Abstract
Alignment of the mitotic spindle with the axis of cell division is an essential process in Saccharomyces cerevisiae that is mediated by interactions between cytoplasmic microtubules and the cell cortex. We found that a cortical protein, the yeast formin Bni1p, was required for spindle orientation. Two striking abnormalities were observed in bni1Δ cells. First, the initial movement of the spindle pole body (SPB) toward the emerging bud was defective. This phenotype is similar to that previously observed in cells lacking the kinesin Kip3p and, in fact, BNI1 and KIP3 were found to be in the same genetic pathway. Second, abnormal pulling interactions between microtubules and the cortex appeared to cause preanaphase spindles in bni1Δ cells to transit back and forth between the mother and the bud. We therefore propose that Bni1p may localize or alter the function of cortical microtubule-binding sites in the bud. Additionally, we present evidence that other bipolar bud site determinants together with cortical actin are also required for spindle orientation.
Keywords: mitotic spindle apparatus, cell division, microtubule, actin, Saccharomyces cerevisiae
Cell division requires the correct positioning of the spindle within the cell in addition to assembly of the mitotic apparatus and segregation of the chromosomes (Hyman and Karsenti, 1996; Stearns, 1997). Significant progress has been made in characterizing the proteins involved in spindle construction and chromosome movement, but little is known about the proteins that mediate interactions between microtubules and the cell cortex to establish spindle position.
Various strategies to align the spindle with the plane of cell division are used in different organisms (Rhyu and Knoblich, 1995). In budding yeast, the axis of division is determined by the signaling molecules that control bud site selection (Freifelder, 1960; Chant and Pringle, 1995). The polarity of the dividing yeast cell is established first and then the spindle is aligned in response to this polarity to segregate the nuclei. This alignment is postulated to be mediated by interactions between cytoplasmic microtubules and asymmetrically localized cortical proteins (Snyder et al., 1991; Hyman and Stearns, 1992). Spindle positioning in plant cells is similar to yeast in that the spindle appears to be aligned by preestablished cortical cues (Staiger and Lloyd, 1991). By contrast, in animal cells spindle position determines the position of cytokinesis (Cao and Wang, 1996). This is important for development because correct spindle position is essential for generating asymmetric cell divisions. Furthermore, there are several examples in animal cells where spindle position appears to be developmentally regulated (Hyman and White, 1987; Hyman, 1989; Chenn and McConnell, 1995; Rhyu and Knoblich, 1995; Kraut et al., 1996). Despite the cell type differences in the relationship between spindle position, cell polarity, and cytokinesis, many of the molecular components involved in these processes are conserved, suggesting that some of the mechanisms linking microtubules to cortical polarity factors may also be conserved (Drubin and Nelson, 1996).
The establishment of spindle position in Saccharomyces cerevisiae occurs in distinct morphological steps (Yeh et al., 1995; DeZwaan et al., 1997; Carminati and Stearns, 1997; Shaw et al., 1997, 1998). First, the centrosome or spindle pole body (SPB)1 migrates toward the incipient bud site. Around the time of SPB separation and the formation of a bipolar spindle, cytoplasmic microtubules penetrate into the bud, and the spindle assumes a relatively stable position at the neck aligned with the mother–bud axis (Shaw et al., 1997). At anaphase, the spindle is inserted across the bud neck and then elongates, segregating the chromosomes between mother and daughter cells (Yeh et al., 1995). Genetic analysis has identified two partially redundant pathways for spindle orientation. One mediated by the kinesin Kip3p, is necessary for the initial movement of the SPB toward the bud. The other is mediated by cytoplasmic dynein which is necessary for the insertion of the spindle through the bud neck (Cottingham and Hoyt, 1997; DeZwaan et al., 1997; Stearns, 1997). It is not known if these motors act primarily by affecting the dynamics of cytoplasmic microtubules or whether they directly link cytoplasmic microtubules to the cell cortex.
One of the most interesting questions about the mechanism of spindle orientation is the identity and molecular function of the cortical proteins that interact with cytoplasmic microtubules. The existence of microtubule “capture sites,” loosely analogous to those at the kinetochore, has been postulated, but little is known about the molecular composition of these cortical regions (Kaverina et al., 1998; Snyder et al., 1991). One factor that is clearly important for controlling spindle position in diverse organisms is the actin cytoskeleton. In S. cerevisiae, mutations in ACT1, the gene that encodes actin, result in defects in spindle position and nuclear segregation (Palmer et al., 1992; Drubin et al., 1993). In Caenorhabditis elegans, actin is required for the developmentally regulated rotation of the spindle during specific early embryonic cell divisions (Hyman and White, 1987; Hyman, 1989). Actin is also required for centrosome movement in human leukocytes (Euteneuer and Schliwa, 1985). The molecular role of actin in these different systems is not known. Cortical actin and associated proteins might bind microtubules or alternatively, they might be required to localize cortical microtubule-binding proteins. Recent genetic analysis in yeast has identified two novel cortical proteins, Num1p and Kar9p, that are required for spindle orientation. Num1p localizes to the mother cell cortex (Farkasovsky and Küntzel, 1995), whereas Kar9p localizes to a discrete region of the bud cortex and to the tip of the mating projection (shmoo). Although it has not been determined if these proteins bind microtubules, it is interesting that overexpressed Kar9p concentrates at sites of contact between cytoplasmic microtubules and the bud cortex (Miller and Rose, 1998).
Here we report a requirement for the conserved protein Bni1p in spindle orientation. Bni1p belongs to a family of proteins called formins, which are required for diverse cellular and developmental functions (Woychik et al., 1990; Castrillon and Wasserman, 1994; Emmons et al., 1995; Petersen et al., 1995; Zahner et al., 1996; Chang et al., 1997; Evangelista et al., 1997; Harris et al., 1997). Formins are characterized by conserved regions called formin homology (FH) domains. There is substantial evidence to suggest that Bni1p and other formins function as molecular scaffolds that link Rho-type GTPases with components of the actin cytoskeleton. The NH2-terminal domain of Bni1p interacts with the activated forms of G proteins, Cdc42p and Rho1p, which regulate organization of the actin cytoskeleton (Kohno et al., 1996; Evangelista et al., 1997). The proline-rich FH1 domain of Bni1p binds profilin and this interaction may regulate actin polymerization at cortical sites (Evangelista et al., 1997; Imamura et al., 1997). The COOH-terminal domain of Bni1p interacts with another putative actin-binding protein, Aip3p/Bud6p (Amberg et al., 1997; Evangelista et al., 1997). In S. cerevisiae, a second formin, Bnr1p, has at least partial functional overlap with Bni1p (Imamura et al., 1997). Whether formins link polarity signals to other cytoskeletal systems, such as the microtubule cytoskeleton, has not been established.
Bni1p has been implicated in actin function during polarized morphogenesis and the establishment of cell polarity. Bni1p is localized in a cap-like structure (Jansen et al., 1996; Evangelista et al., 1997; Fujiwara et al., 1998), similar to that observed for Cdc42p and other polarity determinants (Ziman et al., 1993) at sites of polarized cell growth: the bud tip early in the cell cycle, the septal region late in the cell cycle, and to the shmoo tip. bni1Δ mutant cells contain normal cortical actin patches and cables, but are defective for the reorganization of these structures required for various aspects of cell polarization (Evangelista et al., 1997). bni1Δ cells are normal for bud emergence, but they are defective for polarized bud growth and appear to construct buds through isotropic growth, generating cells with a rounded shape. bni1Δ cells are also defective in shmoo formation, pseudohyphal growth, localization of ASH1 mRNA to the bud, cytokinesis, and bipolar bud site selection (Jansen et al., 1996; Zahner et al., 1996; Evangelista et al., 1997; Mösch and Fink, 1997). The role of Bni1p in bipolar bud site selection is notable because Bni1p interacts with two other proteins, Spa2p and Bud6p, required for this process (Evangelista et al., 1997; Fujiwara et al., 1998). The molecular links between Bni1p and actin may also be relevant to bipolar bud site selection because many actin mutants also affect this process (Yang et al., 1997).
We have now found that Bni1p is required for specific steps in establishing the position of the mitotic spindle before anaphase. By fluorescence time-lapse microscopy, we observed that Bni1p was required for the initial movement of the SPB toward the incipient bud site. In addition, a strikingly abnormal movement of preanaphase spindles was observed in bni1Δ cells: short spindles were pulled back and forth between the mother cell and the bud. In wild-type cells, apparent pulling forces on the spindle were observed toward the bud, but not toward the mother cell. These findings suggest that in the absence of Bni1p, microtubule-binding sites at the cell cortex are either mislocalized or functionally altered, resulting in the loss of polarization of preanaphase spindle movement. Finally, analysis of mutations affecting the actin cytoskeleton suggest that actin structures involved in bipolar budding are also required for spindle orientation.
Materials and Methods
Microbial Techniques
Media and genetic techniques were as described (Rose et al., 1990). The mating pheromone, α-factor, was added to log phase cultures at 6 μg/ml. Latrunculin-A (Lat-A) (from P. Crews, University of California, Santa Cruz, CA) was added to cultures at 400 mM. Benomyl sensitivity (a gift from E.I. du Pont de Nemours, Wilmington, DE) was tested in rich (YPD) medium. Geneticin (GIBCO BRL) was used at a concentration of 0.2 μg/ ml in YPD. Lists of the strains and plasmids used in this study are provided in Tables I and II. Except where indicated, all strains are congenic to W3031a.
Table I.
Strains Used
| Strains | Genotype | Source | ||
|---|---|---|---|---|
| PY1746 | MAT a ase1::URA3 ade2-100 ade3 leu2-3,112 lys2-801 trp1-1 ura3-1 | D. Pellman | ||
| PY1994 | MAT a ase1::kanR ade2-100 ade3 leu2-3,112 lys2-801 trp1-1 ura3-52 {2μ ASE1 ADE3 TRP1} | This study | ||
| PY1996 | MATα ase1::kanR ade2-100 ade3 leu2-3,112 lys2-801 trp1-1 ura3-52 {2μ ASE1 ADE3 TRP1} | This study | ||
| PY2400 | MAT a kip3::HIS3 ade2-100 leu2-3,112 his3-11,15 trp1-1 ura3-1 | This study | ||
| PY2401 | MAT a kip3::HIS3 bni1::kanR ura3-1 ade2-100 his3-11,15 trp1-1 leu2-3,112 | This study | ||
| PY2403 | MAT a bni1::kanR ade2-100 leu2-3,112 his3-11,15 ura3-1 trp1-1 ade3 | This study | ||
| PY2409 | MAT a kip2::URA3 ura3-1 leu2-3,112 his3-11,15 ade2-100 trp1-1 | This study | ||
| PY2513 | MAT a bni1::kanR ade2-100 ade3 leu2-3,112 his3-11,15 trp1-1 ura3-1 | This study | ||
| PY2515 | MAT a ase1::URA3 ade2-100 leu2-3,112 lys2 his3-11,15 ura3-1 trp1-1 | This study | ||
| PY2516 | MAT a dyn1::URA3 ade3 leu2-3,112 ura3-1 trp1-1 | This study | ||
| PY2581 | MAT a ase1::URA3 bni1::kanR ade2-100 leu2-3,112 lys2 his3-11,15 ura3-1 trp1-1 | This study | ||
| PY2582 | MATα dyn1::URA3 ade2-100 leu2-3,112 his3-11,15 trp1-1 ura3-1 | This study | ||
| PY2583 | MATα dyn1::URA3 bni1::kanR leu2-3,112 his3-11,15 trp1-1 ura3-1 | This study | ||
| PY2595 | MAT a ade2-101 leu2-3,112 his3Δ200 ura3-52 can1 cry1 NUF2::GFP::URA3 | This study | ||
| PY2596 | MAT a ade2-101 leu2-3,112 his3-Δ200 ura3-52 can1 cry1 NUF2::GFP::URA3 act1-116::HIS3 | This study | ||
| PY2608 | MAT a bud6::TRP1 leu2Δ1 lys2-801 his3Δ200 trp1Δ63 ura3-52 NUF2::GFP::URA3 | This study | ||
| PY2611 | MAT a leu2Δ1 lys2-801 his3Δ200 trp1Δ63 ura3-52 NUF2::GFP::URA3 | This study | ||
| AFS306 | MAT a kip1::HIS3 trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 | A. Murray (University of California, San Francisco, CA) | ||
| AFS462 | MAT a kar3::TRP1 ade2-1 ura3-1 his3-11,15 leu2-3,112 trp1-1 | A. Murray | ||
| DBY3357 | MAT a ade2-101 leu2-3,112 his3Δ200 ura3-52 can1 cry1 | D. Botstein (Stanford University, Stanford, CA) | ||
| KAY0227 | MAT a sla1::URA3 his3Δ200 ura3-52 {HIS3 CEN SLA1} | D. Drubin (University of California, Berkeley, CA) | ||
| KAY0228 | MAT a sla1::URA3 his3Δ200 ura3-52 {HIS3 CEN sla1ΔSH3#3} | D. Drubin | ||
| MAY2058 | MAT a cin8::LEU2 ade2-101 leu2-3,112 lys2-801 his3Δ200 ura3-52 | M.A. Hoyt (The Johns Hopkins University, Baltimore, MD) | ||
| PY2061 | MATα smy1::LEU2 leu2 ura3 his4 trp1 | S. Brown (University of Michigan Medical School, Ann Arbor, MI) | ||
| Y66 | MATα far1-c ade2-100 leu2-3,112 lys1 his3-11,15 trp1-1 ura3-1 | C. Boone | ||
| Y1353 | MAT a/MATα bni1::HIS3/bni1::HIS3 leu2Δ1/leu2Δ1 his3Δ200/his3Δ200 trp1Δ63/trp1Δ63lys2-801/lys2-801 | C. Boone | ||
| Y1445 | MAT a bni1::kanR bar1::LEU2 ade2-100 ade3 leu2-3,112 his3-11,15 trp1-1 ura3-1 | C. Boone |
Table II.
Plasmids Used
| Plasmid | Markers | Source | ||
|---|---|---|---|---|
| PB1046 | bni1-CTΔ1 LEU2 | This study | ||
| pFA-KanMX2 | kanr | P. Philippsen (University of Basel, Basel, Switzerland) | ||
| pJIL67 | NUF2::GFP URA3 | P. Silver (Dana-Farber Cancer Institute/ Harvard Medical School, Boston, MA) | ||
| pAFS125 | GFP::TUB1 URA3 | A. Murray | ||
| p182 | BNI1 URA3 CEN ARS | C. Boone | ||
| p491 | bni1-CTΔ1 URA3 CEN ARS | C. Boone | ||
| p1832 | BNI1 TRP1 CEN ARS | C. Boone | ||
| p2029 | bni1-CTΔ1 TRP1 CEN ARS | C. Boone | ||
| pRB1537 | act1-116::HIS3 | D. Botstein |
Strain and Plasmid Construction
A complete deletion of the BNI1 coding sequence was created by one-step gene replacement with the selectable marker kanr, which confers Geneticin resistance to yeast (Wach et al., 1994). The bni1Δ::kanr containing DNA sequence was generated by PCR using oligonucleotide primers: 5′ TATCTATCTTCTGTATTGAGGAGAAACATTTTACTCAAGC - AGCTGAAGCTTCGTACGC and 5′GAGTAAAGATGAATGTAA AGTGTATCATAAGTGATCTATAGCATAGGCCACTAGTGGAT- CTG. Deletion of the BNI1 locus was confirmed by Southern blot analysis of Geneticin-resistant transformants. This BNI1 deletion strain, hereafter referred to as bni1Δ, was used for all subsequent analyses unless specified otherwise. The deletion of ASE1 was also created by one-step gene replacement (Pellman et al., 1995).
The centromere-based (CEN) plasmid carrying BNI1 (p182) has been described (Boone et al., 1992). This genomic sequence was also cloned into a CEN ARS TRP1 plasmid (pRS314) (Sikorski and Hieter, 1989) to create p1832. The COOH-terminal deletion mutant, bni1-CTΔ1, lacks the coding sequence for amino acids 1,749–1,953 of Bni1p, and was created by the following strategy. p182 was digested with SstII which cuts BNI1 at bp 5,249 and in the polylinker, and ligated to an oligonucleotide (5′GTAGGCGGCCGCCTACGC). This created a stop codon followed by a NotI site adjacent to the SstII site in BNI1. A CEN plasmid (p2029) derived from pRS314 and an integrating plasmid (PB1046) derived from pRS305 (Sikorski and Hieter, 1989) contain the bni1-CTΔ1 sequence on a 6.3-kb BamHI/NotI restriction fragment. The strain containing PB1046 was used for genetic crosses, and the strain containing p2029 was used for morphological studies.
The act1-116 and sla1ΔSH3#3 strains and their isogenic wild-type controls are in the S288c genetic background. To construct PY2596, the ACT1 locus was replaced by act1-116::HIS3 from pRB1537 by homologous recombination in a strain containing NUF2::GFP (PB1000). This was done because previously constructed act1-116 strains contain a linked TUB2 mutation. Integration was confirmed by PCR analysis.
Genetic Analysis
The screen to identify mutations that were lethal in combination with an ASE1-null allele was performed using an ADE3 sectoring assay as described (Bender and Pringle, 1991). The starting strains MATα and MAT a ase1::kanr ade2 ade3 leu2 lys2 trp1 ura3 {2μ ASE1 ADE3 TRP1} (PY1994 and PY1996) were mutagenized by transformation with a genomic library containing random insertions of a mini-Tn3lacZ-LEU2 transposon (Burns et al., 1994). Candidate mutants were backcrossed to demonstrate that the mutation represented a single genetic locus and to confirm that the mutation cosegregated with the LEU2 marker. The genomic site of transposon insertion was identified by direct sequencing of PCR products spanning the insertion site. These PCR products were generated by an arbitrary primed PCR strategy (Di Rienzo et al., 1996). The bni1::Tn3::LEU2 allele has an insertion of the transposon at nucleotide 5,006 of the BNI1-coding sequence. This allele has an identical phenotype to the bni1Δ::kanr-null allele described above. In addition to BNI1, genes that affect many aspects of microtubule function in S. cerevisiae were identified by this screen (to be described elsewhere).
For the bni1Δ × ase1Δ cross, 37 tetrads were dissected and 41 double mutants were recovered, all of which were temperature sensitive for growth. For the bni1Δ × dyn1Δ cross, 19 tetrads were dissected and 11 temperature-sensitive double mutants were recovered. For the bni1Δ × kar3Δ cross, 27 tetrads were dissected and 20 viable double mutants were recovered. bni1Δ kar3Δ double mutants formed smaller colonies than the kar3Δ single mutant at 23°C and were inviable at 30°C. For the bni1Δ × cin8Δ cross, 35 tetrads were dissected and 28 double mutants were recovered. For the crosses of bni1Δ with kip1Δ and smy1Δ, 23 and 17 tetrads were dissected respectively, and the double mutants were recovered at the expected frequency.
Quantitative mating assays (Sprague, 1991) were performed using a MAT a bni1::kanr bar1::LEU2 strain (Y1445), carrying different alleles of BNI1 on a CEN plasmid. The mating partner used for the mating assays was a MATα far1-c strain (Y66) which was itself defective in mating and thus increased the sensitivity of the assay. The assays were performed in triplicate.
The strain background used for our initial genetic experiments and for our time-lapse analysis, W3031a, has a mixed random/bipolar budding pattern (Cvrckova et al., 1995). Therefore, in experiments where the movement of the SPB after telophase toward the bud was followed by fluorescence microscopy, the distance between the SPB and the bud was determined by overlaying images from each time point with a differential interference contrast (DIC) image obtained after bud emergence. The spindle orientation defect of bni1Δ that we observed is not specific to this strain background because we observed a similar spindle orientation defect in a YEF473-derived bni1Δ strain (Bi and Pringle, 1996) that has normal axial and bipolar bud site selection (data not shown).
Microscopic Analysis of Cells
Nuclear morphology was observed by fluorescence microscopy in cells fixed and stained with 4′,6′-diamidino-2-phenylindole (DAPI) as described (Rose et al., 1990). To preserve green fluorescent protein (GFP) fluorescence for the analysis of spindle orientation, cells were fixed in culture medium with 3.7% formaldehyde for 30 min, washed, and then applied to slides. Indirect immunofluorescence was performed as described (Pellman et al., 1995). Experiments to measure nuclear or spindle position were performed at least twice.
Budding pattern was assayed after staining bud scars with calcofluor (Pringle et al., 1989). The strain used was a bni1::HIS3/bni1::HIS3 YEF473 diploid (Y1353), containing alleles of BNI1 on a CEN plasmid. Results from these strains were compared with results from an isogenic BNI1/BNI1 diploid. The fact that BNI1 carried on a CEN plasmid does not fully complement bni1::HIS3/bni1::HIS3 suggests that the gene dosage of BNI1 may be important for bipolar bud site selection. Mating projection assays using a W3031a MATa bni1::kanr bar1::LEU2 strain (Y1445) containing the bni1 alleles were performed in triplicate (Evangelista et al., 1997).
For live cell fluorescence microscopy, cells were applied to a microscope slide (DeZwaan et al., 1997) and observed at 23°C. The doubling time of cells in liquid culture at this temperature was 150 min, compared with the average doubling time of 176 min for cells observed with our time-lapse fluorescence protocol. A microscope (model ECLIPSE E600; Nikon) equipped with a 100×/1.4 NA Planapochromat objective, a 100-W mercury arc illuminator, a Z-axis focus motor (Biopoint Z-axis focus motor; Ludl Electronics), a fluorescence illumination shutter (Ludl Electronics), and an Endow GFP filter set (excitation 450–490 nm, dichroic 495, emission 500 nm LP; Chroma Optics) was used. Images were acquired with a cooled charge-coupled device camera (Hamamatsu 4742-95, Hamamatsu Photonics), containing a 1280 × 1024 Sony Interline chip (Hamamatsu Photonics). At each time point a series of fluorescence images in six focal planes spaced 0.5 μm apart were collected along with a DIC image. To minimize photo bleaching and photo damage of the cells, the light from the mercury lamp was attenuated to one-eighth of maximal intensity with neutral density filters and exposure times were limited to 200–400 ms. Images were analyzed using Openlabs Software (Improvision), and all statistical analysis was performed using StatView software (Abacus Concepts).
Results
BNI1 Has a Role in Microtubule Function
An allele of BNI1 was unexpectedly isolated in a genetic screen to identify genes involved in microtubule function. The screen was for mutations that were lethal when combined with an ase1Δ allele (see Materials and Methods). ASE1 encodes a nonmotor microtubule-associated protein that localizes to the spindle midzone and promotes spindle elongation (anaphase B) (Pellman et al., 1995; Juang et al., 1997). The genetic interaction between BNI1 and ASE1 suggested that BNI1 might have a role in maintaining the integrity of the microtubule cytoskeleton.
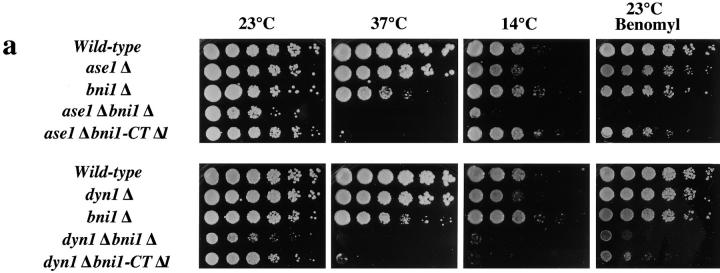
A deletion allele of BNI1 was created (see Materials and Methods). The bni1Δ ase1Δ double mutant strain displayed a marked temperature-sensitive growth defect (Fig. 1 a). Because this strain grew at a near normal rate at 23°C, we were able to test its sensitivity to the microtubule depolymerizing agent, benomyl. In contrast to the control strains, the bni1Δ ase1Δ strain had a strikingly increased sensitivity to benomyl (5 μg/ml). bni1Δ and ase1Δ mutations therefore have an additive detrimental effect on microtubule function.
Figure 1.
Growth phenotypes and nuclear segregation in bni1Δ and double mutant strains. (a) Growth phenotypes. The indicated strains were grown at 23°C to saturation, diluted to the same starting concentration, and then spotted onto medium in fivefold serial dilutions. Plates were incubated for 48 h, except 14°C plates which were incubated for 7 d. Benomyl sensitivity was tested at 5 μg/ml benomyl. Wild-type cells were sensitive to benomyl at 20 μg/ml. (b) Nuclear segregation. Strains were grown to early log phase at 23°C, shifted to the indicated temperatures for 3 h, and then stained with DAPI. At least 300 cells were counted for each sample.
bni1Δ Cells Have a Defect in Spindle Orientation
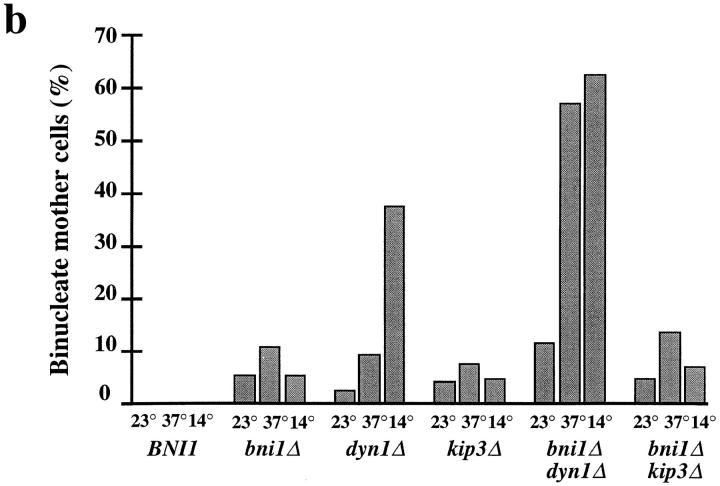
Because Bni1p is localized at the bud cortex and septum of mitotic cells and the shmoo tip in mating cells (Jansen et al., 1996; Evangelista et al., 1997; Fujiwara et al., 1998) (Evangelista, M., and C. Boone, unpublished results) we considered the possibility that BNI1 affects microtubules through a role in spindle orientation and/or nuclear positioning. Here, defects in the location of the nucleus within the mother cell or defects in nuclear segregation that result in binucleate mother cells will be referred to as “nuclear position defects.” Defects in the alignment of the spindle with the mother–bud axis will be described as “spindle orientation defects.” The role of Bni1p in nuclear positioning was initially tested by examining bni1Δ and control cells stained with the DNA-binding dye, DAPI. Whereas mitosis in wild-type cells results in the faithful distribution of nuclei to mother and daughter cells (Fig. 1 b), mitosis in a significant fraction of bni1Δ cells was abnormal, resulting in two nuclei in the mother cell. The fraction of binucleate mother cells in bni1Δ strains increased at 37°C (Fig. 1 b).
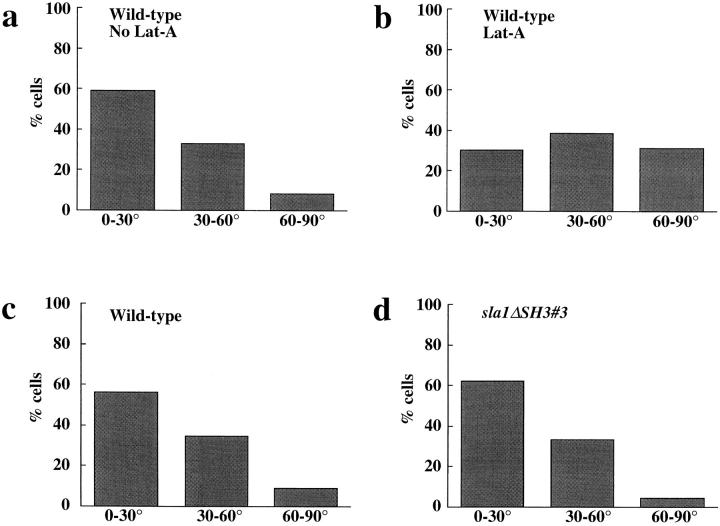
We next tested whether the binucleate phenotype of bni1Δ cells was due to a defect in spindle orientation. bni1Δ and control strains were grown to early logarithmic stage and the angle between the spindle and the mother– bud axis was measured (Fig. 2). A chimeric gene expressing a fusion between Nuf2p, a spindle pole body protein, and GFP was introduced into both strains, allowing visualization of the poles by fluorescence microscopy (Kahana et al., 1995; Kahana and Silver, 1996). In wild-type cells, more than 60% of cells had spindles oriented within 30 degrees of the mother–bud axis (Fig. 2 a). By contrast, spindle orientation in bni1Δ cells appeared to be random (Fig. 2 b). Furthermore, the distance between the neck-proximal SPB and the bud neck was significantly greater in bni1Δ than in BNI1 cells: 1.0 and 0.7 μm, respectively (t test, P < 0.0001, n = 180). Thus, bni1Δ has a defect in spindle orientation.
Figure 2.
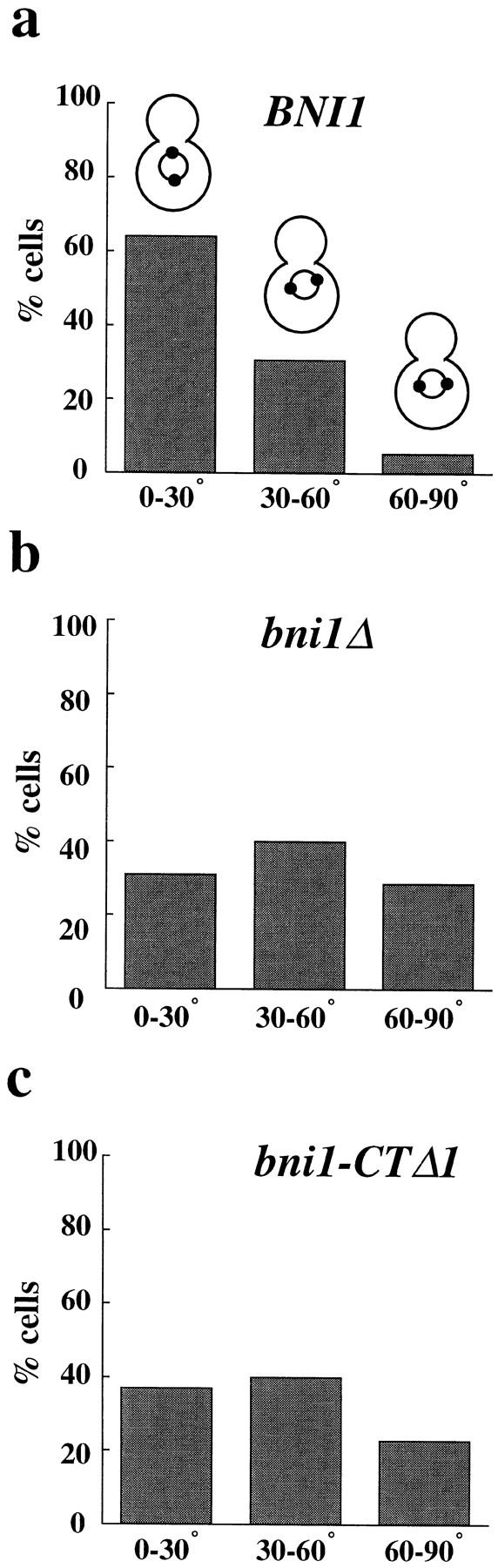
Spindle orientation in BNI1 (a), bni1Δ (b), and bni1-CTΔ1 (c) strains. Early log phase cultures were incubated at either 23° or 37°C for 3 h. The results shown are from the 37°C samples. The histograms show the percentages of cells with spindles oriented to the mother–bud axis at angles of 0°–30°, 30°– 60°, and 60°–90°. 180 cells were examined for each sample. The mean spindle orientation angles are 23.7° (BNI1), 42.8° (bni1Δ), and 39.5° (bni1-CTΔ1) and the mean SPB-neck distances are 0.7, 1.0, and 1.1 μm, respectively. The mean spindle orientation angle of the bni1Δ and the bni1-CTΔ1 strains were significantly different from that of the control (t test, P < 0.0001 for both). For samples maintained at 23°C, the mean spindle orientation angles were 25.2° (BNI1), 39.1° (bni1Δ), and 33.3° (bni1-CTΔ1). The mean angles of the bni1Δ and the bni1-CTΔ1 strains at 23°C were also significantly different from the control strain (t test, P < 0.0001 and P = 0.0005, respectively).
Genetic Interactions Suggest that BNI1 and KIP3 Are in the Same Pathway
The entire set of Saccharomyces cerevisiae microtubule-based motor proteins has been defined. The genome contains a single gene encoding a dynein heavy chain, DYN1, and six genes encoding kinesin-related proteins, KIP1, KIP2, KIP3, KAR3, CIN8, and SMY1. Strains lacking any one of these genes are viable; however, combinations of different null alleles result in lethality. Only Dyn1p, Kip2p, Kip3p, and Kar3p are implicated in cytoplasmic microtubule function (Stearns, 1997; Winsor and Schiebel, 1997). Single mutant kip3Δ and dyn1Δ strains have the most striking effect on nuclear position. kip3Δ and dyn1Δ affect distinct steps in nuclear positioning: kip3Δ cells have a spindle orientation defect, whereas dyn1Δ cells have a defect in the insertion of the spindle across the bud neck. The fact that kip3Δ dyn1Δ double mutant strains are not viable suggests that there are two partially redundant pathways for spindle orientation (Cottingham and Hoyt, 1997; De Zwaan et al., 1997; Miller et al., 1998).
Double mutants were constructed to test if BNI1 was in the KIP3 pathway. Consistent with the similar spindle orientation defect observed in bni1Δ and kip3Δ strains, the bni1Δ kip3Δ strain was viable and grew indistinguishably from single mutant strains (Table III). The percentage of binucleate mother cells in the bni1Δ kip3Δ strain was also not increased relative to either single mutant strain (Fig. 1 b). By contrast, bni1Δ produced a striking synthetic interaction when combined with dyn1Δ. The bni1Δ dyn1Δ strain had a growth defect at 23°C and was inviable at 14° and 37°C (Fig. 1 a and Table III). Also, the bni1Δ dyn1Δ strain had a marked increase in binucleate mother cells relative to the single mutant strains (Fig. 1 b). Like the bni1Δ ase1Δ strain, the bni1Δ dyn1Δ strain had increased sensitivity to benomyl (Fig. 1 a).
Table III.
Genetic Interactions between bni1Δ and Motor Mutations
| 23°C | 37°C | 14°C | 30°C | |||||
|---|---|---|---|---|---|---|---|---|
| Growth phenotype of | ||||||||
| single mutants | ||||||||
| bni1Δ | +++ | ++ | +++ | ND | ||||
| kip3Δ | +++ | +++ | +++ | ND | ||||
| dyn1Δ | +++ | +++ | +++ | ND | ||||
| kar3Δ | + | − | + | + | ||||
| kip1Δ | +++ | +++ | +++ | ND | ||||
| kip2Δ | +++ | +++ | +++ | ND | ||||
| cin8Δ | +++ | +/− | +++ | ND | ||||
| smy1Δ | +++ | +++ | +++ | ND | ||||
| Growth phenotype of | ||||||||
| double mutants | ||||||||
| bni1Δ kip3Δ | +++ | ++ | +++ | ND | ||||
| bni1Δ dyn1Δ | ++ | − | − | ND | ||||
| bni1Δ kar3Δ | + | − | + | − | ||||
| bni1Δ kip1Δ | +++ | ++ | +++ | ND | ||||
| bni1Δ kip2Δ | +++ | ++ | +++ | ND | ||||
| bni1Δ cin8Δ | +++ | − | +++ | ND | ||||
| bni1Δ smy1Δ | +++ | ++ | +++ | ND |
Each strain was struck on a YPD plate and incubated at the appropriate temperatures. For 23°, 30°, and 37°C test, plates were scored after 48 h. For 14°C test, plates were scored after 7 d.
The genetic interactions we observed between bni1Δ and the other motor gene deletions paralleled those observed with kip3Δ (Table III). Like kip3Δ, bni1Δ is lethal when combined with kar3Δ. bni1Δ did not show a significant synthetic interaction with either kip1, cin8Δ, or smy1Δ. This pattern of genetic interactions observed with bni1Δ suggests that BNI1 and KIP3 function in the same pathway.
Defective Movement of the SPB to the Incipient Bud in bni1Δ Cells
Because of the spindle orientation defect in bni1Δ and kip3Δ strains, we determined whether movement of the SPB to the incipient bud site was defective in bni1Δ cells as previously observed in kip3Δ cells. Time-lapse fluorescence microscopy of live cells expressing either Nuf2p– GFP (Kahana et al., 1995; Kahana and Silver, 1996) or GFP–Tub1p (the major α tubulin gene in S. cerevisiae) was used to examine SPB movement early in the cell cycle in both bni1Δ and control cells (see Materials and Methods).
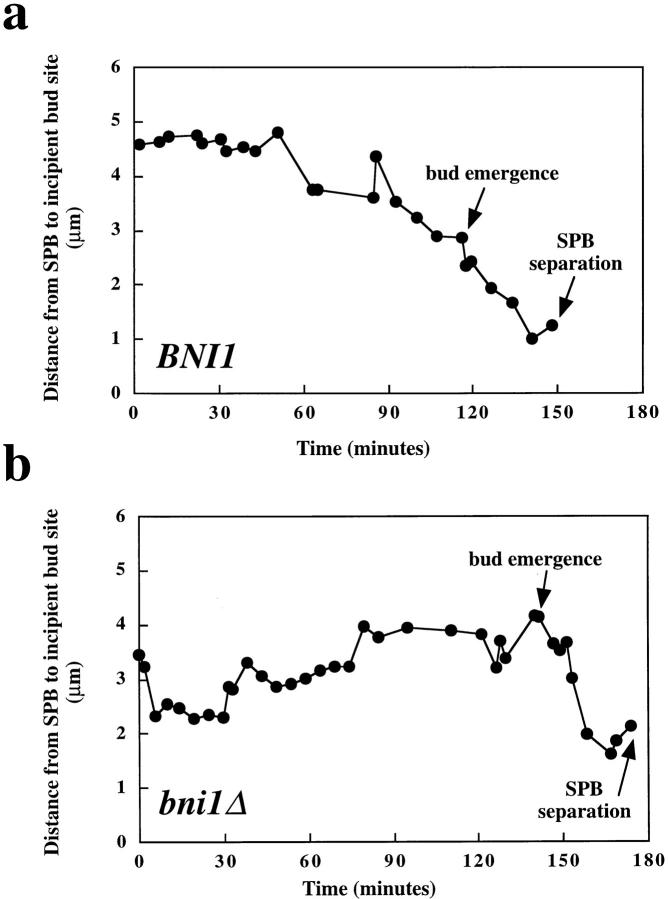
SPB movements in wild-type haploid cells were observed from the end of mitosis (telophase) through bud emergence and SPB separation (Fig. 3 a). At the end of mitosis (Fig. 3 a, 1.7 min, mother cell indicated by white arrow) the poles were extended 8–10 μm and located near the cell periphery. After spindle disassembly, the poles usually moved ∼1–2 μm toward the center of the cell (Fig. 3 a and Fig. 4 a). In the wild-type strain, this initial movement of the SPB was followed by a concerted movement of the pole toward the site of the incipient bud. This movement of the SPB toward the bud occurred on average within 100 min (n = 6), and once the SPB had moved toward the bud its position became relatively stable. SPB separation usually occurred after the old SPB was positioned near the bud. Although the SPB underwent some oscillations as it moved toward the bud, large movements away from the bud were not observed in wild-type cells (Fig. 4 a).
Figure 3.
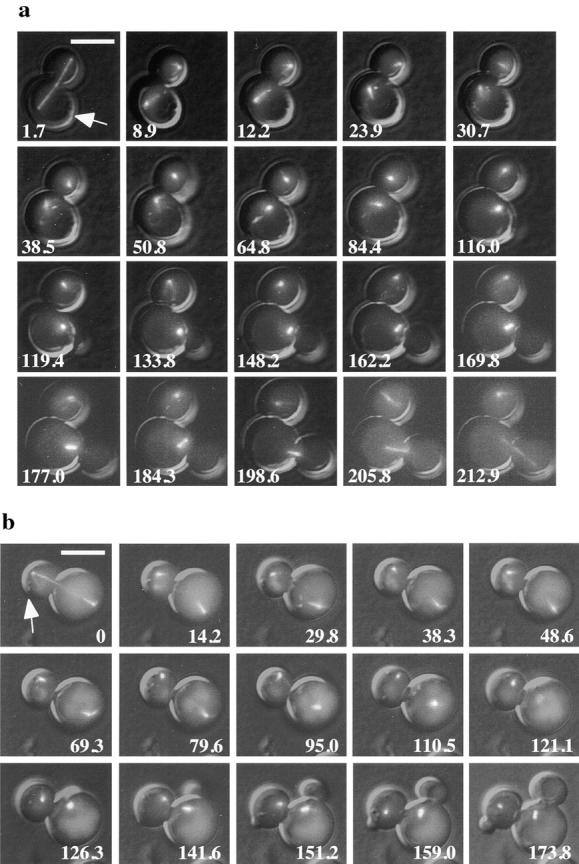
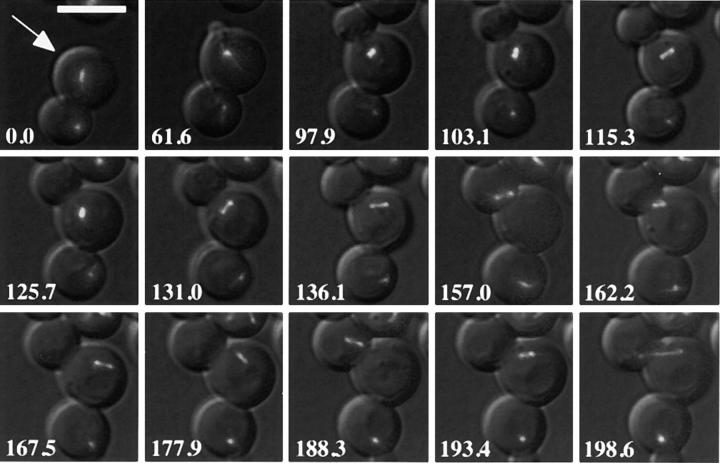
SPB movement toward the incipient bud in posttelophase wild-type and bni1Δ cells. Representative images from time-lapse series of BNI1 (a) and bni1Δ (b) cells. Microtubules are labeled in vivo with GFP–Tub1. Cells were imaged from the time of maximal anaphase spindle elongation until SPB separation or longer. At each time indicated a DIC image and six fluorescent images separated by 0.5 μm in the z focal plane were acquired and overlaid. White arrow, the incipient bud site of the cell of interest. In the BNI1 strain, spindle disassembly is at 8.9 min, bud emergence is at 116 min, SPB separation is at 162.6 min, spindle insertion is at 198.6 min, and spindle elongation is at 212.9 min. In the bni1Δ strain, spindle disassembly is at 0 min, bud emergence is at 141.6 min, and SPB separation is at 173.8 min. The complete data set for each cell is plotted in Fig. 4. Bar, 5 μm.
Figure 4.
Spindle pole body migration in BNI1 (a) and bni1Δ (b) cells from spindle disassembly to SPB duplication. The distance between the SPB and the incipient bud site is measured at each time point. At time points before bud emergence the position of the presumptive bud site was determined by comparison to later time points. Images for a subset of the time points acquired are shown in Fig. 3.
In a significant fraction of posttelophase bni1Δ cells, prolonged movement of the SPB away from the bud was observed (Fig. 3 b and Fig. 4 b). In Fig. 3 b, the daughter cell (white arrow) shows this abnormal movement of the SPB (for example, 14.2–121.1 min). In the cells that exhibited the phenotype, the SPB spent almost as much time moving away from the bud as toward the bud. The more random direction of movement of the SPB with respect to the incipient bud in bni1Δ cells was similar to the abnormal SPB movements previously observed in kip3Δ cells (DeZwaan et al., 1997). This phenotype was detected in four out of 10 cells and was observed in both mother and daughter cells. The mother cell in Fig. 3 b is an example of a bni1Δ cell with relatively normal SPB movement. As in kip3Δ cells, the SPB eventually assumed a relatively stable position near the bud, suggesting that other factors may stabilize the interaction of the SPB with the bud neck.
Abnormal Spindle Rotation and Transiting of Short Mitotic Spindles in bni1Δ Cells
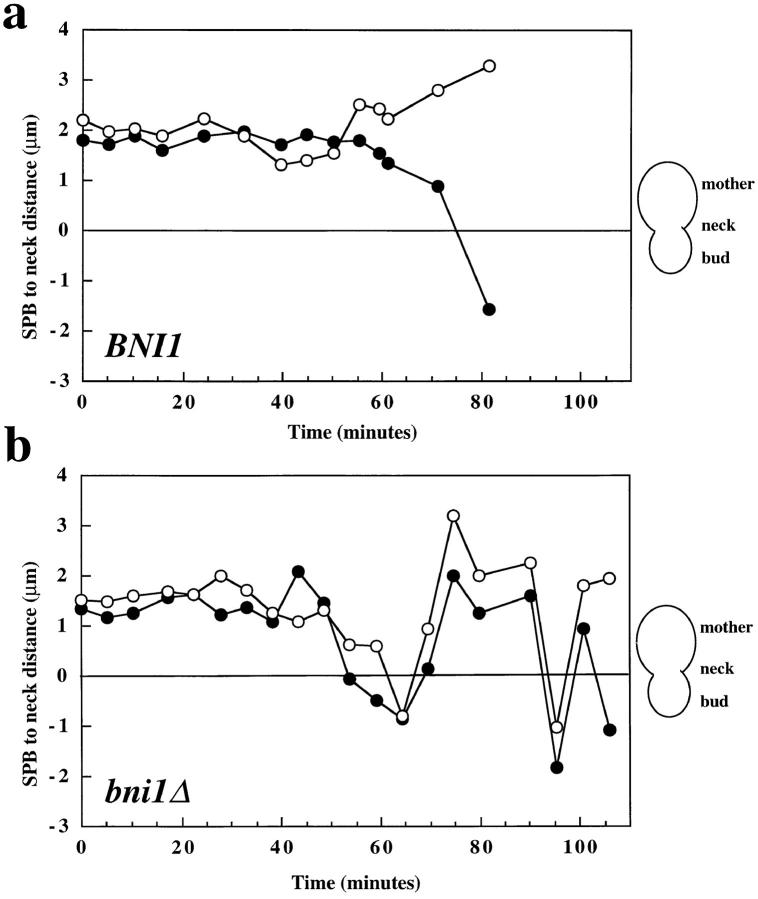
In addition to the defect in the movement of the unduplicated SPB toward the bud, we observed strikingly abnormal movements of short (1.5 μm) spindles that were unique to bni1Δ cells. In another series of experiments, spindle movements before anaphase were observed in bni1Δ and wild-type cells. In wild-type cells, short spindles aligned along the mother–bud axis as expected from our analysis of fixed cells and previous imaging of live cells (Yeh et al., 1995). These short spindles displayed some oscillations relative to the bud neck. However, short spindle movement in these cells was limited and only rarely were large rotations (90–180°) of the spindle observed. At anaphase, the spindles in wild-type cells were correctly inserted into the bud neck, and spindle elongation occurred through the bud neck (Fig. 3 a and Fig. 5 a).
Figure 5.
Preanaphase spindle movement in BNI1 and bni1Δ cells. The distance between each SPB and the bud neck is measured at 5–10 min intervals. Cells were observed from SPB separation through spindle insertion. Images were acquired as in Fig. 3. Closed circles represent the SPB that is initially proximal to the bud and open circles represent the other SPB. The bud neck is at 0 μm, positive numbers are distances from the neck in the mother cell, and negative numbers are distances from the neck in the daughter cell. 0 min in Fig. 5 b corresponds to 97.9 min in Fig. 6.
By contrast, the spindles of bni1Δ cells underwent much more noticeable oscillations, and large rotations of the spindle were frequently observed. In the example shown in Fig. 5 b the spindle underwent four 90–180° rotations in a 40-min time period before anaphase (crossing of lines with open and filled circles). Furthermore, in bni1Δ cells the short spindle frequently underwent what we refer to as transiting: movements of the entire short spindle into the bud or back into the mother cell. In the example shown in Fig. 5 b and Fig. 6, transiting of short spindles back and forth between the bud and mother cell was observed four times in the interval before anaphase (Fig. 6, 157.0, 167.5, 188.3, and 193.4 min). Like the defect in the initial SPB movement toward the bud observed in our other series of experiments, this phenotype was not completely penetrant (four out of 10 cells).
Figure 6.
Defective preanaphase spindle movement in a bni1Δ cell: transiting of the short spindle between the mother and daughter cell. White arrow, the cell of interest. Images were collected as in Fig. 3. The images shown are a subset of the time points acquired and plotted in Fig. 5 b. Bud emergence and SPB separation occur at 61.6 and 97.9 min, respectively. Transiting of the preanaphase spindle into the bud is observed at 157.0 and 188.3 min, respectively, whereas transiting into the mother is observed at 167.5 and 193.4 min, respectively. Bar, 5 μm.
Abnormal Interactions between Microtubules and the Cortex in bni1Δ Cells
Recent live-cell fluorescence microscopy experiments suggest that spindle or SPB movement is driven by cytoplasmic microtubule contacts with the cell cortex (Carminati and Stearns, 1997; Shaw et al., 1997). Three types of contacts between cytoplasmic microtubules and the cell cortex have been correlated with changes in the position of the poles or spindles. Apparent pushing forces are generated when cytoplasmic microtubules remain in contact with the cortex while polymerizing. Apparent pulling forces are generated when they depolymerize while maintaining contact with the cortex. Finally, lateral movements of the SPB or spindles are seen as microtubules sweep across the cortex (Carminati and Stearns, 1997; Shaw et al., 1997).
To determine if the spindle movements in bni1Δ cells were due to abnormal interactions of cytoplasmic microtubules with cortical sites, image sets of wild-type and bni1Δ cells expressing GFP::Tub1p were acquired over short time intervals (30–60 s) using exposures that enabled us to clearly visualize cytoplasmic microtubules. In the wild-type cells with short spindles, the pattern of microtubule interactions with the cell cortex and the associated pattern of SPB and/or spindle movement were similar to that described above. Most notably, pulling events in our wild-type strain were observed in cells with short spindles when microtubules interacted with the bud cortex (five events observed over 140 min in five cells); however, such pulling events toward the mother cell were not observed (0 events over 140 min in five cells). Furthermore, in different experiments, we did not observe transiting of preanaphase spindles in seven wild-type cells, observed for an average time of 57 min per cell, over longer time intervals (data not shown). Although we did not observe spindle transits in our wild-type strain, low frequency spindle transits have been reported in some wild-type strains (Palmer et al., 1989).
We next examined microtubule interactions with the cell cortex in bni1Δ cells (Fig. 7). Unlike the control strain, pulling events between the cortex and the SPB in both the bud and mother cell were frequently observed (14 events toward bud and 10 events toward mother observed over 147 min in four out of 12 cells). An example of such interactions is shown in Fig. 7. In this cell, from time points 8.9– 9.8 min, a microtubule end remained at a fixed position at the mother cell cortex whereas the microtubule shrank from 5.1 to 0 μm. A concomitant 5.6-μm movement of the spindle toward the mother cell cortex was observed. Again, from 15.8–16.6 min, a microtubule with one end at the mother cell cortex shrank from 4.2 to 1.7 μm, whereas the spindle moved 2.1 μm toward the cortex. Overall, we observed a strong correlation between pulling events and the abnormal transiting of short spindles between the mother and the bud in bni1Δ cells. Therefore, although microtubules in the bni1Δ cells still interact with the cortex, the inability to restrict pulling interactions toward the bud suggests that the microtubule cytoskeleton of the bni1Δ strain fails to sense the cell's polarity.
Figure 7.
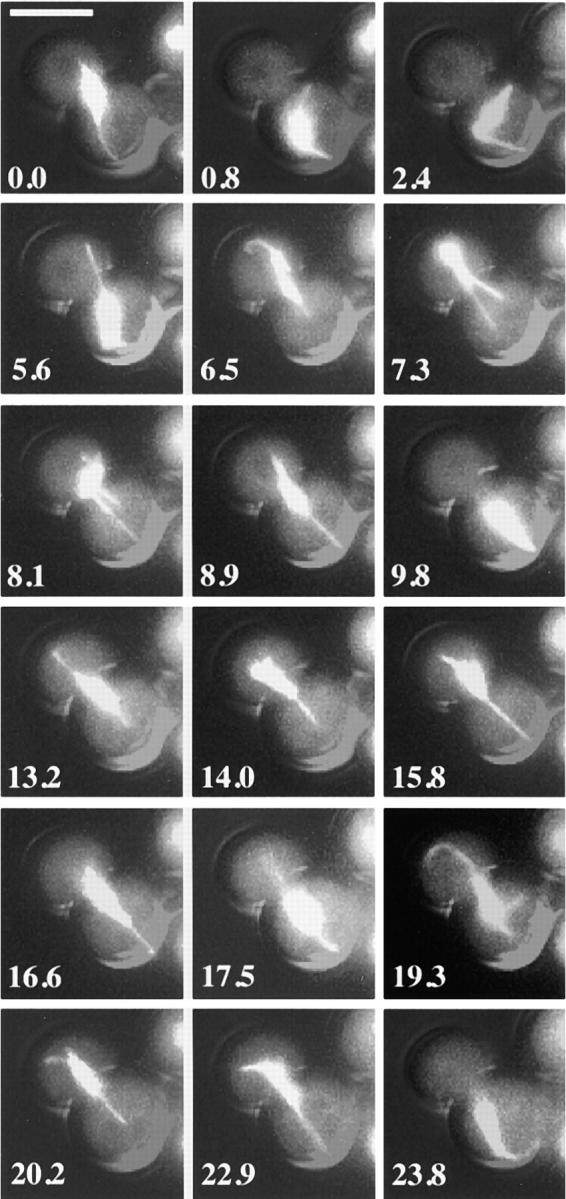
Relationship between preanaphase spindle transiting and cytoplasmic microtubules interactions with the cell cortex: time-lapse series of a preanaphase bni1Δ cell. At each time indicated a DIC image and eight to 10 fluorescent images separated by 0.3 μm in the z focal plane were acquired and overlaid. To enhance the visualization of cytoplasmic microtubules, images were collected using 2 × 2 binning and 400–700-ms exposures. The light from the mercury lamp was attenuated to one-eighth of the maximal intensity. This resulted in overexposure of the spindle microtubules. Spindle transits into the mother cell were observed at 0.8, 9.8, 17.5, and 23.8 min, and transits into the bud were observed at 7.3, 14.0, and 20.2 min. In all cases, spindle transit was correlated with shortening of the cytoplasmic microtubules which maintained contact with the cell cortex. Bar, 5 μm.
Bni1p Is Not Required for Anaphase Spindle Elongation or Cell Cycle Progression
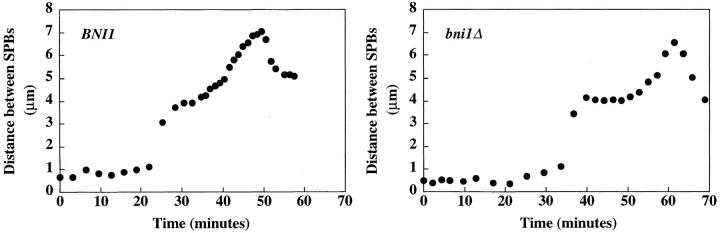
Because loss of BNI1 affects cytoplasmic microtubule function, we next tested whether nuclear microtubule function was affected by measuring the kinetics of anaphase spindle elongation. Two major stages of anaphase have been observed in S. cerevisiae (Kahana et al., 1995; Yeh et al., 1995; Straight et al., 1997). There is an initial fast stage of elongation from an ∼2-μm spindle to an ∼4-μm spindle. The rate of elongation then slows as the spindle reaches maximal extension at 8–10 μm. Anaphase kinetics was measured in bni1Δ and wild-type cells (Fig. 8). Fast elongation was 0.55 μm/min ± 0.14 (n = 7) in the bni1Δ strain and 0.48 μm/min ± 0.10 (n = 5) in the wild-type strain. Slow elongation was 0.14 μm/min ± 0.04 (n = 7) in the bni1Δ strain and 0.12 μm/min ± 0.04 (n = 5) in the wild-type strain. Thus, loss of BNI1 does not have a significant effect on anaphase spindle elongation.
Figure 8.
Anaphase kinetics in BNI1 and bni1Δ cells. Time-lapse series of BNI1 and bni1Δ cells collected from before anaphase through the completion of anaphase. Images were collected as in Fig. 3. The distance between the two SPBs was measured at each time point and plotted versus time.
Because the bni1Δ mutant could affect spindle orientation either through a direct effect on the cytoskeleton or indirectly by perturbing the cell cycle, we next tested whether bni1Δ strains were delayed in any specific stage of the cell cycle. bni1Δ strains grew slightly slower than control strains at 23°C. The distribution of cells throughout the cell cycle was first characterized by examining the distribution of cell morphologies in cultures of logarithmically growing cells (Table IV). The numbers of unbudded G1 cells, budded cells with undivided nuclei, and postanaphase cells were nearly identical in bni1Δ and the wild-type strain. The ratio of cells in G1 and G2 cell cycle phases were also similar in both strains by FACS® analysis (data not shown). Finally, we analyzed our time-lapse experiments to measure the intervals between spindle disassembly and the initiation and completion of budding in bni1Δ and control cells. After completion of anaphase, the bni1Δ and control strains initiated budding on average at 87 and 89 min, respectively. Budding was completed on average at 138 and 150 min, respectively, in these strains.
Table IV.
Cell Cycle Distribution of BNI1 and bni1Δ Cells
|
Percent unbudded | Percent budded (preanaphase) | Percent anaphase | |||
|---|---|---|---|---|---|---|
|
|
||||||
| BNI1 | 46.8 | 43.1 | 10.1 | |||
| bni1Δ | 48.7 | 41.4 | 9.9 |
BNI1 and bni1Δ cells expressing GFP–Tub1p were grown in liquid culture at 23°C and observed by fluorescence microscopy. n = 1,200.
Therefore, although we cannot rule out a subtle effect on cell cycle progression in bni1Δ cells, our analysis shows that the cells do not exhibit a prominent delay at a specific stage of the cell cycle. This is consistent with the idea that loss of BNI1 function affects spindle orientation through direct effects on the cytoskeleton rather than indirect effects on the cell cycle.
Bud6p and the Bud6p-binding Domain of Bni1p Are Required for Spindle Orientation and Bipolar Bud Site Selection
Bni1p associates with other proteins that are required for bipolar bud site selection (Amberg et al., 1997; Evangelista et al., 1997; Fujiwara et al., 1998). As an initial test to determine if these associations are required for spindle orientation, we characterized a BNI1 allele (bni1-CTΔ1) lacking the COOH-terminal domain of Bni1p that interacts with the actin-associated protein Bud6p (Amberg et al., 1997; Evangelista et al., 1997). The bni1-CTΔ1 strain is proficient both for shmoo formation and mating (Table V). By contrast, bni1-CTΔ1 did not complement the bud site selection defect of a bni1Δ/bni1Δ diploid strain (Table V). Furthermore, bni1-CTΔ1 cells had a significant defect in spindle orientation (Fig. 2 c). Like bni1Δ cells, the spindles in bni1-CTΔ1 cells were also located farther from the bud neck and were highly variable in position (mean distance of 1.1 μm in bni1-CTΔ1 versus 0.7 μm in BNI1, t test, P < 0.0001, n = 180) and, like the bni1 null allele, the bni1-CTΔ1 allele produced a striking temperature-sensitive growth defect when combined with ase1Δ or dyn1Δ (Fig. 1 a). bni1-CTΔ1 ase1Δ and bni1-CTΔ1 dyn1Δ strains were also more sensitive to benomyl, although the effect was less pronounced in the bni1-CTΔ1 ase1Δ strain (Fig. 1 a).
Table V.
Mating Projection Formation, Quantitative Mating Assay, and Bipolar Bud Site Selection
| Mating projection formation | Mating efficiency | Bipolar bud site selection | ||||
|---|---|---|---|---|---|---|
| % | % | % | ||||
| bni1::kanr {BNI1} | 83.4 ± 5.5 | 0.17 ± 0.041 | — | |||
| bni1::kanr {bni1-CTΔ1} | 77.2 ± 5.2 | 0.095 ± 0.013 | — | |||
| bni1::kanr {CEN URA3} | 0 | 0.001 ± 0.0006 | — | |||
| BNI1/BNI1 | — | — | 4 ± 1.8 | |||
| bni1Δ/bni1Δ {BNI1} | — | — | 31 ± 2.3 | |||
| bni1Δ/bni1Δ {bni1-CTΔ1} | — | — | 92 ± 3.5 | |||
| bni1Δ/bni1Δ {CEN URA3} | — | — | 93 ± 2.1 |
The mating projection assay measured the percentage of cells with mating projections after 3 h in α factor (n = 300). The quantitative mating assay (Sprague, 1991) measured the percentage of diploids formed after a 4-h mating with the far1-c tester strain. The result of the bipolar bud site selection assay (Pringle et al., 1989) shows the percentage of cells with a random budding pattern.
Spindle orientation was also affected in cells lacking Bud6p. At 23°C, the mean spindle orientation angle in a bud6Δ strain was 24.0°, compared with 17.8° in an isogenic wild-type (t test, P < 0.0001, n = 290). At 37°C, the mean angle in bud6Δ was 28.3° in contrast to 20.6° in the wild-type (t test, P < 0.0001, n = 330). These results demonstrate that Bud6p and the Bud6p interaction domain of Bni1p are required for spindle orientation and bipolar bud site selection, but suggest that Bud6p is less important for spindle orientation than Bni1p.
The Role of Actin in Spindle Orientation and Bipolar Bud Site Selection
Because Bni1p and Bud6p are both thought to be regulators of actin function, we next characterized the nature of actin's role in spindle orientation and nuclear positioning. Previous studies demonstrated that actin mutations interfere with normal nuclear position (Palmer et al., 1992; Drubin et al., 1993). To determine if actin is required for spindle orientation, we treated logarithmically growing wild-type cells with Lat-A for a brief period (5 min), fixed the cells, and then measured spindle orientation (Fig. 9 b). Staining with rhodamine-phalloidin revealed a complete loss of polymerized actin after treatment with Lat-A (data not shown). By contrast with mock-treated cultures (Fig. 9 a), the 5-min treatment with Lat-A–randomized spindle orientation. Thus, actin is required for spindle orientation in normal growing cells. The rapid effect on spindle orientation suggests that the role of actin in controlling spindle orientation may be direct.
Figure 9.
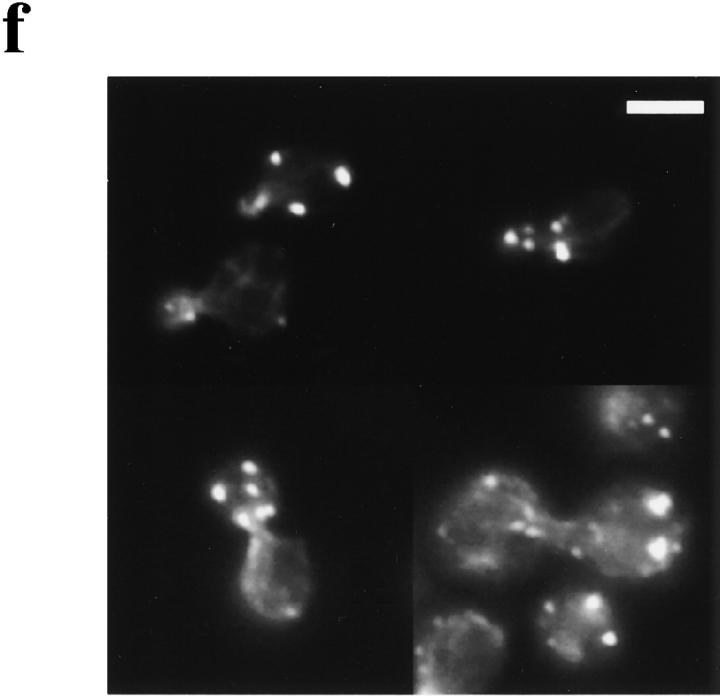
The role of actin in spindle orientation. Early log phase wild-type cells expressing Nuf2p–GFP were treated with DMSO (a) or Lat-A (b) for 5 min. Cells were fixed and spindle orientation was measured as in Fig. 2. Spindle orientation angles are shown on the x axis. A sample from each culture was stained with rhodamine-phalloidin to visualize actin. Spindle orientation angles were measured in sla1ΔSH3#3 (d) and isogenic wild-type strains (c). Spindles were visualized by indirect immunofluorescence using the anti-tubulin mAb, YOL1/34. sla1ΔSH3#3 (f) and isogenic wild-type strains (e) were stained with rhodamine-phalloidin to visualize actin structures. Bar, 3 μm.
Genetic experiments have provided evidence for a specialized actin structure that mediates bipolar bud site selection (Yang et al., 1997). Many, but not all, mutations that affect the actin cytoskeleton affect bipolar bud site selection. A sla1ΔSH3#3 strain with abnormal cortical actin structure did not have a defect in bipolar bud site selection (Yang et al., 1997). Strains with this sla1 allele accumulated abnormal cortical actin aggregates (Fig. 9 f) and like sla1Δ strains, were temperature-sensitive for growth. Like sla1Δ, these strains were also supersensitive to osmotic stress at 23°C (data not shown). On the other hand, two “pseudo-wild-type” actin alleles, act1-116 and act1-117, exhibited normal growth and cortical actin structures but had a defect in bipolar bud site selection (Yang et al., 1997). The reciprocal phenotypes of these mutants suggested that a specific actin structure might mediate bipolar bud site selection. This postulated structure (or substructure) might also contain bipolar bud site determinants since these proteins are known to interact with actin.
The work by Yang et al. (1997) together with our analysis of bni1Δ and bni1-CTΔ1 prompted us to test the hypothesis that similar actin structure(s) required for bipolar budding are required for spindle orientation. We first examined spindle orientation in the sla1ΔSH3#3 strain. As shown in Fig. 9, c and d, spindle orientation in sla1ΔSH3#3 cells was indistinguishable from that of the isogenic wild-type strain. This demonstrates that alterations in the actin cytoskeleton per se do not affect spindle orientation and supports the idea that a specific actin structure may be required for both spindle orientation and bipolar bud site selection.
We next examined spindle orientation in the act1-116 strain. For our analysis we chose act1-116 of the two pseudo-wild-type actin alleles previously studied because it has the most noticeable effect on bipolar bud site selection (Yang et al., 1997). The mean angle for spindle orientation in the act1-116 strain was significantly greater than that for the isogenic wild-type strain (25.8° and 19.1° respectively, t test, P = 0.0002, n = 360). The percentage of cells with obviously misoriented spindles was also greater in the act1-116 strain (10.4% with spindle orientation between 60° and 90° compared with 3.4% for the wild-type strain). Although the spindle orientation defect in act1-116 was small, it was consistent with the magnitude of the defect in bipolar bud site selection pattern (35% of act1-116/ act1-116 diploid cells had a random budding pattern; data not shown). The fact that a mutant strain with no detectable cortical actin defect (act1-116) disrupts spindle orientation and bipolar bud site selection, whereas another mutant strain (sla1ΔSH3#3) with abnormal cortical actin does not, supports the idea that spindle orientation and bipolar bud site selection may be mediated by a similar actin-containing structure.
Discussion
The Role of Bni1p in Spindle Orientation
The formins are important for cytokinesis and aspects of development in many organisms (Frazier and Field, 1997). On the molecular level, formins are thought to be regulators of the actin cytoskeleton, but their exact role in actin function is not defined. Here we report that the yeast formin, Bni1p, is required for orientation of preanaphase mitotic spindles in S. cerevisiae. Our analysis of the spindle positioning defect in bni1Δ cells suggests that Bni1p is required to assemble or transport cortical microtubule-binding sites into the bud.
The idea that Bni1p has a role in spindle orientation was prompted by genetic interactions we observed between bni1Δ and mutations in genes encoding components of the microtubule cytoskeleton. The localization of Bni1p to the bud cortex early in mitosis and to the septum late in mitosis (Jansen et al., 1996; Evangelista et al., 1997; Fujiwara et al., 1998) (Evangelista, M., and C. Boone, unpublished observation) suggested that Bni1p would most likely regulate cytoplasmic microtubules. Indeed, we observed that in bni1Δ cells the orientation of preanaphase spindles was random with respect to the mother–bud axis.
Our real-time analysis demonstrated two striking phenotypes in bni1Δ cells. First, Bni1p was required for the initial directed movement of the SPB toward the incipient bud site. Unlike wild-type cells, where the SPB makes a concerted (but not absolutely direct) movement toward the incipient bud site, the SPB in bni1Δ cells spent almost as much time moving away from the incipient bud site as toward it. Consistent with the idea that a partially redundant pathway is able to compensate for the loss of BNI1 (in some cells better than others), this defect was observed in four out of 10 cells. This phenotype is very similar to that previously observed in kip3Δ cells (DeZwaan et al., 1997), and parallels the common spindle orientation defect observed in fixed populations of bni1Δ and kip3Δ cells.
The polarity of the cell is established before bud emergence. Bni1p and other proteins are localized to the incipient bud before bud emergence (Chant, 1996; Ayscough et al., 1997). Initial studies of fixed populations of cells detected interactions between cytoplasmic microtubules and the incipient bud site in unbudded cells (Snyder et al., 1991). These observations suggested that the microtubule cytoskeleton must be able to sense and respond to polarity cues even before the bud emerges. This study and previous fluorescence time-lapse microscopy studies of wild-type cells demonstrate that the SPB does move toward the bud site before bud emergence (DeZwaan et al., 1997). The finding that Bni1p was required for this initial movement of the SPB suggests that Bni1p is either directly involved in the physical interactions between cytoplasmic microtubules and the cortex, or that it regulates these interactions.
It is important to note that, although there was a profound delay in movement of the SPB in bni1Δ cells toward the incipient bud, the SPB eventually assumed a position close to the bud before SPB separation. Therefore it is possible that other proteins can also mediate the cortical interaction, but that they assemble at cortical sites later than Bni1p. Alternatively, there may be a Bni1p-independent pathway for spindle orientation. Because of the genetic evidence of functional overlap between BNI1 and DYN1, cytoplasmic dynein is one candidate to mediate an alternate mechanism for spindle orientation.
A second striking phenotype that was unique to bni1Δ cells was the abnormal rotations of the preanaphase spindle and transiting of these spindles between the mother and the bud. The transiting motions of the spindle were correlated with shortening of the cytoplasmic microtubules that maintain contact with the cell cortex. We interpret these movements to be the result of pulling forces between the cortex and the spindle, because the shortening microtubule consistently maintained contact with the cell cortex. These spindle movements did not appear to result from pushing forces, because we did not observe consistent contact between the cortex and cytoplasmic microtubules from the distal SPB.
What was most remarkable about the bni1Δ cells was that these apparent pulling movements were observed with approximately equal frequency toward the mother cell body as toward the bud. Apparent pulling interactions between cytoplasmic microtubules and the bud cortex were observed in wild-type cells during spindle orientation. However, apparent pulling movements of the spindle into the mother cell were not observed in our wild-type samples.
The simplest explanation for the high frequency of transiting of preanaphase spindles in bni1Δ cells is that Bni1p is required to concentrate microtubule-binding sites to the bud cortex while spindle orientation occurs. In the absence of Bni1p, these microtubule-binding sites would no longer be asymmetrically localized and would be distributed evenly between the mother and the bud. Bni1p could participate in the assembly or transport of microtubule-binding sites into the bud. Other models are also possible. For examples, cortical attachment sites might be evenly distributed throughout the cell, but Bni1p might specifically alter those in the bud, therefore biasing pulling movements toward the bud. Interestingly, we have found that the second formin in yeast, Bnr1p, is not required for spindle orientation (Lee, L., and D. Pellman, unpublished results). This is consistent with Bnr1p's specific localization to the septal regions and not the bud cortex (Kamei et al., 1998).
The Genetic Pathways Controlling Spindle Orientation
Two major pathways for spindle positioning have been identified. As discussed above, the kinesin Kip3p is required for the initial migration of the SPB toward the incipient bud site before SPB duplication (DeZwaan et al., 1997). Cytoplasmic dynein is required for insertion of the preanaphase spindle into the bud neck, but not for the initial alignment of the spindle with the axis of division (Yeh et al., 1995; DeZwaan et al., 1997). These pathways are at least partially overlapping, because cells lacking both KIP3 and DYN1 are inviable (Cottingham and Hoyt, 1997; DeZwaan et al., 1997; Miller et al., 1998). The pattern of genetic interactions that we observed between bni1Δ and null alleles of different motor genes was remarkably similar to the pattern of genetic interactions previously observed with kip3Δ. These findings together with the fact that bni1Δ kip3Δ double mutant cells were viable and did not have an exacerbated defect in nuclear segregation relative to the single mutant cells, place KIP3 and BNI1 in the same genetic pathway.
There are several molecular models that can explain the relationship between BNI1 and KIP3. One possibility is that Kip3p is a minus end–directed motor, and that the pulling of the spindle toward the bud is mediated by Kip3p tethered to Bni1p. Arguing against this idea is the fact that Kip3p is not detected at the cell cortex (DeZwaan et al., 1997; Miller et al., 1998). Also, Kip3p and Bni1p do not coimmunoprecipitate under a variety of conditions, and the localization of Kip3p is not altered in a bni1Δ mutant (Ellingson, E., L. Lee, and D. Pellman, unpublished results). However, it remains possible that there is a physical interaction that is transient or of low affinity.
Another explanation for the similar phenotypes and genetic interactions observed with bni1Δ and kip3Δ strains is that Kip3p may promote microtubule dynamics that are required for cortical microtubule capture. Dynamic microtubules could be required to increase the number of encounters between the ends of the cytoplasmic microtubules and the cortex, thereby increasing the likelihood that the microtubules will find a binding site. Finally, Kip3p might mediate microtubule depolymerization after the cytoplasmic microtubule is tethered to the bud cortex through interactions with another protein. Microtubule depolymerization in this manner would generate a pulling force on the spindle. Consistent with the idea that Kip3p has an important role in controlling microtubule dynamics, kip3Δ cells have longer cytoplasmic microtubules and are resistant to microtubule-depolymerizing agents (Cottingham and Hoyt, 1997; DeZwaan et al., 1997). Interestingly, we have noted that bni1Δ cells also have longer cytoplasmic microtubules than control cells, but we have not characterized microtubule dynamics in bni1Δ cells in detail (Lee, L., and D. Pellman, unpublished observation).
Bipolar Bud Site Determinants, Actin, and Cortical Microtubule Capture
Several lines of evidence suggest that other bipolar budding determinants in addition to Bni1p may be involved in spindle orientation. First, several genes required for bipolar budding appear to define a functional group based on similar mutant phenotypes, similar genetic interactions, and similar localization of the encoded proteins (Snyder, 1989; Valtz and Herskowitz, 1996; Zahner et al., 1996; Amberg et al., 1997; Winsor and Schiebel, 1997). Furthermore, there is evidence for physical interactions among at least four of these proteins. Bni1p and Spa2p interact by two-hybrid analysis and in vitro, and the localization of Bni1p is dependent upon Spa2p function (Fujiwara et al., 1998). There is also evidence for physical interactions between Spa2p, Pea2p, and Bud6p (Sheu et al., 1998). Second, we found that bud6Δ cells exhibit a defect in spindle orientation. Third, cells expressing Bni1-CTΔ1p, which lack the Bud6p-interacting domain of Bni1p, are defective in bipolar budding and spindle orientation but not Bni1p mating functions. Although Bud6p is necessary for normal spindle positioning, our results demonstrate that it is not as important as Bni1p. This suggests that the biochemical mechanism for cortical microtubule capture may be complex, perhaps involving several partially redundant factors.
The actin cytoskeleton is also required for both spindle orientation and bipolar budding (Palmer et al., 1992; Yang et al., 1997). We have extended previous studies by demonstrating that spindle orientation in normal logarithmically growing cells becomes random within 5 min of depolymerizing cellular actin with Lat-A. The rapid effect of Lat-A on spindle orientation argues that the role of actin may be direct.
Our findings are also consistent with the idea that actin structures postulated to be required for bipolar budding (Yang et al., 1997), are also required for spindle orientation. The supposition that a specific actin structure is required for both bipolar budding and spindle orientation is based on several lines of evidence. First, a mutation that results in abnormal cortical actin (sla1ΔSH3#3) does not affect bipolar budding or spindle orientation, demonstrating that alterations in the actin cytoskeleton per se do not perturb these two processes. Second, an act1-116 allele that does not alter cortical actin or cell growth is defective for both bipolar budding and spindle orientation. Third, both bni1Δ and bni1-CTΔ1 cells have normal appearing actin structures (Lee, L., and D. Pellman, unpublished data) but are defective for spindle orientation and bipolar bud site selection. Together, these findings are consistent with a model where Bni1p is a component of an actin complex that mediates bipolar bud site selection and spindle orientation.
Although our results suggest that Bni1p is necessary to concentrate microtubule/cortical interaction sites into the bud, the molecular nature of this postulated microtubule-binding site remains to be defined. As discussed above, one possibility is that the interaction is mediated by transient association of a microtubule motor such as Kip3p with the cortex. Another possibility is that cytoplasmic microtubules are captured by microtubule-binding proteins located at cortical sites. One candidate cortical protein that might have microtubule-binding activity is Kar9p (Miller and Rose, 1998). The kar9Δ phenotype partially overlaps with that of bni1Δ. Furthermore, Kar9p is mislocalized in bni1Δ cells, although not obviously to cortical sites in the mother cell (see accompanying paper by Miller et al., 1999). Another candidate for a cortical interaction site is coronin (Heil-Chapdelaine et al., 1998; Goode et al., 1999). Coronin colocalizes with actin patches in vivo and, interestingly, binds to both microtubules and actin in vitro. Since most of the proteins required for spindle orientation are not essential, it seems likely that there may be several cortical proteins that interact with microtubules.
Whatever the nature of the cortical microtubule-binding sites, it is likely that their activity is regulated during the cell cycle. One plausible mechanism to achieve this would be through Rho-type GTPases. Because Bni1p links Rho-type GTPases to the actin cytoskeleton (Evangelista et al., 1997; Imamura et al., 1997; Fujiwara et al., 1998), it is interesting to note that genetic evidence suggests a role for Rho2p and the GDP/GTP exchange factor Rom2p in microtubule function (Manning et al., 1997). Whether the interaction between Bni1p and the activated forms of GTPases is required to maintain spindle orientation remains to be determined.
The results described here define an important role for an S. cerevisiae formin, Bni1p, in regulating spindle position. Whether formins in other organisms have similar functions is unknown. However, we note that mutations in the genes encoding the Drosophila formins, cappuccino and diaphanous, result in abnormal microtubule structures (Emmons et al., 1995; Giansanti et al., 1998). cappuccino mutant oocytes contain long and thick microtubule bundles around the cell cortex. During male meiosis in diaphanous mutants the structure of both the actin contractile ring and the central spindle is disrupted. These findings raise the possibility that formins have a conserved role in microtubule function.
Acknowledgments
The authors wish to thank K. Salisbury and S. Randall of Improvision (Coventry, UK) for assistance with the imaging system and software, K. Ayscough (University of California, Berkeley, CA), K. Bloom (University of North Carolina, Chapel Hill, NC), S. Brown, D. Drubin, M.A. Hoyt, J. Kahana (Dana-Farber Cancer Institute/Harvard Medical School, Boston, MA), R. Miller, A. Murray, P. Silver, and A. Straight (Harvard Medical School) for plasmids and strains, A. Datta (Ludwig Institute for Cancer Research, La Jolla, CA) for help with cloning by PCR, N. Lee (Massachusetts Institute of Technology, Lexington, MA) for mathematical advice, K. Bloom, D. Drubin, D. Lew (Duke University, Durham, NC), R. Miller (Princeton University, Princeton, NJ), and M. Rose (Princeton University, Princeton, NJ) for helpful discussion, M. Christman (University of Virginia, Charlottesville, VA), R. Li (Harvard Medical School), M. McLaughlin (Brigham and Women's Hospital, Harvard Medical School), D. Roof (University of Pennsylvania, Philadelphia, PA), S. Schuyler (Harvard Medical School), and J. Tirnauer (Dana-Farber Cancer Institute) for comments on the manuscript, and members of the Pellman lab for stimulating discussions.
This work was supported by the National Institutes of Health (GM55772) and a Kimmel Scholar Award to D. Pellman, grants from the Natural Sciences and Engineering Research Council of Canada and the National Cancer Institute of Canada to C. Boone, and a Natural Sciences and Engineering Research Council of Canada postgraduate fellowship to M. Evangelista.
Abbreviations used in this paper: CEN
centromere-based
- DAPI
4′,6′-diamidino-2-phenylindole
- DIC
differential interference contrast
- GFP
green fluorescent protein
- Lat-A
Latrunculin-A
- SPB
spindle pole body
References
- Amberg DC, Zahner JE, Mulholland JW, Pringle JR, Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor Latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Pringle JR. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. . Mol Cell Biol. 1991;11:1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Pringle JR. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. . Mol Cell Biol. 1996;10:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone C, Clark KL, Sprague GFJ. Identification of a tRNAGln ochre suppressor in Saccharomyces cerevisiae. . Nucleic Acids Res. 1992;20:4661. doi: 10.1093/nar/20.17.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns N, Grimwade B, Ross-Macdonald PB, Choi E-Y, Finberg K, Roeder GS, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. . Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- Cao L-G, Wang Y-L. Signals from the spindle midzone are required for the stimulation of cytokinesis in cultured epithelial cells. Mol Biol Cell. 1996;7:225–232. doi: 10.1091/mbc.7.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillon DH, Wasserman SA. diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformitygene. Development (Camb) 1994;120:3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J. Generation of cell polarity in yeast. Curr Opin Cell Biol. 1996;8:557–565. doi: 10.1016/s0955-0674(96)80035-4. [DOI] [PubMed] [Google Scholar]
- Chant J, Pringle JR. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. . J Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiaeis accomplished by antagonistically acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrckova F, De Virgilio C, Manser E, Pringle JR, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin-related KIP3 of Saccharomyces cerevisiaeis required for a distinct step in nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzo A, Peterson AC, Freimer NB. Amplification with arbitrary primers. Methods Mol Biol. 1996;54:123–129. doi: 10.1385/0-89603-313-9:123. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Jones HD, Wertman KF. Actin structure and function: roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol Biol Cell. 1993;4:1277–1294. doi: 10.1091/mbc.4.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Emmons S, Phan H, Calley J, Chen W, James B, Manseau L. cappuccino, a Drosophila maternal effect gene required for polarity of the egg and embryo, is related to the vertebrate limb deformitylocus. Genes Dev. 1995;9:2482–2494. doi: 10.1101/gad.9.20.2482. [DOI] [PubMed] [Google Scholar]
- Euteneuer U, Schliwa M. Evidence for an involvement of actin in the positioning and motility of centrosomes. J Cell Biol. 1985;101:96–103. doi: 10.1083/jcb.101.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Farkasovsky I, Küntzel H. Yeast Num1p associates with the mother cell cortex during S/G2 phase and affects microtubular functions. J Cell Biol. 1995;131:1003–1014. doi: 10.1083/jcb.131.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier JA, Field CM. Actin cytoskeleton: are FH proteins local organizers? . Curr Biol. 1997;7:R414–R417. doi: 10.1016/s0960-9822(06)00205-3. [DOI] [PubMed] [Google Scholar]
- Freifelder D. Bud formation in Saccharomyces cerevisiae. . J Bacteriol. 1960;80:567–568. doi: 10.1128/jb.80.4.567-568.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Tanaka K, Mino A, Kikyo M, Takahashi K, Shimizu K, Takai Y. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. . Mol Biol Cell. 1998;9:1221–1233. doi: 10.1091/mbc.9.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti MG, Bonaccorsi S, Williams B, Williams EV, Santolamazza C, Goldberg ML, Gatti M. Cooperative interactions between the central spindle and the contractile ring during Drosophilacytokinesis. Genes Dev. 1998;12:396–410. doi: 10.1101/gad.12.3.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Wong JJ, Butty A, Peter M, McCormack AL, Yates JR, Drubin DG, Barnes G. Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J Cell Biol. 1999;144:83–98. doi: 10.1083/jcb.144.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SD, Hamer L, Sharpless KE, Hamer JE. The Aspergillus nidulans sepAgene encodes an FH1/2 protein involved in cytokinesis and the maintenance of cellular polarity. EMBO (Eur Mol Biol Organ) J. 1997;16:3474–3483. doi: 10.1093/emboj/16.12.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil-Chapdelaine RA, Tran NK, Cooper JA. The role of Saccharomyces cerevisiaecoronin in the actin and microtubule cytoskeletons. Curr Biol. 1998;8:1281–1284. doi: 10.1016/s0960-9822(07)00539-8. [DOI] [PubMed] [Google Scholar]
- Hyman AA. Centrosome movement in the early divisions of Caenorhabditis elegans: a cortical site determining centrosome position. J Cell Biol. 1989;109:1185–1193. doi: 10.1083/jcb.109.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, White JG. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. . J Cell Biol. 1987;105:2123–2135. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Stearns T. Spindle positioning and cell polarity. Curr Biol. 1992;2:469–471. doi: 10.1016/0960-9822(92)90660-3. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Karsenti E. Morphogenic properties of microtubules and mitotic spindle assembly. Cell. 1996;84:401–410. doi: 10.1016/s0092-8674(00)81285-4. [DOI] [PubMed] [Google Scholar]
- Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R-P, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HOexpression in budding yeast depends on the unconventional myosin Myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- Juang Y-L, Huang J, Peters J-M, McLaughlin ME, Tai C-Y, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- Kahana, J.A., and P.A. Silver. 1996. Use of the A. victoria green fluorescent protein to study dynamics in vivo. In Current Protocols in Molecular Biology. F.M. Ausubel, R. Brent, R.E. Kingston, D.E. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, editors. John Wiley and Sons, New York, NY. 9.6.13–9.6.19.
- Kahana JA, Schnapp BJ, Silver PA. Kinetics of spindle pole body separation in budding yeast. Proc Natl Acad Sci USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei T, Tanaka K, Hihara T, Umikawa M, Imamura H, Kikyo M, Ozaki K, Takai Y. Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. J Biol Chem. 1998;273:28341–28345. doi: 10.1074/jbc.273.43.28341. [DOI] [PubMed] [Google Scholar]
- Kaverina I, Rottner K, Small JV. Targeting, capture, and stabilization of microtubules at early focal adhesions. J Cell Biol. 1998;142:181–190. doi: 10.1083/jcb.142.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno H, Tanaka K, Mino A, Umikawa M, Imamura H, Fujiwara T, Fujita Y, Hotta K, Qadota H, Watanabe T, Ohya Y, Takai Y. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1996;15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Kraut R, Chia W, Jan LY, Knoblich JA. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. . Nature. 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- Manning BD, Padmanabha R, Snyder M. The Rho-GEF Rom2p localizes to sites of polarized cell growth and participates in cytoskeletal functions in Saccharomyces cerevisiae. . Mol Biol Cell. 1997;10:1829–1844. doi: 10.1091/mbc.8.10.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R.K., D. Matheos, and M.D. Rose. 1999. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J. Cell Biol. 963–975. [DOI] [PMC free article] [PubMed]
- Miller RK, Rose MD. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol. 1998;140:377–390. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Heller KK, Frisèn L, Wallack DL, Loayza D, Gammie AE, Rose MD. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol Biol Cell. 1998;9:2051–2068. doi: 10.1091/mbc.9.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch H-U, Fink GR. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. . Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RE, Koval M, Koshland D. The dynamics of chromosomal movement in the budding yeast Saccharomyces cerevisiae. . J Cell Biol. 1989;109:3355–3366. doi: 10.1083/jcb.109.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellman D, Bagget M, Tu H, Fink GR. Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. . J Cell Biol. 1995;130:1373–1385. doi: 10.1083/jcb.130.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Weilguny D, Egel R, Nielsen O. Characterization of fus1 of Schizosaccharomyces pombe: a developmentally controlled function needed for conjugation. Mol Cell Biol. 1995;15:3697–3707. doi: 10.1128/mcb.15.7.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AEM, Stearns T, Drubin DG, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Rhyu MS, Knoblich JA. Spindle orientation and asymmetric cell fate. Cell. 1995;82:523–526. doi: 10.1016/0092-8674(95)90022-5. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., F. Winston, and P. Hieter. 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 198 pp.
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Maddox P, Skibbens RV, Yeh E, Salmon ED, Bloom K. Nuclear and spindle dynamics in budding yeast. Mol Biol Cell. 1998;9:1627–1631. doi: 10.1091/mbc.9.7.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu Y-J, Santos B, Fortin N, Costigan C, Snyder M. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol Cell Biol. 1998;18:4053–4069. doi: 10.1128/mcb.18.7.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. The SPA2protein of yeast localizes to sites of cell growth. J Cell Biol. 1989;108:1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M, Gehrung S, Page BD. Studies concerning the temporal and genetic control of cell polarity in Saccharomyces cerevisiae. . J Cell Biol. 1991;114:515–532. doi: 10.1083/jcb.114.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague GF., Jr Assay of yeast mating reaction. Methods Enzymol. 1991;194:77–93. doi: 10.1016/0076-6879(91)94008-z. [DOI] [PubMed] [Google Scholar]
- Staiger CJ, Lloyd CW. The plant cytoskeleton. Curr Opin Cell Biol. 1991;3:33–42. doi: 10.1016/0955-0674(91)90163-s. [DOI] [PubMed] [Google Scholar]
- Stearns T. Motoring to the finish: kinesin and dynein work together to orient the yeast mitotic spindle. J Cell Biol. 1997;138:957–960. doi: 10.1083/jcb.138.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Valtz N, Herskowitz I. Pea2 protein of yeast is localized to sites of polarized growth and is required for efficient mating and bipolar budding. J Cell Biol. 1996;135:725–739. doi: 10.1083/jcb.135.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. . Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Winsor B, Schiebel E. Review: an overview of the Saccharomyces cerevisiaemicrotubule and microfilament cytoskeleton. Yeast. 1997;13:399–434. doi: 10.1002/(SICI)1097-0061(199704)13:5<399::AID-YEA126>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Woychik RP, Maas RL, Zeller R, Vogt TF, Leder P. “Formins”: proteins deduced from the alternative transcripts of the limb deformitygene. Nature. 1990;346:850–853. doi: 10.1038/346850a0. [DOI] [PubMed] [Google Scholar]
- Yang S, Ayscough KR, Drubin DG. A role for the actin cytoskeleton of Saccharomyces cerevisiaein bipolar bud-site selection. J Cell Biol. 1997;136:111–123. doi: 10.1083/jcb.136.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner JE, Harkins HA, Pringle JR. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. . Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M, Preuss D, Mulholland J, O'Brien JM, Botstein D, Johnson DI. Subcellular localization of Cdc42p, a Sacchamromyces cerevisiaeGTP-binding protein involved in the control of cell polarity. Mol Biol Cell. 1993;4:1307–1316. doi: 10.1091/mbc.4.12.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]