Abstract
To identify novel components required for cell division processes in complex eukaryotes, we have undertaken an extensive mutational analysis in the one cell stage Caenorhabditis elegans embryo. The large size and optical properties of this cell permit observation of cell division processes with great detail in live specimens by simple differential interference contrast (DIC) microscopy. We have screened an extensive collection of maternal-effect embryonic lethal mutations on chromosome III with time-lapse DIC video microscopy. Using this assay, we have identified 48 mutations in 34 loci which are required for specific cell division processes in the one cell stage embryo. We show that mutations fall into distinct phenotypic classes which correspond, among others, to the processes of pronuclear migration, rotation of centrosomes and associated pronuclei, spindle assembly, chromosome segregation, anaphase spindle positioning, and cytokinesis. We have further analyzed pronuclear migration mutants by indirect immunofluorescence microscopy using antibodies against tubulin and ZYG-9, a centrosomal marker. This analysis revealed that two pronuclear migration loci are required for generating normal microtubule arrays and four for centrosome separation. All 34 loci have been mapped by deficiencies to distinct regions of chromosome III, thus paving the way for their rapid molecular characterization. Our work contributes to establishing the one cell stage C. elegans embryo as a powerful metazoan model system for dissecting cell division processes.
Keywords: genetics, video microscopy, mitosis, centrosomes, microtubules
Proper cell division requires the faithful distribution of chromosomes and cytoplasmic material to daughter cells. In eukaryotes, this is achieved by a series of coordinated cytoskeletal processes which include spindle assembly, chromosome segregation, spindle positioning and cytokinesis. Although the mechanisms governing such processes have been investigated in a number of systems, they remain incompletely understood, and there persists a need for identifying novel participating molecules, especially in complex eukaryotes.
Mutational analysis has played an important role in achieving our current understanding of cell division processes, owing in particular to extensive genetic screens conducted in yeast and other simple eukaryotes (Hartwell et al., 1974; Morris, 1975; Nurse et al., 1976; Hoyt et al., 1990). However, some aspects of cell division are not conserved between simple and complex eukaryotes. For instance, the nuclear envelope does not break down during mitosis in yeast, in contrast to the situation in complex eukaryotes. Moreover, components required for specific cell division processes in simple eukaryotes do not necessarily govern solely the analogous process in complex eukaryotes. For example, whereas cytoplasmic dynein appears to be essential only for spindle positioning in Saccharomyces cerevisiae (Eshel et al., 1993; Li et al., 1993), it is also required for other processes in mammalian cells (reviewed in Hirokawa et al., 1998), including proper spindle assembly (Vaisberg et al., 1993; Heald et al., 1996, 1997; Gaglio et al., 1997). Such differences underscore the importance of performing a mutational analysis of cell division processes directly in a complex eukaryote.
Drosophila has proven especially valuable in this regard. Indeed, genetic and cytological analysis of meiotic and mitotic mutants has led to the identification of many loci required for aspects of cell division (Baker and Carpenter, 1972; Gatti and Baker, 1989; Castrillon et al., 1993; reviewed in Gatti and Goldberg, 1991; Fuller, 1993; Hawley et al., 1993). For instance, the minus-end–directed kinesin ncd was originally discovered as a locus required for proper chromosome segregation during female meiosis (Baker and Carpenter, 1972; Zhang et al., 1990). However, these primary screens have not always allowed the identification of the exact cell division process affected in mutant animals, because they relied on the analysis of terminal phenotypes. In general, detailed observations in live cells appear better suited for analyzing defects in cell division processes. Thus, in the syncytial Drosophila embryo, injection of fluorescent tracer molecules has greatly enhanced the resolution with which these processes can be studied (reviewed in Sullivan and Theurkauf, 1995). For instance, time-lapse observations of daughterless-abo-like mutant embryos injected with labeled tubulin and histone revealed specific defects in centrosome separation (Sullivan et al., 1990). However, although appropriate for analyzing select mutants, such techniques are probably too cumbersome to become the basis of a large scale primary screen.
The one cell stage Caenorhabditis elegans embryo offers an attractive alternative for undertaking a mutational analysis of cell division processes in a complex eukaryote for a number of reasons. First, division of the one cell stage C. elegans embryo resembles that of most other complex eukaryotic cells. The cell cycle is mitotic, not meiotic, and cytokinesis is conventional, unlike, for instance, that seen in the syncytial Drosophila embryo. Thus, homologues of genes required for cell division processes in the one cell stage C. elegans embryo will likely play similar roles in other metazoans. Second, cell division processes can be observed in real time with great detail by differential interference contrast (DIC)1 microscopy in this 50-μm-long cell (Nigon et al., 1960; Sulston et al., 1983). Thus, a simple visual screen can allow the identification of the exact cell division process affected in a given mutant strain. Importantly, a large number of strains can be analyzed by such a straightforward primary screen. Third, the early C. elegans embryo is amenable to experimental manipulations, including the use of cytoskeletal drugs and localized laser irradiations (Strome and Wood, 1983; Hyman and White, 1987; Hyman, 1989). In combination with mutant analysis, this offers a unique potential for a thorough analysis of cell division processes. Fourth, the recent completion of the genome sequence allied with the powerful genetics available in C. elegans will greatly facilitate the molecular characterization of loci identified by mutational analysis. This will be significantly aided by the advent of RNA-mediated interference (RNAi), whereby expression of a given gene can be silenced via microinjection of a corresponding fragment of double-stranded RNA (Fire et al., 1998). Thus, candidate genes can be rapidly tested by RNAi to determine whether they correspond to loci identified by mutational analysis.
Because of these advantages, we have screened an extensive collection of C. elegans maternal-effect embryonic lethal strains on chromosome III using time-lapse DIC video microscopy. Maternal-effect strains were analyzed since events in the one cell stage are thought to rely exclusively on the maternal genome (Edgar et al., 1994; Seydoux and Fire, 1994). We have identified and mapped 34 loci which are required for specific cell division processes in the one cell stage embryo, including spindle assembly, chromosome segregation, anaphase spindle positioning, and cytokinesis. By carefully analyzing mutant phenotypes in live and fixed specimens, we have gained novel mechanistic insights into several aspects of cell division in this metazoan model system.
Materials and Methods
Culture Conditions and Strains
Basic methods of C. elegans culture and handling were as previously described (Brenner, 1974; Wood, 1988). Strains carrying the following mutations were used: unc-32(e189), qC1 dpy-19(e1259ts) glp-1(q339), him-3(e1147), zyg-8(b235) (Wood et al., 1980), zyg-11(b2) (Wood et al., 1980), cyk-1 (or36) (Swan et al., 1998), mel-27(e2561) and mel-28(e2567) (Ahringer, J., personal communication), let-733(s2621), let-748(s2578) and let-771(s2442) (Stewart et al., 1998), emb-30(g53) (Cassada et al., 1981), par-3(it62) (Kemphues et al., 1988b), and emb-8(hc69) (Schierenberg et al., 1980). The following deficiencies were used: sDf121 and sDf125 (Stewart et al., 1998), sDf110 (from the Genetic Toolkit Project), nDf16, nDf20, nDf40, tDf2, tDf5, tDf6, tDf7, and tDf9. Information on these strains and deficiencies can be found in the C. elegans database Acedb which is available by anonymous FTP from ncbi.nlm.nih.gov, directory repository/ acedb or can be accessed online at the following Internet address: http://www.sanger.ac.uk/Projects/C_elegans/webace_front_end.shtml
Mutant Collection and Deficiency Mapping
Mutations were induced on an unc-32(e189) labeled third chromosome. L4 hermaphrodites of genotype unc-32(e189)/qC1 III; him-3(e1147) IV were mutagenized with 20 mM ethyl methane sulfonate according to standard procedures (Brenner, 1974). F1 progeny were placed on individual plates and several F2 Unc L4 hermaphrodites were picked to a separate plate and placed at 25°C. Strains giving rise to adult Unc hermaphrodites which produced eggs that did not hatch were kept for further analysis. 254 strains carrying parental-effect embryonic lethal mutations were thus recovered from 15,600 mutagenized genomes (Schnabel, H., T. Kaletta, and R. Schnabel, manuscript in preparation). The mutations were outcrossed twice, and a male stock of each was established. This was facilitated by the him-3 mutation which causes 3.5% male production (Hodgkin et al., 1979).
The 254 strains were subdivided into sets by deficiency mapping (see Fig. 1). Complementation analysis was performed among mutations within each set, and with loci previously mapped to each of the corresponding regions. Six strains failed to complement more than one deficiency, or members of more than one complementation group, and are thus considered to carry two mutations, although no attempt has been made to separate them. In some cases, this behavior could be due to the presence of a small deficiency, rather than a double mutation. Thus, the collection comprised 260 mutations, which fell into 125 complementation groups, 81 of which had a single allele, and 44 of which had 2 or more alleles (see Tables II and III; H. Schnabel, T. Kaletta, and R. Schnabel, manuscript in preparation).
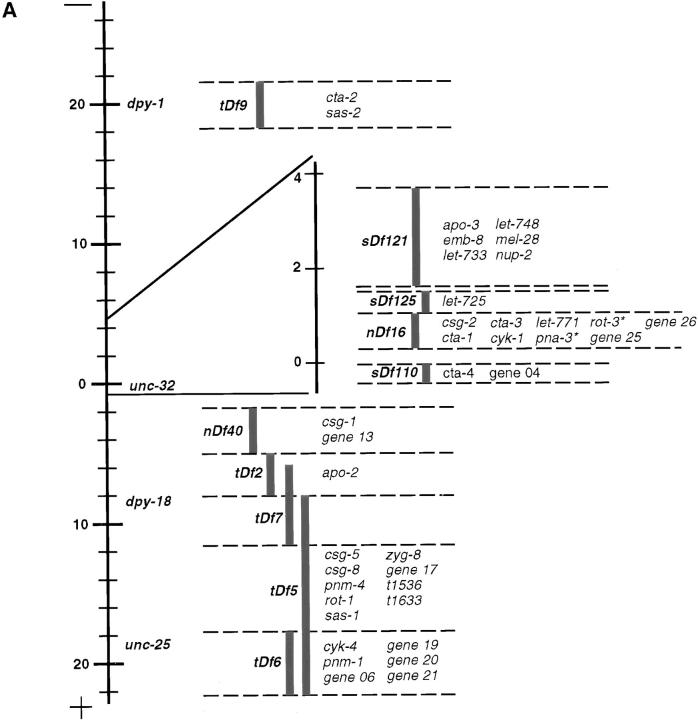
Figure 1.
Deficiency mapping. (A) Failure to complement 1 or several of 11 tested deficiencies (tDf9, sDf121, sDf125, nDf16, nDf20, sDf110, nDf40, tDf2, tDf7, tDf5, or tDf6) led to the assignment of a majority of loci to distinct intervals on chromosome III (see Materials and Methods). The positions of the visible markers dpy-1, unc-32, dpy-18, and unc-25 are indicated for reference. The gene-rich region towards the center of the chromosome III is magnified. *pna-3 and rot-3 map under both nDf16 and nDf20, which overlaps with nDf16 but is not shown on the figure. (B) Loci which complemented all 11 tested deficiencies.
Table II.
Mutations Affecting One of Six Major Cell Division Processes
| Phenotypic class | Class phenotypic description | Locus* | Alleles‡ | Alleles examined§ | Comments ¶ | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pronuclear migration | Male pronucleus remains at posterior | pnm-1 | 2 | t1550 (s), t1698 (p) | No spindle | |||||
| (pnm) (see Fig. 4) | cortex | |||||||||
| Female pronucleus usually stays in the | pnm-2 | 1 | t1445 (s) | No spindle in most embryos; | ||||||
| anterior | small spindle in some | |||||||||
| In some mutants, spindle is absent or | pnm-3 | 1 | t1458 (s) | Small spindle | ||||||
| barely detectable (“no spindle” in | ||||||||||
| comments) | pnm-4 | 1 | t1491 (nd) | No spindle | ||||||
| In some mutants, spindle is small and | ||||||||||
| sets up at the posterior cortex, transverse | ||||||||||
| to the longitudinal axis (“small spindle” | pnm-5 | 1 | t1543 (s) | Small spindle | ||||||
| in comments) | ||||||||||
| Sometimes more than one female | pnm-6 | 1 | t1586 (s) | No spindle; large areas | ||||||
| pronucleus | lacking yolk granules | |||||||||
| Rotation | Centrosomes and associated pronuclei | rot-1 | 1 | t1599 (s) | No centration | |||||
| (rot) (see Fig. 5) | fail to rotate | |||||||||
| In some mutants, there is no centration | rot-2 | 1 | t1623 (s) | No centration; female | ||||||
| and spindle sets up at 70% egg length, | pronucleus does not always | |||||||||
| transverse to the longitudinal axis (“no | migrate | |||||||||
| concentration” in comments) | ||||||||||
| In one mutant (t1640), centration takes | ||||||||||
| place and spindle sets up at 30–40% | rot-3 | 1 | t1640 (s) | |||||||
| egg length, transverse to the longitudinal | ||||||||||
| axis | ||||||||||
| Spindle usually reorients along the | ||||||||||
| longitudinal axis by the end of anaphase | ||||||||||
| Often more than one nucleus in | ||||||||||
| daughter cells | ||||||||||
| Spindle assembly | Bipolar spindle fails to assemble | sas-1 | 3 | t1476 (p), t1521 (s) | ||||||
| (sas) (see Fig. 6) | Cleavage furrow absent | sas-2 | 2 | t1539 (p), t1595 (z) | t1539 is on a double-mutant | |||||
| Many small nuclei form in resulting | chromosome; rotation often | |||||||||
| single cell | incomplete in t1595 | |||||||||
| Sometimes centrosomal-pronuclear | ||||||||||
| complex fails to center and rotate | sas-3 | 1 | t1465 (s) | No fast phase of female | ||||||
| pronuclear migration | ||||||||||
| Phenotypic class | Class phenotypic description | Locus* | Alleles‡ | Alleles examined§ | Comments ¶ | |||||
| Anaphase spindle | In some mutants, anaphase spindle | zyg-8 | 7 | t1547 (s), t1650 (s), | Allelic to zyg-8 (Wood et al., | |||||
| positioning (apo) | moves strongly towards the posterior | t1518 (s) | 1980); strong | |||||||
| (see Fig. 7) | (“strong” in comments) | |||||||||
| In some mutants, anaphase spindle | apo-1 | 2 | t1638 (s), t1441 (s) | Strong | ||||||
| drifts towards the posterior or lateral | ||||||||||
| cortex (“mild drift” in comments) | apo-2 | 1 | t1454 (p) | Mild drift | ||||||
| Sometimes centrosomes and associated | ||||||||||
| pronuclei fail to rotate, in which case | let-771 | 1 | t1554 (ns**) | Mild drift; allelic to let-771 | ||||||
| spindle sets up transverse to the | (Stewart et al., 1998) | |||||||||
| longitudinal axis, drifting anteriorly | ||||||||||
| during anaphase | apo-3 | 1 | t1613 (s) | Mild drift | ||||||
| Aberrant placement of cleavage furrow | apo-4 | 1 | t1707 (p) | Mild drift | ||||||
| in all embryos | ||||||||||
| Chromosome | Karyomeres (i.e., > 1 nucleus) | csg-1 | 6 | t1501 (s), t1519 (s) | eos; t1463 is on a double- | |||||
| segregation (csg) | in AB and/or P1 | t1463 (s) | mutant chromosome; t1501 | |||||||
| (see Fig. 8) | First division spindle usually | and t1519; sometimes | ||||||||
| indistinguishable from wild-type | multiple female pronuclei | |||||||||
| Some strains have unusual looking | csg-2 | 4 | t1637 (p), t1587 (p), | t1637: only 2/5 embryos | ||||||
| first division spindle (as indicated in | t1433 (s) | have karyomeres; t1587: | ||||||||
| comments) | weak central spindle | |||||||||
| Some embryos in some strains have | and sometimes multiple | |||||||||
| more than one female pronucleus (as | female pronuclei | |||||||||
| indicated in comments) | csg-3 | 2 | t1685 (s), t1708 (s) | eos; t1708: poorly visible | ||||||
| spindle | ||||||||||
| csg-4 | 1 | t1467 (nd‖) | Small; sometimes multiple | |||||||
| female pronuclei | ||||||||||
| csg-5 | 1 | t1545 (s) | ||||||||
| csg-6 | 1 | t1556 (s) | eos; sometimes multiple | |||||||
| female pronuclei | ||||||||||
| csg-7 | 1 | t1612 (s) | Sometimes multiple female | |||||||
| pronuclei; no | ||||||||||
| pseudocleavage furrow | ||||||||||
| csg-8 | 1 | t1714 (s) | Large areas lacking yolk | |||||||
| granules; sometimes | ||||||||||
| multiple female pronuclei; | ||||||||||
| many embryos are eos | ||||||||||
| (gene 04) | 9 | t1504 (s), t1712 (s) | t1504: eos and sometimes | |||||||
| multiple female pronuclei; | ||||||||||
| t1712: spindle bent | ||||||||||
| towards the end of anaphase | ||||||||||
| (gene 39) | 2 | t1653 (s) | Sometimes multiple female | |||||||
| (gene 06) | 8 | t1669 (nd) | pronuclei | |||||||
| (gene 31) | 2 | t1717 (s) | ||||||||
| Cytokinesis (cyk) | In some mutants, cleavage furrow is | cyk-1 | 2 | t1568 (s), t1611 (s) | Furrow regresses; allelic to | |||||
| (see Fig. 9) | not specified (as indicated in comments) | cyk-1 (Swan et al., 1998) | ||||||||
| In some mutants, cleavage furrow | ||||||||||
| regresses (as indicated in comments) | cyk-3 | 2 | t1525 (s), t1535 (s) | eos; furrow not specified | ||||||
| Daughter nuclei appose in resulting | ||||||||||
| single cell | cyk-4 | 1 | t1689ts†† (nd) | Furrow regresses | ||||||
| Additional nuclei, probably containing | (gene 17) | 2 | t1603 (p) | eos; furrow regresses; | ||||||
| non-extruded polar body material, come | spindle usually poorly | |||||||||
| from the anterior to join daughter nuclei | defined | |||||||||
| Phenotypic class | Class phenotypic description | Locus* | Alleles‡ | Alleles examined§ | Comments ¶ | |||||
| Fast phase of female | Female pronucleus does not accelerate | faf-1 | 1 | t1678 (s) | ||||||
| pronuclear | or pauses during migration | |||||||||
| migration (faf) | ||||||||||
| (gene 34) | 3 | t1502 (s) | Only one allele examined, | |||||||
| second allele is on a double | ||||||||||
| mutant chromosome with an | ||||||||||
| earlier phenotype, third one is | ||||||||||
| semisterile | ||||||||||
| Pronuclear meeting | Pronuclei tend to meet centrally, | (gene 26) | 6 | t1447 (s), t1472 (s) | ||||||
| seemingly due to premature mi- | ||||||||||
| gration of male pronucleus | ||||||||||
| Sometimes rotation of centrosomes and | ||||||||||
| associated pronuclei fails to take | ||||||||||
| place | ||||||||||
| Progression through | In some eos strains, variable processes | gene 21 | 2 | t1614ts** (s), t1674 | eos | |||||
| one cell stage | are affected in different embryos: | (p) | ||||||||
| pronuclei can be very slow to appear; | ||||||||||
| centration/rotation or pronuclear | emb-30 | 7 | t1446 (s), t1497 (s), | eos; allelic to emb-30 (Cassada | ||||||
| envelope breakdown can be very | t1600 (s) | et al., 1981) | ||||||||
| slow; the cleavage furrow is | ||||||||||
| sometimes not specified or regresses | 1 | t1536 (s) | eos | |||||||
| Sometimes many embryos appear | 1 | t1633 (s) | eos | |||||||
| unfertilized | ||||||||||
| (gene 28) | 4 | t1461 (s) | eos | |||||||
| Other unique | For sake of compactness, strains with | gene 13 | 2 | t1642 (s), t1676 (s) | Events from pronuclear | |||||
| phenotypes | phenotypes not displayed by mutations at any other locus | migration onwards until spindle set up are slow; |
||||||||
| are grouped here | division of P1 delayed with | |||||||||
| The specific phenotypes are given | respect to that of AB | |||||||||
| under comments | ||||||||||
| gene 20 | 2 | t1644 (s), t1664 (s) | Complex phenotype: slow to | |||||||
| form pronuclei, multiple | ||||||||||
| pronuclei, absence of | ||||||||||
| centration/rotation; karyomeres | ||||||||||
| in daughter blastomeres; | ||||||||||
| reminiscent of phenotypes | ||||||||||
| observed in zyg-11 mutant | ||||||||||
| embryos (Kemphues et al., | ||||||||||
| 1986) | ||||||||||
| par-3 | 3 | t1591 (s), t1688 (s) | Symmetric first division; allelic | |||||||
| to par-3 (Kemphues et al., | ||||||||||
| 1988) | ||||||||||
| (emb-8) | 6 | t1462 (s) | eos; poorly visible spindle, but | |||||||
| normal daughter nuclei; allelic | ||||||||||
| to emb-8 (Miwa et al., 1980) | ||||||||||
| (gene 35) | 2 | t1470 (s) | No male pronucleus; only one | |||||||
| allele could be examined, | ||||||||||
| because the other is semisterile | ||||||||||
| 1 | t1648 (p) | No or very small female | ||||||||
| pronucleus |
Loci are named according to phenotypic classes. Multiple allele loci which display allele-specific phenotypes are not given name designations. Instead, they are referred to by the gene number utilized during generation of the mutant collection.
The total number of alleles in the entire chromosome III collection is given for each locus.
s, strict; p, nonstrict, paternal rescue; z, nonstrict, zygotic rescue (see Materials and Methods); nd, strict test and paternal test not conducted.
Strains that gave rise to small or eos embryos are indicated. Indicated are also additional phenotypes that are apparent in some strains.
Leaky: homozygous mutant hermaphrodites produce some live progeny.
Nonstrict, paternal test not conducted.
Homozygous mutant hermaphrodites produce live progeny at 15°C. Refer to Materials and Methods for additional information about strain classification.
Table III.
Mutations Affecting Other Processes
| Phenotypic class | Class phenotypic description | Locus* | Alleles‡ | Alleles examined§ | Comments ¶ | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cytoplasmic | Abnormal distribution of yolk granules | let-725 | 3 | t1440 (nd), t1661 | Sparse yolk granules | |||||
| appearance (cta) | (as specified in comments) | (nd) | throughout; allelic to let-725 | |||||||
| (Stewart et al., 1998) and mel-27 | ||||||||||
| (J. Ahringer, personal | ||||||||||
| communication) | ||||||||||
| cta-1 | 4 | t1618 (s), t1704 (nd), | Large areas lacking yolk | |||||||
| t1630 (ns‖) | granules | |||||||||
| cta-2 | 3 | t1701 (s), t1710 (nd) | Large areas lacking yolk granules | |||||||
| let-748 | 1 | t1452 (s) | Large areas lacking yolk granules; | |||||||
| division of P1 delayed with | ||||||||||
| respect to AB; allelic to let-748 | ||||||||||
| (Stewart et al., 1998) | ||||||||||
| cta-3 | 1 | t1492 (p) | eos; large areas lacking yolk | |||||||
| granules | ||||||||||
| cta-4 | 1 | t1562 (s) | Large areas lacking yolk granules, | |||||||
| especially around asters | ||||||||||
| cta-5 | 1 | t1627 (p) | Small; sparse yolk granules | |||||||
| throughout | ||||||||||
| (gene 25) | 4 | t1439 (p) | Some aggregated yolk granules | |||||||
| (gene 04) | 9 | t1517 (s) | Sparse yolk granules | |||||||
| throughout; more than one | ||||||||||
| female pronucleus | ||||||||||
| (gene 27) | 7 | t1575 (ns‖) | Sparse yolk granules | |||||||
| throughout | ||||||||||
| Pronuclear | No or poorly visible pronuclei, as well | mel-28 | 10 | t1579 (s), t1578 (s), | Allelic to mel-28 (J. Ahringer, | |||||
| appearance (pna) | as AB and P1 nuclei | t1589 (s) | personal communication) | |||||||
| Usually poorly visible spindle | ||||||||||
| Division of P1 delayed with respect to | pna-1 | 2 | t1639 (s), t1499 (s) | |||||||
| that of AB | pna-2 | 1 | t1434 (s) | |||||||
| pna-3 | 1 | t1598ts** (s) | ||||||||
| Pronuclear/nuclear | Unusual location of either male | nup-1 | 1 | t1482 (nd††) | Central male pronucleus | |||||
| position (nup) | pronucleus, female pronucleus, or | |||||||||
| AB and P1 nuclei (as specified in | nup-2 | 1 | t1574 (s) | Central/posterior female | ||||||
| comments) | pronucleus | |||||||||
| let-733 | 1 | t1683 (s) | AB and P1 nuclei at central | |||||||
| cortex; allelic to let-733 | ||||||||||
| Stewart et al., 1998) | ||||||||||
| (gene 06) | 8 | t1580 (s) | Central/posterior female | |||||||
| pronucleus | ||||||||||
| (gene 19) | 2 | t1675 (nd) | Central male pronucleus |
Loci are named according to phenotypic classes. Multiple allele loci which display allele-specific phenotypes are not given name designations. Instead, the gene numbers utilized during generation of the mutant collection are given in parentheses. In addition, strains that fell either in the “progression through the one cell stage” or “unique phenotypes” class were not given new name designations, as further analysis seems required to identify the primary defect in many of these strains.
The total number of alleles in the entire chromosome III collection is given for each locus.
s, strict; p, nonstrict, paternal rescue; z, nonstrict, zygotic rescue (see Materials and Methods); nd, strict test and paternal test not conducted.
Strains that gave rise to small or eos embryos are indicated. Indicated are also additional phenotypes that are apparent in some strains.
Nonstrict, paternal test not conducted.
Homozygous mutant hermaphrodites produce live progeny at 15°C.
Leaky: homozygous mutant hermaphrodites produce some live progeny. Refer to Materials and Methods for additional information about strain classification.
Strict and Paternal Tests
The “strict” test was conducted to determine whether there is a strict requirement for maternal contribution, or, alternatively, whether a wild-type paternal allele is sufficient to rescue lethality. Five homozygous mutant hermaphrodites (mut unc-32/mut unc-32) were crossed with plg-1 males at 25°C. After mating with plg-1 males, hermaphrodites have a visible plug in the vulva, indicating that mating took place (Barker, 1994). As a control, five homozygous mutant hermaphrodites were allowed to self fertilize without males. If only nonviable eggs were produced in the cross and at least some animals showed a mating plug, the mutation was called strict (see “s” in Tables II and III). In contrast, if live cross-progeny was produced, the mutation was deemed nonstrict.
The “paternal” test was conducted on nonstrict strains to determine whether zygotic expression or, alternatively, sperm cytoplasm can rescue lethality. Five homozygous mutant hermaphrodites (mut unc-32/mut unc-32) were crossed with heterozygous animals from the same strain (mut unc-32/qC1). If only non-Unc progeny (mut unc-32/qC1) were produced, then homozygous mutant progeny (mut unc-32/mut unc-32) must have died, and zygotic expression is needed to rescue lethality. Such strains are marked “z” for zygotic rescue (see Tables II and III). In contrast, if both non-Unc progeny (mut unc-32/qC1) and Unc progeny were produced, then the rescue is independent of the genotype of the embryo, and sperm cytoplasm rescues lethality. Such strains are marked “p” for paternal rescue (see Tables II and III).
Number of Strains Analyzed
We attempted to analyze each single-allele locus of the collection, two independent alleles for loci represented by two or three alleles, and three independent alleles for loci represented by four or more alleles. However, 13 strains representing mutations in 10 loci could not be analyzed for the following reasons. First, in two strains, including a double-mutant chromosome, adult Unc hermaphrodites could no longer be found in the population at the time of analysis, even though they were present when the strains were initially derived. Second, in seven strains, including a double-mutant chromosome, homozygous mutant hermaphrodites gave rise to no or very little progeny, precluding analysis of events in one cell stage embryos. Third, for two double-mutant chromosomes, the phenotype was identical to that observed with single-mutant alleles at one of the two loci and may have precluded scoring the phenotype of the mutation at the other locus. Fourth, for one double-mutant chromosome, other single-mutant alleles at both loci could not be examined, making it impossible to ascribe the phenotype observed in the double-mutant combination to either locus with certainty. Fifth, we found during preparation of the manuscript that one strain carried two mutations; although this double-mutant chromosome had a pronuclear meeting phenotype (see Table III), it is not included in this study because no attempt has been made as of yet to determine which of the two mutations is responsible for the phenotype. In addition, 10 strains representing 10 loci were not included in the data set because they gave rise to unfertilized embryos (3 strains) or were extremely slow to progress through the one cell stage (7 strains).
All together the phenotypes in one cell stage embryos were analyzed for 160 mutations in 106 loci. 2 of the 160 mutations (t1463, t1539) were on double-mutant chromosomes (as indicated in Tables II and III).
Sample Preparation and Recordings
Worms were grown at 16°C, picked as L4s, and shifted to 25°C for a minimum of 12 h. Gravid hermaphrodites were then dissected in M9 medium (Brenner, 1974). Their embryos were collected with a mouth pipette and mounted on a 2% agarose pad. A coverslip was placed onto the pad and sealed with melted Vaseline (Sulston et al., 1983).
Embryos were analyzed at 25°C ± 1°C with a 100× (1.25 NA) Achrostigmat objective lens and standard DIC optics on a Zeiss Axioskop microscope. Under these conditions, pronuclei, asters, spindles, and nuclei can be observed in living C. elegans embryos. Early embryonic events were recorded with time-lapse video microscopy (one image every 5 s), typically from the time of pronuclear formation to the four cell stage, using a monochrome CCD camera (Hamamatsu C5405 or Sony CCD-IRIS).
In Utero Observations
In a few strains, the embryos lost structural integrity upon dissection or mounting on the agarose pad. These strains were dubbed egg-osmotic sensitive (eos). For eos strains, gravid hermaphrodites were anaesthetized for 10 min in M9 medium containing 0.1% tricaine and 0.01% tetramisole, and mounted intact on an agarose pad to observe embryogenesis in utero (Kirby et al., 1990). Although early events could usually be scored with confidence in this manner, the resolution achieved by viewing through the body wall does not equal that achieved by observing dissected embryos. Therefore, subtle phenotypic manifestations may have been missed in eos strains.
Phenotypic Classes
One cell stage embryos were scored for the following: number and position of female and male pronuclei, migration of pronuclei, position of pronuclear meeting, centration and rotation of centrosomes and associated pronuclei, spindle assembly, anaphase spindle positioning, and cytokinesis. The number and position of daughter nuclei in AB and P1 were also determined. Moreover, the size of the embryo, the appearance of its cytoplasm, and the general timing of events were noted.
Five or more embryos were analyzed for each strain, and a given phenotype was ascribed to the allele if it was observed in at least three out of the first five embryos analyzed. The observed phenotypes fell into different classes; mutations that affect one of the six cell division processes which are the focus of this report are found in Table II, mutations that affect other processes are in Table III.
The vast majority of strains (157/160) fell into only one phenotypic class. However, four strains (t1586, t1465, t1517, and t1714) displayed characteristics of two phenotypic classes. These strains have been classified according to their phenotype in one of the six cell division processes (see Table II), with a mention of the other phenotypic characteristic in the comments column.
Loci were named according to phenotypic classes (see abbreviations in Tables II and III). Multiple allele loci which displayed allele-specific phenotypes were not given name designations. Instead, they are referred to by gene numbers used during generation of the mutant collection. In addition, strains that fell in the “progression through the one cell stage” or “unique phenotypes” classes were not given name designations, as further analysis is required to identify the primary defect in most of these cases.
Other Deviations from Wild-Type
Some embryos were significantly smaller than wild-type. Being small, like being eos, was not considered as a phenotypic class per se; both traits are nevertheless indicated for the relevant strains in Tables II and III.
Aspects that were sometimes variable in wild-type and often variable in many mutant strains were not included in the phenotypic classification. These aspects include the position and the strength of the pseudocleavage furrow, the completeness of the rotation of the centrosomes and associated pronuclei, the extent of rocking of the posterior spindle pole during anaphase displacement, or the extent of cortical contractions in the AB blastomere after cytokinesis.
Other apparent phenotypic deviations from wild-type seemed too subtle to justify a placement in a phenotypic class at this stage of the analysis. These included a minor lack of yolk granules, a slightly smaller size of pronuclei, a hesitant rotation of the centrosomes and associated pronuclei, a delay for the AB or P1 nucleus to reach their appropriate positions, or an unstable AB nucleus.
Indirect Immunofluorescence
Indirect immunofluorescence was carried out with slight modifications of standard procedures (Strome and Wood, 1983; Albertson, 1984). Slides were coated with 1% poly-l-lysine (Sigma, P 1524) in PBS. 2 μl H2O was pipetted onto the slide, and five gravid hermaphrodites were placed into the drop and cut open with a scalpel to release embryos. An 18-mm2 coverslip was placed onto the drop and excess fluid was wicked away with 3MM Whatman paper. The slide was frozen on a block of metal precooled on packed dry ice. After a few minutes, the coverslip was flicked off with a razor blade, and the slide was plunged into −20°C methanol for 15 min or more. Slides were rehydrated in PBS for 5 min and incubated with 30 μl of primary antibody in PBS for 45 min at room temperature in a wet chamber. The following primary antibodies were used: mouse anti-tubulin (clone DM1A, used at 1:400; Sigma) and rabbit anti–ZYG-9 (used at 1:100; Matthews et al., 1998). Slides were washed for 5 min in PBT (PBS-0.05% Tween 20), 5 min in PBS, and incubated as described above for 45 min with the following secondary antibodies: 1:800 goat anti–mouse Alexa™ 488 (Molecular Probes) and 1:1,000 donkey anti–rabbit Cy3 (Dianova). Slides were placed for 5 min in PBT containing 1 μg/ml Hoechst 33258 (Sigma), washed 5 min in PBT, 5 min in PBS, and mounted in 6 μl of 90% glycerol containing 4% n-propyl-gallate in PBS. An 18-mm2 coverslip was placed onto the sample and sealed with nail polish. Immunofluorescence microscopy was performed with a Zeiss Confocal Microscope (LSM 510), and images were processed with Adobe Photoshop 4.0.
Results
Screening of an Extensive Mutant Collection Leads to the Identification of 34 Loci Required for Cell Division Processes
We set out to identify novel components required for cell division processes in a complex eukaryote by mutational analysis, using the one cell stage C. elegans embryo as a model system. To this end, we screened an extensive collection of maternal-effect embryonic lethal mutations on chromosome III with time-lapse DIC video microscopy (see Materials and Methods). 160 mutations in 106 loci were analyzed; an overview of the screen is given in Table I.
Table I.
Summary of the Screen
| Overview | ||||
| Genomes mutagenized | 15,600 | |||
| Maternal-effect embryonic lethal strains | 254 | |||
| Complementation groups | 125 | |||
| Loci analyzed in this study* | 106 | |||
| Mutations analyzed in this study‡ | 160 | |||
| Loci with phenotype in one cell stage§ | 65 | |||
| Mutations with phenotype in one cell stage | 95 | |||
| Breakdown of phenotypes | Loci§¶ | Alleles‖ | ||
| Cell division processes | 34 | 48 | ||
| Pronuclear migration | 6 | 7 | ||
| Rotation | 3 | 3 | ||
| Spindle assembly | 3 | 5 | ||
| Anaphase spindle positioning | 6 | 9 | ||
| Chromosome segregation | 12 | 18 | ||
| Cytokinesis | 4 | 6 | ||
| Other phenotypes | 34 | 47 | ||
| Cytoplasmic appearance | 10 | 14 | ||
| Pronuclear appearance | 4 | 7 | ||
| Pronuclear/nuclear position | 5 | 5 | ||
| Fast phase of female pronuclear migration | 2 | 2 | ||
| Pronuclear meeting | 1 | 2 | ||
| Progression through one cell stage | 5 | 8 | ||
| Other unique phenotypes** | 6 | 9 | ||
| No phenotype in one cell stage | 51 | 65 |
Mutations at 19 loci could not be analyzed (see Materials and Methods).
See Materials and Methods for how many alleles were examined at each locus.
Includes loci for which only some alleles display a phenotype in the one cell stage embryo.
Loci are listed twice if different alleles display different phenotypic characteristics.
Alleles analyzed in this study; additional alleles are available in the collection for certain loci (see Tables II and III).
These phenotypes are not displayed by mutations at any other locus; these strains are grouped in this class solely for sake of compactness; details about particular phenotypes are found in Table III.
We identified 48 mutations in 34 loci affecting one of six major processes which normally contribute to proper distribution of chromosomes and cytoplasmic material at cell division. These 48 strains are listed by phenotypic class in Table II. Each of the six phenotypic classes is described in the main text and is illustrated by a figure (see Figs. 4–9), with the top three panels (A–C) representing the relevant wild-type sequence, and the bottom three panels (D–F) the corresponding mutant sequence. In addition, Quicktime movies corresponding to these figures can be viewed on the Hyman lab web site (http://embl-heidelberg.de/ ExternalInfo/hyman/Data.htm). We also identified 47 mutations in 34 loci which had other phenotypic manifestations detectable by DIC microscopy in the one cell stage embryo. These 47 strains are listed by phenotypic class in Table III. Finally, 65 mutations in 51 loci had either no phenotype or no penetrant phenotype in the one cell stage embryo (data not shown).
Figure 4.
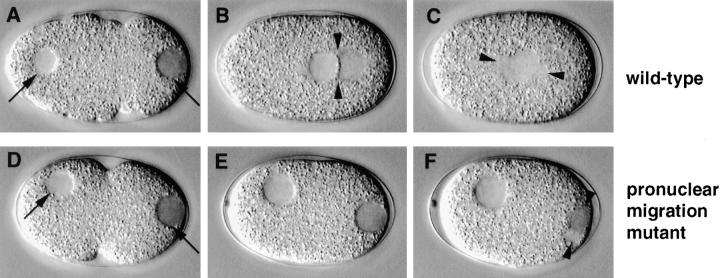
Pronuclear migration mutant. Relevant sequence of events in wild-type (N2; A–C) and corresponding sequence in a pronuclear migration mutant (t1543; D–F). Arrowheads in B, C, and F point to centrosomes and spindle poles. (A and D) In both wild-type and mutant embryo, the male pronucleus is tightly apposed to the posterior cortex (right arrow); the female pronucleus is slightly off the anterior cortex (left arrow). (B and E) While in wild-type the male and female pronuclei migrate to meet at 70% egg length (B), neither male nor female pronucleus migrates in the mutant (E). (C and F) As a consequence, while in wild-type the spindle sets up in the cell center and along the longitudinal axis (C), the spindle sets up at the very posterior and perpendicular to the longitudinal axis in the mutant (F). Note that the female pronuclear membrane is still intact after the male pronuclear membrane broke down (F).
Figure 9.
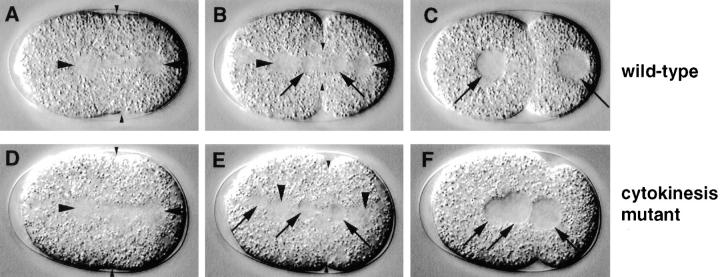
Cytokinesis mutant. Relevant sequence of events in wild-type (N2; A–C) and corresponding sequence in a cytokinesis mutant (t1568; D–F). Larger arrowheads point to spindle poles in all panels. (A and D) In both wild-type and mutant embryo, the cleavage furrow is specified at a slightly asymmetric position along the longitudinal axis (small arrowheads). (B and E) In both wild-type and mutant embryo, the cleavage furrow starts to ingress (small arrowheads). Note that an additional nucleus (E, left-most arrow) is visible towards the anterior of the mutant embryo; this most likely corresponds to nonextruded polar body material reincorporated in the embryo proper. (C and F) While in wild-type the cleavage furrow has fully ingressed, thus splitting the one cell stage embryo into two daughter blastomeres (C), the cleavage furrow has regressed in the mutant (F). As a consequence, both daughter nuclei (F, two right-most arrows) and the nucleus coming from the anterior (F, left arrow) appose in the undivided mutant cell.
All 106 loci analyzed were mapped by deficiencies to distinct regions of chromosome III (see Materials and Methods). The location of the loci reported in Tables II and III is given in Fig. 1.
Time-Lapse DIC Video Microscopy Reveals Six Major Classes of Mutants in Cell Division Processes
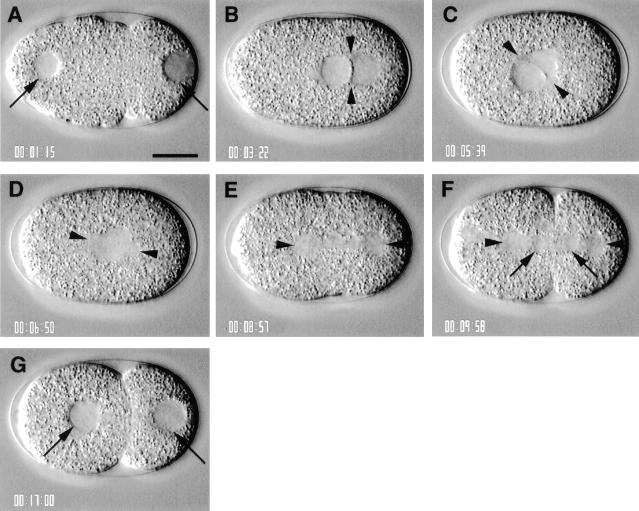
Analysis of one cell stage embryos typically started ∼20 min after fertilization. In wild-type at this stage, the male and female pronuclei are clearly visible by DIC microscopy at opposite poles of the embryo (Fig. 2 A, arrows). The male pronucleus is tightly apposed to the cortex (Fig. 2 A, right arrow), while the female pronucleus lies slightly off the cortex (Fig. 2 A, left arrow). Since the position of the male pronucleus defines the future posterior of the embryo (Goldstein and Hird, 1996), the location of the male and female pronuclei serve to orient the embryo, which is shown with anterior to the left and posterior to the right in all figures.
Figure 2.
Wild-type; sequence of events in one cell stage embryo. Time elapsed since the beginning of the recording is indicated at the bottom left of each panel in hours, minutes, and seconds. All panels in this figure and in Figs. 4–9 are at the same magnification. Bar, 10 μm. (A) Embryo ∼20 min after fertilization. The male pronucleus is tightly apposed to the posterior cortex (right arrow); the female pronucleus is slightly off the anterior cortex (left arrow). Note the pseudocleavage furrow forming towards the middle of the embryo. (B) Embryo at pronuclear meeting. The pronuclei have migrated and have just met at 70% egg length, and the pseudocleavage furrow has regressed. The two centrosomes (arrowheads) lie perpendicular to the longitudinal axis. Note also that a line corresponding to the apposed pronuclear membranes runs between the two centrosomes. (C) Embryo during rotation. The centrosome pair and associated pronuclei have begun to centrate and rotate. The centrosomes (arrowheads) are approaching the longitudinal axis. (D) Embryo just after pronuclear envelope breakdown. Completion of centration/ rotation has aligned the centrosomes (arrowheads) along the longitudinal axis. The pronuclear envelopes have broken down, and a bipolar spindle is assembled. (E) Embryo towards the end of anaphase. The posterior displacement of the posterior spindle pole (right arrowhead) during anaphase has positioned the spindle asymmetrically along the longitudinal axis. (F) Embryo shortly after telophase. The cleavage furrow cleaves the one cell stage embryo into two unequally sized daughters, the larger anterior AB blastomere, and the smaller posterior P1 blastomere. (G) Two cell stage embryo, with centrally located nuclei (arrows).
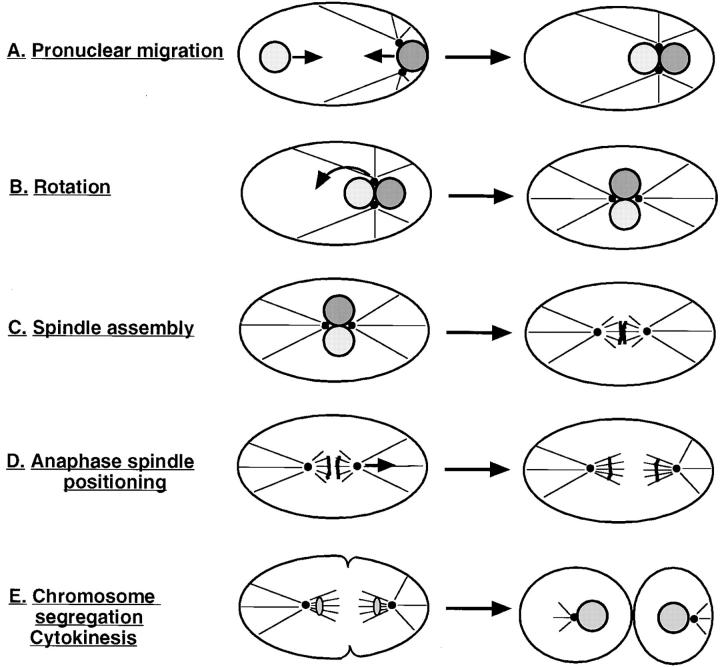
The sperm contributes the single centrosome of the one cell stage embryo (Albertson, 1984). This centrosome duplicates and the two resulting centrosomes then separate, while remaining associated with the male pronucleus (Albertson, 1984; Hyman and White, 1987). The cell division that follows entails a sequence of six major processes which can be monitored by DIC microscopy: pronuclear migration, rotation of centrosomes and associated pronuclei, spindle assembly, anaphase spindle positioning, chromosome segregation, and cytokinesis (Figs. 2 and 3; Nigon et al., 1960; Sulston et al., 1983; Albertson, 1984). Screening of the mutant collection with time-lapse DIC video microscopy led to the identification of the first point in time when a given mutant deviates from the wild-type sequence. Mutant strains affecting cell division processes were thus classified into six phenotypic classes, corresponding to defects in one of the six major processes which take place in wild-type. For each of these processes, we first describe the events in wild-type and then those in the corresponding mutant strains.
Figure 3.
Sequence of processes which contribute to proper distribution of chromosomes and cytoplasmic material to daughters of the first cell division. (A) Pronuclear migration: female pronucleus (left, light shading) and male pronucleus (right, dark shading) migrate (arrows) from their initial location to meet at 70% egg length. (B) Rotation: the centrosome pair and associated pronuclei move to the center of the embryo and rotate by 90° (arrow). (C) Spindle assembly: the bipolar spindle sets up after pronuclear envelope breakdown. (D) Anaphase spindle positioning: the posterior spindle pole is displaced towards the posterior cortex during anaphase (arrow). (E) Chromosome segregation: the spindle segregates one complement of chromosomes towards each spindle pole. While chromosome segregation occurs earlier (see D), it can be monitored by DIC microscopy only after reformation of daughter nuclei. Cytokinesis: the cleavage furrow is first specified and then ingresses, thus dividing the one cell stage embryo into two unequally sized daughters.
Pronuclear Migration Mutants
The fiirst process to be considered is pronuclear migration. In wild-type, the female pronucleus migrates away from the anterior region, slowly at first and faster thereafter; meanwhile, the male pronucleus migrates slightly away from the posterior cortex (Figs. 3 A and 4, A and B). As a result of these migrations, the female and male pronuclei meet at 70% egg length (0%: anterior-most; 100%: posterior-most).
Pronuclear migration was defective in seven mutants representing six loci in the collection. We define as pronuclear migration mutants those strains in which the female and male pronuclei are initially positioned similarly to wild-type (Fig. 4 D, arrows), but fail to migrate towards each other (Fig. 4 E). In two pronuclear migration mutants (see Table II), a spindle which appears smaller than wild-type assembles around the male pronucleus at the very posterior of the embryo and perpendicular to the longitudinal axis (Fig. 4 F, arrowheads). This spindle moves off the posterior cortex by the end of mitosis, and is bisected by a cleavage furrow ingressing from the posterior and along the longitudinal axis. In the remaining five pronuclear migration mutants (see Table II), the spindle is usually barely detectable or not visible at all after breakdown of the male pronucleus. Consistent with the absence of a functional spindle, there is usually no cleavage furrow ingression from the posterior of the embryo. In occasional pronuclear migration mutant embryos from either set, the female pronucleus undergoes part of its migration towards the posterior. In others, there is more than one female pronucleus, indicating that the female meiotic divisions can be affected as well.
Interestingly, in all pronuclear migration mutants, the nuclear envelope of the male pronucleus breaks down ∼1 min before that of the female pronucleus (see Fig. 4 F). This asynchrony in pronuclear envelope breakdown may reflect a wave in cell cycle progression along the longitudinal axis of the embryo (see Discussion).
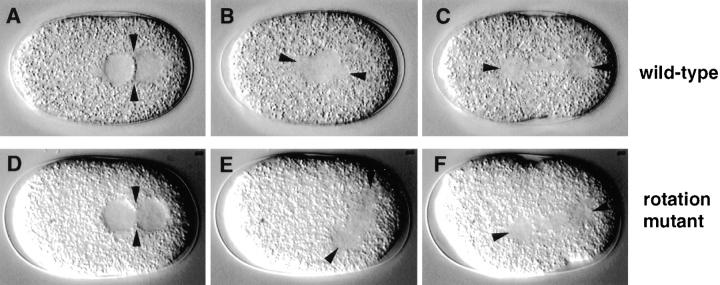
Rotation Mutants
After pronuclear migration in wild-type, the centrosome pair and associated pronuclei move to the embryo center, while undergoing a 90° rotation that aligns the centrosomes along the longitudinal axis (Figs. 3 B and 5, A and B).
Rotation was defective in three mutants representing three loci in the collection. We define as rotation mutants those strains in which the pronuclei migrate as in wild-type (Fig. 5 D), but in which the centrosome pair and associated pronuclei then fail to rotate. In two rotation mutants (see Table II), the centrosome pair and associated pronuclei also fail to center, and the spindle thus assembles at 70% egg length, perpendicular to the longitudinal axis (Fig. 5 E). In the third rotation mutant (see Table II), the centrosome pair and associated pronuclei do center, and in fact move towards the anterior more so than in wild-type; as a result, the spindle assembles at 30–40% egg length, perpendicular to the longitudinal axis. In all three rotation mutants, the spindle is reoriented along the longitudinal axis by the end of anaphase (Fig. 5 F), perhaps because of the physical constraints of the eggshell. As a consequence of this reorientation, roughly normal-sized daughter blastomeres are generated. However, these blastomeres often have more than one nucleus, indicative of problems in chromosome segregation.
Figure 5.
Rotation mutant. Relevant sequence of events in wild-type (N2; A–C) and corresponding sequence in a rotation mutant (t1599; D–F). Arrowheads in all panels point to centrosomes and spindle poles. (A and D) In both wild-type and mutant embryo, the pronuclei migrate to meet at 70% egg length; arrowheads point to centrosomes. (B and E) While in wild-type the centrosome pair and associated pronuclei move towards the center and undergo a 90° rotation (B), this does not happen in the mutant (E). As a consequence, while in wild-type the spindle sets up in the cell center and along the longitudinal axis (B), the spindle sets up at 70% egg length and perpendicular to the longitudinal axis in the mutant (E). (C and F) By the end of anaphase, the spindle in the mutant embryo is reoriented along the longitudinal axis (F), perhaps due to constraints exerted by the egg shell.
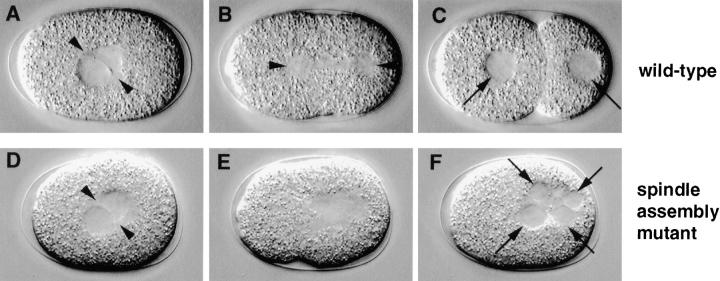
Spindle Assembly Mutants
After rotation in wild-type, the nuclear envelopes of the apposed pronuclei break down and a bipolar spindle is assembled along the longitudinal axis, in the center of the embryo (Figs. 2 D and 3 C).
Spindle assembly was defective in five mutants representing three loci in the collection. We define as spindle assembly mutants those strains in which the pronuclear envelopes break down similarly to wild-type, but in which a bipolar spindle is not apparent thereafter (Fig. 6 E). Although there is an area in the center that appears clear by DIC microscopy, this area lacks bipolarity. Consistent with the absence of a functional spindle, the cleavage furrow does not form and many small nuclei, presumably containing nonsegregated chromosomes, form as the cell returns to interphase (Fig. 6 F, arrows). In a minority of spindle assembly mutant embryos, prior rotation of the centrosome pair and associated pronuclei is also defective.
Figure 6.
Spindle assembly mutant. Relevant sequence of events in wild-type (N2; A–C) and corresponding sequence in a spindle assembly mutant (t1595; D–F). (A and D) In both wild-type and mutant embryo, the centrosome pair (arrowheads) and associated pronuclei are in the cell center before pronuclear envelope breakdown. (B and E) While in wild-type a bipolar spindle is assembled (B, arrowheads point to spindle poles), this is not the case in the mutant (E). (C and F) As a consequence, while in wild-type the bipolar spindle segregates one complement of chromosomes to each daughter nucleus (C, arrows), the lack of functional spindle in the mutant leads to an absence of cell division and to the formation of many small nuclei (F, arrows), presumably reforming around nonsegregated chromosomes as the cell returns to interphase.
Anaphase Spindle Positioning Mutants
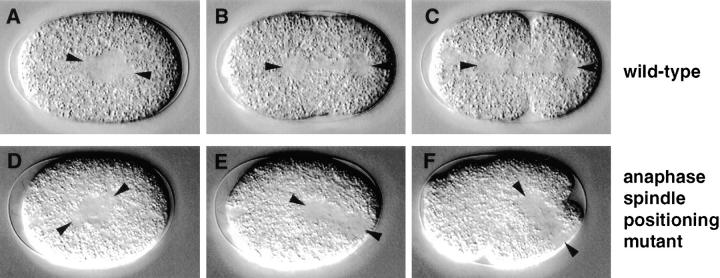
After spindle assembly in wild-type, the posterior spindle pole is displaced towards the posterior during anaphase, resulting in an asymmetric spindle position along the longitudinal axis (Figs. 3 D and 7, A and B). Since the cleavage furrow in animal cells is specified by the end of anaphase to form at equal distance from the spindle poles (reviewed in Rappaport, 1971; Glotzer, 1997; Oegema and Mitchison, 1997), this posterior displacement in the one cell stage C. elegans embryo results in an asymmetric placement of the cleavage furrow (Fig. 7, B and C).
Figure 7.
Anaphase spindle positioning mutant. Relevant sequence of events in wild-type (N2; A–C) and corresponding sequence in an anaphase spindle positioning mutant (t1650; D–F). Arrowheads point to spindle poles in all panels. (A and D) In both wild-type and mutant embryo, the spindle sets up along the longitudinal axis and in the cell center. (B and E) While wild-type embryos exhibit a slight posterior displacement of the posterior spindle pole during anaphase (B), this displacement occurs in an exaggerated manner in the mutant, all the way to the posterior cortex (compare E with B). (C and F) While in wild-type the slightly asymmetric position of the spindle along the longitudinal axis at the end of anaphase ensures proper placement of the cleavage furrow (C), the aberrant position of the spindle at the end of anaphase in the mutant leads to an abnormal first cleavage division (F).
As expected from previous work (Kemphues et al., 1988b), we found that mutations at the par-3 locus abolish posterior anaphase displacement, resulting in a symmetric first cell division (Table III). However, this lack of posterior displacement is probably a secondary consequence of the known requirement for par-3 in setting up antero-posterior embryonic polarity (Kemphues et al., 1988b; reviewed in Kemphues and Strome, 1997). To indicate this fact, par-3 is not included among the loci strictly required for cell division processes which are reported in Table II; instead, par-3 is mentioned in Table III.
We found that anaphase spindle positioning was defective in a novel manner in nine mutants representing six loci in the collection. In these anaphase spindle positioning mutants, the spindle is usually assembled in the cell center similarly to wild-type (Fig. 7 D), but is displaced to aberrant locations on the cortex during anaphase. In five anaphase spindle positioning mutants (see Table II), the spindle exhibits an exaggerated and abrupt movement towards the posterior during anaphase (Fig. 7 E). The spindle thus comes in contact with the posterior cortex and is reoriented perpendicular to the longitudinal axis by the end of anaphase, resulting in an aberrant first cleavage (Fig. 7 F). In four other anaphase spindle positioning mutants (see Table II), the spindle drifts slowly during anaphase, and ends up being positioned towards the posterior or lateral cortices. In a minority of anaphase spindle positioning mutant embryos from either set, prior rotation of the centrosome pair and associated pronuclei is defective, in which case the spindle sets up perpendicular to the longitudinal axis and tends to drift towards the anterior of the embryo.
Chromosome Segregation Mutants
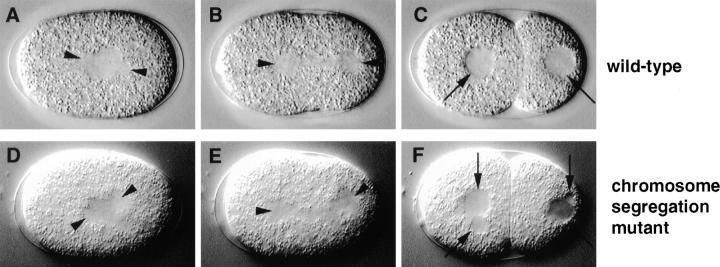
In wild-type, the spindle segregates one chromosome complement to each daughter of the first cell division (Fig. 3, D and E). In contrast to the other processes described here, chromosome segregation cannot be directly observed by DIC microscopy in C. elegans embryos because individual chromosomes cannot be seen. However, proper chromosome segregation can be inferred from the presence of two equally sized daughter nuclei, one in each daughter blastomere (Fig. 8 C, arrows).
Figure 8.
Chromosome segregation mutant. Relevant sequence of events in wild-type (N2; A–C) and corresponding sequence in a chromosome segregation mutant (t1712; D–F). Arrowheads point to spindle poles in all panels. (A and D) In both wild-type and mutant embryo, a bipolar spindle is set up. (B and E) The anaphase spindle in this particular chromosome segregation mutant appears less visible, especially towards the center of the spindle, and is bent (compare E with B); the spindle of most chromosome segregation mutants is indistinguishable by DIC microscopy from that of wild-type (see Table II). (C and F) In wild-type, the segregation of one complement of chromosomes towards each spindle pole results in the formation of two equally sized daughter nuclei, one in each daughter blastomere (C, arrows). In the mutant embryo, more than one nucleus is formed in each daughter of the first cell division (F, arrows), perhaps because chromosomes which were lagging during anaphase have not been incorporated in the main nucleus of daughter cells.
According to this criterion, chromosome segregation was defective in 18 mutants representing 12 loci, comprising the largest phenotypic class in the collection. We define as chromosome segregation mutants those strains in which a bipolar spindle is assembled similarly to wild-type (Fig. 8 D), but generates more than one nucleus in daughter blastomeres (Fig. 8 F, arrows). This indicates that the spindle is unable to properly segregate the entire complement of chromosomes to daughter cells. Perhaps some chromosomes are lagging during anaphase, and are thus not incorporated in the larger nucleus that reforms after telophase. In 15 chromosome segregation mutant strains, the first division spindle looks indistinguishable from wild-type by DIC microscopy, while it has a somewhat unusual appearance in the other three (see Table II; compare Fig. 8, B and E). In seven chromosome segregation mutant strains (see Table II), some embryos have more than one female pronucleus, indicative of defects during the female meiotic divisions.
Cytokinesis Mutants
Cytokinesis is the last cell division process to be observed in the one cell stage embryo. In wild-type (Fig. 3 E), the cleavage furrow is first specified at equal distance from the spindle poles (Fig. 9 A, small arrowheads), then ingresses (Fig. 9 B, small arrowheads) and finally cleaves the cell into two unequally sized blastomeres (Fig. 9 C).
Cytokinesis was defective in six mutants representing four loci in the collection. We define as cytokinesis mutants those strains in which the spindle is positioned by the end of anaphase similarly to wild-type (Fig. 9 D), but in which the cell fails to cleave in two. In two cytokinesis mutants (see Table II), the cleavage furrow is not visible by DIC optics and may thus not be properly specified. In the remaining four cytokinesis mutants (see Table II), the cleavage furrow is specified in the correct location (Fig. 9 D, small arrowheads), starts to ingress (Fig. 9 E, small arrowheads), but then regresses before going to completion (Fig. 9 F). In both sets of cytokinesis mutant embryos, daughter nuclei then appose in the undivided cell (see Fig. 9 F, two right-most arrows). Cytokinesis during the female meiotic divisions seems also affected: additional nuclei, most likely corresponding to nonextruded polar body material, join the daughter nuclei from the anterior of the embryo (Fig. 9, E and F, left-most arrow).
Immunofluorescence Microscopy Reveals that Two Pronuclear Migration Loci Are Required for Generating Normal Microtubule Arrays and Four for Centrosome Separation
Secondary screens customized for each phenotypic class will enhance understanding of the underlying cellular defect. While it is beyond the scope of this study to perform secondary screens on every phenotypic class, we wanted to examine pronuclear migration mutants further to elucidate the basis of a subdivision that was apparent from our observations with time-lapse DIC video microscopy. We had noted that a small but detectable spindle assembled around the male pronucleus and moved off the posterior cortex during mitosis in two pronuclear migration mutants. In contrast, the spindle was barely detectable or seemed altogether absent in five mutants of this class. To better understand the basis for this subdivision, we analyzed fixed wild-type and pronuclear migration mutant embryos with anti-tubulin and anti–ZYG-9 antibodies, as well as with the DNA dye Hoechst 33258.
We examined the distribution of microtubules because migration of both male and female pronuclei is abolished in the presence of the microtubule-depolymerizing agent nocodazole (Strome and Wood, 1983), as well as in zyg-9 mutant embryos, which exhibit short astral microtubules (Albertson, 1984). Therefore, short or otherwise abnormal microtubules may be causing the phenotype in some pronuclear migration mutants identified here. In addition, we simultaneously examined the distribution of ZYG-9 protein, a convenient centrosomal marker which maintains this localization even after nocodazole treatment (Matthews et al., 1998).
We examined embryos both during prophase, when pronuclear migration normally takes place, and during mitosis, when a potential difference in spindle morphology may be apparent. In wild-type, the single centrosome lies initially between the male pronucleus and the posterior cortex (Albertson, 1984; Hyman and White, 1987). After centrosome duplication, the two resulting centrosomes separate and move towards the anterior, seemingly along the surface of the male pronucleus. As a result, both centrosomes lie on the anterior of the male pronucleus, facing towards the female pronucleus (Fig. 10 C, arrows). The cell is in prophase at this stage, as can be seen from condensing DNA in both male (Fig. 10 A, arrow) and female (Fig. 10 A, arrowhead) pronuclei. During prophase, astral microtubules emanating from the centrosomes become numerous and some of them are fairly long (Fig. 10 B, arrows). The dense mesh of cortical microtubules which is predominant in earlier stages (Albertson, 1984) is still present during prophase, but begins to disappear (Fig. 10 B). During anaphase (Fig. 10 D), an extensive array of astral microtubules extends towards the anterior and posterior cortices (Fig. 10 E, arrows), while spindle microtubules extend centrally towards the chromosomes (Fig. 10 E, arrowhead).
Figure 10.
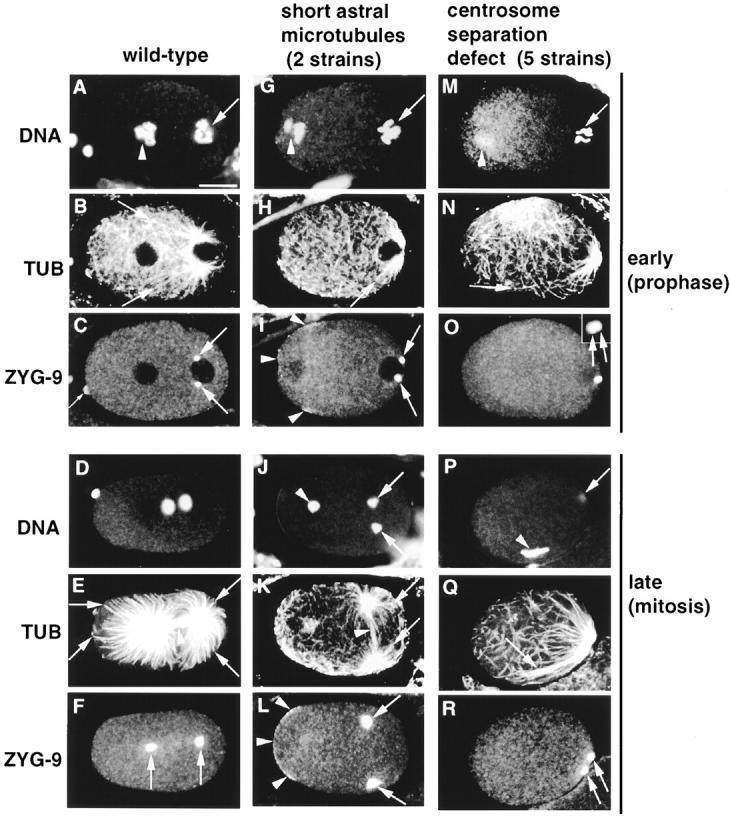
Wild-type (first column), pronuclear migration mutant with short astral microtubules (second column, t1543, G–I, t1458, J–L) and pronuclear migration mutant with defective centrosome separation (third column, t1550) stained with antitubulin (TUB) and anti–ZYG-9 antibodies, and counterstained with Hoechst 33258 to reveal DNA. (Top) Prophase, just before pronuclear migration stage. (Bottom) Mitosis. Images are 1–1.2-μm confocal slices; in some cases, the stage was refocused slightly between channels. I is a projection of two confocal slices 2.4 μm apart. Bar in A, 10 μm. All panels are at the same magnification. Inset in O is 3.6 μm2. (A–F) Wild-type (A) prophase; DNA of male (arrow) and female (arrowhead) pronuclei is condensing; (B) numerous astral microtubules emanate from the centrosomes; arrows point to the ends of two of the longest astral microtubules; a mesh of cortical microtubules is also visible; (C) ZYG-9 marks centrosomes, which are separated and anterior of the male pronucleus; ZYG-9 is also present throughout the cytoplasm and in the polar body (small arrow); (D) anaphase; the two sets of chromosomes segregate towards the spindle poles; DNA at the very anterior of the embryo corresponds to polar body material; (E) numerous and long astral microtubules (arrows) extend towards the anterior and posterior cortices, while spindle microtubules (arrowhead) extend centrally towards chromosomes; (F) ZYG-9 signal at spindle poles is stronger during anaphase than in earlier stages. (G–L) Pronuclear migration mutant with short astral microtubules. (G) Prophase; DNA of male (arrow) and female (arrowhead) pronuclei is condensing; (H) astral microtubules are shorter than in wild-type; arrow points to the end of the longest astral microtubule; (I) centrosomes have separated but are positioned more posteriorly than in wild-type; ZYG-9 is also found at the anterior cortex (arrowheads); in this particular embryo, ZYG-9 distribution at the anterior cortex is patchy; (J) anaphase; chromosomes from the male pronucleus (arrows) are found on the spindle, in the posterior of the embryo, whereas chromosomes from the female pronucleus are left in the anterior (arrowhead); (K) a spindle (arrowhead) which contains less microtubules than in wild-type is present in the posterior of the embryo, perpendicular to the longitudinal axis; arrows point to the ends of two of the longest astral microtubules, which are significantly shorter than in wild-type; note how cortical microtubules are denser at the anterior; (L) ZYG-9 signal at the spindle poles (arrows) is stronger than in wild-type; in addition, ZYG-9 is present at the anterior cortex (arrowheads). (M–R) Pronuclear mutant defective in centrosome separation. (M) Prophase; DNA of male (arrow) and female (arrowhead) pronuclei is condensing; (N) some astral microtubules are fairly long; arrow points to the end of a particularly long one, which is not in focus throughout its length in this focal plane; (O) centrosomes have failed to separate (inset, arrows) and are posterior of the male pronucleus; (P) mitosis; condensed chromosomes from the female pronucleus (arrowhead); chromosomes from the male pronucleus are in a different focal plane (arrow); (Q) absence of bipolar spindle assembly; astral microtubules are fairly long and seem to grow preferentially or are stabilized in the vicinity of chromosomes (arrow); (R) ZYG-9 signal confirms that centrosomes (arrows) are still in close proximity to one another.
We found that astral microtubules were short in the first set of two pronuclear migration mutants, representing two loci (see Table II). During prophase, astral microtubules emanating from the centrosomes were consistently shorter than in wild-type (Fig. 10 H, arrow, compare with 10 B, arrows). This was still the case during anaphase (Fig. 10 K, arrows, compare with 10 E, arrows). At this stage, a spindle which contained less microtubules than in wild-type was apparent (Fig. 10 K, arrowhead, compare with 10 E, arrowhead). This attenuated spindle was perpendicular to the longitudinal axis and located slightly off the posterior cortex, as had been suggested by the DIC observations. Chromosomes coming from the male pronucleus were found to lie on this attenuated spindle in 19 out of 20 embryos examined for the two mutants combined (Fig. 10 J, arrows). In contrast, chromosomes coming from the female pronucleus were left towards the anterior of the embryo (Fig. 10 J, arrowhead).
Interestingly, ZYG-9 protein distribution was affected in these two mutants, in a similar manner. First, ZYG-9 was localized to the anterior cortex (Fig. 10, I and L, arrowheads), where it is not detected in wild-type (see Fig. 10, C and F). In some embryos, ZYG-9 was present at the anterior cortex in patches (see Fig. 10 I, arrowheads). Second, compared with wild-type, ZYG-9 signal appeared stronger at centrosomes and spindle poles (for example, compare Fig. 10 L, arrows, and 10 F, arrows). Taken together, these observations suggest that these two pronuclear migration loci are required for generating normal microtubule arrays and for proper subcellular localization of ZYG-9 protein.
Somewhat unexpectedly, we found that centrosomes failed to separate in the second set of five pronuclear migration mutants, representing four loci (see Table II). In prophase, the centrosome had duplicated, but the resulting daughter centrosomes failed to separate and remained positioned posterior of the male pronucleus (Fig. 10 O, arrows). During mitosis (Fig. 10 P), the two centrosomes were still in close proximity to one another and located at the very posterior of the embryo (Fig. 10 R, arrows). Probably as a consequence of this failure in centrosome separation, no bipolar spindle was assembled, as had been suggested by the DIC observations. Chromosomes were not located between the spindle poles in 42 out of 43 embryos examined during mitosis for the five mutants combined (see for example Fig. 10 P, arrowhead, and 10 R, arrows). Microtubules seemed to grow preferentially or to be stabilized in the vicinity of chromosomes (Fig. 10 P, arrowhead), and were occasionally fairly long (Fig. 10 Q, arrow). In some mutant embryos, centrosomes were separated to a greater extent towards the end of mitosis (data not shown). Taken together, these observations suggest that these four pronuclear migration loci are required for centrosome separation.
Discussion
Large Scale Mutational Analysis Reveals Spectrum of Phenotypic Classes Affecting Cell Division Processes in the One Cell Stage Embryo
Mutational analysis has been successfully used to dissect a number of biological processes. Often, success has relied on having a mutant collection sufficiently extensive to recognize the full spectrum of phenotypic classes. Thus, analysis of a large number of cell division cycle mutants in S. cerevisiae was instrumental for unraveling the dependence between successive events in the cell cycle (Hartwell et al., 1974). In Drosophila, a large scale screen revealed that a limited number of gene classes pattern the early embryo (Nüsslein-Volhard and Wieschaus, 1980).
The availability of an extensive mutant collection was also of utmost importance in this work. This allowed us to recognize that mutations affecting cell division processes in the one cell stage C. elegans embryo fall into a limited number of characteristic phenotypic classes when analyzed by DIC microscopy. The spectrum of phenotypic classes was not apparent from earlier studies relying on smaller mutant collections. Maternal-effect embryonic lethal mutations affecting aspects of C. elegans development have been identified previously, notably among three sets of temperature-sensitive mutant collections (Schierenberg et al., 1980; Wood et al., 1980; Isnenghi et al., 1983), and one set of nonconditional mutants on chromosome II (Kemphues et al., 1988a). However, these studies did not focus solely on the one cell stage embryo nor on cell division processes, in contrast to what was done here. More recently, a screen specifically designed to identify mutants in cell division processes has generated 19 temperature-sensitive mutants, 10 of which display defects in early embryos (O'Connell et al., 1998). However, it is clear that this screen was far from saturation, as all 19 mutations were single alleles of different loci. In contrast, 39 loci of the chromosome III collection analyzed here are represented by multiple alleles, arguing that this collection is significantly closer to saturation. Thus, analyzing the most extensive mutant collection to date has allowed us to build on earlier studies and recognize a well-defined spectrum of phenotypic classes affecting cell division processes in the one cell stage embryo.
100–400 True Maternal-Effect Genes May Be Required Genome-wide for Cell Division Processes in the One Cell Stage Embryo
Screening an extensive mutant collection has also allowed us to estimate the number of true maternal-effect genes that are required genome-wide for cell division processes in the one cell stage embryo. We found that mutations in 34 loci on chromosome III affect one of these processes. These mutations probably alter two different sets of genes; the first of these corresponds to true maternal-effect genes, which encode components required solely in the early embryo. Such genes should mutate to a maternal-effect lethal phenotype with reasonable frequency. It follows that many of the 39 multiple-allele loci analyzed in the chromosome III collection may correspond to true maternal-effect genes. Mutations in 15 (38%) of these multiple-allele loci affect a cell division process in the one cell stage embryo. The number of genes in C. elegans which mutate with reasonable frequency to maternal-effect embryonic lethality is postulated to be 300–1,000 (Feichtinger, 1995). Assuming an even distribution of true maternal-effect genes in the genome, one can extrapolate that some 100–400 of them may be required for cell division processes genome-wide.
The second set comprises genes which encode components required not only in the early embryo but also later during development. The null phenotype in such genes is usually zygotic lethal. However, rare mutations in these genes may still be isolated as maternal-effect alleles if, for instance, they generate sufficient gene product to sustain a given process in the small larval and adult cells, but not in the larger embryonic blastomeres. Thus, several single- allele loci in maternal-effect mutant collections turned out to be hypomorphic alleles of essential genes (Perrimon et al., 1986; Kemphues et al., 1988a; Schüpbach and Wieschaus, 1989). Such rare mutations are drawn from a very large pool of genes, whose size cannot be estimated with accuracy.
Time-Lapse DIC Video Microscopy Permits Identification of Mutations Affecting Specific Cell Division Processes
Detailed time-lapse analysis was crucial during our screen for determining the first deviation from the wild-type sequence of events, thus identifying the earliest cell division process for which a given component may be required. It is worth noting that an early phenotypic manifestation may mask a potential requirement in a later process. For example, if a mutation results in a failure in pronuclear migration, a requirement for the corresponding locus in subsequent rotation may not be assessed. Importantly, how early the first deviation from wild-type occurs can yield clues about the type of component encoded by the corresponding locus. Indeed, one might expect that mutations affecting fundamental aspects of cytoskeletal function, such as microtubule integrity, would interfere with many cell division processes, and, thus, exhibit a very early defect. Conversely, mutations affecting components more specifically required for a later cell division process should not exhibit a very early defect.
In fact, our analysis demonstrates that the vast majority of mutations do not exhibit a very early defect, and may thus identify loci required for specific cell division processes. Importantly, we note that for 29 of 39 (74%) multiple allele loci, all alleles examined fall in the same phenotypic class, indicating that the encoded genes may indeed be primarily required for the corresponding cell division process. However, whether this will truly be the case remains to be determined for each locus. Indeed, some of the mutations are likely not null, and additional alleles may reveal that some loci are required for other processes as well. Moreover, in some cases, the function of a molecule in a given process may be difficult to unravel by mutational analysis. For instance, a small protein domain required for a given process may be rarely hit by mutagen.
Even in the event that a mutation is not null, the availability of mutations that specifically disrupt a given process can provide important insights. For example, in Drosophila, although the null phenotype of mutations in the actin binding protein profilin is embryonic lethality, female-sterile alleles at this locus revealed the role of the actin cytoskeleton in regulating transport of cytoplasmic material during oogenesis (Cooley et al., 1992; Verheyen and Cooley, 1994).
Pronuclear Migration: An Early Process Which Relies on Fundamental Aspects of Cytoskeletal Function
The earliest defect in a cell division process that we uncovered is a failure in both male and female pronuclear migration. Analysis of this phenotypic class demonstrates that the corresponding genes are required, as expected, for fundamental aspects of cytoskeletal function. In addition, analysis of the timing of nuclear envelope breakdown in this mutant class reveals the existence of a wave in cell cycle progression in the one cell stage embryo.
When newly fertilized embryos are treated with nocodazole, migration of both male and female pronuclei is abolished (Strome and Wood, 1983). Therefore, this phenotype is the first deviation from wild-type expected for mutations in loci generally required for microtubule structure or function. Accordingly, our immunofluorescence data show that astral microtubules are shorter than wild-type in mutations at two of the pronuclear migration loci identified here. Short astral microtubules likely prevent migration of the male and female pronuclei in different ways, as distinct mechanisms are thought to normally govern these processes. In wild-type, migration of the male pronucleus away from the posterior cortex may result from the movement of the associated centrosomes (Albertson, 1984). Polymerization forces from astral microtubules would act against the posterior cortex to push centrosomes and associated male pronucleus away from it. In contrast, migration of the female pronucleus is postulated to result from its interaction with astral microtubules emanating from the centrosomes. Minus-end–directed motors anchored on the female pronuclear membrane would power movement along microtubules towards the centrosomes, and, thus, towards the male pronucleus (reviewed in Reinsch and Gönczy, 1998). Based on these models, the failure in male pronuclear migration in the two mutants with short astral microtubules likely results from insufficient polymerization forces acting against the cortex to push centrosomes and associated male pronucleus away from it. Meanwhile, the lack of female pronuclear migration may be due to astral microtubules not reaching all the way to the female pronuclear membrane.
Intriguingly, our immunofluorescence data raise the possibility that ZYG-9 protein mislocalization is involved in generating the short astral microtubule phenotype in these two pronuclear migration mutants. Short astral microtubules are also observed in zyg-9 mutant embryos, and ZYG-9 is a member of an evolutionarily conserved family of microtubule-binding proteins which promote microtubule growth in vitro (Albertson, 1984; Gard and Kirschner, 1987; Matthews et al., 1998). Here, we found that ZYG-9 protein localization is abnormal in these two mutant strains. Most notably, ZYG-9 protein is present at the anterior cortex, where it is not detected in wild-type. Possibly ZYG-9 protein trapped at this location promotes abnormal microtubule growth. Consistent with this view, cortical microtubules appear denser than in wild-type at the anterior cortex in these mutant embryos (see Fig. 10 K). This may in turn result in less efficient microtubule growth in the cytoplasm, perhaps because a rate-limiting component is trapped at the anterior cortex along with ZYG-9. Further analysis is needed to determine whether ZYG-9 mislocalization indeed causes the short astral microtubule phenotype. In the meantime, however, the present observations demonstrate that these two pronuclear migration loci are required for proper ZYG-9 subcellular localization.
Our immunofluorescence analysis also led to the unexpected discovery that centrosome separation is a prerequisite for the migration of both male and female pronuclei, as separation is defective in mutations at the remaining four pronuclear migration loci. The unseparated centrosomes do not leave the vicinity of the posterior cortex, despite fairly long astral microtubules. Perhaps the overlap of astral microtubules emanating from two centrosomes in close proximity to one another causes steric hindrance which prevents polymerization forces from efficiently acting against the posterior cortex. Alternatively, the lack of centrosome separation may signal to prevent the subsequent migration of centrosomes away from the posterior cortex.
Asynchronous Breakdown of Pronuclei in Pronuclear Migration Mutant Embryos Reveals Possible Regional Differences in Cell Cycle Progression
Time-lapse observations revealed a remarkable property of pronuclear migration mutants. In all mutant embryos of this class, the nuclear envelope of the male pronucleus breaks down ∼1 min before that of the female pronucleus. A similar asynchrony had been noted in zyg-9 mutant embryos (Albertson, 1984), and is observed in embryos treated with nocodazole (Gönczy, P., unpublished observations). The generality of this phenomenon suggests that it reflects a fundamental feature of the one cell stage C. elegans embryo, rather than a peculiar behavior of particular mutant strains.
In sea urchin zygotes in which pronuclear fusion has been prevented, the female pronucleus always breaks down earlier than the male pronucleus (Sluder and Rieder, 1985; Sluder et al., 1994). In this case, nuclear envelope breakdown is controlled at the level of each individual pronucleus, rather than by regional differences of cell cycle progression, as the female pronucleus breaks down earlier even if it is found by chance in the immediate vicinity of the male pronucleus (Sluder et al., 1995). In contrast, in C. elegans, both pronuclei appear to undergo nuclear envelope breakdown synchronously when they happen to be juxtaposed in rare pronuclear migration mutant embryos (Gönczy, P., unpublished observations). Therefore, the asynchrony observed under conditions where the two pronuclei are distant from one another most likely results from regional differences in cell cycle progression.
One possibility to explain such regional differences is that par genes, which set up polarity along the antero-posterior axis shortly after fertilization (reviewed in Kemphues and Strome, 1997), simultaneously set up a wave of cell cycle progression emanating from the posterior pole. Another possibility is that centrosomes, which remain at the posterior of pronuclear migration mutant embryos, are preferred sites of cdc2 activation. In this context, it might be interesting to note that cyclin B1, a component of cdc2 kinase, is associated with centrosomes in HeLa cells (Bailly et al., 1992). Perhaps a component or an activator of cdc2 kinase similarly localizes to centrosomes in the one cell stage C. elegans embryo to start a wave of cdc2 kinase activity.
Centration and Rotation of Centrosomes Can Be Separated by Mutational Analysis
It has been proposed that centration and rotation of centrosomes result from capture and pulling of astral microtubules by the anterior cortex (Hyman and White, 1987; Hyman, 1989). One centrosome would be pulled preferentially, thus generating a torque which induces rotation of the entire centrosome–pronuclear complex. However, to what extent centration and rotation are separable processes was previously unknown.
During this screen, we have identified two kinds of rotation mutants. In the first kind, represented by two mutant strains, both centration and rotation are defective. Possibly, these mutations correspond to weak alleles of loci more generally required for microtubule structure and function, since astral microtubules are required for centration and rotation (Hyman and White, 1987). Compatible with this view, pronuclear migration is sometimes defective in one of these two strains (see Table II). Alternatively, however, these mutations may identify components required more specifically for generating anteriorly directed pulling forces. A similar centration and rotation phenotype has been described for let-99 mutant embryos (Rose and Kemphues, 1998), as well as for embryos in which the C. elegans homologues of the dynactin components p50/dynamitin or p150Glued have been inactivated by RNAi (Skop and White, 1998). This latter finding has reinforced the view that the minus-end–directed motor cytoplasmic dynein may power rotation by pulling astral microtubules towards the anterior cortex.
In the second kind of rotation mutant identified here, represented by one mutant strain, rotation alone is defective. Centrosomes centrate and move all the way towards the anterior without ever undergoing rotation, suggesting that anteriorly pulling forces are still active. Perhaps the lack of rotation in this mutant is due to the fact that forces are equally pulling on both centrosomes, and that no torque is generated under these conditions. Thus, centration and rotation can be separated by mutational analysis and, thus, likely correspond to distinct, though coupled, processes.
Components Governing Spindle Assembly and Function
Upon entry into mitosis, the interphase microtubule network reorganizes into a bipolar array to form the mitotic spindle. Studies using primarily Xenopus egg extracts have shown that this reorganization involves participation of microtubule motors and appropriate modulation of microtubule dynamics (reviewed in Hyman and Karsenti, 1996). However, it remains to be determined to what extent such factors govern spindle assembly in vivo.
We have identified mutations at four loci in which microtubules do not assemble a bipolar spindle following pronuclear envelope breakdown. These spindle assembly mutants undergo earlier interphasic microtubule-dependent processes such as pronuclear migration. Therefore, the corresponding loci may encode motor proteins or regulators of microtubule dynamics which are specifically required during mitosis. Alternatively, components distinct from these may be identified by such an unbiased mutational analysis, and thus define novel mechanisms governing spindle assembly in vivo.
Analysis of spindle assembly mutants revealed that the spindle assembly checkpoint is not active in the one cell stage C. elegans embryo. We noted that, despite the absence of a bipolar spindle, all mutant embryos of this class exit mitosis and progress into the next cell cycle without significant delay. Therefore, the spindle assembly checkpoint, which prevents most eukaryotic cells with an unassembled spindle from progressing into anaphase (reviewed in Straight and Murray, 1997), must be inactive in the early C. elegans embryo. This parallels observations in Xenopus where in vitro experiments have demonstrated that the checkpoint machinery is inactive in the early embryo due to a low nucleus/cytoplasmic volume ratio (Minshull et al., 1994).
We have also identified mutations in 12 loci which yield daughter blastomeres with more than one nucleus, despite the presence of a bipolar spindle during the first division. This phenotype most likely results from having nonclustered chromosomes at telophase, when chromatin induces nuclear envelope reformation. These chromosome segregation mutants are analogous to mutants in the fidelity of chromosome transmission in S. cerevisiae (Hartwell and Smith, 1985; Hieter et al., 1985; Spencer et al., 1990). Detailed analysis of the yeast mutants demonstrated that they affect distinct processes contributing to proper chromosome transmission (reviewed in Brown et al., 1991). Similarly, appropriate secondary screens will undoubtedly break down C. elegans chromosome segregation mutants into subclasses, corresponding to distinct subprocesses, including aspects of kinetochore function. Direct visualization of individual chromosome behavior in these mutants should be particularly revealing in this regard.
A Balance of Forces May Act to Properly Position the First Division Spindle
Asymmetric cell divisions play a central role in the generation of cell diversity in a number of organisms (reviewed in Gönczy and Hyman, 1996; Strome and White, 1996; Jan and Jan, 1998). In the wild-type one cell stage C. elegans embryo, the mitotic spindle becomes asymmetrically positioned following a posterior displacement of the posterior spindle pole during anaphase. Since the cleavage furrow is specified to form equidistant from the spindle poles (reviewed in Rappaport, 1971; Glotzer, 1997; Oegema and Mitchison, 1997), this results in an asymmetric division which yields a larger anterior blastomere and a smaller posterior one. The mechanisms which mediate posterior anaphase displacement are not known. While displacement is abolished in par mutant embryos (Kemphues et al., 1988b), this is probably a secondary consequence of earlier defects in antero-posterior polarity (reviewed in Kemphues and Strome, 1997). Components downstream of the par genes must exist which translate embryonic polarity into posterior displacement. The identity of such components remains to be determined, and except for par-3 alleles, we did not isolate mutations which result in the absence of posterior displacement.
However, we did uncover a novel phenotypic class which affects posterior displacement. In anaphase spindle positioning mutant embryos, the entire anaphase spindle is usually displaced in an exaggerated manner towards the posterior. This raises the possibility that mechanisms exist in wild-type to restrict the extent of posterior displacement, and that these mechanisms are affected in anaphase spindle positioning mutants. Thus, asymmetric division of the one cell stage embryo may normally be achieved by a balance of forces: par-dependent forces which promote posterior displacement during anaphase, and forces dependent on anaphase spindle positioning loci which counteract this movement.
Mutations Specific for Distinct Steps of Cytokinesis
Cytokinesis involves at least two distinct steps: specification of the cleavage furrow at the cell cortex, and ingression of the furrow to cleave the cell in two. We have identified cytokinesis mutants affecting each of these two steps. In mutations at one locus, the cleavage furrow is not detectable by DIC microscopy, despite the spindle being present, suggesting a defect in cleavage furrow specification. The molecular basis of this process is still poorly understood (reviewed in Rappaport, 1971; Glotzer, 1997; Oegema and Mitchison, 1997), and the identification of components required for cleavage furrow specification is thus of great general interest.
In mutations at three other loci, the cleavage furrow is correctly specified, but regresses before going to completion. One of the loci which we have identified as mutating to this phenotype encodes cyk-1 (Swan et al., 1998), a member of the FH family of genes, some of which are also required for cytokinesis in other species (reviewed in Frazier and Field, 1997). CYK-1 protein localizes to the leading edge of the cleavage furrow late in cytokinesis, where it may act by bridging actin from the contractile ring with midzone spindle microtubules (Swan et al., 1998). Similar cleavage furrow regression occurs in embryos lacking zen-4/CeMKLP1 function, which encodes a kinesin of the CHO family localizing to midzone spindle microtubules (Powers et al., 1998; Raich et al., 1998). CHO can bridge antiparallel microtubules in vitro (Nislow et al., 1992), and zen-4/CeMKLP1 may thus act to stabilize midzone spindle microtubules in C. elegans (Powers et al., 1998; Raich et al., 1998). The molecular characterization of the two other loci identified here which are required for a late step in cytokinesis may help clarify the role of midzone spindle microtubules in cleavage furrow ingression.
Prospects
In conclusion, our work demonstrates that cell division processes in the one cell stage C. elegans embryo can be systematically dissected by mutational analysis. Collections similar to the one analyzed here exist for chromosomes II, IV, and V, corresponding to >50% of the genome in total (Schnabel, H., and R. Schnabel, unpublished observations). All of the loci identified here have been mapped by deficiencies, thus facilitating their future molecular characterization. This should be especially valuable in combination with the recently completed genome sequence and the use of RNAi to silence gene function (Fire et al., 1998). Thus, candidate genes in defined regions of chromosome III could be directly tested by RNAi to see if they correspond to the loci identified here by mutational analysis. This approach should lead to the rapid identification of numerous novel components required for cell division processes in metazoans.
Acknowledgments
We thank Elke Stenvers and Matthew Kirkham for expert technical assistance and Lisa Matthews for providing anti–ZYG-9 antibodies. For help in improving the manuscript, we thank especially Chris Echeverri, as well as Steve DiNardo, Michael Glotzer, Karen Oegema, Silke Pichler, and Sigrid Reinsch.
Abbreviations used in this paper
- DIC
differential interference contrast
- eos
egg-osmotic sensitive
- RNAi
RNA-mediated interference
Footnotes
Address correspondence to Pierre Gönczy, European Molecular Biology Laboratory, 1, Meyerhofstrasse, D-69117 Heidelberg, Germany. Tel.: 49-6221-387-337. Fax: 49-6221-387-512. E-mail: gonczy@EMBL- Heidelberg.de
Some strains used in this work were obtained from the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources (NIH NCRR). Other strains were provided by the C. elegans Genetic Toolkit Project, which is funded by a grant from the NIH NCRR to Drs. Ann Rose, David L. Baillie, and Donald L. Riddle. Part of this project was supported by the Deutsche Forschungsgemeinschaft and the EMBL (European Molecular Biology Laboratory). Pierre Gönczy was a fellow from the European Molecular Biology Organization (ATLF 787-1995), the Human Frontier Science Program (LT-202/96), and the Swiss National Science Foundation (TMR 83EU-045376) during parts of this work.
Heinke Schnabel and Ralf Schnabel's present address is Technische Universität Braunschweig, Institut für Genetik, Spielmannstrasse 7, D-38106 Braunschweig, Germany. Titus Kaletta's present address is deVGen, Technologiepark 9, B-9052 Gent-Zwijnaarde, Belgium.
References
- Albertson D. Formation of the first cleavage spindle in nematode embryos. Dev Biol. 1984;101:61–72. doi: 10.1016/0012-1606(84)90117-9. [DOI] [PubMed] [Google Scholar]
- Bailly E, Pines J, Hunter T, Bornens M. Cytoplasmic accumulation of cyclin B1 in human cells: association with a detergent-resistant compartment and with the centrosome. J Cell Sci. 1992;101:529–545. doi: 10.1242/jcs.101.3.529. [DOI] [PubMed] [Google Scholar]
- Baker BS, Carpenter AT. Genetic analysis of sex chromosomal meiotic mutants in Drosophila melanogaster. . Genetics. 1972;71:255–286. doi: 10.1093/genetics/71.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DM. Copulatory plugs and paternity assurance in the nematode Caenorhabditis elegans. . Anim Behav. 1994;48:147–156. [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M, Garvick B, Hartwell L, Kadyk L, Seeley T, Weinert T. Fidelity of mitotic chromosome transmission. Cold Spring Harbor Symp Quant Biol. 1991;56:359–365. doi: 10.1101/sqb.1991.056.01.043. [DOI] [PubMed] [Google Scholar]
- Cassada R, Isnenghi E, Culotti M, von Ehrenstein G. Genetic analysis of temperature-sensitive embryogenesis mutants in Caenorhabditis elegans. . Dev Biol. 1981;84:193–205. doi: 10.1016/0012-1606(81)90383-3. [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Gönczy P, Alexander S, Rawson R, Eberhart CG, Viswanathan S, DiNardo S, Wasserman SA. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics. 1993;135:489–505. doi: 10.1093/genetics/135.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L, Verheyen E, Ayers K. chickadee encodes a profilin required for intercellular cytoplasm transport during Drosophilaoogenesis. Cell. 1992;69:173–184. doi: 10.1016/0092-8674(92)90128-y. [DOI] [PubMed] [Google Scholar]
- Edgar LG, Wolf N, Wood WB. Early transcription in Caenorhabditis elegansembryos. Development. 1994;120:443–451. doi: 10.1242/dev.120.2.443. [DOI] [PubMed] [Google Scholar]
- Eshel D, Urrestarazu LA, Vissers S, Jauniaux JC, van Vliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feichtinger, R.E. 1995. Quantitative analysis of maternal gene function of Caenorhabditis elegans. Ph.D. thesis. Universität Wien, Wien, Austria. 71 pp.
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Frazier JA, Field CM. Actin cytoskeleton: are FH proteins local organizers? . Curr Biol. 1997;7:414–417. doi: 10.1016/s0960-9822(06)00205-3. [DOI] [PubMed] [Google Scholar]
- Fuller, M.T. 1993. Spermatogenesis. In The Development of Drosophila melanogaster: Cold Spring Harbor Press, Cold Spring Harbor, NY. 71–147.
- Gaglio T, Dionne MA, Compton DA. Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J Cell Biol. 1997;138:1055–1066. doi: 10.1083/jcb.138.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Kirschner MW. A microtubule-associated protein from Xenopuseggs that specifically promotes assembly at the plus-end. J Cell Biol. 1987;105:2203–2215. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M, Baker BS. Genes controlling essential cell-cycle functions in Drosophila melanogaster. . Genes Dev. 1989;3:438–453. doi: 10.1101/gad.3.4.438. [DOI] [PubMed] [Google Scholar]
- Gatti, M., and M.L. Goldberg. 1991. Mutations affecting cell division in Drosophila. In Methods in Cell Biology. Academic Press, Inc., New York. 543– 586. [DOI] [PubMed]
- Glotzer M. The mechanism and control of cytokinesis. Curr Opin Cell Biol. 1997;9:815–823. doi: 10.1016/s0955-0674(97)80082-8. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Hird SN. Specification of the anteroposterior axis in Caenorhabditis elegans. . Development. 1996;122:1467–1474. doi: 10.1242/dev.122.5.1467. [DOI] [PubMed] [Google Scholar]
- Gönczy P, Hyman AA. Cortical domains and the mechanisms of asymmetric cell division. Trends Cell Biol. 1996;6:382–387. doi: 10.1016/0962-8924(96)10035-0. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. . Genetics. 1985;110:381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- Hawley RS, McKim KS, Arbel T. Meiotic segregation in Drosophila melanogasterfemales: molecules, mechanisms, and myths. Annu Rev Genet. 1993;27:281–317. doi: 10.1146/annurev.ge.27.120193.001433. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopusegg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A. Spindle assembly in Xenopusegg extracts: respective roles of centrosomes and microtubule self-organization. J Cell Biol. 1997;138:615–628. doi: 10.1083/jcb.138.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P, Mann C, Snyder M, Davis RW. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985;40:381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Okada Y. Kinesin and dynein superfamily proteins in organelle transport and cell division. Curr Opin Cell Biol. 1998;10:60–73. doi: 10.1016/s0955-0674(98)80087-2. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Horvitz HR, Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. . Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Stearns T, Botstein D. Chromosome instability mutants of Saccharomyces cerevisiaethat are defective in microtubule-mediated processes. Mol Cell Biol. 1990;10:223–234. doi: 10.1128/mcb.10.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA. Centrosome movement in the early divisions of Caenorhabditis elegans: a cortical site determining centrosome position. J Cell Biol. 1989;109:1185–1193. doi: 10.1083/jcb.109.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Karsenti E. Morphogenetic properties of microtubules and mitotic spindle assembly. Cell. 1996;84:401–411. doi: 10.1016/s0092-8674(00)81285-4. [DOI] [PubMed] [Google Scholar]
- Hyman AA, White JG. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. . J Cell Biol. 1987;105:2123–2135. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isnenghi E, Cassada R, Smith K, Denich K, Radnia K, von Ehrenstein G. Maternal effects and temperature-sensitive period of mutations affecting embryogenesis in Caenorhabditis elegans. . Dev Biol. 1983;98:465–480. doi: 10.1016/0012-1606(83)90376-7. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. Asymmetric cell division. Nature. 1998;392:775–778. doi: 10.1038/33854. [DOI] [PubMed] [Google Scholar]
- Kemphues, K.J., and S. Strome. 1997. Fertilization and establishment of polarity in the embryo. In C. elegans II. D.L. Riddle, T. Blumenthal, B.J. Meyer, and J.R. Priess, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 335–359. [PubMed]
- Kemphues KJ, Wolf N, Wood WB, Hirsch D. Two loci required for cytoplasmic organization in early embryos of Caenorhabditis elegans. . Dev Biol. 1986;113:449–460. doi: 10.1016/0012-1606(86)90180-6. [DOI] [PubMed] [Google Scholar]
- Kemphues KJ, Kusch M, Wolf N. Maternal-effect lethal mutations on linkage group II of Caenorhabditis elegans. . Genetics. 1988a;120:977–986. doi: 10.1093/genetics/120.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng N. Identification of genes required for cytoplasmic localization in early C. elegansembryos. Cell. 1988b;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Kirby C, Kusch M, Kemphues K. Mutations in the par genes of Caenorhabditis elegansaffect cytoplasmic reorganization during the first cell cycle. Dev Biol. 1990;142:203–215. doi: 10.1016/0012-1606(90)90164-e. [DOI] [PubMed] [Google Scholar]
- Li YY, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews LR, Carter P, Thierry MD, Kemphues K. ZYG-9, a Caenorhabditis elegansprotein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. J Cell Biol. 1998;141:1159–1168. doi: 10.1083/jcb.141.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull J, Sun H, Tonks NK, Murray AW. A MAP kinase-dependent spindle assembly checkpoint in Xenopusegg extracts. Cell. 1994;79:475–486. doi: 10.1016/0092-8674(94)90256-9. [DOI] [PubMed] [Google Scholar]
- Miwa J, Schierenberg E, Miwa S, von Ehrenstein G. Genetics and mode of expression of temperature-sensitive mutations arresting embryonic development in Caenorhabditis elegans. . Dev Biol. 1980;76:160–174. doi: 10.1016/0012-1606(80)90369-3. [DOI] [PubMed] [Google Scholar]
- Morris NR. Mitotic mutants of Aspergillus nidulans. . Genet Res. 1975;26:237–254. doi: 10.1017/s0016672300016049. [DOI] [PubMed] [Google Scholar]
- Nigon V, Guerrier P, Monin H. L'architecture polaire de l'oeuf et les mouvements des constituants cellulaires au cours des premières étapes du développement chez quelques nématodes. Bull Biol Fr Belg. 1960;93:131–202. [Google Scholar]
- Nislow C, Lombillo VA, Kuriyama R, McIntosh JR. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 1992;359:543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. . Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. . Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- O'Connell KF, Leys CM, White JG. A genetic screen for temperature-sensitive cell-division mutants of Caenorhabditis elegans. . Genetics. 1998;149:1303–1321. doi: 10.1093/genetics/149.3.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K, Mitchison TJ. Rappaport rules: cleavage furrow induction in animal cells. Proc Natl Acad Sci USA. 1997;94:4817–4820. doi: 10.1073/pnas.94.10.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N, Mohler D, Engstrom L, Mahowald AP. X-linked female-sterile loci in Drosophila melanogaster. . Genetics. 1986;113:695–712. doi: 10.1093/genetics/113.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J, Bossinger O, Rose D, Strome S, Saxton W. A nematode kinesin required for cleavage furrow advancement. Curr Biol. 1998;8:1133–1136. doi: 10.1016/s0960-9822(98)70470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raich WB, Moran AN, Rothman JH, Hardin J. Cytokinesis and midzone microtubule organization in Caenorhabditis elegansrequire the kinesin-like protein ZEN-4. Mol Biol Cell. 1998;9:2037–2049. doi: 10.1091/mbc.9.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport R. Cytokinesis in animal cells. Int Rev Cytol. 1971;31:169–213. doi: 10.1016/s0074-7696(08)60059-5. [DOI] [PubMed] [Google Scholar]
- Reinsch S, Gönczy P. Mechanisms of nuclear positioning. J Cell Sci. 1998;111:2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- Rose LS, Kemphues K. The let-99 gene is required for proper spindle orientation during cleavage of the C. elegansembryo. Development. 1998;125:1337–1346. doi: 10.1242/dev.125.7.1337. [DOI] [PubMed] [Google Scholar]
- Schierenberg E, Miwa J, von Ehrenstein G. Cell lineages and developmental defects of temperature-sensitive embryonic arrest mutants in Caenorhabditis elegans. . Dev Biol. 1980;76:141–159. doi: 10.1016/0012-1606(80)90368-1. [DOI] [PubMed] [Google Scholar]
- Schüpbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics. 1989;121:101–117. doi: 10.1093/genetics/121.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Fire A. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. . Development. 1994;120:2823–2834. doi: 10.1242/dev.120.10.2823. [DOI] [PubMed] [Google Scholar]
- Skop AR, White JG. The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegansembryos. Curr Biol. 1998;8:1110–1115. doi: 10.1016/s0960-9822(98)70465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder G, Rieder CL. Experimental separation of pronuclei in fertilized sea urchin eggs: chromosomes do not organize a spindle in the absence of centrosomes. J Cell Biol. 1985;100:897–903. doi: 10.1083/jcb.100.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder G, Miller FJ, Thompson EA, Wolf DE. Feedback control of the metaphase-anaphase transition in sea urchin zygotes: role of maloriented chromosomes. J Cell Biol. 1994;126:189–198. doi: 10.1083/jcb.126.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder G, Thompson EA, Rieder CL, Miller FJ. Nuclear envelope breakdown is under nuclear not cytoplasmic control in sea urchin zygotes. J Cell Biol. 1995;129:1447–1458. doi: 10.1083/jcb.129.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. . Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart HI, O'Neil NJ, Janke DL, Franz NW, Chamberlin HM, Howell AM, Gilchrist EJ, Ha TT, Kuervers LM, Vatcher GP, et al. Lethal mutations defining 112 complementation groups in a 4.5 Mb sequenced region of Caenorhabditis eleganschromosome III. Mol Gen Genet. 1998;260:280–288. doi: 10.1007/pl00013816. [DOI] [PubMed] [Google Scholar]
- Straight AF, Murray AW. The spindle assembly checkpoint in budding yeast. Methods Enzymol. 1997;283:425–440. doi: 10.1016/s0076-6879(97)83035-2. [DOI] [PubMed] [Google Scholar]
- Strome S, White J. Cleavage plane specification. Cell. 1996;84:195–198. doi: 10.1016/s0092-8674(00)80974-5. [DOI] [PubMed] [Google Scholar]
- Strome S, Wood WB. Generation of asymmetry and segregation of germ-line granules in early C. elegansembryos. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- Sullivan W, Theurkauf WE. The cytoskeleton and morphogenesis of the early Drosophilaembryo. Curr Opin Cell Biol. 1995;7:18–22. doi: 10.1016/0955-0674(95)80040-9. [DOI] [PubMed] [Google Scholar]
- Sullivan W, Minden JS, Alberts BM. daughterless-abo-like, a Drosophilamaternal-effect mutation that exhibits abnormal centrosome separation during the late blastoderm divisions. Development. 1990;110:311–323. doi: 10.1242/dev.110.2.311. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. . Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Swan KA, Severson AF, Carter JC, Martin PR, Schnabel H, Schnabel R, Bowerman B. cyk-1: a C. elegansFH gene required for a late step in embryonic cytokinesis. J Cell Sci. 1998;111:2017–2027. doi: 10.1242/jcs.111.14.2017. [DOI] [PubMed] [Google Scholar]
- Vaisberg EA, Koonce MP, McIntosh JR. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen EM, Cooley L. Profilin mutations disrupt multiple actin-dependent processes during Drosophiladevelopment. Development. 1994;120:717–728. doi: 10.1242/dev.120.4.717. [DOI] [PubMed] [Google Scholar]
- Wood, W.B. 1988. Introduction to C. elegans biology. In The Nematode Caenorhabditis elegans. W.B. Wood, editor. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 1–16.
- Wood WB, Hecht R, Carr S, Vanderslice R, Wolf N, Hirsh D. Parental effects and phenotypic characterization of mutations that affect early development in Caenorhabditis elegans. . Dev Biol. 1980;74:446–469. doi: 10.1016/0012-1606(80)90445-5. [DOI] [PubMed] [Google Scholar]
- Zhang P, Knowles BA, Goldstein LS, Hawley RS. A kinesin-like protein required for distributive chromosome segregation in Drosophila. . Cell. 1990;62:1053–1062. doi: 10.1016/0092-8674(90)90383-p. [DOI] [PubMed] [Google Scholar]