution was performed with PBS containing 20 mM glutathione.
Antibodies
A rabbit antiserum against ponsin was raised against GST–ponsin-2-SH3 (1) (the first SH3 domain). This antiserum was affinity-purified with GST– ponsin-2-SH3(1) covalently coupled to NHS-activated Sepharose beads (Amersham-Pharmacia Biotechnology) and used as a polyclonal antiponsin antibody. A mouse monoclonal anti–l-afadin antibody was prepared as described (Sakisaka et al., 1999). A rabbit polyclonal anti–l- and s-afadin antibody, which recognized both l- and s-afadins, was prepared as described (Mandai et al., 1997). A mouse monoclonal anti-vinculin antibody was purchased from Sigma Chemical Co. Rat monoclonal anti– E-cadherin (ECCD2) and anti–P-cadherin (PCD1) antibodies were purchased from TAKARA Shuzo, Inc. (Shiga).
Immunofluorescence and Immunoelectron Microscopy
Immunofluorescence microscopy of cultured cells and frozen sections of various rat tissues was done as described (Itoh et al., 1991; Mandai et al., 1997). Immunoelectron microscopy using the silver-enhancement technique was done as described (Mizoguchi et al., 1994; Mandai et al. 1997).
Other Procedures
125I-Labeled F-actin blot overlay and F-actin cosedimentation were done as described (Mandai et al., 1997; Satoh et al., 1998). Native vinculin was purified from chicken gizzard as described (O'Halloran et al., 1986). COS7, mouse mammary tumor MTD-1A, and rat 3Y1 cells were maintained in DME containing 10% FCS. Protein concentrations were determined with BSA as a reference protein (Bradford, 1976). SDS-PAGE was done as described (Laemmli, 1970). Fluorescence in situ hybridization (FISH) analysis was performed as described (Matsuda et al., 1992; Matsuda and Chapman, 1995).
Results
Purification of Ponsin and Molecular Cloning of its cDNA
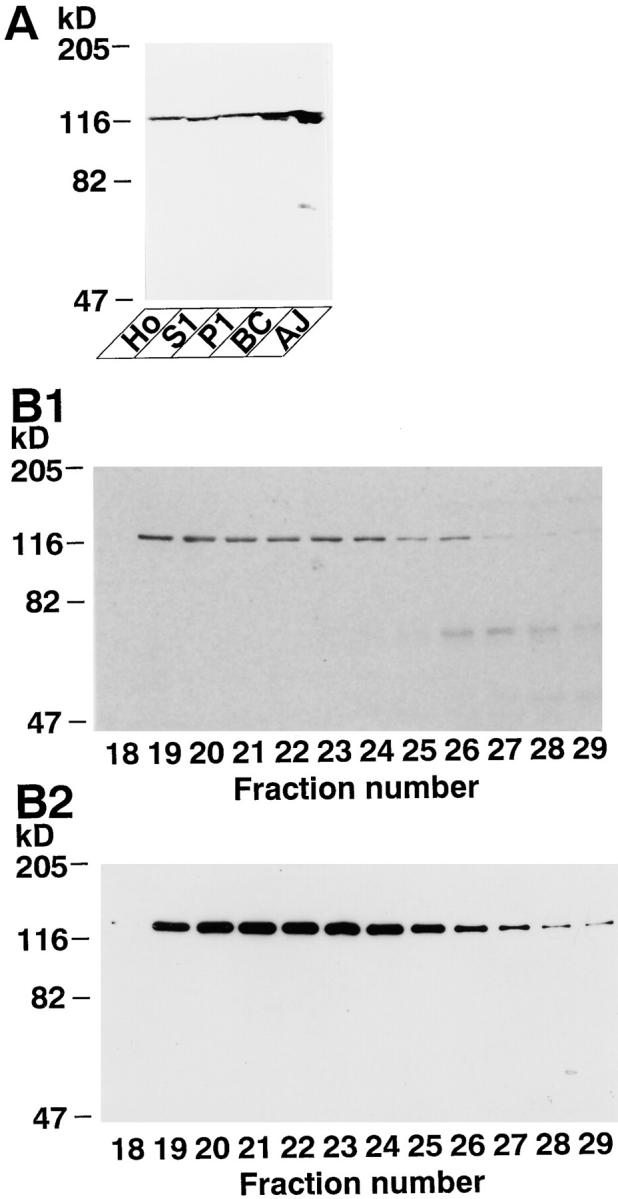
To identify an l-afadin–binding protein(s), we used a blot overlay method with 35S-labeled l-afadin. When each subcellular fraction of rat liver was subjected to SDS-PAGE, followed by the blot overlay, a radioactive band of ∼93 kD (p93) was detected and enriched in the AJ-enriched fraction (Fig. 1 A). p93 was a major binding partner for l-afadin in the AJ-enriched fraction. p93 was solubilized from this fraction with a combination of urea and Triton X-100, and highly purified by successive chromatographies of Mono Q, hydroxyapatite, and Mono S columns. On the final Mono S column chromatography, the radioactive band with l-afadin–binding activity well coincided with a protein of ∼93 kD (Fig. 1 B). p93 was purified with two other radioactive bands of ∼95 (p95) and 70 (p70) kD. They also coincided with proteins of ∼95 and 70 kD that were identified by protein staining with Coomassie brilliant blue. In addition to these proteins, a radioactive band of ∼55 kD (p55) was detected, but this band remained to be characterized.
These protein bands were separately cut out from the gel and digested with a lysyl endopeptidase. The digested peptides were separated by HPLC, and peptide maps of these proteins were obtained. Their maps were similar to each other, but the maps of p95 and p93 had additional peptide peaks when compared with that of p70 (data not shown). When two peptide peaks of each of p95, p93, and p70, both of which were common among these proteins, were separately sequenced, each of the three proteins included the following peptide sequences: peptide-1, LNRDDDSDLHSPRYSFSEDTK; peptide-2, CDDGWFVGTSRRTK. BLAST search of the GenBank database indicated that these sequences were almost identical to that of the mouse SH3P12 gene product (U58883; GenBank/EMBL/DDBJ), which had originally been identified as an SH3-containing protein (Sparks et al., 1996). These results suggest that p95, p93, and p70 are related to SH3P12.
We performed PCR to amplify the cDNA of SH3P12 using a mouse brain cDNA as a template, and isolated at least 12 clones whose restriction maps were similar to but slightly different from each other (data not shown). We determined the nucleotide sequences of two of them, clones 2 and 17. Each clone contained an in-frame stop codon upstream of the predicted initiation codon. The protein encoded by clone 2 had 724 aa with a calculated molecular weight of 81,217 (AF078666; GenBank/EMBL/ DDBJ) and the protein encoded by clone 17 had 714 aa with a calculated molecular weight of 79,492 (AF078667; GenBank/EMBL/DDBJ) (Fig. 2 A). Both the proteins included the aa sequences identical to peptide-1 and -2, except that L (at the 9th aa residue) and E (at the 18th aa residue) of peptide-1 were replaced with V and D, respectively, and K (at the 14th aa residue) of peptide-2 was replaced with R. This difference in the aa residues is likely to be due to the species difference between mouse and rat. When the aa sequences of clones 2, 17, and SH3P12 were aligned, they were highly homologous (92% aa identity between clone 2 and SH3P12 and 96% aa identity between clone 17 and SH3P12) but their differences were clustered and not distributed throughout the sequences (Fig. 2 A). To examine whether clones 2 and 17 were derived from the same locus, we made the chromosomal assignment by the FISH analysis using the cDNAs as probes. Clones 2 and 17 were located at the same locus (data not shown). These results suggest that clones 2, 17, and SH3P12 were splicing variants derived from the same gene.
Figure 2.
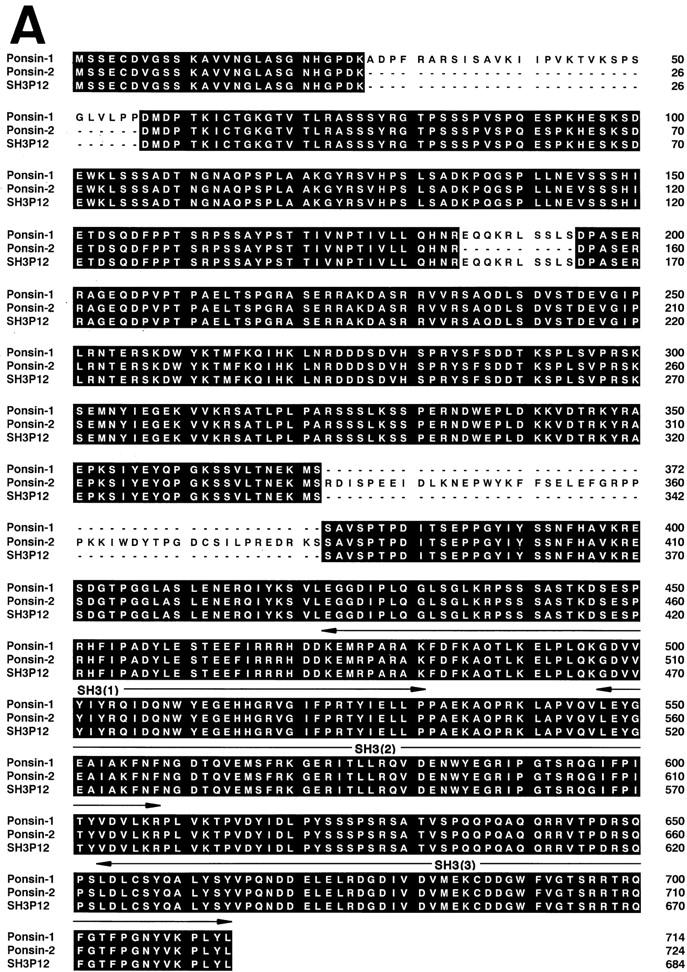
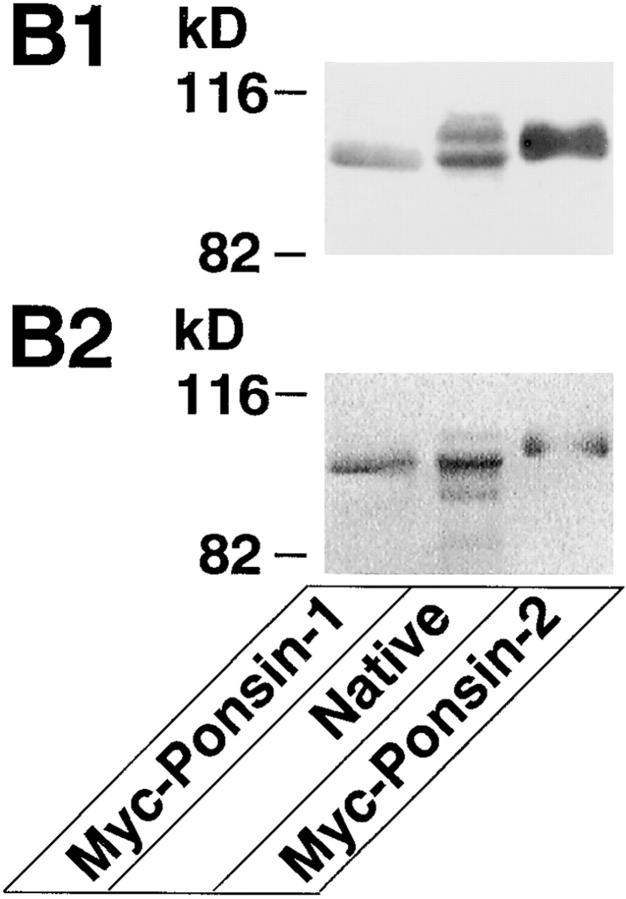
Molecular characterization of ponsin-1 and -2. (A) Deduced amino acid sequences of ponsin-1, -2, and SH3P12. Identity is highlighted. Signaling domain organization was determined with the Web-based tool, SMART (simple modular architecture research tool) program (Schultz et al., 1998). (B) l-Afadin–binding activity of ponsin-1 and -2. Myc– ponsin-1 (the full length) or -2 (the full length) was expressed in COS7 cells by transfection with pCMV-Myc– ponsin-1 or -2, respectively. The Myc-tagged protein was immunoprecipitated with the anti–Myc antibody, and then subjected to SDS-PAGE (8% polyacrylamide gel), followed by 35S-labeled l-afadin blot overlay or protein staining with Coomassie brilliant blue. An aliquot (2 μl) of fraction 10 in the Mono S column chromatography was also subjected to SDS-PAGE, followed by 35S-labeled l-afadin blot overlay or by protein staining. (B1) 35S-Labeled l-afadin blot overlay, and (B2) protein staining. The results are representative of three independent experiments.
To examine whether the protein encoded by clone 2 or 17 was the mouse counterpart of p95, p93, or p70, we constructed a Myc-tagged mammalian expression vector with each DNA, and expressed the protein in COS7 cells. The Myc-tagged recombinant protein was immunoprecipitated with the anti–Myc antibody and subjected to SDS-PAGE, followed by 35S-labeled l-afadin blot overlay. The protein encoded by clone 17 showed a mobility similar to that of native p93 on SDS-PAGE and the 35S-labeled l-afadin– binding activity (Fig. 2 B). On the basis of these observations, we concluded that clone 17 encoded the full-length cDNA of the mouse counterpart of p93 and named it ponsin-1, although we did not know the reason for the difference between the molecular mass value calculated from the predicted aa sequence and that estimated by SDS-PAGE. On the other hand, the protein encoded by clone 2 had the 35S-labeled l-afadin–binding activity, but showed a mobility between p93 and p95 on SDS-PAGE. Thus, we could not conclude whether p95 was encoded by clone 2. The protein encoded by clone 2 was named ponsin-2. Ponsin-2 appeared to bind 35S-labeled l-afadin more than ponsin-1 under the conditions used here. Hydrophilicity analysis of ponsin-1 and -2 predicted that these proteins had no transmembrane region (data not shown). Computer analysis of domain organization indicated that ponsin-1 and -2 had three SH3 domains at their COOH-terminal regions and two sorbin-like regions at their NH2-terminal regions, but did not have other known signaling domains (Fig. 2 A).
Binding of l-Afadin to Ponsin
We confirmed the binding of l-afadin to ponsin by an immunoprecipitation method. Myc–l-afadin (the full length) and FLAG–ponsin (the full length) were transiently coexpressed in COS7 cells. When FLAG–ponsin-2 was immunoprecipitated with the anti–FLAG antibody, Myc– l-afadin was coprecipitated (Fig. 3 A). Conversely, when Myc–l-afadin was immunoprecipitated with the anti–Myc antibody, FLAG–ponsin-2 was coprecipitated. Similarly, FLAG–ponsin-2 and tag-free l-afadin (the full length) or s-afadin (the full length) were transiently coexpressed in COS7 cells. When FLAG–ponsin-2 was immunoprecipitated with the anti–FLAG antibody, most of l-afadin was coprecipitated, but s-afadin was coprecipitated to a small extent and mostly recovered in the supernatant fraction (Fig. 3 B). The essentially same results were obtained with ponsin-1 (data not shown). These results indicate that ponsin binds mainly l-afadin and slightly s-afadin.
Figure 3.
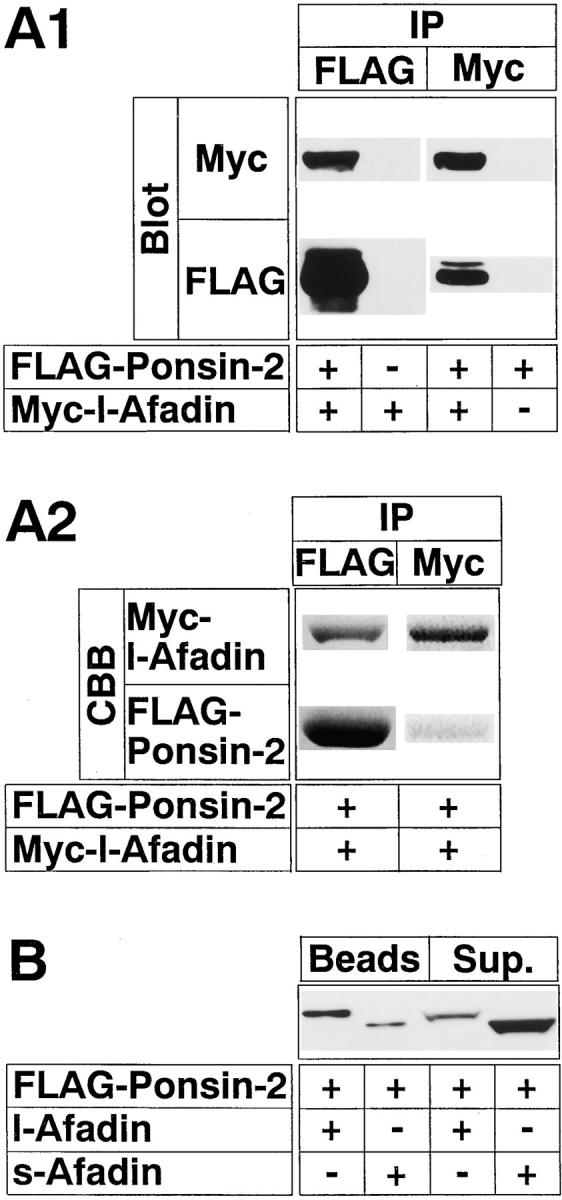
Binding of l-afadin to ponsin in vivo. (A) Immunoprecipitation analysis of l-afadin and ponsin-2. FLAG–ponsin-2 (the full length) and Myc–l-afadin (the full length) were coexpressed in COS7 cells in various combinations by transfection with pFLAG-CMV2–ponsin-2 and pCMV-Myc–l-afadin, respectively. Each cell extract was subjected to immunoprecipitation with the anti–FLAG or anti–Myc antibody. The precipitate was then subjected to SDS-PAGE (8% polyacrylamide gel), followed by Western blot analysis using the anti–FLAG or anti–Myc antibody, or by protein staining with Coomassie brilliant blue. (A1) Western blot analysis, and (A2) protein staining. (IP) Immunoprecipitation, and (CBB) staining with Coomassie brilliant blue. (B) Immunoprecipitation analysis of s-afadin and ponsin-2. Both FLAG–ponsin-2 and tag-free l-afadin (the full length) or both FLAG–ponsin-2 and tag-free s-afadin (the full length) were coexpressed in COS7 cells by transfection with both pFLAG-CMV2– ponsin-2 and pCMV5–l-afadin or both pFLAG-CMV2–ponsin-2 and pCMV5–s-afadin, respectively. Each cell extract was incubated with the anti–FLAG antibody and protein G–Sepharose beads, followed by centrifugation. The beads and the supernatant were separately subjected to SDS-PAGE (8% polyacrylamide gel), followed by Western blot analysis using the anti–l- and s-afadin antibody. (Sup) supernatant. The results are representative of three independent experiments.
To examine the possibility that ponsin was an F-actin– binding protein and bound l-afadin through F-actin, we performed 125I-labeled F-actin blot overlay and F-actin cosedimentation using the recombinant proteins as described (Mandai et al., 1997; Satoh et al., 1998). GST–ponsin-2-F (the full length) did not bind to F-actin under the conditions where l-afadin bound to F-actin (data not shown).
Binding Regions of l-Afadin and Ponsin
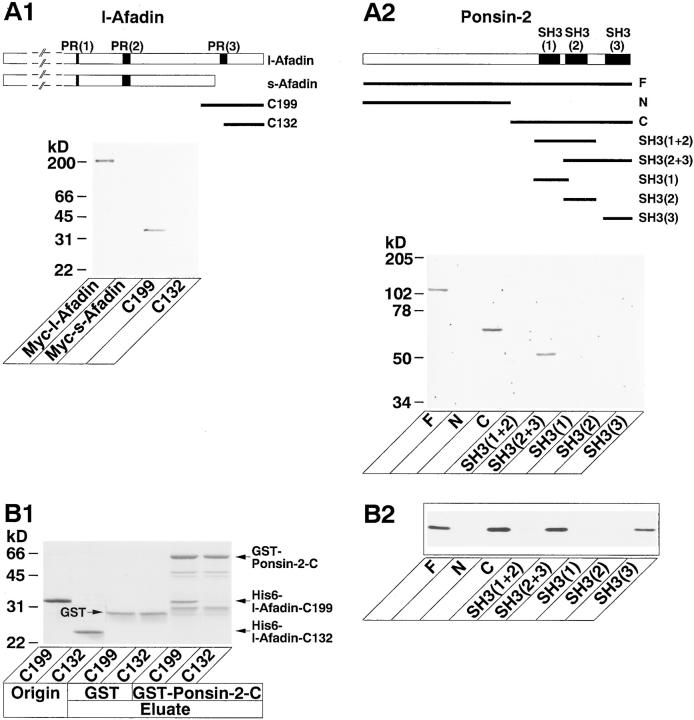
We first determined the ponsin-binding region of l-afadin. 35S-labeled ponsin-2 blot overlay analysis revealed that Myc–l-afadin (the full length), but not Myc–s-afadin (the full length) that lacked the third proline-rich region, bound to ponsin-2 (Fig. 4 A1). His6–l-Afadin-C199 (the COOH-terminal 199 aa region including the third proline-rich region), but not His6–l-afadin-C132 (the COOH-terminal 132 aa region lacking the aa residues [PPLP] of the third proline-rich region), bound to ponsin-2. These results suggest that the third proline-rich region of l-afadin binds to ponsin.
Figure 4.
Binding regions of l-afadin and ponsin. (A) Blot overlay. (A1) 35S-Labeled ponsin-2 blot overlay. Myc–l-Afadin (the full length), Myc–s-afadin (the full length), His6–l-afadin-C199 (the COOH-terminal 199 aa region including the third proline-rich region), and His6– l-afadin-C132 (the COOH-terminal 132 aa region lacking the aa residues [PPLP] of the third proline-rich domain) (5 pmol each) were subjected to SDS-PAGE (8 and 12% discontinuous polyacrylamide gel), followed by 35S-labeled ponsin-2 blot overlay. (A2) 35S-Labeled l-afadin blot overlay. Various GST-fused proteins of ponsin-2 (5 pmol each) were subjected to SDS-PAGE (8% polyacrylamide gel), followed by 35S-labeled l-afadin blot overlay. (B) Affinity chromatography. (B1) Binding region of l-afadin. His6–l-Afadin-C199 or His6–l-afadin-C132 was incubated with GST–ponsin-2-C (the COOH-terminal half) or GST alone immobilized on glutathione-Sepharose beads. Each original sample and eluate were subjected to SDS-PAGE (15% polyacrylamide gel), followed by protein staining with Coomassie brilliant blue. (B2) Binding region of ponsin. His6– l-afadin-C199 was incubated with various GST-fused proteins of ponsin-2 immobilized on glutathione-Sepharose beads. Each eluate was subjected to SDS-PAGE (12% polyacrylamide gel), followed by Western blot analysis using the anti–l-afadin antibody. [PR(1)] the first proline-rich region (aa 1219–1229), [PR(2)] the second proline-rich region (aa 1372–1399), [PR(3)] the third proline-rich region (aa 1691–1713), (C199) His6–l-afadin-C199, (C132) His6–l-afadin-C132, (F) GST–ponsin-2-F (the full length), (N) GST–ponsin-2-N (the NH2-terminal half), (C) GST–ponsin-2-C, [SH3(1+2)] GST– ponsin-2-SH3(1+2) (the first and second SH3 domains), [SH3(2+3)] GST–ponsin-2-SH3(2+3) (the second and third SH3 domains), [SH3(1)] GST–ponsin-2-SH3(1) (the first SH3 domain), [SH3(2)] GST–ponsin-2-SH3(2) (the second SH3 domain), and [SH3(3)] GST– ponsin-2-SH3(3) (the third SH3 domain). The results are representative of three independent experiments.
We next determined the l-afadin–binding region of ponsin. 35S-Labeled l-afadin blot overlay analysis revealed that GST–ponsin-2-F (the full length) and GST–ponsin-2-C (the COOH-terminal half) bound to l-afadin (Fig. 4 A2). In contrast, GST–ponsin-2-N (the NH2-terminal half) did not show this activity. In the COOH-terminal half of ponsin-2, GST–ponsin-2-SH3(2+3) (the second and third SH3 domains) showed this activity, but GST–ponsin-2-SH3 (1+2) (the first and second SH3 domains) did not. When each of the three SH3 domains was used, neither GST– ponsin-2-SH3(1) (the first SH3 domain), GST–ponsin-2-SH3 (2) (the second SH3 domain), nor GST–ponsin-2-SH3(3) (the third SH3 domain) bound to l-afadin. These results suggest that the region containing the second and third SH3 domains binds to l-afadin.
We furthermore examined the binding regions of l-afadin and ponsin by an affinity chromatography method using the recombinant proteins. Consistent with the results by the blot overlay method, His6–l-afadin-C199 bound to GST–ponsin-2-C immobilized on glutathione-Sepharose beads, whereas His6–l-afadin-C132 did not (Fig. 4 B1). His6–l-afadin-C199 bound to GST–ponsin-2-F, GST–ponsin-2-C, and GST–ponsin-2-SH3(2+3), but not to GST– ponsin-2-N, GST–ponsin-2-SH3(1+2), GST–ponsin-2-SH3 (1), or GST–ponsin-2-SH3(2) (Fig. 4 B2). In contrast, His6–l-afadin-C199 slightly bound to GST–ponsin-2-SH3 (3). This inconsistency may be due to the sensitivity difference between the two methods.
Binding of Ponsin to Vinculin
Because ponsin-1 and -2 were predicted not to be integral membrane proteins as described above, these proteins by themselves would not determine the specific localization of l-afadin at ZA in epithelial cells and at cell–cell AJ in nonepithelial cells. To explore another molecule(s) through which l-afadin was localized there, we furthermore attempted to identify a ponsin-binding protein(s). To identify the protein(s), we performed a blot overlay method with 35S-labeled ponsin-2. When each subcellular fraction of rat liver was subjected to SDS-PAGE, followed by the blot overlay, a radioactive band of ∼120 kD was detected and relatively enriched in the AJ-enriched fraction (Fig. 5 A). Each fraction from the Mono Q column chromatography, which was used for the purification of ponsin, was subjected to 35S-labeled ponsin-2 blot overlay. The radioactive band of ∼120 kD was detected in fractions 19–29 (Fig. 5 B1). The radioactive band well coincided with a protein band of ∼120 kD that was identified by protein staining with Coomassie brilliant blue (data not shown). This protein was recognized by the anti-vinculin antibody, suggesting that ponsin directly binds vinculin (Fig. 5 B2).
Figure 5.
Binding of ponsin to vinculin in vitro. (A) Subcellular distribution of 35S-labeled ponsin-2–binding proteins in rat liver. An aliquot (60 μg protein) of each subcellular fraction was subjected to SDS-PAGE (8% polyacrylamide gel), followed by 35S-labeled ponsin-2 blot overlay. (Ho) homogenate, (S1) soluble fraction, (P1) pellet fraction, (BC) fraction rich in bile canaliculi, and (AJ) fraction rich in AJ. (B) Mono Q column chromatography of 35S-labeled ponsin-2–binding proteins. (B1) 35S-labeled ponsin-2 blot overlay. An aliquot (10 μl) of each fraction was subjected to SDS-PAGE (8% polyacrylamide gel), followed by 35S-labeled ponsin-2 blot overlay. (B2) Western blot analysis. An aliquot (4 μl) of each fraction was subjected to SDS-PAGE (8% polyacrylamide gel), followed by Western blot analysis using the anti-vinculin antibody. The results are representative of three independent experiments.
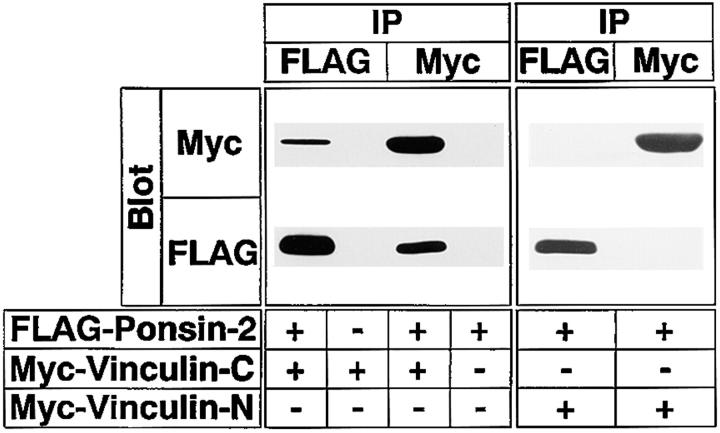
We confirmed the binding of ponsin to vinculin by an immunoprecipitation method. Because vinculin shows the intramolecular association of its NH2- and COOH-terminal halves (Johnson and Craig, 1995), we used the COOH- and NH2-terminal halves of vinculin. FLAG–ponsin-2 (the full length) and Myc–vinculin-C (the COOH-terminal half) were transiently coexpressed in COS7 cells. When FLAG–ponsin-2 was immunoprecipitated with the anti– FLAG antibody, Myc–vinculin-C was coprecipitated (Fig. 6). Conversely, when Myc–vinculin-C was immunoprecipitated with the anti–Myc antibody, FLAG–ponsin-2 was coprecipitated. When Myc–vinculin-N (the NH2-terminal half) was used instead of Myc–vinculin-C, FLAG–ponsin-2 or Myc–vinculin-N was not coimmunoprecipitated. The essentially same results were obtained with ponsin-1 (data not shown). These results indicate that ponsin binds not only l-afadin but also vinculin both in cell-free and intact cell systems.
Figure 6.
Immunoprecipitation analysis of ponsin-2 and vinculin. FLAG–ponsin-2 (the full length), Myc–vinculin-N (the NH2-terminal half), and Myc–vinculin-C (the COOH-terminal half) were expressed in COS7 cells in various combinations by transfection with pFLAG-CMV2–ponsin-2, pCMV-Myc–vinculin-N, and pCMV-Myc–vinculin-C, respectively. Each cell extract was subjected to immunoprecipitation with the anti–FLAG or anti–Myc antibody. The precipitate was then subjected to SDS-PAGE (8% polyacrylamide gel), followed by Western blot analysis using the anti–FLAG or anti–Myc antibody. (IP) Immunoprecipitation. The results are representative of three independent experiments.
Binding Regions of Ponsin and Vinculin
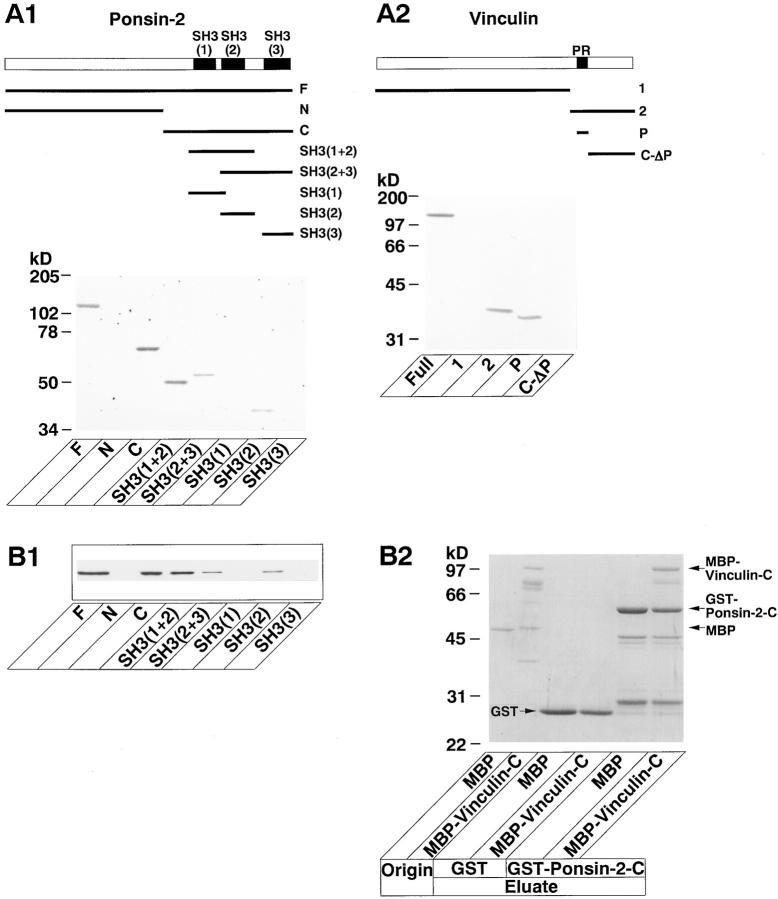
We first examined the vinculin-binding region of ponsin. 35S-labeled vinculin blot overlay analysis revealed that GST–ponsin-2-F (the full length) and GST–ponsin-2-C (the COOH-terminal half) bound to vinculin (Fig. 7 A1). In contrast, GST–ponsin-2-N (the NH2-terminal half) did not show this activity. In the COOH-terminal half of ponsin, GST–ponsin-2-SH3(1+2) (the first and second SH3 domains) bound to vinculin, whereas neither GST–ponsin-2-SH3(1) (the first SH3 domain) nor GST–ponsin-2-SH3 (3) (the third SH3 domain) bound. GST–ponsin-2-SH3 (2+3) (the second and third SH3 domains) and GST–ponsin-2-SH3(2) (the second SH3 domain) slightly bound to vinculin. These results suggest that the region containing the first and second SH3 domains binds to vinculin.
Figure 7.
Binding regions of ponsin and vinculin. (A) Blot overlay. (A1) 35S-Labeled vinculin blot overlay. Various GST-fused proteins of ponsin-2 (5 pmol each) were subjected to SDS-PAGE (8% polyacrylamide gel), followed by 35S-labeled vinculin blot overlay. (A2) 35S- Labeled ponsin-2 blot overlay. Full-length native vinculin and various recombinant proteins of vinculin (5 pmol each) were subjected to SDS-PAGE (8 and 12% discontinuous polyacrylamide gel), followed by 35S- labeled ponsin blot overlay. (B) Affinity chromatography. (B1) Binding region of ponsin. MBP– vinculin-C (the COOH-terminal half) was incubated with various GST-fused proteins of ponsin-2 immobilized on glutathione-Sepharose beads. Each eluate was subjected to SDS-PAGE (10% polyacrylamide gel), followed by Western blot analysis using the anti-vinculin antibody. (B2) Binding region of vinculin. MBP–vinculin-C or MBP alone was incubated with GST–ponsin-2-C (the COOH-terminal half) or GST alone immobilized on glutathione-Sepharose beads. Each original sample and eluate were subjected to SDS-PAGE (12% polyacrylamide gel), followed by protein staining with Coomassie brilliant blue. (F) GST– ponsin-2-F (the full length), (N) GST–ponsin-2-N (the NH2-terminal half), (C) GST–ponsin-2-C, [SH3(1+2)] GST–ponsin-2-SH3(1+2) (the first and second SH3 domains), [SH3(2+3)] GST–ponsin-2-SH3(2+3) (the second and third SH3 domains), [SH3(1)] GST–ponsin-2-SH3(1) (the first SH3 domain), [SH3(2)] GST–ponsin-2-SH3(2) (the second SH3 domain), [SH3(3)] GST–ponsin-2-SH3(3) (the third SH3 domain), [PR] the proline-rich region of vinculin (aa 837–878), (1) Myc–vinculin-1 (the NH2-terminal region, aa 1–800), (2) Myc–vinculin-2 (the COOH-terminal region, aa 801–1066); (P) GST–vinculin-P (the proline-rich region), and (C-ΔP) GST–vinculin-C-ΔP (the COOH-terminal region lacking the proline-rich region). The results are representative of three independent experiments.
We next examined the ponsin-binding region of vinculin. Consistent with the results by the immunoprecipitation method, 35S-labeled ponsin-2 blot overlay analysis revealed that native vinculin and Myc–vinculin-2 (the COOH-terminal region, aa 801–1066), but not Myc–vinculin-1 (the NH2-terminal region, aa 1–800), bound to ponsin-2 (Fig. 7 A2). GST–vinculin-P (the proline-rich region), but not GST–vinculin-C-ΔP (the COOH-terminal region lacking the proline-rich region), bound to ponsin-2. These results suggest that the proline-rich region of vinculin binds to ponsin. The proline-rich region has two proline-rich sequences, one of which binds to the cytoskeletal protein, vasodilator-stimulated phosphoprotein (VASP; Brindle et al., 1996; Niebuhr et al., 1997). It remains to be clarified which proline-rich sequence of vinculin is responsible for the binding to ponsin.
We furthermore examined the binding regions of ponsin and vinculin by an affinity chromatography method using the recombinant proteins. Consistent with the results by the immunoprecipitation and blot overlay methods, MBP– vinculin-C (the COOH-terminal half) bound to GST–ponsin-2-F, GST–ponsin-2-C, and GST–ponsin-2-SH3(1+2), but not to GST–ponsin-2-SH3(1) or GST–ponsin-2-SH3 (3) (Fig. 7 B1). MBP–vinculin-C slightly bound to GST– ponsin-2-SH3(2+3) and GST–ponsin-2-SH3(2). When MBP alone was used instead of MBP–vinculin-C, MBP alone did not bind to GST–ponsin-2-C (Fig. 7 B2).
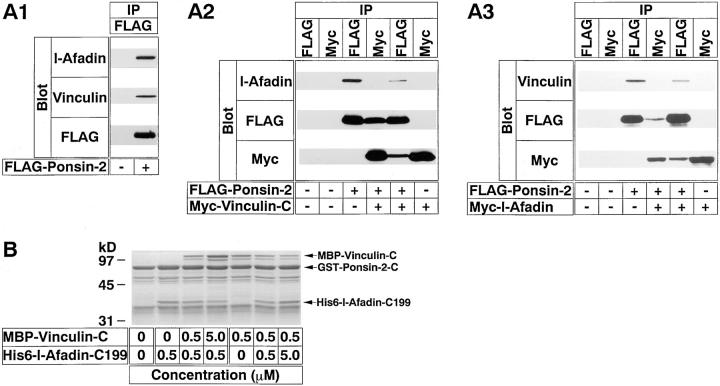
Inability of Ponsin to Form a Ternary Complex with l-Afadin and Vinculin
The above result that the l-afadin– and vinculin-binding regions of ponsin are partly overlapped suggests that l-afadin and vinculin bind to ponsin in a competitive manner and that these three proteins do not form a ternary complex. We examined this possibility by two different methods, immunoprecipitation and affinity chromatography. When FLAG–ponsin-2 (the full length) alone was transiently expressed in COS7 cells and immunoprecipitated with the anti–FLAG antibody, both endogenous l-afadin and vinculin were coprecipitated (Fig. 8 A1). However, it was not clear from this result whether a ternary complex of ponsin, l-afadin, and vinculin, or a mixture of binary complexes of ponsin and l-afadin and of ponsin and vinculin was coprecipitated. To address this issue, we examined whether ponsin and l-afadin or ponsin and vinculin were coprecipitated when vinculin or l-afadin was immunoprecipitated, respectively. Both FLAG–ponsin-2 and Myc– vinculin-C (the COOH-terminal half) were transiently coexpressed in COS7 cells. When Myc–vinculin-C was immunoprecipitated with the anti–Myc antibody, FLAG– ponsin-2, but not endogenous l-afadin, was coprecipitated (Fig. 8 A2). When FLAG–ponsin-2 was immunoprecipitated with the anti–FLAG antibody, both Myc–vinculin-C and endogenous l-afadin were coprecipitated, but the amount of the precipitated l-afadin was reduced, compared with that from COS7 cells expressing FLAG–ponsin-2 alone. Next, both FLAG–ponsin-2 and Myc–l-afadin (the full length) were transiently coexpressed in COS7 cells. When Myc–l-afadin was immunoprecipitated with the anti–Myc antibody, FLAG–ponsin-2, but not endogenous vinculin, was coprecipitated (Fig. 8 A3). When FLAG–ponsin-2 was immunoprecipitated with the anti– FLAG antibody, both Myc–l-afadin and endogenous vinculin were coprecipitated, but the amount of the precipitated vinculin was reduced compared with that from COS7 cells expressing FLAG–ponsin-2 alone. These results suggest that l-afadin and vinculin bind to ponsin in a competitive manner, and that l-afadin, ponsin, and vinculin hardly form a ternary complex.
Figure 8.
Inability of ponsin to form a ternary complex with l-afadin and vinculin. (A) Immunoprecipitation from COS7 cells. Myc– l-afadin (the full length), FLAG–ponsin-2 (full length), and Myc–vinculin-C (the COOH-terminal half) were expressed in COS7 cells in various combinations by transfection with pCMV-Myc–l-afadin, pFLAG-CMV2–ponsin-2, and pCMV-Myc–vinculin-C, respectively. Each cell extract was subjected to immunoprecipitation with the anti–Myc or anti–FLAG antibody. The precipitate was then subjected to SDS-PAGE (8% polyacrylamide gel), followed by Western blot analysis using the anti–Myc, anti–FLAG, anti–l-afadin, or anti-vinculin antibody. (A1) COS7 cells expressing FLAG–ponsin-2 alone, (A2) COS7 cells expressing both FLAG–ponsin-2 and Myc–vinculin-C, (A3) COS7 cells expressing both Myc–l-afadin and FLAG–ponsin-2. (IP) Immunoprecipitation. (B) Affinity chromatography using ponsin-immobilized beads. Various doses of His6–l-afadin-C199 (the COOH-terminal 199 aa region including the third proline-rich region) and MBP–vinculin-C (the COOH-terminal half) were mixed and incubated with GST–ponsin-2-C (the COOH-terminal half) immobilized on glutathione-Sepharose beads. Each eluate was subjected to SDS-PAGE (15% polyacrylamide gel), followed by protein staining with Coomassie brilliant blue. The results are representative of three independent experiments.
We confirmed this conclusion by an affinity chromatography method using the recombinant proteins. His6–l-Afadin-C199 (the COOH-terminal 199 aa region including the third proline-rich region) was mixed with MBP–vinculin-C (the COOH-terminal half) and incubated with GST–ponsin-2-C (the COOH-terminal half) immobilized on glutathione-Sepharose beads. When the concentration of His6–l-afadin-C199 was fixed and the concentration of MBP–vinculin-C was increased, the amount of the bound His6–l-afadin-C199 was decreased, whereas the amount of the bound MBP–vinculin-C was increased (Fig. 8 B). Conversely, when the concentration of His6–l-afadin-C199 was increased and the concentration of MBP–vinculin-C was fixed, the amount of the bound His6–l-afadin-C199 was increased, whereas the amount of the bound MBP–vinculin-C was decreased. The exact affinities of l-afadin and vinculin to ponsin were not examined, but their affinities were apparently similar.
We confirmed that l-afadin and vinculin did not bind to each other. Myc–l-afadin or Myc–vinculin-C alone was transiently expressed in COS7 cells. When Myc–l-afadin was immunoprecipitated with the anti–Myc antibody, endogenous vinculin was not precipitated (Fig. 8 A3). Similarly, when Myc–vinculin-C was immunoprecipitated with the anti–Myc antibody, endogenous l-afadin was not precipitated (Fig. 8 A2).
Tissue Distribution and Intracellular Localization of Ponsin
In the next series of experiments, we analyzed the tissue distribution and intracellular localization of ponsin. Northern blot analysis using ponsin-2 as a probe showed that ponsin was ubiquitously expressed in all the mouse tissues examined, including heart, brain, spleen, lung, liver, skeletal muscle, kidney, and testis (Fig. 9 A). In heart, brain, liver, and skeletal muscle, several bands were detected, consistent with the above result that ponsin had several splicing variants. Western blot analysis using the anti-ponsin antibody showed that ponsin as well as l-afadin was concentrated in the AJ-enriched fraction of liver (Fig. 9 B). This antibody specifically recognized p93 (ponsin-1), p95, and p70. Vinculin was detected in all the fractions and relatively concentrated in the AJ-enriched fraction (Fig. 9 B).
Figure 9.
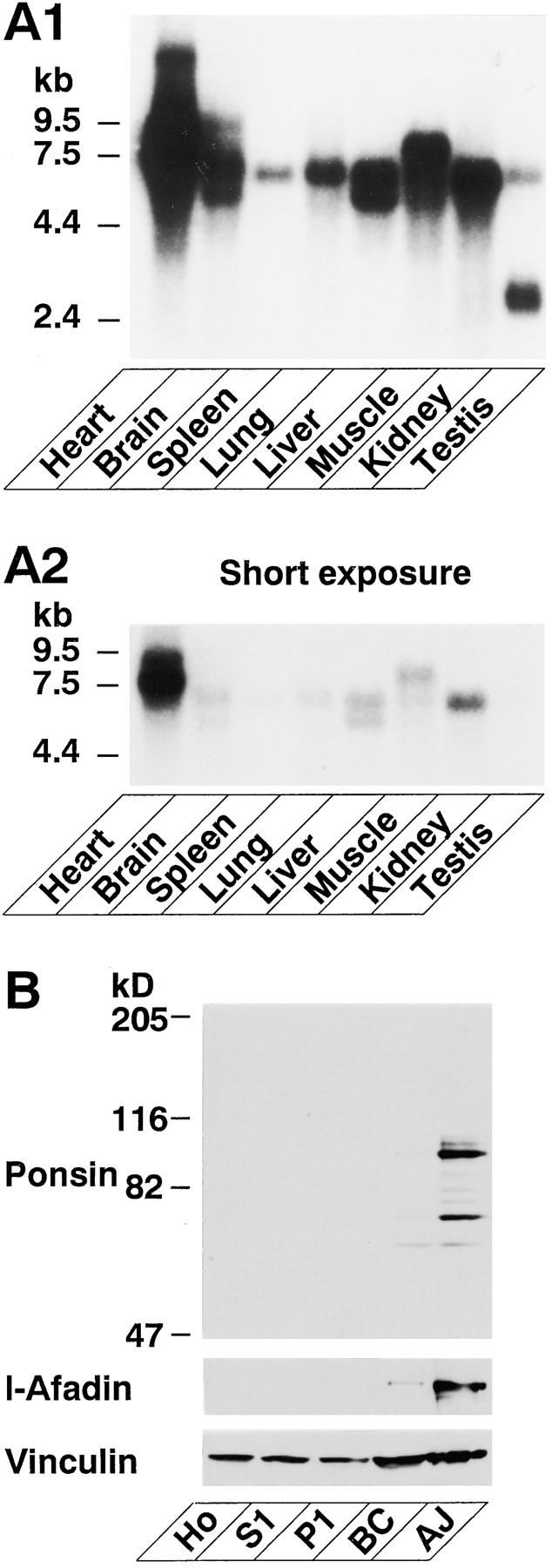
Tissue and subcellular distribution of ponsin. (A) Northern blot analysis. A mouse RNA blot membrane (Clontech) was hybridized with the 32P-labeled full-length cDNA of ponsin-2, followed by autoradiography with 48- or 8-h exposure. (A1) 48-h exposure, and (A2) 8-h exposure. (B) Subcellular distribution in rat liver. An aliquot (15 μg protein) of each subcellular fraction was subjected to SDS-PAGE (8% polyacrylamide gel), followed by Western blot analysis using the anti-ponsin, anti–l-afadin, or anti-vinculin antibody. (Ho) homogenate, (S1) soluble fraction, (P1) pellet fraction, (BC) fraction rich in bile canaliculi, and (AJ) fraction rich in AJ.
To determine the intracellular localization of ponsin, we performed immunofluorescence microscopy of cultured cells and various rat tissues. At the junctional level of mouse mammary tumor MTD-1A cells, ponsin was colocalized with l-afadin and vinculin at cell–cell AJ (Fig. 10). At the basal level, ponsin was colocalized with vinculin at cell–matrix AJ where l-afadin was not localized. In rat 3Y1 fibroblasts, ponsin was colocalized with l-afadin, vinculin, and P-cadherin at cell–cell AJ and with vinculin at cell– matrix AJ (Fig. 11). When the frozen sections of rat small intestine were doubly stained with the monoclonal anti– E-cadherin antibody, ponsin was concentrated with E-cadherin at the junctional complex region of absorptive epithelial cells, but ponsin was more highly concentrated at the junctional complex region than E-cadherin was (Fig. 12 A). When the frozen sections of rat heart were doubly stained with the monoclonal anti-vinculin antibody, ponsin was colocalized with vinculin at the intercalated disc. They were also periodically located along the lateral borders of cardiac muscle cells (costamere) (Fig. 12 B). In liver, ponsin showed the beltlike localization along the bile canaliculi (Fig. 12 C). This localization was consistent with its subcellular distribution in liver (see Fig. 9 B). These results indicate that ponsin is localized at both cell–cell and cell–matrix AJs, although l-afadin is localized only at cell–cell AJ, and that ponsin is colocalized with l-afadin at cell–cell AJ.
Figure 10.
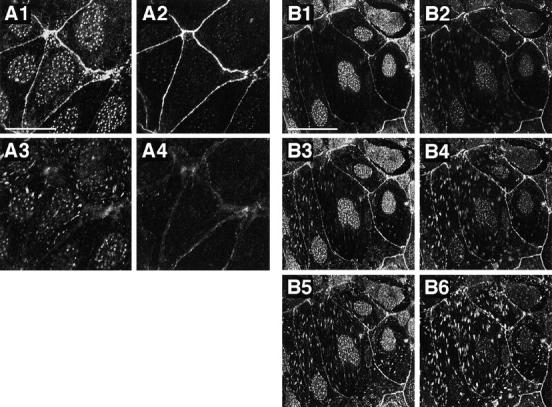
Localization of l-afadin, ponsin, and vinculin in cultured mouse mammary tumor MTD-1A cells. The cells were doubly stained with the rabbit anti-ponsin and mouse anti–l-afadin antibodies, or with the rabbit anti-ponsin and mouse anti-vinculin antibodies. There were nuclear stainings with this anti-ponsin antibody, but its significance was not clear. (A) The double staining with the antiponsin and anti–l-afadin antibodies. (A1 and A3) Ponsin, (A2 and A4) l-afadin, (A1 and A2) junctional level, and (A3 and A4) basal level. (B) The double staining with the anti-ponsin and anti-vinculin antibodies. (B1, B3, and B5) Ponsin, (B2, B4, and B6) vinculin, (B1 and B2) junctional level, (B3 and B4) middle level, and (B5 and B6) basal level. Bars: (A) 25 μm and (B) 50 μm. The results are representative of three independent experiments.
Figure 11.
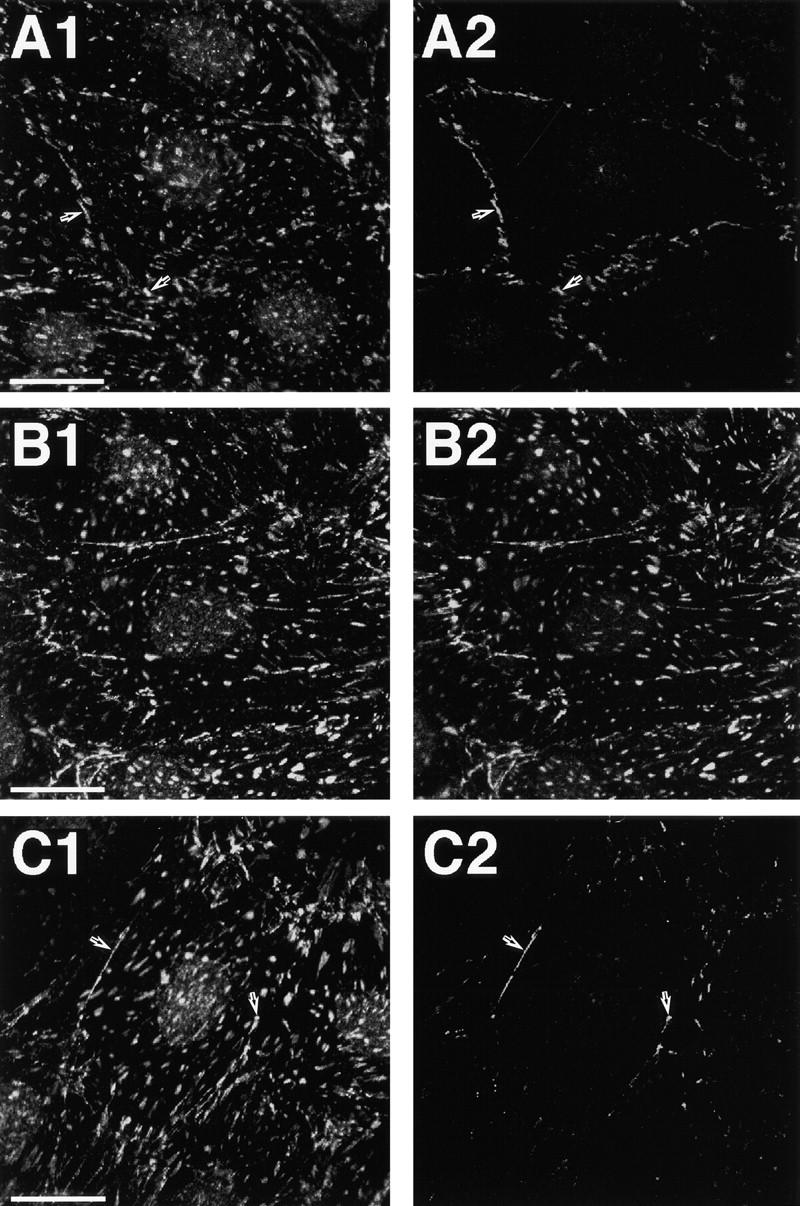
Localization of l-afadin, ponsin, vinculin, and P-cadherin in cultured rat 3Y1 fibroblasts. (A) The double staining with the anti-ponsin and anti–l-afadin antibodies. (A1) Ponsin and (A2) l-afadin. (B) The double staining with the anti-ponsin and anti-vinculin antibodies. (B1) Ponsin and (B2) vinculin. (C) The double staining with the anti-ponsin and anti–P-cadherin antibodies. (C1) Ponsin and (C2) P-cadherin. (Arrows) Cell–cell AJ. Bars: 25 μm. The results are representative of three independent experiments.
Figure 12.
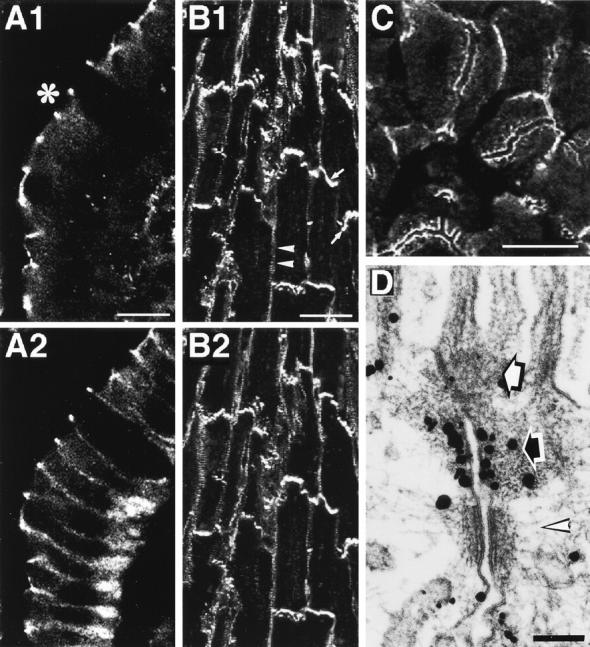
Localization of ponsin, E-cadherin, and vinculin in various rat tissues. (A) The double staining of small intestine absorptive epithelial cells with the antiponsin and anti–E-cadherin antibodies. (A1) Ponsin and (A2) E-cadherin. *Apical side. (B) The double staining of heart with the anti-ponsin and anti-vinculin antibodies. (B1) Ponsin and (B2) vinculin. (Arrow) Intercalated disc and (arrowhead) costamere. (C) The staining of liver with the anti-ponsin antibody. (D) Ultrastructural localization of ponsin in small intestine absorptive epithelial cells. Intestine absorptive epithelial cells were labeled with the anti-ponsin antibody. (Open arrow) ZO, (closed arrow) ZA, and (open arrowhead) desmosome. Bars: (A and C) 10, (B) 50, and (D) 0.1 μm. The results are representative of three independent experiments.
To examine the precise localization of ponsin in the junctional complex region, we performed immunoelectron microscopy of rat small intestine absorptive epithelial cells using the silver enhancement technique. Ponsin was associated with the undercoat of ZA where l-afadin is localized (Mandai et al., 1997) (Fig. 12 D).
Discussion
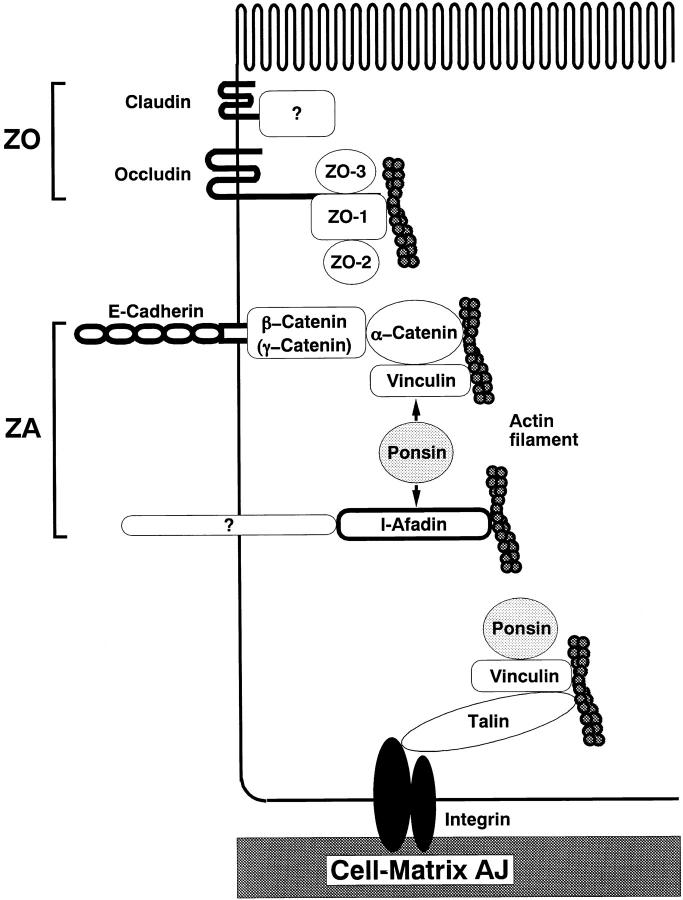
In this study, we isolated an l-afadin– and vinculin-binding protein, named ponsin, which was ubiquitously expressed and colocalized with vinculin at ZA in epithelial cells, at cell–cell AJ in nonepithelial cells, and at cell–matrix AJ in both types of cells (Fig. 13). We first purified three 35S-l-afadin–binding proteins, p95, p93, and p70, from the AJ-enriched fraction of rat liver. We then analyzed the peptide maps and sequences of these proteins and found that they were related to each other. Based on their peptide sequences, we obtained the mouse cDNAs of ponsin-1 and -2. Ponsin-1 was likely a mouse counterpart of rat p93. However, ponsin-2 did not show the same mobility to that of p95 or p70 on SDS-PAGE. During the molecular cloning of ponsin, we obtained at least 12 independent clones, all of which had the similar restriction maps. Northern blot analysis indicated that ponsin had several mRNA bands in heart, brain, liver, and skeletal muscle. When the aa sequences of ponsin-1 and -2 were aligned together with that of SH3P12, they were highly homologous but their differences were clustered and not distributed throughout the sequences. Moreover, the FISH analysis indicated that ponsin-1 and -2 were located at the same locus (data not shown). We have determined neither the genetic locus of SH3P12 nor the clones other than ponsin-1 and -2, but ponsin/SH3P12 had many splicing variants, and all of these variants might be derived from the same gene. The exact relationship of the 35S-l-afadin–binding proteins (p95 and p70) and ponsin-1 and -2 has not been clarified here, but there are two possibilities: (a) p95 and p70 are splicing variants of ponsin other than ponsin-1 and -2, and (b) they are posttranslationally modified forms of ponsin-1 or -2, such as the phosphorylated or proteolytic form. The physiological significance of the presence of many splicing variants of ponsin is unknown at present.
Figure 13.
Schematic diagram of ZO, ZA, and cell–matrix AJ.
We presented the evidence that l-afadin bound to ponsin by four different methods: (a) blot overlay, (b) affinity chromatography, (c) immunoprecipitation, and (d) immunofluorescence and immunoelectron microscopy. Blot overlay analysis indicated that ponsin-2 appeared to bind l-afadin more than ponsin-1, but their exact binding affinities remain to be clarified. We moreover showed that the binding regions of l-afadin and ponsin were the third proline-rich region and the region containing the second and third SH3 domains, respectively, consistent with the earlier observation that proline-rich sequences interact with either profilin or SH3 domains (Tanaka and Shibata, 1985; Yu et al., 1994). Affinity chromatography analysis on each SH3 domain indicated that the third SH3 domain bound to l-afadin, whereas neither the first nor second SH3 domain bound, suggesting that the third SH3 domain is the minimal l-afadin–binding region. However, the l-afadin–binding activity of the third SH3 domain alone was less than that of the region containing the second and third SH3 domains. It remains to be clarified why the l-afadin–binding activity of ponsin is reduced when the second and third SH3 domains are separated from each other, but the highly ordered structure of the region containing the second and third SH3 domains may be necessary for the more efficient binding of ponsin to l-afadin. In contrast to l-afadin, s-afadin, which has the first and second proline-rich regions but lacks the third proline-rich region, hardly bound to ponsin. Neurabin-II, which also has three proline-rich regions (Satoh et al., 1998), did not bind to ponsin either (data not shown). These results suggest that the SH3 domains of ponsin recognize a specific proline-rich region.
Computer analysis indicated that ponsin-1 and -2 had three SH3 domains and two sorbin-like regions, but no transmembrane region or other known signaling domains. However, ponsin was localized at ZA in epithelial cells, at cell–cell AJ in nonepithelial cells, and at cell–matrix AJ in both types of cells (Fig. 13). These results suggest that there is another protein that determines the specific localization of l-afadin at these specific sites of cells. Along this line, we found here that ponsin-1 and -2 bound vinculin, which interacts with α-catenin at cell–cell AJ (Watabe-Uchida et al., 1998; Weiss et al., 1998) and with talin at cell–matrix AJ (Burridge and Mangeat, 1984). α-Catenin interacts with cadherin through β-catenin (Aberle et al., 1994), whereas talin directly interacts with integrin (Horwitz et al., 1986). We presented here the evidence that ponsin directly bound vinculin by four different methods: (a) blot overlay, (b) affinity chromatography, (c) immunoprecipitation, and (d) immunofluorescence and immunoelectron microscopy. Moreover, we showed that the binding regions of ponsin and vinculin were the region containing the first and second SH3 domains and the proline-rich region, respectively. The proline-rich region of vinculin is divided into at least two proline-rich sequences, the first of which binds to vasodilator-stimulated phosphoprotein (VASP) (Brindle et al., 1996; Niebuhr et al., 1997). We have not determined which proline-rich sequence is responsible for the binding to ponsin. Blot overlay and affinity chromatography analyses on each SH3 domain indicated that the second SH3 domain bound to vinculin, whereas neither the first nor third SH3 domain bound, suggesting that the second SH3 domain is the minimal vinculin-binding region. However, the vinculin-binding activities of the second SH3 domain alone and the region containing the second and third SH3 domains were less than that of the region containing the first and second SH3 domains. It remains to be clarified why the vinculin-binding activity of ponsin is reduced when the first and second SH3 domains are separated from each other. The highly ordered structure of the region containing the first and second SH3 domains may be necessary for the more efficient binding of ponsin to vinculin.
The result (the l-afadin– and vinculin-binding regions of ponsin were partly overlapped) suggested that l-afadin and vinculin bind to ponsin in a competitive manner and that these three proteins do not form a ternary complex. Consistently, we found that l-afadin and vinculin bound to ponsin in a competitive manner and that the affinities of l-afadin and vinculin to ponsin were apparently similar. We found here by immunoprecipitation and affinity chromatography analyses that ponsin formed a binary complex with either l-afadin or vinculin, but hardly formed a ternary complex with l-afadin and vinculin. This result suggests that ponsin does not serve as a direct linker between l-afadin and vinculin. The physiological significance of the result that ponsin forms a binary complex with either l-afadin or vinculin at the same time but not a ternary complex with the two F-actin–binding proteins is currently unknown, but ponsin may independently regulate the function of each protein, or may regulate the linkage between l-afadin and vinculin in cooperation with another still unidentified factor. If the latter is the case, l-afadin may be localized at ZA in epithelial cells and at cell–cell AJ in nonepithelial cells by interacting with the cadherin-catenin system through the ponsin-vinculin system (Fig. 13).
ZO and ZA in the junctional complex of polarized epithelial cells are closely aligned from the apical side to the basal side, suggesting that these two junction structures have their molecular interactions. ZO is also linked to the actin cytoskeleton. At ZO, multiple integral membrane proteins with four transmembrane regions, the claudin family members and occludin, constitute tight junction strands (Furuse et al., 1993, 1998; Ando-Akatsuka et al., 1996). The cytoplasmic region of occludin binds ZO-1 (Furuse et al., 1994), which interacts with F-actin (Itoh et al., 1997). The cytoplasmic regions of the claudin family members may also directly or indirectly bind ZO-1 (Furuse et al., 1998). Evidence is accumulating that the cadherin-catenin system plays essential roles for the assembly of the junctional complex (Gumbiner and Simons, 1986; Gumbiner et al., 1988; Watabe et al., 1994). It has recently been shown by use of an α-catenin–deficient colon carcinoma cell line that the binding of vinculin to α-catenin is required for the organization of ZO (Watabe-Uchida et al., 1998). It has furthermore been shown that the junctional organization is impaired in vinculin-null F9 cells (Watabe-Uchida et al., 1998). It should be noted from the present and previous (Mandai et al., 1997) results that l-afadin and ponsin as well as vinculin are more highly concentrated at ZA than cadherin. The afadin-ponsin system may be involved in the assembly of the junctional complex by interacting with vinculin.
We recently found that l-afadin and E-cadherin showed different behavior during the formation and destruction of cell–cell AJ in MDCK and L cells (Sakisaka et al., 1999). Dissociation of MDCK cells by culturing the cells at 2 μM Ca2+ caused rapid endocytosis of E-cadherin, but not that of l-afadin or ZO-1. Addition of phorbol 12-myristate 13-acetate to these dissociated cells formed a ZO-like structure where ZO-1 and l-afadin, but not E-cadherin, accumulated. Even in cadherin-deficient L cells, l-afadin was mainly localized at cell–cell contacts, whereas ZO-1 was mainly localized at the tip area of cell processes. l-Afadin did not directly bind to α-, β-catenin, E-cadherin, ZO-1, or occludin. All of the results thus far available suggest that there is an integral membrane protein that is specifically localized at ZA in epithelial cells and at cell–cell AJ in nonepithelial cells and interacts with the afadin–ponsin– vinculin system (Fig. 13). It would be of crucial importance to identify this protein for our understanding of the physiological significance of l-afadin and ponsin.
Ponsin-1 and -2 were splicing variants of SH3P12. SH3P12 was suddenly renamed CAP in the GenBank database during the revision of this manuscript. CAP was originally isolated as a c-Cbl–binding protein (Ribon et al., 1998a). c-Cbl is a proto-oncogene product involved in T cell antigen receptor–mediated signaling (Thien and Langdon, 1998). In this CAP paper, no nucleotide or amino acid sequence information was available. Homology searches of DNA and protein databases furthermore revealed another protein structurally related to ponsin/ SH3P12, ArgBP2. ArgBP2 was isolated as an Arg- and Abl-binding protein (Wang et al., 1997). Arg and Abl represent the members of the Aberson family of tyrosine kinase. During the revision of this manuscript, Kioka et al. (1999) isolated a protein structurally related to ponsin/ SH3P12, named vinexin, as a vinculin-binding protein. All of these proteins have three SH3 domains and one or two sorbin-like regions. These results suggest that ponsin/ SH3P12/CAP, ArgBP2, and vinexin constitute a family. Structural comparison of these proteins suggest that ponsin/SH3P12/CAP, ArgBP2, and vinexin are derived from three different genes and constitute three subfamilies. Vinexin as well as ponsin/SH3P12 is localized at both cell– cell and cell–matrix AJs (Kioka et al., 1999), whereas both SH3P12/CAP and ArgBP2 are associated with actin stress fibers (Wang et al., 1997; Ribon et al., 1998b). Vinexin enhances the actin cytoskeleton reorganization and cell spreading (Kioka et al., 1999). SH3P12/CAP enhances actin stress fiber formation and focal adhesions and is associated with signaling molecules such as the insulin receptor, focal adhesion kinase (FAK), and SOS, a Ras small G protein exchanger (Ribon et al., 1998a,b). Thus, the members of the family that has three SH3 domains and one or two sorbin-like regions show similar and different subcellular localization and association with other proteins, suggesting their related but diverse functions.
Acknowledgments
We thank Drs. Sh. Tsukita and M. Itoh (Kyoto University, Kyoto, Japan) for helpful discussions and for providing us with MTD-1A cells and 3Y1 cells. We also thank Drs. Y. Imamura, A. Nagafuchi, and Sh. Tsukita (Kyoto University) for providing us with the vinculin cDNA, and Dr. Y. Matsuda (Nagoya University, Nagoya, Japan) for the FISH analysis of ponsin-1 and -2.
Abbreviations used in this paper
- aa
amino acid
- AJ
adherens junction
- F-actin
actin filament
- FISH
fluorescence in situ hybridization
- GST
glutathione S-transferase
- His6
six histidine residues
- MBP
maltose-binding protein
- MDCK cells
Madin-Darby canine kidney cells
- SH3
Src homology 3
- ZA
zonula adherens
- ZO
zonula occludens
References
- Aberle H, Butz S, Stappert J, Weissig H, Kemler R, Hoschuetzky H. Assembly of the cadherin–catenin complex in vitro with recombinant proteins. J Cell Sci. 1994;107:3655–3663. doi: 10.1242/jcs.107.12.3655. [DOI] [PubMed] [Google Scholar]
- Ando-Akatsuka Y, Saitou M, Hirase T, Kishi M, Sakakibara A, Itoh M, Yonemura S, Furuse M, Tsukita S. Interspecies diversity of the occludin sequence: cDNA cloning of human, mouse, dog, and rat-kangaroo homologues. J Cell Biol. 1996;133:43–47. doi: 10.1083/jcb.133.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brindle NP, Holt MR, Davies JE, Price CJ, Critchley DR. The focal-adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem J. 1996;318:753–757. doi: 10.1042/bj3180753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K, Mangeat P. An interaction between vinculin and talin. Nature. 1984;308:744–746. doi: 10.1038/308744a0. [DOI] [PubMed] [Google Scholar]
- Chevesich J, Kreuz AJ, Montell C. Requirement for the PDZ domain protein, INAD, for localization of the TRP store-operated channel to a signaling complex. Neuron. 1997;18:95–105. doi: 10.1016/s0896-6273(01)80049-0. [DOI] [PubMed] [Google Scholar]
- Cimino G, Moir DT, Canaani O, Williams K, Crist WM, Katzav S, Cannizzaro L, Lange B, Nowell PC, Croce CM, Canaani E. Cloning of ALL-1, the locus involved in leukemias with the t(4;11)(q21;q23), t(9; 11)(p22;q23), and t(11;19)(q23;p13) chromosome translocations. Cancer Res. 1991;51:6712–6714. [PubMed] [Google Scholar]
- Cohen GB, Ren R, Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol. 1993;123:1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Itoh M, Hirase T, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J Cell Biol. 1994;127:1617–1626. doi: 10.1083/jcb.127.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol. 1998;141:1539–1550. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B, Dutton AH, Kokuyasu KT, Singer SJ. Immunoelectron microscopic studies of membrane-microfilament interactions: distribution of α-actinin, tropomyosin, and vinculin in intestinal epithelial brush border and chicken gizzard smooth muscle cells. J Cell Biol. 1981;91:614–628. doi: 10.1083/jcb.91.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B, Ginsberg D. The cytoplasmic domain of adherens-type junctions. Cell Motil Cytoskelet. 1991;20:1–6. doi: 10.1002/cm.970200102. [DOI] [PubMed] [Google Scholar]
- Gumbiner B, Simons K. A functional assay for proteins involved in establishing an epithelial occluding barrier: identification of a uvomorulin-like polypeptide. J Cell Biol. 1986;102:457–468. doi: 10.1083/jcb.102.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner BM, Stevenson B, Grimaldi A. The role of the cell adhesion molecule uvomorulin in the formation and maintenance of the epithelial junctional complex. J Cell Biol. 1988;107:1575–1587. doi: 10.1083/jcb.107.4.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- Hata Y, Südhof TC. A novel ubiquitous form of Munc-18 interacts with multiple syntaxins. Use of the yeast two-hybrid system to study interactions between proteins involved in membrane traffic. J Biol Chem. 1995;270:13022–13028. doi: 10.1074/jbc.270.22.13022. [DOI] [PubMed] [Google Scholar]
- Horwitz A, Duggan K, Buck C, Beckerle MC, Burridge K. Interaction of plasma membrane fibronectin receptor with talin-a transmembrane linkage. Nature. 1986;320:531–533. doi: 10.1038/320531a0. [DOI] [PubMed] [Google Scholar]
- Imazumi K, Sasaki T, Takahashi K, Takai Y. Identification of a rabphilin-3A-interacting protein as GTP cyclohydrolase I in PC12 cells. Biochem Biophys Res Commun. 1994;205:1409–1416. doi: 10.1006/bbrc.1994.2822. [DOI] [PubMed] [Google Scholar]
- Itoh M, Yonemura S, Nagafuchi A, Tsukita S, Tsukita S. A 220-kD undercoat-constitutive protein: its specific localization at cadherin-based cell–cell adhesion sites. J Cell Biol. 1991;115:1449–1462. doi: 10.1083/jcb.115.5.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Nagafuchi A, Moroi S, Tsukita S. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to a catenin and actin filament. J Cell Biol. 1997;138:181–192. doi: 10.1083/jcb.138.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch BM, Bubeck P, Giehl K, Kroemker M, Moschner J, Rothkegel M, Rüdiger M, Schluter K, Stanke G, Winkler J. The molecular architecture of focal adhesions. Annu Rev Cell Dev Biol. 1995;11:379–416. doi: 10.1146/annurev.cb.11.110195.002115. [DOI] [PubMed] [Google Scholar]
- Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995;373:261–264. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- Kioka N, Sakata S, Kawauchi T, Amachi T, Akiyama SK, Okazaki K, Yaen C, Yamada KM, Aota S. Vinexin: a novel vinculin-binding protein with multiple SH3 domains enhances actin cytoskeletal organization. J Cell Biol. 1999;144:59–69. doi: 10.1083/jcb.144.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen KA, Soler AP, Johnson KR, Wheelock MJ. Interaction of α-actinin with the cadherin/catenin cell–cell adhesion complex via α-catenin. J Cell Biol. 1995;130:67–77. doi: 10.1083/jcb.130.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Luna EJ, Hitt AL. Cytoskeleton-plasma membrane interactions. Science. 1992;258:955–964. doi: 10.1126/science.1439807. [DOI] [PubMed] [Google Scholar]
- Mandai K, Nakanishi H, Satoh A, Obaishi H, Wada M, Nishioka H, Itoh M, Mizoguchi A, Aoki T, Fujimoto T, et al. Afadin: a novel actin filament-binding protein with one PDZ domain localized at cadherin-based cell-to-cell adherens junction. J Cell Biol. 1997;139:517–528. doi: 10.1083/jcb.139.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Harada YN, Natsuume-Sakai S, Lee K, Shiomi T, Chapman VM. Location of the mouse complement factor H gene(cfh) by FISH analysis and replication R-banding. Cytogenet Cell Genet. 1992;61:282–285. doi: 10.1159/000133423. [DOI] [PubMed] [Google Scholar]
- Matsuda Y, Chapman VM. Application of fluorescence in situ hybridization in genome analysis of the mouse. Electrophoresis. 1995;16:261–272. doi: 10.1002/elps.1150160142. [DOI] [PubMed] [Google Scholar]
- Mizoguchi A, Yano Y, Hamaguchi H, Yanagida H, Ide C, Zahraoui A, Shirataki H, Sasaki T, Takai Y. Localization of Rabphilin-3A on the synaptic vesicle. Biochem Biophys Res Commun. 1994;202:1235–1243. doi: 10.1006/bbrc.1994.2063. [DOI] [PubMed] [Google Scholar]
- Nagafuchi A, Takeichi M, Tsukita S. The 102 kd cadherin-associated protein: similarity to vinculin and posttranscriptional regulation of expression. Cell. 1991;65:849–857. doi: 10.1016/0092-8674(91)90392-c. [DOI] [PubMed] [Google Scholar]
- Niebuhr K, Ebel F, Frank R, Reinhard M, Domann E, Carl UD, Walter U, Gertler FB, Wehland J, Chakraborty T. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO (Eur Mol Biol Organ) J. 1997;16:5433–5444. doi: 10.1093/emboj/16.17.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Halloran T, Morony L, Burridge K. Purification and assay of vinculin, metavinculin, and talin. Methods Enzymol. 1986;134:69–77. doi: 10.1016/0076-6879(86)34076-x. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Nakanishi H, Ikeda W, Satoh A, Momose Y, Nishioka H, Takai Y. Nexilin: a novel actin filament-binding protein localized at cell-matrix adherens junction. J Cell Biol. 1998;143:1227–1238. doi: 10.1083/jcb.143.5.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otey CA, Pavalko FM, Burridge K. An interaction between α-actinin and the β1 integrin subunit in vitro. J Cell Biol. 1990;111:721–729. doi: 10.1083/jcb.111.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO (Eur Mol Biol Organ) J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansu D, Vagne M, Bosshard A, Mutt V. Sorbin, a peptide contained in porcine upper small intestine which induces the absorption of water and sodium in the rat duodenum. Scand J Gastroenterol. 1981;16:193–199. doi: 10.3109/00365528109181955. [DOI] [PubMed] [Google Scholar]
- Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- Prasad R, Gu Y, Alder H, Nakamura T, Canaani O, Saito H, Huebner K, Gale RP, Nowell PC, Kuriyama K, et al. Cloning of the ALL-1 fusion partner, the AF-6gene, involved in acute myeloid leukemias with the t(6;11) chromosome translocation. Cancer Res. 1993;53:5624–5628. [PubMed] [Google Scholar]
- Ribon V, Printen JA, Hoffman NG, Kay BK, Saltiel AR. A novel, multifunctional c-Cbl binding protein in insulin receptor signaling in 3T3-L1 adipocytes. Mol Cell Biol. 1998a;18:872–879. doi: 10.1128/mcb.18.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribon V, Herrera R, Kay BK, Saltiel AR. A role for CAP, a novel, multifunctional Src homology 3 domain-containing protein in formation of actin stress fibers and focal adhesions. J Biol Chem. 1998b;273:4073–4080. doi: 10.1074/jbc.273.7.4073. [DOI] [PubMed] [Google Scholar]
- Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. a1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci USA. 1995;92:8813–8817. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakisaka, T., H. Nakanishi, K. Takahashi, K. Mandai, M. Miyahara, A. Satoh, K. Takaishi, and Y. Takai. 1999. Different behavior of l-afadin and neurabin-II during the formation and destruction of cell–cell adherens junction. Oncogene. In press. [DOI] [PubMed]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. 2nd edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 1.1–1.110.
- Satoh A, Nakanishi H, Obaishi H, Wada M, Takahashi K, Satoh K, Hirao K, Nishioka H, Hata Y, Mizoguchi A, Takai Y. Neurabin-II/ spinophilin. An actin filament-binding protein with one PDZ domain localized at cadherin-based cell–cell adhesion sites. J Biol Chem. 1998;273:3470–3475. doi: 10.1074/jbc.273.6.3470. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh BH, Zhu MY. Regulation of the TRP Ca2+ channel by INAD in Drosophilaphotoreceptors. Neuron. 1996;16:991–998. doi: 10.1016/s0896-6273(00)80122-1. [DOI] [PubMed] [Google Scholar]
- Sparks AB, Hoffman NG, McConnell SJ, Fowlkes DM, Kay BK. Cloning of ligand targets: systematic isolation of SH3 domain-containing proteins. Nat Biotechnol. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- Takaishi K, Sasaki T, Kameyama T, Tsukita S, Tsukita S, Takai Y. Translocation of activated Rho from the cytoplasm to membrane ruffling area, cell–cell adhesion sites and cleavage furrows. Oncogene. 1995;11:39–48. [PubMed] [Google Scholar]
- Takeichi M. The cadherins: cell–cell adhesion molecules controlling animal morphogenesis. Development (Camb) 1988;102:639–655. doi: 10.1242/dev.102.4.639. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Shibata H. Poly(l-proline)-binding proteins from chick embryos are a profilin and a profilactin. Eur J Biochem. 1985;151:291–297. doi: 10.1111/j.1432-1033.1985.tb09099.x. [DOI] [PubMed] [Google Scholar]
- Thien CB, Langdon WY. c-Cbl: a regulator of T cell receptor-mediated signalling. Immunol Cell Biol. 1998;76:473–482. doi: 10.1046/j.1440-1711.1998.00768.x. [DOI] [PubMed] [Google Scholar]
- Tsukita S, Tsukita S. Isolation of cell-to-cell adherens junctions from rat liver. J Cell Biol. 1989;108:31–41. doi: 10.1083/jcb.108.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S, Tsukita S, Nagafuchi A, Yonemura S. Molecular linkage between cadherins and actin filaments in cell–cell adherens junctions. Curr Opin Cell Biol. 1992;4:834–839. doi: 10.1016/0955-0674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- Wang B, Golemis EA, Kruh GD. ArgBP2, a multiple Src homology 3 domain-containing, Arg/Abl-interacting protein, is phosphorylated in v-Abl-transformed cells and localized in stress fibers and cardiocyte Z-disks. J Biol Chem. 1997;272:17542–17550. doi: 10.1074/jbc.272.28.17542. [DOI] [PubMed] [Google Scholar]
- Watabe M, Nagafuchi A, Tsukita S, Takeichi M. Induction of polarized cell–cell association and retardation of growth by activation of the E-cadherin-catenin adhesion system in a dispersed carcinoma line. J Cell Biol. 1994;127:247–256. doi: 10.1083/jcb.127.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M, Uchida N, Imamura Y, Nagafuchi A, Fujimoto K, Uemura T, Vermeulen S, van Roy F, Adamson ED, Takeichi M. α-Catenin–vinculin interaction functions to organize the apical junctional complex in epithelial cells. J Cell Biol. 1998;142:847–857. doi: 10.1083/jcb.142.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EE, Kroemker M, Rüdiger A-H, Jockusch BM, Rüdiger M. Vinculin is part of the cadherin-catenin junctional complex: complex formation between α-catenin and vinculin. J Cell Biol. 1998;141:755–764. doi: 10.1083/jcb.141.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Harada N, Kano K, Taya S, Canaani E, Matsuura Y, Mizoguchi A, Ide C, Kaibuchi K. The Ras target AF-6 interacts with ZO-1 and serves as a peripheral component of tight junctions in epithelial cells. J Cell Biol. 1997;139:785–795. doi: 10.1083/jcb.139.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S, Itoh M, Nagafuchi A, Tsukita S. Cell-to-cell adherens junction formation and actin filament organization: similarities and differences between non-polarized fibroblasts and polarized epithelial cells. J Cell Sci. 1995;108:127–142. doi: 10.1242/jcs.108.1.127. [DOI] [PubMed] [Google Scholar]
- Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]