Abstract
A genetic synthetic dosage lethality (SDL) screen using CTF13 encoding a known kinetochore protein as the overexpressed reference gene identified two chromosome transmission fidelity (ctf) mutants, YCTF58 and YCTF26. These mutant strains carry independent alleles of a novel gene, which we have designated CTF19. In light of its potential role in kinetochore function, we have cloned and characterized the CTF19 gene in detail. CTF19 encodes a nonessential 369–amino acid protein. ctf19 mutant strains display a severe chromosome missegregation phenotype, are hypersensitive to benomyl, and accumulate at G2/M in cycling cells. CTF19 genetically interacts with kinetochore structural mutants and mitotic checkpoint mutants. In addition, ctf19 mutants show a defect in the ability of centromeres on minichromosomes to bind microtubules in an in vitro assay. In vivo cross-linking and chromatin immunoprecipitation demonstrates that Ctf19p specifically interacts with CEN DNA. Furthermore, Ctf19-HAp localizes to the nuclear face of the spindle pole body and genetically interacts with a spindle-associated protein. We propose that Ctf19p is part of a macromolecular kinetochore complex, which may func- tion as a link between the kinetochore and the mitotic spindle.
Keywords: centromere, kinetochores, chromosome segregation, mitotic spindle apparatus, Saccharomyces cerevisiae
Mitotic cell division is a process which ensures that a cell's chromosomes are faithfully replicated and equally segregated to two daughter cells. The kinetochore (centromere DNA and associated proteins) is essential to the high fidelity of chromosome transmission. The kinetochore mediates attachment of the chromosomes to the spindle microtubules (MTs)1 and directs chromosome movement during mitosis and meiosis (reviewed in Bloom et al., 1989; Koshland, 1994). The kinetochore–MT interaction is monitored by at least one centromere-based mitotic checkpoint system which delays onset of anaphase until stable bipolar attachment is achieved (Gorbsky, 1995; Rudner and Murray, 1996). Thus, kinetochores play both important structural and regulatory roles in ensuring faithful chromosome segregation during mitosis.
The kinetochores of Saccharomyces cerevisiae are relatively simple compared with the large trilaminar structures seen in multicellular eukaryotes (Rieder, 1982; Pluta et al., 1990). Major advances in defining components necessary for a functional kinetochore in yeast have implications for understanding the corresponding elements in more complex eukaryotes. The minimal functional centromere of S. cerevisiae (∼125 bp) is comprised of three conserved centromere DNA elements (CDEs). The central CDEII element consists of 78–86 bp of AT-rich DNA, and is flanked by two highly conserved palindromic motifs, CDEI (8 bp) and CDEIII (25 bp). Of these, CDEIII is absolutely essential for centromere function. In vivo, a unique nuclease resistant chromatin structure encompassing 160–220 bp is associated with CEN DNA and presumably corresponds to the yeast kinetochore (Bloom and Carbon, 1982; Funk et al., 1989). Seven genes that encode centromere proteins have been identified. Four of these, NDC10/CTF14/CBF2 (Goh and Kilmartin, 1993; Jiang et al., 1993), CEP3/CBF3 (Lechner, 1994; Strunnikov et al., 1995), CTF13 (Doheny et al., 1993), and SKP1 (Connelly and Hieter, 1996; Stemmann and Lechner, 1996) are essential for viability and encode the components of a multisubunit complex, CBF3, that binds to CDEIII DNA in vitro (Lechner and Carbon, 1991). CBF1/CEP1/CPF1 encodes a nonessential basic helix-loop-helix protein which binds to CDEI (Baker and Masison, 1990; Cai and Davis, 1990; Mellor et al., 1990). In addition, MIF2, a homologue of mammalian CENP-C, has been shown to interact with CEN DNA in a CDEIII- and CDEII-dependent manner (Meluh and Koshland, 1995, 1997), and CSE4, a homologue of mammalian CENP-A, is suggested to be involved in a specialized centromeric nucleosome (Meluh et al., 1998). Further analysis of the architecture of the CBF3 complex using DNA protein cross-linking (Espelin et al., 1997) suggests that three subunits of CBF3, Ndc10p, Cep3p, and Ctf13p, are in direct contact with CDEIII over a region spanning 80 bp of DNA.
The binding of the CBF3 complex to CDEIII is necessary, but not sufficient for attachment of chromosomes to MTs. Several studies indicate the existence of other factors which are required to link the CBF3-DNA complex to polymerized MTs (Kingsbury and Koshland, 1991; Sorger et al., 1994). The CBF3 subcomplex is thought to act as the nucleation site onto which these as yet unidentified components assemble and form active MT-binding complexes. A rich resource for identification of these additional kinetochore components is the chromosome transmission fidelity (ctf) mutant collection, which was isolated by the sole criterion of an increased rate of missegregation of a nonessential chromosome (Spencer et al., 1990). Previously, two secondary in vivo genetic screens, the centromere transcriptional readthrough assay and the dicentric chromosome stabilization assay, were used to detect potential kinetochore mutants from this collection (Doheny et al., 1993). A subset of 12 ctf mutants screened positive for the transcriptional readthrough assay. Three of these also tested positive for the dicentric stabilization assay. Two mutants were characterized in further detail, ctf13-30 and ctf14-42, and were shown to correspond to CTF13 and CTF14 (NDC10/CBF2), genes which encode essential kinetochore proteins (Doheny et al., 1993).
To identify other proteins involved in kinetochore structure or function, a third in vivo genetic assay was developed as a tertiary screen. This novel screen, which we have called synthetic dosage lethality (SDL; Kroll et al., 1996), involves overexpressing a wild-type reference gene in a collection of target mutants. CTF13, encoding a known kinetochore protein (Doheny et al., 1993), was used as the inducibly overexpressed reference gene to screen for synthetic interactions within a subset of 12 potential kinetochore ctf mutants. Overexpression of CTF13 caused SDL in combination with four ctf mutants; one corresponded to a known kinetochore component, (CTF14/NDC10), and two were independent alleles of a previously uncharacterized gene, CTF19. We report here the cloning and detailed characterization of the CTF19 gene. The genetic and biochemical results presented here indicate that Ctf19p represents a new kinetochore protein which may play a role as a molecular link between kinetochores and the mitotic spindle.
Materials and Methods
Yeast Strains and Media
Table I lists the genotypes of all yeast strains used in this study. Media for yeast growth and sporulation were as described (Rose et al., 1990), except as otherwise indicated. For experiments monitoring the loss of a nonessential chromosome fragment, adenine was added to 6 μg/ml minimal media to enhance the development of the red pigment in ade2-101 strains. For galactose inductions, strains were grown on solid medium containing 2% raffinose as the sole carbon source, and then transferred to solid medium containing 2% galactose, either with or without additional 2% raffinose. For experiments using liquid cultures, strains were grown in liquid medium containing 2% raffinose overnight, and galactose was added to a final concentration of 2% for the induction times indicated. To inhibit MT function in liquid cultures, nocodazole (NZ; Sigma Chemical Co.) was added to 20 μg/ml and cultures incubated at 25°C for 2 h, or to 15 μg/ml at 30°C for 90 min. To inhibit MT function on solid medium, benomyl (DuPont) was added at 5, 10, and 20 μg/ ml as indicated. DMSO alone was added to the media as a control. All yeast transformations were done by the method of Ito et al. (1983).
Table I.
List of Yeast Strains Used in this Study
| Strains | Genotype | Source | ||
|---|---|---|---|---|
| YPH277 | MATa ura3-52 lys2-801 ade2-101 trp1Δ1 leu2Δ1 CFVII (RAD2d. YPH277) URA3 SUP11 | F. Spencer and P. Hieter | ||
| YPH500 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 | P. Hieter | ||
| YPH501 | MATA/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3Δ200/his3Δ200 | P. Hieter | ||
| trp1Δ63/trp1Δ63 leu2Δ1/leu2Δ1 | ||||
| YCTF58 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 CFIII (CEN3L. YPH278) URA3 SUP11 ctf19-58 | Kroll et al., 1996; | ||
| F. Spencer and P. Hieter | ||||
| YCTF26 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 CFIII (CEN3L. YPH278) URA3 SUP11 ctf19-26 | Kroll et al., 1996; | ||
| F. Spencer and P. Hieter | ||||
| YPH877 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ1 CFVII (RAD2d. YPH877) TRP1 SUP11 | P. Hieter | ||
| YPH982 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3Δ200/his3Δ200 | P. Hieter | ||
| trp1Δ63/trp1Δ63 leu2Δ1/leu2Δ1 CFIII (CEN3L. YPH982) URA3 SUP11 | ||||
| YPH1314 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ1 leu2Δ1 ctf19Δ1:HIS3 | This study | ||
| YPH1315 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ1 leu2Δ1 ctf19Δ1:HIS3 | This study | ||
| YPH1125 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 CFIII (CEN3L. YPH278) URA3 SUP11 | This study | ||
| YPH1316 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 ctf19Δ1:TRP1 | This study | ||
| YPH1317 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 ctf19Δ1:TRP1 | This study | ||
| YPH1318 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 ctf19Δ1:TRP1 | This study | ||
| CFIII (CEN3L. YPH278) URA3 SUP11 | ||||
| YPH1319 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 ctf19Δ1:TRP1 | This study | ||
| CFIII (CEN3L. YPH278) URA3 SUP11 | ||||
| YPH1320 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3Δ200/his3Δ200 | This study | ||
| trp1Δ63/trp1Δ63 leu2Δ1/leu2Δ1 ctf19Δ1:TRP1/ctf19Δ1:TRP1 CFIII (CEN3L. YPH278) URA3 SUP11 | ||||
| YPH1321 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3Δ200/his3Δ200 | This study | ||
| leu2Δ1/leu2Δ1 ctf19-58/ctf19-58 CFVII (RAD2d. YPH277) URA3 SUP11 | ||||
| YPH1322 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 HIS3/his3Δ200 | This study | ||
| trp1Δ1/TRP1 leu2Δ1/leu2Δ1 ctf19-26/ctf19-26 CFVII (RAD2d. YPH277) URA3 SUP11 | ||||
| YPH1329 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3Δ200/his3Δ200 | This study | ||
| trp1Δ1/trp1Δ1 leu2Δ1/leu2Δ1 ctf13-30/ctf13-30 | ||||
| YPH1330 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3Δ200/his3Δ200 | This study | ||
| trp1Δ1/trp1Δ1 leu2Δ1/leu2Δ1 ctf19Δ1:HIS3/ctf19Δ1:HIS3 | ||||
| YPH1331 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3Δ200/his3Δ200 | This study | ||
| trp1Δ1/trp1Δ1 leu2Δ1/leu2Δ1 ctf13-30/ctf13-30 ctf19Δ1:HIS3/ctf19Δ1:HIS3 | ||||
| YPH1332 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3Δ200/his3Δ200 | This study | ||
| trp1Δ1/TRP1 leu2Δ1/leu2Δ1 ctf13-30/ctf13-30 ctf19Δ1:HIS3/ctf19Δ1:HIS3 | ||||
| YPH1327 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 ctf19Δ1:TRP1 leu2Δ1::CTF19-3HA,HIS3 | This study | ||
| YPH1328 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3Δ200/his3Δ200 trp1Δ63/ | This study | ||
| trp1Δ63 leu2Δ1/leu2Δ1 ctf19Δ1:TRP1/ctf19Δ1:TRP1 leu2Δ1::CTF19-3HA/leu2Δ1::CTF19-3HA | ||||
| YPH1323 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 ctf19Δ1:TRP1 | This study | ||
| CFIII (CEN3L. YPH278)URA3 SUP11 [YKH 65] + pKH7(CTF19-CEN-LEU2; pRS315) | ||||
| YPH1324 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 ctf19Δ1:TRP1 | This study | ||
| CFIII (CEN3L. YPH278)URA3 SUP11 [YKH 65] + pKH32(CTF19-3HA-CEN-LEU2; pRS315) | ||||
| YPH1325 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 ctf19Δ1/:TRP1 | This study | ||
| CFIII (CEN3L. YPH278)URA3 SUP11 [YKH 65] + pKH14(CTF19-CEN-HIS3; pRS 313) | ||||
| YPH1326 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 ctf19Δ1:TRP1 | This study | ||
| CFIII (CEN3L. YPH278)URA3 SUP11 [YKH 65] + pKH31(CTF19-3HA-CEN-HIS3; pRS313) | ||||
| YCTF30 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 CFIII (CEN3L. YPH278) URA3 SUP11 ctf13-30 | Spencer et al., 1990; | ||
| Kroll et al., 1996 | ||||
| YCTF42 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 CFIII (CEN3L. YPH278) URA3 SUP11 ndc10-42 | Spencer et al., 1990; | ||
| Kroll et al., 1996 | ||||
| YPH971 | MATa ura3 leu2 trp1 lys2 ndc10-1 | J. Kilmartin | ||
| YPH1027 | MATa ura3-52 leu2-3,112 ndc10-2 | T. Huffaker | ||
| YPH1234 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 skp1-3::LEU2 | C. Connelly and P. Hieter | ||
| YPH1235 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 skp1-4::LEU2 | C. Connelly and P. Hieter | ||
| YAS281 | MATa ura3 lys2 ade2 his3 trp1 leu2 cep3-1 | Strunnikov et al., 1995 | ||
| YAS282 | MATa ura3 lys2 ade2 his3 trp1 leu2 cep3-2 | Strunnikov et al., 1995 | ||
| YFP2 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 CFIII (CEN3L. YPH278) URA3 SUP11 | F. Pangilinan and | ||
| bub1Δ::LEU2 | F. Spencer | |||
| YFS822 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 CFIII (CEN3L. YPH278) URA3 SUP11 | F. Pangilinan and | ||
| bub2Δ::LEU2 | F. Spencer | |||
| YFP74 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 CFIII (CEN3L. YPH278) URA3 SUP11 | F. Pangilinan and | ||
| bub3Δ::LEU2 | F. Spencer | |||
| YPH1238 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 mad2Δ::HIS3 | M. Mayer and P. Hieter | ||
| YFS722 | MATα ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ1 leu2Δ1 rad9Δ::HIS3 | F. Spencer | ||
| YPH311 | MATa ura3-52 ade2(ochre) his3Δ200 leu2-3,112 tub1-1 | A. Hoyt and D. Botstein | ||
| YPH312 | MATα ura3-52 ade2(ochre) tub2-104 | A. Hoyt and D. Botstein | ||
| YNN421 | MATa ura3 ade2 his3 lys2 trp1 cbf1::TRP1 | Cai and Davis, 1990 | ||
| SBD520-8c | MATα ura3-52 leu2-3 ade2-101 trp1Δ 901 his3-11,15 lys2-801 CEN3 cse4-1::RSCL1-1H(HIS3) | Stoler et al., 1995 | ||
| PMY1002-3A | MATα ura3 leu2 trp1 mif2-3 | P. Meluh | ||
| SMY6-4b | MATα ura3-52 his3Δ200 leu2,3-112 mps2-1 | Winey et al., 1991 | ||
| HC10-42D | MATα ura3-52 ade2 ndc1-1 | Winey et al., 1993 | ||
| ELW65-9d | MATa ura3-52 trp1-1 cdc31-2 + pCDC31;URA3 | M. Winey | ||
| TSY498 | MATα ura3-52 lys2-801 his3Δ200 leu2-3,112 tub4-32 | Marschall et al., 1996 | ||
| TSY502 | MATa ra3-52 lys2-801 his3Δ200 leu2-3,112 tub4-34 | Marschall et al., 1996 | ||
| Yspc42-10 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 spc42Δ::LEU2 spc42-10,TRP1::trp1-1 | Donaldson and | ||
| Kilmartin, 1996 | ||||
| Yspc110-1 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 spc110Δ::LEU2 spc110-1,TRP1::trp1-1 | J.V. Kilmartin | ||
| Yndc80-1 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ndc80-1 | Wigge et al., 1998 | ||
| Yndc80-2 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ndc80-2 | Wigge et al., 1998 | ||
Genetic Analysis
Synthetic Lethality (SL) Studies.
Strains containing mutations in the gene of interest were mated to a ctf19 null strain to create the heterozygous diploids. These strains were sporulated, dissected, and cultured at 25°C. At least two independent diploid clones were analyzed for each mating. To test for conditional interactions, all double mutant spores recovered at 25°C were assayed for growth at higher temperatures, initially at 30°C and 37°C. Other temperatures were tested if there was an indication of a conditional effect at a permissive temperature to determine the lowest temperature at which lethality was observed. All synthetic lethal interactions indicated by the tetrad data were confirmed by plasmid shuffle. CBF3 mutant strains which demonstrated potential SL with a ctf19 null were mated to a ctf19Δ1::TRP1 or ctf19Δ1::HIS3 strain containing a wild-type copy of CTF19 on a URA3-CEN plasmid (pKH7), sporulated and dissected. The bub (budding uninhibited by benzimidazole) and mad (mitotic arrest deficient) deletion strains were transformed with the appropriate wild-type gene (BUB1, BUB2, BUB3, or MAD2) on a URA3-CEN plasmid, mated back to the ctf19 null strain, sporulated, and dissected. Double mutant spores recovered containing a URA3-marked plasmid were streaked on plates containing 5-FOA, which does not allow growth of cells expressing URA3 (Boeke et al., 1987). Therefore, only viable double mutants can grow on 5-FOA, and true synthetic lethals can not.
SDL Studies.
Methods are as described previously (Kroll et al., 1996). In brief, mutant strains were transformed under noninducing conditions, with overexpressing constructs containing either CTF13 (pKF88) or CTF19 (pKH21) reference genes under transcriptional control of a GAL1 promoter, and the corresponding vector, p415GEU2. Expression of the reference gene was induced on solid medium containing 2% galactose. Growth was compared directly for each mutant containing either an overexpression plasmid or the respective vector alone. To test for conditional effects, transformants viable at 25°C were also assayed at 30°C and 37°C.
Molecular Cloning and Characterization of CTF19
The chromosome missegregation phenotype of ctf19-58 was confirmed to be due to a single genetic mutation through genetic analysis after backcrossing this mutant strain to its wild-type parent (YPH277). The CTF19 gene was cloned by complementation of the ctf sectoring phenotype from a library containing 10–12-kb fragments of yeast genomic DNA inserted into a LEU2-CEN vector (Spencer, F., and P. Hieter, unpublished results). Appropriate restriction fragments were used for subcloning the genomic DNA into pRS based vectors (Sikorski and Hieter, 1989). A 2-kb MluI-NsiI fragment rescued the sectoring phenotype and was shown to contain the CTF19 gene by genetic linkage analysis. CTF19 was physically mapped to chromosome XVI by hybridization of a [32P] labeled SalI-NsiI fragment to filters containing overlapping lambda and cosmid clones of the S. cerevisiae genome (Link and Olson, 1991). This placed CTF19 on the right arm of chromosome XVI, 40–50 kb from CEN6, distal to RAD1.
A complete deletion of the CTF19 open reading frame (ORF) was generated using PCR-mediated gene disruption (Lorenz et al., 1995). Oligonucleotides for PCR were synthesized as follows. OKH1 (5′-GTGTGATCTTGTT GATAC TAGGT CGCAAAGAACGCAAATAGATTG- TACTGAGAGTGCAC-3′) has a 40-bp homology to the sense strand upstream of the CTF19 ATG, followed by a 20-bp sequence from the plus strand of pRS vectors (Sikorski and Hieter, 1989) adjacent to the vector selectable marker. OKH2 (5′-GTTTAAGCAAGCCGTCCAGTTGGCAATGGCAAATGGAACACTGTGCGGTATTTCACACCG-3′) has a 40-bp homology to the antisense strand downstream of the stop codon followed by a 20-bp sequence from the minus strand of pRS vectors adjacent to the vector selectable marker. OKH1 and OKH2 can be used to incorporate any one of the markers from the pRS vectors (URA3, HIS3, LEU2, or TRP1) by PCR. These oligonucleotides were used to amplify either HIS3 from pRS303 or TRP1 from pRS304. The HIS3 PCR product was transformed into the haploid strain YPH877 and the diploid strain YJP57. The TRP1 PCR product was transformed in the haploid strain YPH1125 and the diploid strain YPH982 (Table I). Gene replacement was confirmed in each case by Southern blot analysis.
Plasmids and Epitope-tagged Constructs
Plasmids containing CTF19 fused to the GAL1 promoter, either with or without an in frame E1 epitope tag, were constructed from the plasmid p415GEU1 and p415GEU2, respectively (Connelly and Hieter, 1996; Kroll et al., 1996). A PCR strategy was used to place the second codon of CTF19 in frame with the ATG of p415GEU1 and p415GEU2, using a 5′ engineered Xho1 cloning site and a downstream engineered HindIII site (Fig. 1 b). To avoid possible PCR errors, a wild-type PstI (bp 566 in ORF) HindIII (in multiple cloning site) genomic fragment was used to replace the 3′ end of the PCR-generated sequence in pKH20. When transformed into a ctf19 deletion strain containing pKF88 (CTF13 overexpression construct; Kroll et al., 1996), both pKH20 and pKH21 were able to rescue the SDL phenotype. pKH20 and pKH21 were also able to stabilize a chromosome fragment in the ctf19 deletion strain when compared with the vector alone control on galactose containing media.
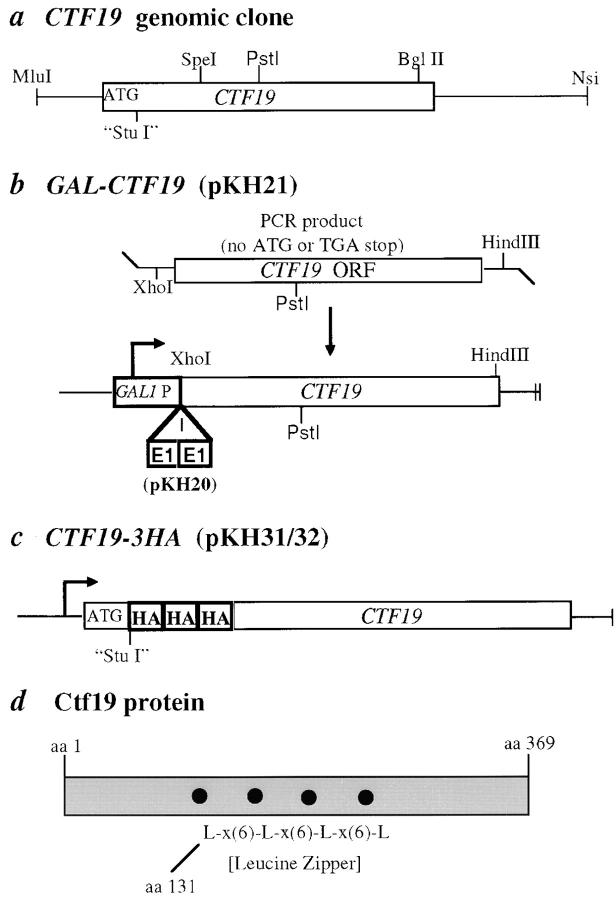
Figure 1.
CTF19 genomic clone, overexpression construct, epitope tag fusion construct, and schematic of protein. (a) A simple restriction map of the 2-kb genomic clone. (b) Overexpression and E1 epitope-tagged constructs with CTF19 under control of a GAL1 promoter. (c) NH2-terminal 3XHA epitope-tagged Ctf19p fusion protein. Three HA tags were inserted in tandem at a StuI site which was generated by PCR based site-directed mutagenesis directly after the ATG. (d) Schematic of Ctf19p, showing putative leucine zipper.
To make an HA epitope-tagged construct, a PCR based site-directed mutagenesis method which utilizes Pfu polymerase was used to engineer a StuI site directly 3′ to the ATG of CTF19 in pKH6 (protocol from QuickChange™ site-directed mutagenesis kit; Stratagene), creating pKH27. Three tandem copies of the 9–amino acid HA epitope tag (Field et al., 1988) were amplified by PCR from the vector pSM492 (a gift from Susan Michaelis) with StuI sites engineered on the 5′ and 3′ ends. The triple HA tag was ligated into the StuI site engineered at the NH2 terminus of CTF19, and this epitope-tagged fusion construct was subcloned into pRS313 and pRS315, generating pKH31 and pKH32, respectively (Fig. 1 c). These constructs were transformed into ctf19Δ1::TRP1 (YPH1319) and shown to rescue the chromosome missegregation phenotype. The CTF19-3HA fusion was integrated into the genome by gamma integration (Sikorski and Hieter, 1989). A 1.4-kb XhoI-SmaI fragment containing CTF19-3HA from pKH32 was subcloned into a HIS3 marked gamma integration vector, p679 (Doheny et al., 1993), which is designed to direct an integration event at the leu2Δ1 locus on chromosome III. The resulting integrating construct, pKH35, was linearized at the NotI site, between the targeting sequences to the left (5′) and right (3′) of the leu2Δ1 locus, and transformed into a ctf19Δ1::TRP1 deletion strain (YPH1317), creating leu2Δ1::CTF19-HA3 (YPH1327). Integration at the leu2Δ1 locus was confirmed by colony PCR analysis, using primers OMB-leu2Δ (5′-GTGTAGAATTGCAGATTCCC-3′, provided by M. Basrai) and T7. An integration event targeted correctly to the genomic leu2Δ1 locus results in a 550-bp product. Both the plasmid based and genome integrated versions of the CTF19-3HA epitope-tagged fusion produce the same 48-kD band by Western blotting and probing with anti-HA antibody, which is not seen in control strains containing a wild-type untagged copy of CTF19.
Quantitation of Chromosome Missegregation
Colony color half sector analysis was performed as previously described (Koshland and Hieter, 1987; Gerring et al., 1990). In brief, homozygous diploid strains (YPH982, CTF19/CTF19; YPH1320, ctf19Δ::TRP1/ctf19Δ:: TRP1; YPH1321, ctf19-58/ctf19-58; and YPH1322, ctf19-26/ctf19-26) containing a single SUP11-marked chromosome fragment were plated to single colonies on solid media containing a limiting amount of adenine (6 μg/ml), and grown at 30°C. The red pigment was allowed to develop at 4°C before scoring the sectoring phenotypes. Colonies scored as half-sectored were ≥50% red. A 1:0 missegregation event (chromosome loss) in the first division results in pink/red half-sectored colonies, whereas a 2:0 missegregation event (nondisjunction; chromosome gain) results in white/ red half-sectored colonies.
Flow Cytometry and Cytological Analysis
Cells were processed for flow cytometry as previously described (Gerring et al., 1990), with a few modifications. In brief, logarithmically growing cells were pelleted, resuspended in 0.2 M Tris, pH 7.5, containing 70% ethanol, and fixed overnight at 4°C. Cells were treated with 1 mg/ml RNase A at 37°C for 1 h. Proteinase K was added, and the cell suspension incubated at 55°C for 1 h. Cells were stained overnight at 4°C in 0.2 M Tris containing 3 μg/ml propidium iodide (Sigma Chemical Co.). Before being subjected to flow cytometry, cells were sonicated, using a Branson sonifier 450 on a setting of two for 2–5 s. Aliquots of cells fixed with 3.7% formaldehyde, as described for immunofluorescence, were stained with 300 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) to analyze nuclear morphology, and anti–α-tubulin decorated with fluorescein isocyanate, to analyze spindle morphology.
For experiments comparing ctf13-30 and ctf19Δ1 single mutants to ctf13-30 ctf19Δ1 double mutants, cultures of wild-type (YPH501), ctf13-30/ctf13-30 (YPH1329), ctf19Δ1::HIS3/ctf19Δ1::HIS3 (YPH1330), and ctf19Δ1::HIS3/ctf19Δ1::HIS3 ctf13-30/ctf13-30 (YPH1331, YPH1332) were grown at 25°C to early log stage (∼106 cells/ml), then split at time = 0, keeping half of the culture at 25°C and shifting the other half to 37°C. Samples were taken at the start of the experiment (time = 0), and at 3 and 5 h after the temperature shift. For each time point, samples were assayed for viability, nuclear and spindle morphology, and cell DNA content by flow cytometry.
Isolation of Minichromosomes and MT-Binding Assay for Yeast Centromeres
Isolation of minichromosomes from yeast cells and assaying their ability to bind to MTs were performed as previously described (Kingsbury and Koshland, 1991).
In Vivo Cross-Linking and Chromatin Immunoprecipitation
The methods used for chromatin immunoprecipitation and the PCR primers used to amplify both centromeric and noncentromeric test loci were as described in Meluh and Koshland (1997). Purified HA mAb 12CA5 (Berkeley Antibody Co.) was added at either a 1:250 dilution (resulting in a 4 μg/ml final concentration), or 50 μl of HA antibody pre–cross-linked to CNBr–Sepharose beads was added to the chromatin solution. Crude anti-Mif2p rabbit antisera, provided by Pam Meluh (Carnegie Institute of Washington, Baltimore, MD), was used at a 1:250 dilution and served as a positive control for this assay. For NZ treatment experiments, NZ was added to logarithmically growing cells at 15 or 20 μg/ml final concentration, and incubated for 90 min at 30°C to completely depolymerize MTs before fixation.
Immunofluorescence
Ctf19p was localized by indirect immunofluorescence microscopy as described by Pringle et al. (1989), with a few modifications. To visualize MTs, cells were fixed with 3.7% formaldehyde for 90 min at 30°C. To visualize Ctf19-HAp or Tub4p, cells were fixed for 60 min at 25°C. Primary antibodies were diluted in block solution (4% milk, 2% BSA, and 0.1% Tween in 1× PBS) as follows: 1:50 for anti–α-tubulin (YOL1/34, Serra Lab), 1:5,000 for anti-HA antibody (12CA5, Boehringer Mannheim Corp.), and 1:500 for anti-Tub4p (provided by L. Marschall; see Marschall et al., 1996). Affinity-purified secondary antibodies (Cappel Research Products) were used at 1:1,000 dilution in block solution. YOL1/34 was detected with rhodamine conjugated goat anti–rat antibodies, anti-HA antibody was detected with fluorescein conjugated goat anti–mouse antibodies, and anti-Tub4p was detected with CY3 conjugated goat anti–rabbit antibodies (Jackson ImmunoResearch Laboratories, Inc.). When costaining for Ctf19-HAp and either α-tubulin or Tub4p, primary and secondary antibodies were applied in rounds, with anti-HA and goat anti–mouse applied first, to avoid background from secondary antibody cross-reactivity. Cells were examined using fluorescence microscopy with a Zeiss Axioskop fitted with UV, fluorescein isothiocyanate, and rhodamine/CY3 optics and a 100× objective. Digital images were captured using a cooled charged-coupled device (CCD) camera (Photometrix) and IPLabSpectrum software (Signal Analytics).
Results
Isolation and Characterization of the CTF19 Gene
An SDL screen (Kroll et al., 1996) was performed in which CTF13 was inducibly overexpressed in a subset of 12 potential kinetochore ctf mutants, which tested positive for the centromere transcription readthrough assay (Doheny et al., 1993). Overexpression of CTF13 caused SDL in combination with four mutants within this set: ndc10-42, ctf17-61, ctf19-26 (YCTF26), and ctf19-58 (YCTF58). The chromosome missegregation ctf phenotype (visualized by the formation of red sectors in a white colony) of ctf19-58 was confirmed by genetic analysis to be due to a single nuclear mutation, and the corresponding gene was cloned by complementation of this phenotype (see Materials and Methods; Fig. 1 a). CTF19 was localized to the right arm of chromosome XVI using physical mapping methods. Nucleotide sequencing of the 2-kb CTF19 clone revealed a previously unidentified 1.2-kb ORF that encodes a protein of 369 amino acids with a predicted molecular mass of 40 kD. Database searching revealed no significant overall homology at the amino acid level. Protein motif searching (ProSite) revealed a putative leucine zipper beginning at amino acid 131 (Fig. 1 d). The sequence and mapping data of CTF19 (YPL018W) was corroborated upon release of the complete sequence of the S. cerevisiae genome (Goffeau et al., 1996).
ctf19-26 was confirmed to be an independent allele of CTF19 using four lines of evidence: first, the cloned CTF19 DNA complements the ctf19-26 sectoring phenotype; second, the ctf19-58/ctf19-26 diploid exhibits a chromosome missegregation phenotype; third, when this diploid is sporulated, all spores which bear a chromosome fragment display the sectoring phenotype (Basrai, M., personal communication); and fourth, when ctf19-26 is crossed to a wild-type strain with the LEU2-marked genomic CTF19 locus (YPH1313), the ctf phenotype always segregated away from the LEU2 marker.
A complete deletion of the CTF19 ORF was generated using PCR-mediated gene disruption (Lorenz et al., 1995). The CTF19 ORF was replaced with two different marker genes, creating ctf19Δ1::HIS3 and ctf19Δ1::TRP1. The deletion strains are viable, display no temperature conditional phenotypes, and exhibit a chromosome missegregation phenotype similar to that seen in strains containing either of the original mutant alleles, ctf19-58 or ctf19-26. Upon overexpression of CTF13 under the control of a GAL1 promoter, SDL was observed in the deletion strain, comparable to that seen with ctf19-58 and ctf19-26, albeit somewhat more pronounced.
ctf19 Mutants Display Phenotypes Consistent with a Role in Kinetochore Function
Strains carrying the ctf19Δ1, ctf19-58, or ctf19-26 mutations were tested for sensitivity to benzimidazole compounds. These agents cause depolymerization of MTs, and it has been observed that kinetochore mutants, as well as mitotic checkpoint mutants, are sensitive to compounds such as benomyl (Spencer et al., 1990; Hoyt et al., 1991; Li and Murray, 1991). All three strains are highly sensitive to 10 μg/ml of benomyl at 25°C, and slightly sensitive at the level of 5 μg/ml (Fig. 2 a). Isogenic wild-type strains grow well with 10 μg/ml of benomyl at 25°C, and are sensitive to 20 μg/ml. As an interpretive note, the bub (Hoyt et al., 1991) and mad (Li and Murray, 1991; Hardwick and Murray, 1995) mutants are also sensitive to 10 μg/ml of benomyl.
Figure 2.
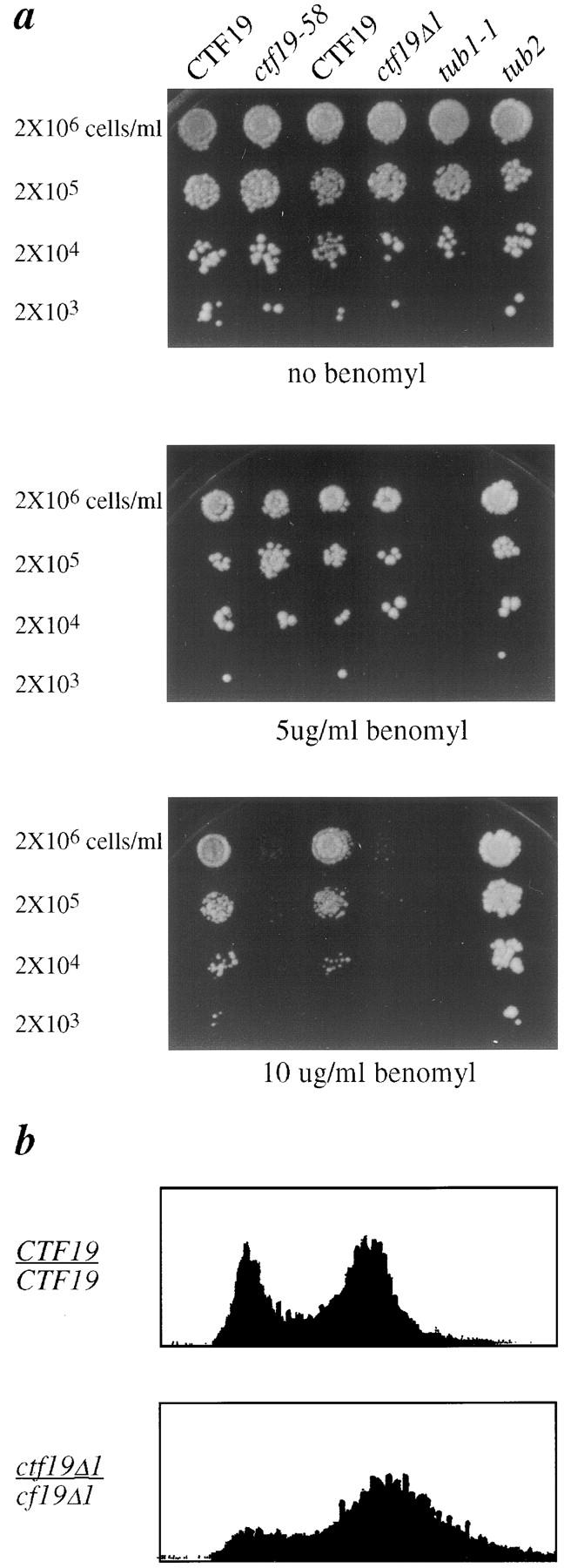
Phenotypes of ctf19 mutants. (a) Benomyl sensitivity; serial dilutions of yeast cells (YPH278, YCTF58, YPH1125, YPH1319, YPH311, and YPH312) were spotted onto YPD plates containing the indicated concentrations of benomyl, and were photographed after 4 d at 25°C. (b) Flow cytometry profiles for homozygous diploid cultures of wild-type CTF19 (YPH501) and ctf19Δ1 (YPH1330) strains at 25°C.
Examination of possible cell cycle defects revealed that a ctf19Δ1/ctf19Δ1 mutant shows a G2/M accumulation with 2C DNA content in a logarithmically growing culture (Fig. 2 b). Cytological analysis of these cells reveals an accumulation of large budded cells with the nucleus at or near the neck (29% in ctf19Δ1 versus 6% in wild-type). A similar G2 delay is seen in ctf13-30, ndc10-42 (Doheny et al., 1993), cep3-1/-2 (Strunnikov et al., 1995), and skp1-4 (Connelly and Hieter, 1996) kinetochore structural mutants at their nonpermissive temperatures. While there is an accumulation of large budded uninucleate cells, the number of telophase cells in the ctf19 null mutant remains similar to that seen for wild-type, 17% versus 16%, respectively. In contrast, the percentage of telophase cells in a ctf13-30 mutant at the nonpermissive temperature drops to 3%. This is consistent with the notion that ctf13-30 cells experience a strong arrest at G2/M at the nonpermissive temperature, whereas ctf19 null cells experience a shorter delay, most likely because the lesion is less severe.
Chromosome missegregation in the ctf19 mutants was quantitated in homozygous diploid strains using colony color half sector analysis (Koshland and Hieter, 1987). Chromosome loss (1:0 segregation) results in a pink/red half-sectored colony, whereas nondisjunction (2:0 segregation) results in a white/red half-sectored colony. The rate of chromosome loss in a ctf19Δ1/ctf19Δ1 mutant is ∼100 times greater than that of wild-type, and the rate of nondisjunction is 60-fold higher than wild-type (Table II). For ctf19 mutant alleles, the rates of chromosome loss and nondisjunction are 77- and 17-fold higher than wild-type for ctf19-58/ctf19-58, and 37- and 13-fold higher for ctf19-26/ctf19-26, respectively. Thus, both chromosome loss and gain are occurring in ctf19 mutant diploids, and the deletion strain is affected the most severely. Similar rates of chromosome loss and nondisjunction have been reported for essential kinetochore mutants ctf13-30 (Doheny et al., 1993) and skp1-4 (Connelly and Hieter, 1996).
Table II.
Rates of Chromosome Missegregation Events in ctf19 Mutants
| Genotype* | 1:0 Events‡ | 2:0 Events§ | Total # colonies | |||
|---|---|---|---|---|---|---|
| ctf19Δ::TRP1 | ||||||
| ctf19Δ::TRP1 | 2.8 | 1.8 | 2,737 | |||
| ctf19-58 | ||||||
| ctf19-58 | 2.3 | 0.5 | 2,533 | |||
| ctf19-26 | ||||||
| ctf19-26 | 1.1 | 0.4 | 3,180 | |||
| CTF19 ‖ | ||||||
| CTF19 | 0.03 | 0.03 | 29,046 |
Mutants of the above genotypes were plated to single colonies and visual sectoring phenotypes were scored. Colonies scored as half-sectored were ≥50% red (Koshland and Hieter, 1987; Gerring et al., 1990).
Strains used were YKH67, YKH588, YKH590, and YPH279
.
1:0 events (chromosome loss) were scored as colonies that are half red, half pink. Rates are shown as events per 100 cells.
2:0 events (nondisjunction) were scored as above for half red, half white colonies. Rates are shown as events per 100 cells.
∥ Rates for wild-type YPH279 are as reported by Gerring et al. (1990).
Ctf19p Interacts Genetically with Components of the Kinetochore and the Mitotic Spindle Checkpoint
To explore a potential role of Ctf19p in kinetochore function, genetic analysis was used to look for synthetic phenotypes between ctf19 mutants and known kinetochore mutants. As our first genetic test, we used the SDL screen (Kroll et al., 1996) to assay the effect of overexpression of CTF19 in a wild-type background, and in strains containing mutations in each of the four subunits of the CBF3 kinetochore complex. CTF19, the reference gene, was placed under a GAL1 promoter (pKH21, Fig. 1 b) and this plasmid was transformed into a set of target mutants (ndc10, cep3, ctf13, and skp1). Growth was assessed upon galactose induction, as previously described (Kroll et al., 1996). Results of this dosage study are summarized in Table III. SDL was seen when CTF19 was overexpressed in the background of two independent mutant alleles of NDC10, ndc10-42, and ndc10-1, thus providing another genetic link between CTF19 and the kinetochore. Moreover, overexpression of NDC10 results in lethality in ctf19-26 and ctf19-58 mutants. The plasmid expressing CTF13 under a GAL1 promoter (pKF88) was also transformed into these mutant strains and viability assessed. Overexpression of CTF13 results in SDL in ndc10-42 (as previously described), and also in ndc10-1 at elevated temperature (30°C), as well as cep3-1 and cep3-2 (Table III).
Table III.
Dosage Studies with Kinetochore Mutants
| Mutant strain | Gene overexpressed* | |||
|---|---|---|---|---|
| GAL-CTF13 | GAL-CTF19 | |||
| ctf13-30 | + | + | ||
| ndc10-42 | SDL | SDL | ||
| ndc10-1 | SDL‡ | SDL‡ | ||
| ndc10-2 | + | + | ||
| skp1-3 | + | + | ||
| skp1-4 | DS | + | ||
| cep3-1 | SDL | + | ||
| cep3-2 | SDL | + | ||
| ctf19Δ1::TRP1 | SDL | + | ||
| ctf19-58 | SDL | + | ||
| ctf19-26 ‡ | SDL | + | ||
+, normal growth; SDL, synthetic dosage lethality; DS, dosage suppression (i.e., rescue of viability at the nonpermissive temperature).
Phenotypes are at 25°C, unless otherwise indicated.
Temperature ≥30°C.
To look for additional genetic interactions between CTF19 and CBF3 kinetochore mutations, double mutants were made between a ctf19 null mutant and the same CBF3 subunit mutations used in the dosage studies above. The heterozygous diploids were sporulated at 25°C and tetrads analyzed (Table IV). To detect any conditional interactions, all double mutants recovered at 25°C were assayed for growth at successively higher semipermissive temperatures. The ctf19Δ1 ctf13-30 double mutant showed conditional SL at 30°C, which is lower than the nonpermissive temperature of ctf13-30 alone (35.5°C; Doheny et al., 1993). ctf19Δ1 ndc10-42 double mutants could not be recovered at 25°C, although ctf19Δ1 ndc10-1 double mutants were viable at all permissive temperatures tested, demonstrating allele specific SL. ctf19Δ1 skp1-4 double mutants were conditionally synthetic lethal at 28°C, whereas ctf19Δ1 skp1-3 double mutants were viable at all permissive temperatures. This allele specificity is significant since the skp1-4 allele arrests in G2, and the skp1-3 allele arrests in G1 at the respective nonpermissive temperatures (Connelly and Hieter, 1996), suggesting that the interaction detected between CTF19 and SKP1 is within the G2/M phase of the cell cycle. Similarly, a conditional synthetic lethal interaction was detected between ctf19Δ1 and sgt1-3, but not with sgt1-5 (SGT1 is a suppressor of skp1-4; Kitagawa, K., personal communication). sgt1-3 arrests at G2/M, whereas sgt1-5 arrests at G1/S. Therefore, this allele specificity is analogous to the ctf19Δ1-skp1-4 interaction. Finally, both mutant alleles of CEP3 tested, cep3-1 and cep3-2, were synthetically lethal with ctf19Δ1 at 25°C. Thus, CTF19 genetically interacts with all four components of the CBF3 CDEIII-binding complex.
Table IV.
Synthetic Lethal Interactions between ctf19Δ1 and Kinetochore Mutants
| Double mutant genotype | Phenotype | 4-Spore tetrads | 3-Spore tetrads | 2-Spore tetrads | 1-Spore tetrads | Total tetrads | Viable spores* | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # | % | |||||||||||||
| ctf19Δ::HIS3 ctf13-30 | CSL‡ | 19 | 3 | 0 | 0 | 22 | 97 | |||||||
| ctf19Δ::HIS3 ndc10-42 | SL | 4 | 15 | 9 | 2 | 30 | 68 | |||||||
| ctf19Δ::TRP1 ndc10-1 | Viable | 21 | 4 | 0 | 0 | 25 | 96 | |||||||
| ctf19Δ::TRP1 cep3-1 | SL | 7 | 15 | 5 | 2 | 29 | 72 | |||||||
| ctf19Δ::TRP1 cep3-2 | SL | 5 | 11 | 4 | 0 | 20 | 76 | |||||||
| ctf19Δ::TRP1 skp1-3 | Viable | 14 | 2 | 0 | 0 | 16 | 97 | |||||||
| ctf19Δ::TRP1 skp1-4 | CSL§ | 15 | 2 | 1 | 0 | 18 | 94 | |||||||
| ctf19Δ::TRP1 sgt1-3 | CSL‖ | 18 | 2 | 8 | 2 | 30 | 80 | |||||||
| ctf19Δ::TRP1 sgt1-5 | Viable | 24 | 0 | 2 | 1 | 27 | 94 | |||||||
| ctf19Δ::TRP1 mif2-3 | SL | 5 | 20 | 4 | 1 | 30 | 74 |
Numbers presented are for 25°C. Percent viable spores = number of viable spores from a given mating/total number of possible spores.
ctf19Δ::HIS3 ctf13-30 double mutant spores were inviable at 30°C. The nonpermissive temperature of ctf13-30 is 36°C.
ctf19Δ::TRP1 skp1-4 double mutant spores were inviable at 28°C. The nonpermissive temperature of skp1-4 is 37°C.
ctf19Δ::TRP1 sgt1-3 double mutant spores were inviable at 30°C. The nonpermissive temperature for sgt1-3 is 37°C. SL, synthetic lethality; CSL, conditional synthetic lethality, at temperatures indicated.
To address the specificity of these genetic interactions, a ctf19Δ1 strain was crossed to strains containing cbf1:: TRP1, cse4-1, or mif2-3 mutations, representing other proteins which function at the kinetochore, as well as tub1-1 or tub2 tubulin mutations. No synthetic lethal interactions were detected in any of the corresponding double mutants, except for ctf19Δ1 mif2-3, which is inviable (Table IV). Interestingly, Meluh and Koshland (1997) have shown that Mif2p interacts with CEN DNA primarily through CDEIII. Thus, the genetic interactions identified between CTF19 and the kinetochore appear specific for CBF3 and other CDEIII-associated components.
The ctf19Δ1 ctf13-30 double mutant was a valuable reagent, because double mutants could be recovered at 25°C, but were conditionally synthetic lethal at 30°C. It has been shown that ctf13-30 alone causes initiation of the mitotic checkpoint at the nonpermissive temperature, resulting in delay at the G2/M point of the cell cycle (Doheny et al., 1993). When ctf13-30 is combined with mad2 or bub1 mitotic checkpoint mutants, the cells do not arrest upon shift to the nonpermissive temperature, and rapid death ensues (Wang and Burke, 1995; Pangilinan and Spencer, 1996). In contrast, when ctf13-30 is combined with a ctf19Δ1 mutant, although a rapid loss in viability is observed upon shift to the nonpermissive temperature, the checkpoint appears to be intact judging by a G2/M delay visible in flow cytometry profiles at 25°C and 37°C (data not shown). This G2/M delay in the double mutant is accompanied by an accumulation of large budded uninucleate cells with short spindles. The inability of the ctf19Δ1 ctf13-30 double mutant to recover after exposure to the nonpermissive temperature suggests that there is a more severe structural aberration than in the ctf13-30 mutant alone.
In light of the observations that the mitotic spindle checkpoint responds to impaired kinetochore function (Wang and Burke, 1995; Pangilinan and Spencer, 1996; Wells and Murray, 1996), we asked directly whether CTF19 genetically interacts with BUB1, BUB2, BUB3, or MAD2 mitotic checkpoint genes. To do this, we again used the SDL assay. The CTF19 overexpression plasmid (pKH21) was transformed into mitotic checkpoint target mutants, bub1Δ, bub2Δ, bub3Δ, and mad2Δ. Upon induction of CTF19 on galactose-containing medium, growth was assessed as previously described. SDL was observed in the bub1Δ and bub3Δ strains, but no defect of growth was seen with mad2Δ or bub2Δ. To interpret these results, we also determined the effect of overexpressing CTF13 in the same target mutants, especially since the ctf13-30 mutation triggers the mitotic checkpoint resulting in a G2/M delay, as described above. When CTF13 was overexpressed in combination with bub2Δ, bub3Δ, and mad2Δ, SDL was observed with bub3Δ, but not with mad2Δ or bub2Δ.
To test for synthetic lethal interactions, crosses were made between strains carrying a ctf19 null mutation and strains carrying either bub1Δ, bub2Δ, bub3Δ, or mad2Δ mutations (Table V). A rad9Δ mutation, previously characterized as defective in monitoring incomplete replication and DNA damage (Weinert and Hartwell, 1988; Lydall and Weinert, 1995), was also tested. SL was seen when ctf19Δ1 was combined with bub1Δ, bub3Δ, or mad2Δ, but not with bub2Δ or rad9Δ (Table V). Bub1p, Bub3p, and Mad2p are all necessary for the initiation of the mitotic checkpoint pathway and are sufficient for shorter delays, as seen with low levels of NZ, whereas Bub2p is required for maintaining longer delays, as seen with high levels of NZ (Wang and Burke, 1995; Pangilinan and Spencer, 1996). These synthetic dosage lethal and synthetic lethal genetic interaction results are consistent with the notion that ctf19 mutations invoke a mild premitotic delay, which does not require Bub2p. Taken together, these genetic studies strongly suggest that Ctf19p plays a structural role at the kinetochore.
Table V.
Synthetic Lethal Interactions between ctf19Δ and Mitotic Checkpoint Mutants
| Double mutant genotype | Phenotype | 4-Spore tetrads | 3-Spore tetrads | 2-Spore tetrads | 1-Spore tetrads | Total tetrads | Viable spores* | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # | % | |||||||||||||
| ctf19Δ::HIS3 bub1Δ::LEU2 | SL | 2 | 6 | 8 | 4 | 20 | 58 | |||||||
| ctf19Δ::HIS3 bub2Δ::LEU2 | Viable | 12 | 3 | 3 | 1 | 19 | 84 | |||||||
| ctf19Δ::HIS3 bub3Δ:: | SL‡ | 16 | 8 | 6 | 1 | 31 | 81 | |||||||
| LEU2 + pBUB3-URA3 ‡ | ||||||||||||||
| ctf19Δ::TRP1 mad2Δ::HIS3 | SL | 6 | 10 | 12 | 0 | 28 | 70 | |||||||
| ctf19Δ::TRP1 rad9Δ::HIS3 | Viable | 26 | 1 | 3 | 0 | 30 | 94 |
Numbers presented are for 25°C.
Analysis of spores from this diploid required the inclusion of a plasmid (pBUB3-URA3) before sporulation to achieve sufficient sporulation efficiency and acceptable viability of spore products. Double mutant spores containing pBUD3-URA3 were not viable on FOA plates, thus confirming synthetic lethality between bub3 and ctf19.
Ctf19p Interacts with Centromere DNA In Vivo
Sucrose gradient sedimentation analysis revealed that Ctf19-HAp sediments with an S value of 20 (data not shown). This suggests that Ctf19p may be part of a large protein complex in the cell. There is no evidence that Ctf19p is part of the CBF3 CDEIII-binding complex, as Ctf19p is not required for the normal CEN DNA bandshift in vitro, and antibodies to an epitope-tagged Ctf19p do not result in a supershift of the CBF3-CEN DNA complex (data not shown; Stemmann et al., 1999). In addition, coimmunoprecipitation experiments using crude yeast cell extracts were unable to detect a direct interaction between Ctf19p and CBF3 proteins.
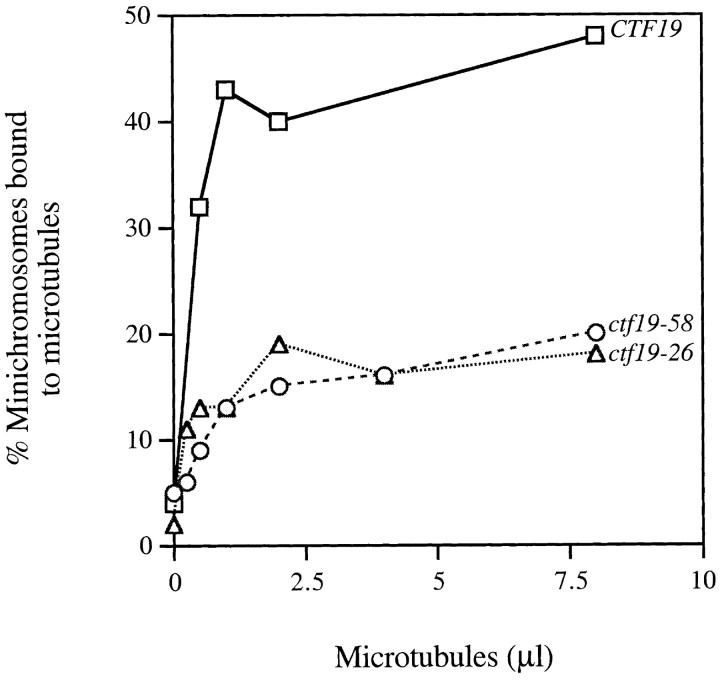
Knowing that Ctf19p is not a part of the CDEIII-binding complex detected by CEN DNA bandshift in vitro, we investigated other ways in which it could be involved in kinetochore function. One possibility is that Ctf19p plays a role in binding of MTs to kinetochores. To test this hypothesis, an in vitro assay was used which measures the ability of yeast centromeres to bind to MTs (Kingsbury and Koshland, 1991). In this assay, minichromosomes containing a centromere are introduced into wild-type or mutant strains, and purified chromatin is prepared. Taxol-stabilized bovine MTs are added to the lysate and sedimented. The supernatant and pellet are separated, DNA from each fraction is extracted, and the relative amount of the minichromosome in each fraction is determined. In wild-type cultures with wild-type centromeres on the minichromosomes, the minichromosomes sediment with the MTs. CEN DNA mutations in CDEII and CDEIII abolish the centromere's ability to bind MTs (Kingsbury and Koshland, 1991). Similarly, trans-acting mutations in CEN-binding proteins also inhibit the ability of the minichromosome to bind MTs, exemplified by the cep3-1 mutant (Strunnikov et al., 1995). Two independent alleles of CTF19 were tested in this assay, ctf19-58 and ctf19-26, and both were shown to exhibit a dramatic defect in centromere-dependent binding of minichromosomes to MTs (Fig. 3). Therefore, knowing that Ctf19p interacts genetically with the CBF3 complex, is important for binding MTs to centromeres of minichromosomes, and sediments with an S value consistent with being part of a protein complex, we next investigated whether Ctf19p is involved in a higher order centromere complex.
Figure 3.
ctf19 mutants exhibit reduced MT binding to minichromosomes. Cleared lysates were prepared from wild-type CTF19 (solid line with squares), ctf19-26 (dotted line with triangles), and ctf19-58 (dashed line with circles) cells containing minichromosomes. Increasing amounts of taxol stabilized bovine MTs were added to the lysates for 15 min at 23°C. MTs were pelleted, and the percentage of minichromosomes that cosedimented with MTs was determined (see Kingsbury and Koshland, 1991).
To examine whether Ctf19p physically interacts with a kinetochore macromolecular complex, we used an in vivo cross-linking and immunoprecipitation strategy (Meluh and Koshland, 1997). Formaldehyde cross-linked chromatin (2 h fixation) prepared from an epitope-tagged CTF19-3HA strain and a wild-type untagged control strain were sonicated to shear the DNA and immunoprecipitated with anti-HA antibody. After reversing the cross-links, the presence of specific DNA sequences in the immunoprecipitates was assessed by PCR analysis. Mif2p, which was previously shown to associate with CEN DNA in vivo (Meluh and Koshland, 1997), served as a positive control for this assay. Two CEN DNA sequences were tested, CEN3 and CEN16, and both were found to be present in the Ctf19-HAp immunoprecipitate, but not in the untagged control strain or in the mock-treated control (Fig. 4 a). The anti-Mif2p immunoprecipitate also contained these CEN DNA sequences. To test if the interaction detected is specific for CEN DNA, two noncentromeric loci, PGK1 and HMRa, which are AT-rich intergenic regions located on yeast chromosome III, were analyzed. Both of these sequences were not found in the Ctf19-HAp immunoprecipitate, as was the case with the Mif2p positive control (Fig. 4 a). Thus, as predicted by the numerous genetic interactions with kinetochore protein components, Ctf19p specifically associates with the centromere in chromatin preparations.
Figure 4.
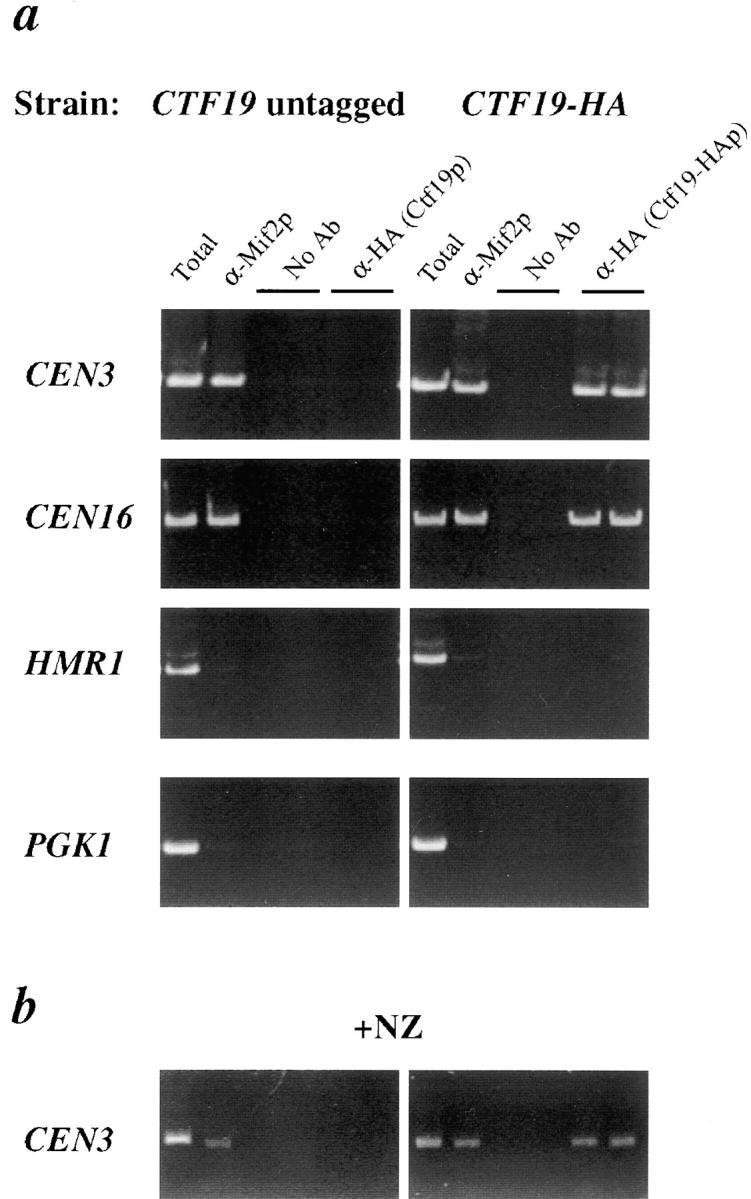
Coimmunoprecipitation of CEN DNA with Ctf19p. (a) Formaldehyde cross-linked chromatin (2 h fixation) prepared from a wild-type CTF19 untagged strain (YPH500) and a CTF19-HA epitope-tagged strain (YPH1327) were immunoprecipitated with anti-HA antibody, anti-Mif2p antiserum, or mock-treated (No Ab). Experimental reactions were prepared in duplicate. Total input material (3 μl chromatin solution, at a 1:5 dilution) and coimmunoprecipitated DNA (equal to ∼30 μl chromatin solution) were analyzed by PCR with primers specific for CEN3 (244 bp), CEN16 (345 bp), or two noncentromeric loci on chromosome III, HMR1 (314 bp) and PGK1 (288 bp). (b) The same wild-type untagged and CTF19-HA tagged strains used in a were treated with NZ, then fixed in formaldehyde, processed, and immunoprecipitated as above. Shown is the CEN3 PCR product. A similar result was seen with primers for CEN16. As in a, no PCR product was seen in the α-mif2 or α-HA immunoprecipitate for the noncentromeric loci tested (not shown).
It is plausible that the association of Ctf19p with the centromere is transient and contact may be made with kinetochore proteins upon MT attachment. To test whether MTs are necessary for Ctf19p to associate with the centromere, the chromatin immunoprecipitation assay was performed with strains grown in the presence of NZ, a MT depolymerizing agent. Results from this experiment show that, as with Mif2p and Ndc10p (not shown), Ctf19p specifically immunoprecipitates CEN DNA, and not noncentromeric loci, even in the presence of NZ (Fig. 4 b). These data further support the conclusion that Ctf19p is in fact part of a centromere–protein complex.
Ctf19p Localizes Near the Spindle Pole Body (SPB)
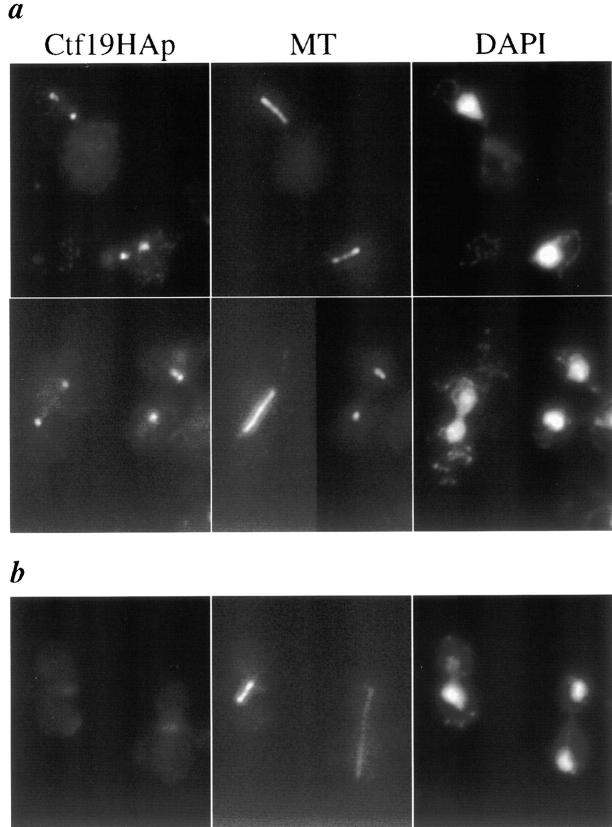
The localization of Ctf19p in yeast cells was determined by indirect immunofluorescence using an HA epitope fusion construct (pKH32). To avoid potential copy number effects, the CTF19-3HA fusion was integrated into the genome at the leu2Δ1 locus (leu2Δ1::CTF19-3HA, YPH1327). For immunofluorescence studies, an asynchronous population of cells from strains containing CTF19-3HA (either plasmid-based or integrated, YPH1324 or YPH1327) or an untagged control (YPH1323 or YPH500) were formaldehyde-fixed, and stained with anti-HA antibody and anti–α-tubulin, which stains spindle MTs. The HA antibody detected a bright dot at the vertex of the MTs in interphase cells, and at the ends of the spindle in mitotic cells (Fig. 5). This staining pattern, reminiscent of SPB staining, is similar to that seen with other centromere proteins, including Ndc10p (Goh and Kilmartin, 1993), Mif2p (Meluh and Koshland, 1997), and Cse4p (Meluh et al., 1998), and is consistent with the notion that centromeres display a nonrandom localization throughout the cell cycle, clustering near the SPB during interphase (G1) and late mitosis (anaphase and telophase; Guacci et al., 1997; Straight et al., 1997). A key difference between the staining pattern observed for Ctf19-HAp and those reported for other centromere proteins occurs during early mitosis. Both Ndc10p and Cse4p are reported to stain the spindle MTs, evident as a short bar, in cells with short spindles. Ctf19-HAp staining, however, resolves into two distinct foci even in cells with short spindles.
Figure 5.
Ctf19p localizes to the SPB region. Immunofluorescence with a CTF19-3HA epitope-tagged strain (YPH1384; a) and an untagged control strain (YPH1323; b). Shown in the first panel of a are two early anaphase cells. The second panel exhibits an anaphase cell, an interphase cell with a single SPB, and a cell with a duplicated SPB. (b) Two representative untagged control cells, one in early anaphase, and one in telophase. No signals were seen with the anti-HA antibody in any untagged control cells analyzed. Ctf19p was detected with anti-HA antibody followed by FITC conjugated goat anti–mouse secondary antibody. Spindle MTs were detected with anti–α-tubulin antibody followed by rhodamine conjugated goat anti–rat secondary antibody. Total DNA was stained with DAPI.
To examine the specificity of this localization, we obtained antibodies against Tub4p (provided by L. Marschall, Stanford, CA), which served as an SPB marker (Sobel and Snyder, 1995; Marschall et al., 1996; Spang et al., 1996). Tub4p, the γ-tubulin homologue in S. cerevisiae, is part of a complex that localizes to the inner and outer plaques of the SPB (Geissler et al., 1996; Knop et al., 1997), where nuclear and cytoplasmic MTs are organized, respectively. Costaining experiments revealed that Ctf19-HAp localizes to the same region as Tub4p (Fig. 6, a and b). Further analysis of cells stained for both Ctf19-HAp and Tub4p revealed that the Ctf19-HAp signal appears to be just interior to (on the nucleoplasmic side of) the Tub4p signal in mid-mitotic cells with short spindles (Fig. 6 d). This observation was examined in more detail by measuring the distance between the Ctf19-HAp signals and Tub4p signals in individual cells. The average distance between Ctf19-HAp signals for 50 mitotic cells was 1.5 μm ± 1.1, whereas the comparable distance between Tub4p signals was 1.8 μm ± 1.1. These measurements correspond to an average distance of 0.2 μm between Ctf19p and Tub4p in mitotic cells. To interpret the significance of this difference, it should be considered that Tub4p resides on both nuclear and cytoplasmic surfaces of the SPB and that the fluorescent signal observed for Tub4p is an average of both signals. Therefore, the distance from the Ctf19-HAp signal to the nuclear side of the SPB may be less than indicated by these measurements. These data suggest that Ctf19p localizes near the SPB, but, as expected, is not likely to be an integral component of the SPB. Consistent with this hypothesis, we were unable to demonstrate a direct interaction between Ctf19-HAp and Tub4p by coimmunoprecipitation (data not shown). It is possible that Ctf19p is part of a complex which resides at the nucleoplasmic surface of the SPB and transiently interacts with kinetochores. However, it is more likely that the staining pattern observed for Ctf19-HAp corresponds to a centromere localization, as noted above for other kinetochore proteins, and is thus compatible with data implicating a kinetochore function for Ctf19p.
Figure 6.
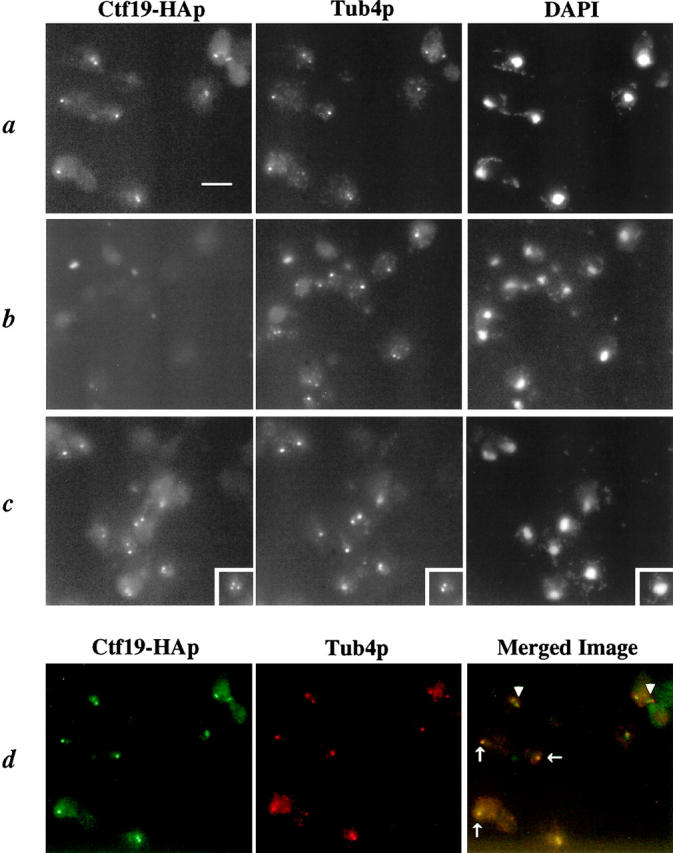
The localization of Ctf19-HAp is similar to Tub4p, a known SPB component. (a) A strain containing CTF19-3HA (integrated into the genome, YPH1327) and (b) a wild-type control strain (YPH500) were fixed and stained with anti-HA antibody to detect Ctf19-HAp, anti-Tub4p, and the DNA stain DAPI. Bar, 5 μm. (c) Cells were treated with 20 μg/ml NZ for 2 h at 25°C to depolymerize the MTs. No MT staining was detected after NZ treatment (not shown). Ctf19-HAp staining remains at the SPB, as does the SPB marker Tub4p. Note that a few cells in this field display an additional Ctf19-HAp signal which does not correspond to a Tub4p signal. The inset clearly demonstrates this phenomenon, which is seen in ∼7% of cells analyzed. (d) A merged image, with Ctf19-HAp stained green (FITC) and Tub4p stained red (Cy3). Note that complete colocalization, observed as yellow spots, is seen in cells with a single SPB or with elongated spindles as in late anaphase or telophase (arrows), whereas in cells with short spindles, the green Ctf19-HAp staining is seen adjacent (interior) to the red Tub4p staining (arrowheads).
To test if we could differentiate between these two hypotheses, we asked whether Ctf19p requires MTs for its localization. Assuming that kinetochores are tethered to the SPB in a MT-dependent manner (Guacci et al., 1997), cells were treated with 15 μg/ml NZ (a concentration that depolymerizes all MTs) and examined by immunofluorescence. For proteins that are known to be integral components of the SPB, localization is unchanged by the presence of NZ (Marschall et al., 1996). Fluorescent in situ hybridization analysis has revealed that centromere localization is, however, changed in the presence of NZ as they are no longer clustered (Guacci et al., 1997). It should be noted that, although NZ and other benzimidazole drugs depolymerize most of the MTs, it is possible that a small amount of the polymer which is undetectable by immunofluorescence remains (Marschall et al., 1996; Jacobs et al., 1988). Interestingly, Ctf19p localization in most cells remained near the SPB, visualized as one (∼66% of cells) or two (∼27% of cells) brightly staining dots (Fig. 6 c). This compares to 69% and 31%, respectively, in cells not treated with NZ. However, ∼7% of the cells (out of 500 cells analyzed) contained three or four discrete signals for Ctf19-HAp. There were also cells observed in which two signals were seen for Ctf19-HAp, but only one signal was seen for Tub4p. These observations indicate that the majority of Ctf19p remains near the SPB in the absence of MTs, however, a portion of the protein in some cells has an altered localization in the presence of NZ. These results may indicate that Ctf19p is present both at the centromere and near the nucleoplasmic surface of the SPB. Whether or not these localization data are suggestive of a function for Ctf19p at the SPB, or merely a consequence of the limitations of the assay, has yet to be determined.
CTF19 Interacts Genetically with a Spindle- and Pole-associated Protein
As a preliminary investigation into a potential role for Ctf19p at the SPB, we tested for genetic interactions between CTF19 and several genes encoding either integral SPB proteins or proteins involved in SPB duplication. No synthetic lethal interactions were detected in any double mutants created between ctf19Δ1 and mps2-1, ndc1-1, cdc31-1, spc42-10, spc110-1, tub4-32, or tub4-34 (data not shown). In comparison to the unequivocal genetic interactions detected between CTF19 and the genes encoding the CBF3 kinetochore components, we conclude that the function of Ctf19p lies at the kinetochore. We also tested for a genetic interaction between CTF19 and NDC80, which encodes a spindle- and pole-associated protein. Interestingly, a conditional synthetic lethal effect was detected in ndc80-1 ctf19Δ1 double mutants at 28°C. This genetic interaction is allele specific, as no conditional SL was detected in ndc80-2 ctf19Δ1 double mutants. NDC80 mutants display phenotypes very similar to a temperature-sensitive mutant of NDC10, which encodes a component of the CBF3 kinetochore complex (Wigge et al., 1998). Although the phenotypes and immunofluorescent staining pattern of Ndc80p are consistent with localization to the kinetochore, no genetic interactions have been detected between NDC80 and CBF3 kinetochore components (Wigge et al., 1998). These authors have concluded that an indirect interaction may exist between Ndc80p and the kinetochore. It is quite possible that such an indirect interaction may occur through Ctf19p.
Discussion
We have identified and characterized the CTF19 gene of S. cerevisiae, and show that it encodes a novel protein which is required for faithful chromosome transmission and efficient binding of centromeres to MTs, associates with a centromere DNA complex in vivo, and localizes to the SPB region. Previously, two uncharacterized mutations, ctf19-58 and ctf19-26, were classified as putative kinetochore mutants by an in vivo centromere transcription readthrough assay (Doheny et al., 1993). ctf19 mutants were further implicated in kinetochore function by our SDL screen (Kroll et al., 1996), in which overexpression of the CTF13 reference gene, encoding an essential CBF3 kinetochore protein, resulted in lethality in the context of the ctf19-58 and ctf19-26 mutations. The phenotypes described here for ctf19 mutants, including chromosome missegregation, increased sensitivity to benomyl, and a G2/M accumulation of logarithmically growing cells, are all consistent with a defect in kinetochore function.
Extensive genetic analysis revealed that CTF19 exhibits strong genetic interactions with both kinetochore structural mutants and mitotic checkpoint mutants. Genetic interactions (SDL, SL, or both) were detected between the ctf19 null mutation and mutant alleles of all four subunits of the CBF3 kinetochore complex, as well as with Mif2p, which has been shown to interact with CEN DNA in a CDEIII-specific manner (Meluh and Koshland, 1997). We also demonstrate that cells defective in, or overexpressing Ctf19p require a functional mitotic checkpoint pathway, as do cells defective in, or overexpressing Ctf13p. The extent and specificity of these interactions strongly implicates Ctf19p as playing a role in kinetochore structure or function.
Ctf19p Functions as Part of a Macromolecular Kinetochore Complex
Consistent with the numerous genetic interactions linking Ctf19p to the kinetochore, as well as the sedimentation data, we demonstrate that Ctf19p associates specifically with centromeric DNA in vivo through formaldehyde cross-linking followed by chromatin immunoprecipitation. Two known kinetochore proteins, Ndc10p and Cbf1p, as well as two implicated kinetochore proteins, Mif2p and Cse4p, have been shown to cross-link wild-type CEN DNA in vivo (Meluh and Koshland, 1997; Meluh et al., 1998). Ndc10p and Mif2p both demonstrate a specific dependence on the integrity of CDEIII (Meluh and Koshland, 1997). A similar specificity of interaction between Ctf19p and CDEIII is described by Lechner and colleagues, who have independently identified Ctf19p as part of a complex of proteins that interacts with CBF3 (Stemmann et al., 1999). These data are consistent with genetic interactions reported here, which are also specific for CBF3 components.
The fact that Ctf19p is able to specifically cross-link CEN DNA does not differentiate between a direct and indirect association. Ctf19p is not a subunit of the CBF3 complex (Stemmann and Lechner, 1996), and is not necessary for the gel mobility shift seen with the assembled kinetochore complex on a CEN DNA fragment (data not shown; Lechner, J., personal communication). Therefore we propose that Ctf19p interacts indirectly with CEN DNA, likely through interactions with the CBF3 complex, or perhaps through a larger macromolecular complex whose assembly is initiated by recruitment of CBF3 to CDEIII (Sorger et al., 1994; Meluh and Koshland, 1997). Given the ability of Ctf19p to cross-link to CEN DNA, and the defect seen in the ability of ctf19 mutants to bind centromeres of minichromosomes to MTs, Ctf19p would be a good candidate for a factor which interacts with CBF3 and is necessary to form active spindle MT-binding complexes. The fact that Ctf19p is able to associate with centromeric DNA in the presence of NZ suggests that MTs are not required for this association, and places Ctf19p at the kinetochore instead of on the spindle. Similar results were observed for Mif2p and Ndc10p. We are currently testing whether Ctf19p is required for attachment of reassembled kinetochore complexes in vitro to polymerized MTs.
Implications of a Kinetochore Protein with SPB Localization
The immunolocalization pattern of Ctf19p is consistent with kinetochore proteins, which is strongly supported by the genetic and biochemical data presented here. Other proteins which have been biochemically placed at the centromere, including Ndc10p, Mif2p, and Cse4p, show immunofluorescent staining patterns similar to Ctf19p. The SPB staining pattern observed in G1 cells and in late mitotic cells could be due to the effect of centromere clustering, a phenomenon seen in higher eukaryotes and demonstrated in yeast by fluorescent in situ hybridization (Guacci et al., 1997). Studies on cell cycle dependent centromere positioning in S. cerevisiae reveal that in G1 cells, centromeres are loosely clustered around the SPB, similar to that reported for fission yeast and mammalian cells (Ferguson and Ward, 1992; Funabiki et al., 1993; Vourc'h et al., 1993). In mitosis before anaphase (mid M), centromeres are centrally localized within the nucleus, away from the SPB, reminiscent of metaphase in larger eukaryotic cells. In anaphase and telophase cells, centromeres are clustered tightly and proximal to the SPBs. One caveat to interpreting the Ctf19p localization as consistent with it being a CEN binding protein is that, unlike Ndc10p (Goh and Kilmartin, 1993) and Cse4p (Meluh et al., 1998), Ctf19p does not display spindle staining, as may be expected for centromere associated proteins when kinetochores are forming bipolar attachments to the spindle MTs. The absence of spindle staining for Ctf19p may reflect limitations of the immunofluorescence assay, as other kinetochore proteins, including Ctf13p and Cep3p, have not been visualized in the cell by traditional immunofluorescent techniques, or it may reflect a unique role for Ctf19p in mitosis, perhaps functioning to tether kinetochores to the SPB. Given the prominent role of anaphase B in sister chromatid separation in budding yeast (Straight et al., 1997), Ctf19p may be important in stabilizing the link between kinetochores and each SPB during spindle pole separation.
The results of immunofluorescence with Ctf19-HAp in the presence of NZ suggest that at least some amount of Ctf19p localizes adjacent to the SPB, as opposed to Ctf19p residing solely at the kinetochores, because the positioning of kinetochores near the SPB is believed to be dependent on MTs. In the presence of NZ, it has been observed that centromeres are no longer clustered, but rather disperse throughout the nucleus (Guacci et al., 1997), presumably because MTs are no longer present for the kinetochores to remain attached to the SPB. However, the procedures used to fix and process nuclei for in situ hybridization in yeast may be more disruptive than that used for immunofluorescence, and theoretically could contribute to an aberrant delocalization of centromeres after treatment with NZ. Although Ctf19-HAp does maintain a discrete localization in the presence of NZ, we did observe a few examples (∼7% of cells analyzed) in which more than two fluorescent signals were seen for Ctf19-HAp which did not correlate with a Tub4p signal. We reason that, because a small polymer of MTs may still be present at the face of the SPB even after NZ treatment (Marschall et al., 1996; Jacobs et al., 1988), Ctf19p may be part of a complex that is peripheral to the SPB. The localization of this complex may be less stable in the presence of MT depolymerizing agents, accounting for the small population of cells that exhibit more than two Ctf19-HAp signals. Wigge et al. (1998) have identified several new spindle pole and spindle-associated proteins through analysis of purified spindle preparations with MALDI mass spectrometry. Ndc80p is of particular interest because it stains spindles as well as poles, and ndc80-1 mutants display phenotypes similar to yeast kinetochore mutants, including chromosome missegregation and anaphase defects. In addition, Ndc80p is a potential homologue of human HEC protein, which localizes to the centromere. However, no genetic interactions were detected between NDC80 and three of the CBF3 components tested. Ndc80p partially copurifies with the factors which bind kinetochores to MTs (Sorger et al., 1994), but is absent from the final fraction (Wigge et al., 1998). Since Ctf19p interacts with the centromere, both genetically and biochemically, and it genetically interacts with Ndc80p, we propose that Ctf19p provides a link between the mitotic spindle and the kinetochore in budding yeast.
Acknowledgments
We thank O. Stemmann, J. Ortiz, S. Rank, and J. Lechner for communicating unpublished results; P. Meluh, J. Kingsbury, S. Strunnikov, D. Koshland, F. Pangilinan, F. Spencer, S. Michaelis, L. Marschall, M. Basrai, and J. Lechner for reagents and advice; M. Winey, J.V. Kilmartin, and T. Stearns for strains; and Hieter lab members for their helpful discussions and support.
This work was supported by a grant from the National Cancer Institute (CA16519) to P. Hieter.
Abbreviations used in this paper: bub
budding uninhibited by benzimidazole
- CDEs
centromere DNA elements
- ctf
chromosome transmission fidelity
- 5-FOA
5-fluoroorotic acid
- mad
mitotic arrest deficient
- MT
microtubule
- NZ
nocodazole
- ORF
open reading frame
- SDL
synthetic dosage lethality
- SL
synthetic lethality
- SPB
spindle pole body
References
- Baker RE, Masison DC. Isolation of the gene encoding the Saccharomyces cerevisiaecentromere-binding protein CP1. Mol Cell Biol. 1990;10:2458–2467. doi: 10.1128/mcb.10.6.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K, Hill A, Kenna M, Saunders M. The structure of a primitive kinetochore. Trends Biochem Sci. 1989;14:223–227. doi: 10.1016/0968-0004(89)90031-5. [DOI] [PubMed] [Google Scholar]
- Bloom KS, Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Cai M, Davis RW. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990;61:437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiaekinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin CW, Kaplan KB, Sorger PK. Probing the architecture of a simple kinetochore using DNA-protein cross-linking. J Cell Biol. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M, Ward DC. Cell cycle dependent chromosomal movement in pre-mitotic human T-lymphocyte nuclei. Chromosoma. 1992;101:557–565. doi: 10.1007/BF00660315. [DOI] [PubMed] [Google Scholar]
- Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson IA, Lerner RA, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiaeby use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk M, Hegemann JH, Philippsen P. Chromatin digestion with restriction endonucleases reveals 150–160 bp of protected DNA in the centromere of chromosome XIV in Saccharomyces cerevisiae. . Mol Gen Genet. 1989;219:153–160. doi: 10.1007/BF00261171. [DOI] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Soues S, Kilmartin J, Schiebel E. The spindle pole body component Spc98p interacts with the gamma-tubulin–like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment [published erratum appears in EMBO (Eur. Mol. Biol. Organ.) J.1996. 15:5124] EMBO (Eur Mol Biol Organ) J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Gerring SL, Spencer F, Hieter P. The CHL 1 (CTF 1) gene product of Saccharomyces cerevisiaeis important for chromosome transmission and normal cell cycle progression in G2/M. EMBO (Eur Mol Biol Organ) J. 1990;9:4347–4358. doi: 10.1002/j.1460-2075.1990.tb07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau, A., B.G. Barrell, H. Bussey, R.W. Davis, B. Dujon, H. Feldmann, F. Galibert, J.D. Hoheisel, C. Jacq, M. Johnston, et al. 1996. Life with 6000 genes. Science. 274:546, 563–567. [DOI] [PubMed]
- Goh PY, Kilmartin JV. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. . J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky GJ. Kinetochores, microtubules and the metaphase checkpoint. Trends Cell Biol. 1995;5:143–148. doi: 10.1016/s0962-8924(00)88968-0. [DOI] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Centromere position in budding yeast: evidence for anaphase A. Mol Biol Cell. 1997;8:957–972. doi: 10.1091/mbc.8.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Murray AW. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiaegenes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs CW, Adams AEM, Szaniszlo PJ, Pringle JR. Functions of microtubules in the Saccharomyces cerevisiaecell cycle. J Cell Biol. 1988;107:1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Lechner J, Carbon J. Isolation and characterization of a gene (CBF2) specifying a protein component of the budding yeast kinetochore. J Cell Biol. 1993;121:513–519. doi: 10.1083/jcb.121.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury J, Koshland D. Centromere-dependent binding of yeast minichromosomes to microtubules in vitro. Cell. 1991;66:483–495. doi: 10.1016/0092-8674(81)90012-x. [DOI] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiaeand functions in microtubule organization and spindle pole body duplication. EMBO (Eur Mol Biol Organ) J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D. Mitosis: back to the basics. Cell. 1994;77:951–954. doi: 10.1016/0092-8674(94)90432-4. [DOI] [PubMed] [Google Scholar]
- Koshland D, Hieter P. Visual assay for chromosome ploidy. Methods Enzymol. 1987;155:351–372. doi: 10.1016/0076-6879(87)55024-8. [DOI] [PubMed] [Google Scholar]
- Kroll ES, Hyland KM, Hieter P, Li JJ. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics. 1996;143:95–102. doi: 10.1093/genetics/143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J. A zinc finger protein, essential for chromosome segregation, constitutes a putative DNA binding subunit of the Saccharomyces cerevisiaekinetochore complex, Cbf3. EMBO (Eur Mol Biol Organ) J. 1994;13:5203–5211. doi: 10.1002/j.1460-2075.1994.tb06851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast [published erratum appears in Cell. 1994. 79:388] Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Link AJ, Olson MV. Physical map of the Saccharomyces cerevisiaegenome at 110-kb resolution. Genetics. 1991;127:681–698. doi: 10.1093/genetics/127.4.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Muir RS, Lim E, McElver J, Weber SC, Heitman J. Gene disruption with PCR products in Saccharomyces cerevisiae. . Gene. 1995;158:113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- Marschall LG, Jeng RL, Mulholland J, Stearns T. Analysis of Tub4p, a yeast gamma-tubulin–like protein: implications for microtubule-organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J, Jiang W, Funk M, Rathjen J, Barnes CA, Hinz T, Hegemann JH, Philippsen P. CPF1, a yeast protein which functions in centromeres and promoters. EMBO (Eur Mol Biol Organ) J. 1990;9:4017–4026. doi: 10.1002/j.1460-2075.1990.tb07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiaeencodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. . Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Pangilinan F, Spencer F. Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol Biol Cell. 1996;7:1195–1208. doi: 10.1091/mbc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta AF, Cooke CA, Earnshaw WC. Structure of the human centromere at metaphase. Trends Biochem Sci. 1990;15:181–185. doi: 10.1016/0968-0004(90)90158-8. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AE, Stearns T, Drubin DG, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Rieder CL. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int Rev Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., F. Winston, and P. Hieter. 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 198 pp.
- Rudner AD, Murray AW. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel SG, Snyder M. A highly divergent gamma-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. . J Cell Biol. 1995;131:1775–1788. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Severin FF, Hyman AA. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J Cell Biol. 1994;127:995–1008. doi: 10.1083/jcb.127.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Geissler S, Grein K, Schiebel E. γ-Tubulin–like Tub4p of Saccharomyces cerevisiaeis associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. . Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmann O, Lechner J. The Saccharomyces cerevisiaekinetochore contains a cyclin-CDK complexing homologue, as identified by in vitro reconstitution. EMBO (Eur Mol Biol Organ) J. 1996;15:3611–3620. [PMC free article] [PubMed] [Google Scholar]
- Stemmann, O., J. Ortiz, S. Rank, and J. Lechner. 1999. A putative complex consisting of Ctf19, Mcm21 and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. In press. [DOI] [PMC free article] [PubMed]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Kingsbury J, Koshland D. CEP3 encodes a centromere protein of Saccharomyces cerevisiae. . J Cell Biol. 1995;128:749–760. doi: 10.1083/jcb.128.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourc'h C, Taruscio D, Boyle AL, Ward DC. Cell cycle-dependent distribution of telomeres, centromeres, and chromosome-specific subsatellite domains in the interphase nucleus of mouse lymphocytes. Exp Cell Res. 1993;205:142–151. doi: 10.1006/excr.1993.1068. [DOI] [PubMed] [Google Scholar]
- Wang Y, Burke DJ. Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. . Mol Cell Biol. 1995;15:6838–6844. doi: 10.1128/mcb.15.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. . Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- Wells WA, Murray AW. Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J Cell Biol. 1996;133:75–84. doi: 10.1083/jcb.133.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV. Analysis of the Saccharomycesspindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]