Abstract
The study of the equine immune system and inflammatory responses, by measuring cytokine expression, can provide important insight into disease pathogenesis in the horse. A set of quantitative real time polymerase chain reaction (QPCR) assays for the equine cytokines IL-1α, IL-1β, IL-6, IL-8 and TNF-α were validated using QPCR primers and probes which were generated for the equine IL-1α, IL-1β, IL-6, IL-8, TNF-α and 18S genes. Amplification efficiency, intra-assay and inter-assay variation were determined using 10-fold dilutions of plasmid for each gene. Under these conditions the amplification efficiencies of the primers and probes ranged from 99% to 101%. The mean coefficient of variation (CV) across five sets of plasmid DNA for both intra-assay and inter-assay variation was 0.63% (range 0.2% to 1.8%). Amplification efficiency was also determined using 2-fold dilutions of cDNA and under these conditions amplification efficiency ranged from 83% to 95%. The specificity of amplification was confirmed by DNA sequencing of reaction products. The QPCR assays were also evaluated using three sets of cDNA from equine monocyte derived macrophages (EMDM) stimulated for 1 hour with lipopolysaccharide (LPS). The general trend was the same for all three samples with IL-1α showing the greatest induction and IL-6 the lowest induction. The range of cytokine induction was greater than has previously been reported with values ranging from 12-fold to 30,000-fold. We present a set of QPCR primers and probes that are suitable for quantitation of expression of a set of equine cytokines. The primers and probes have been rigorously analyzed, and we demonstrate that they are specific for the desired genes, have a high amplification efficiency and the assays are highly reproducible.
Keywords: Cytokines, Macrophage, Horse, QPCR
1. Introduction
The lack of reagents for measuring cytokine expression has limited the study of the equine immune system and the understanding of inflammatory responses and disease pathogenesis in the horse. With the exception of TNF-α (Vick et al., 2007), specific monoclonal antibodies (mAb) and cytokine standards for the equine are very limited, and the expression of equine IL-1β, IL-6, and TGF-β can only be measured using antibodies specific to human or other animal cytokines. This has impeded extensive studies using immunoassays (Billinghurst et al., 1995; Charan et al., 1997; Rodriguez et al., 1996). An alternative to immunoassays for the detection of cytokine induction is the measurement of equine cytokine mRNAs (Cherwinski et al., 1987). Methods that have been used to measure the expression of equine cytokine mRNAs include reverse transcription competitive polymerase chain reaction (RT-cPCR), reverse transcription polymerase chain reaction (RT-PCR), Southern blot techniques, quantitative real time polymerase chain reaction (QPCR), and the ribonuclease protection assay (RPA) (Grünig and Antczak, 1995; Rottman et al., 1996; Byrne et al., 1997; Swiderski et al., 1999; Leutenegger et al., 1999; Giguère and Prescott 1999; Garton et al., 2002; Lim, et al., 2003; Lim et al., 2005; Murphy et al., 2007; Vick et al., 2007). Previous studies that have explored the production of equine cytokines in macrophages using RT-cPCR and QPCR techniques (Giguère and Prescott, 1999; Garton et al., 2002; Fumuso et al., 2003; Murphy et al., 2007; Vick et al., 2007) lack information regarding reproducibility (intra- and inter-assay variability), efficiency and specificity of the assays. While it is likely that the absence of validation data in some reports was due to limited manuscript lengths, it remains difficult to evaluate the utility of individual assays for diverse studies. Therefore the main objective of our study was to develop and clearly evaluate a set of QPCR assays for the equine cytokines IL-1α, IL-1-β, IL-6, IL-8, and TNF-α. We describe primer and probe pairs for the listed cytokines that are sensitive, robust, and highly reproducible. These assays were used to measure the effects of lipopolysaccharide (LPS) stimulation on equine monocyte derived macrophages (EMDM) as a baseline for future studies.
2. Methods
2.1. Primer design
Custom TaqMan® Gene Expression Assays of primer and probe sets for IL-1α, IL-1β, IL-6, IL-8, TNF-α, and 18S were designed using the Applied Biosystem's Assays-by-DesignSM Service for Gene Expression Criteria. Table 1 lists the sequences of the forward and reverse primers and the probe. The accession numbers for each gene are as follows: equine IL-6: U64794, AF041975, AF005227; equine IL-8: AY184956, CD53673, CD536703; equine IL-1α: CD466534, U92480; equine IL-1β: U92481, D42165, D42147; equine TNF-α: M64087, AB035735. Available nucleotide sequences for each cytokine were compared, and those with target regions that were fully conserved, free of base ambiguities, and were not within alternatively spliced regions were selected and submitted to ABI for primer and probe development. All primer pairs were designed to produce amplicons smaller than 150 bp. To prevent the amplification of genomic DNA, the primers were placed in consecutive exons with the probe spanning the junction between exons. The 18S rRNA sequence M10098, AJ311673 was specifically chosen to be as universal as possible (Schmittgen and Zakrajsek, 2000; Aerts et al., 2004; Robinson et al., 2007) and to fit the parameters of the ABI 7500 Sequence Detection System (Applied Biosystems). The 18S rRNA primers and probe were designed using the Applied Biosystem's Assays-by-DesignSM Service, in order to be compatible with the TaqMan FAM/MGB probe format.
Table 1.
Primers and probe combinations for quantitative real-time PCR
| IL-6 f | 5′-GAAAAAGACGGATGCTTCCAATCTG-3′ |
| IL-6 r | 5′-TCCGAAAGACCAGTGGTGATTTT-3′ |
| IL-6 p | 5′-CAGGTCTCCTGATTGAAC-3′ |
| IL-8 f | 5′-GCCACACTGCGAAAACTCA-3′ |
| IL-8 r | 5′-GCACAATAATCTGCACCCACTTTG-3′ |
| IL-8 p | 5′-ACGAGCTTTACAATGATTTC-3′ |
| IL-1α f | 5′-CAATATCTTGCGACTGCTGCATTAA-3′ |
| IL-1α r | 5′-CTCTTCTGATGTATAAGCACCCATGT-3′ |
| IL-1α p | 5′-ACGCAGTGAAATTT-3′ |
| IL-1β f | 5′-TGTACCTGTCTTGTGGGATGAAAG-3′ |
| IL-1β r | 5′-GCTTTTCCATTTTCCTCTTTGGGTAA-3′ |
| IL-1β p | 5′-CCTACAGCTGGAGACAGT-3′ |
| TNF-α f | 5′-TTCTCGAACCCCAAGTGACAAG-3′ |
| TNF-α r | 5′-GCTGCCCCTCGGCTT-3′ |
| TNF-α p | 5′-ATGTTGTAGCAAACCC-3′ |
| 18S f | 5′- AAACGGCTACCACATCCAA-3′ |
| 18S r | 5′-TCGGGAGTGGGTAATTTGC-3′ |
| 18S p | 5′-AAGGCAGCAGGCGC-3′ |
Sense primers are designated by (f), antisense primers by an (r), and the probe is designated by a (p). Probes are labeled with 5′ 6FAM, fluorescent dye 6-carboxyfluorescein, and 3′MGBNFQ, a minor groove binder/non-fluorescent quencher.
2.2. TaqMan primer and probe validation
Primer efficiency was determined using standard curves generated from 10-fold dilutions of plasmids encoding the transcript of each gene of interest. Plasmids containing IL-1α, IL-1β, and IL-8 sequences were obtained from the Pratt Laboratory, Plant Biology Department, University of Georgia (http://www.fungen.org/Projects/Horse/Equus%20Project.htm). IL-6 and TNF-α cDNAs were generated by standard RT-PCR and cloned using mRNA isolated from LPS-stimulated EMDM. Plasmid DNA was isolated using the QIAprep Spin Miniprep Kit (Qiagen Inc. Valencia, CA), and plasmid concentration was determined by spectrophotometry. Dilutions were prepared in water such that the final copy number in the QPCR assay ranged from 10 to 105, and QPCR was preformed in triplicate. QPCR using plasmid standards with the addition of cDNA derived from unstimulated equine macrophages was also performed in order to evaluate primer efficiency in the presence of background cDNA. An additional measurement of efficiency of equine cytokine assays was performed by using 2-fold serial dilutions of cDNA generated from LPS stimulated EMDM. QPCR was performed in triplicate using the five cDNA dilutions to measure the efficiency of the five equine cytokine assays and the 18S housekeeping gene.
Inter-assay variability was measured using three separate QPCR assays performed on three different days. Intra-assay variability was measured by performing a single QPCR assay using 10 replicates of each of the following plasmid copy numbers 1 × 105, 1× 103, 1 × 102, and 1 ×101. Analysis of the results included determination of the coefficient of variation (CV), the mean coefficient of variation, and the amplification efficiency.
The specificity of amplification was determined by performing gel electrophoresis and ethidium bromide staining on the QPCR products for each gene of interest to confirm product size and to detect the presence of any nonspecific amplification products. The amplified DNA obtained from each primer pair was then purified using StrataPrep PCR Purification Kit (Stratagene, La Jolla, CA) and sequenced using BigDye® Terminator v1.1 (Applied Biosystems, Foster City, CA) on an ABI PRISM® 3100 Genetic Analyzer.
2.3. Quantitative real-time polymerase chain reaction assay
PCR reactions contained Custom TaqMan® Gene Expression Assays Mix (containing primers and probe), TaqMan® Universal PCR Master Mix (containing Taq, dNTPs and buffer), No AmpErase® UNG (final concentration 1X), and plasmid DNA in a volume of 20 μl. The samples were placed in 96-well plates and amplified in an automated fluorometer (ABI 7500 Sequence Detection System, Applied Biosystems). Amplification conditions were 2 min at 50°C, 10 min at 95.0°C, followed by 40 cycles of 15s at 95.0°C and 1 min at 60.0°C.
2.4. Isolation of peripheral blood mononuclear cells and establishment of equine monocyte derived macrophage cell cultures
EMDM cultures were established with methods adapted from Raabe et al. (1998). Whole blood in the volume of 500ml was collected from EIAV-negative horses into an anticoagulant citrate dextrose (ACD) solution blood collection bottle (The Metrix Company, Dubuque, Iowa 52002, USA) and centrifuged at 700 × g for 20 minutes to produce buffy coats. EMDM were separated by density gradient centrifugation using HybriMax histopaque (d = 1.077g/cm3) cushion (Sigma, St. Louis, MO). The EMDM were washed four times with Dulbecco's Ca2+ - and Mg2+ - free PBS (Sigma, St. Louis, MO), 5% adult horse serum (endotoxin and EIAV tested, Invitrogen/Gibco, Carlsbad, CA) and 1% penicillin- streptomycin solution (Sigma, St. Louis, MO). The EMDM were diluted in complete Minimum Essential Medium Alpha (MEM α) medium supplemented with 10% adult horse serum (Invitrogen, Carlsbad, CA) to a beginning concentration of 5 × 106 cells/cm2, and five ml aliquots were added to 25 cm2 cell culture flasks and incubated at 37 °C in 5% CO2. The next day (day 2), cells were washed to remove the nonadherent cells. Cells were incubated for an additional seven days to allow macrophages to mature. All reagents used for EMDM cultures were tested by the manufacturer for endotoxin and certified ≤ 0.6 EU/mL endotoxin. In order to confirm the purity of the macrophage cultures, cell differentials were performed on cytospin preparations collected at days 1, 3 and 7 using 100μl samples. Cytospin preparations were stained with Wright's-Giemsa and 100-cell differentials were performed. Based on the differentials the cell cultures were composed of 95% or greater macrophages at days 1, 3 and 7.
2.5. Stimulation of equine monocyte derived macrophage cells with lipopolysaccharide
LPS at 10ng/ml (Chen et al., 2003) or a negative control using complete MEM α medium was added to the cell cultures on day seven of culture and incubated for 1 hour at 37 °C in 5% CO2. Culture medium containing LPS or negative control was decanted, and cells were lysed by the addition of Buffer RLT/ β-mercaptoethanol (Qiagen Inc., Valencia, CA). Lysates were used for RNA preparation and cDNA synthesis to examine differences in cytokine expression between the stimulated and unstimulated cells.
2.6. RNA isolation
RNA was extracted from EMDM cultures using the Qiagen RNeasy® Mini Kit (Qiagen Inc., Valencia, CA) according to manufacturer instructions for isolation of total RNA from animal cells, with the exception that 1200 μl of Buffer RLT/ β-mercaptoethanol (Qiagen Inc., Valencia, CA) was added to 25 cm2 cell culture flasks for cell lysis. All RNA extractions were treated with 1 μl of DNase I Amp Grade (Invitrogen, Carlsbad, CA) for 15 min and then heated for 10 min at 65°C. RNA isolated from the EMDM cultures was used for cDNA synthesis.
2.7. cDNA synthesis
First-strand cDNA was synthesized from total RNA using SuperScript™ III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) according to manufacturer instructions in a 20 μl volume containing 200U/μl SuperScript III RT, 1μg of total RNA, 50ng/μl random hexamers, 10mM dNTP mix, and DEPC-treated water. The reaction was carried out for 10 min at 25°C followed by 50 min at 50°C and was terminated at 85°C for 5 min. cDNA, synthesized from the total RNA, was amplified using the ABI 7500 Real Time PCR System in order to determine cytokine expression using quantitative real time PCR. Relative quantification of the target was done using the signal from 18S M10098, AJ311673, a housekeeping gene, in each sample. Raw data was analyzed using the 2-ΔΔCt method also known as the comparative threshold cycle (Ct) or ΔΔ Ct method (Livak and Schmittgen, 2001; Sabek et al., 2002), and the value for each sample was normalized using 18S as described in Applied Biosystems User Bulletin No. 2 (P/N 4303859) (http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf, 1997).
3. Results
3.1. Assay validation
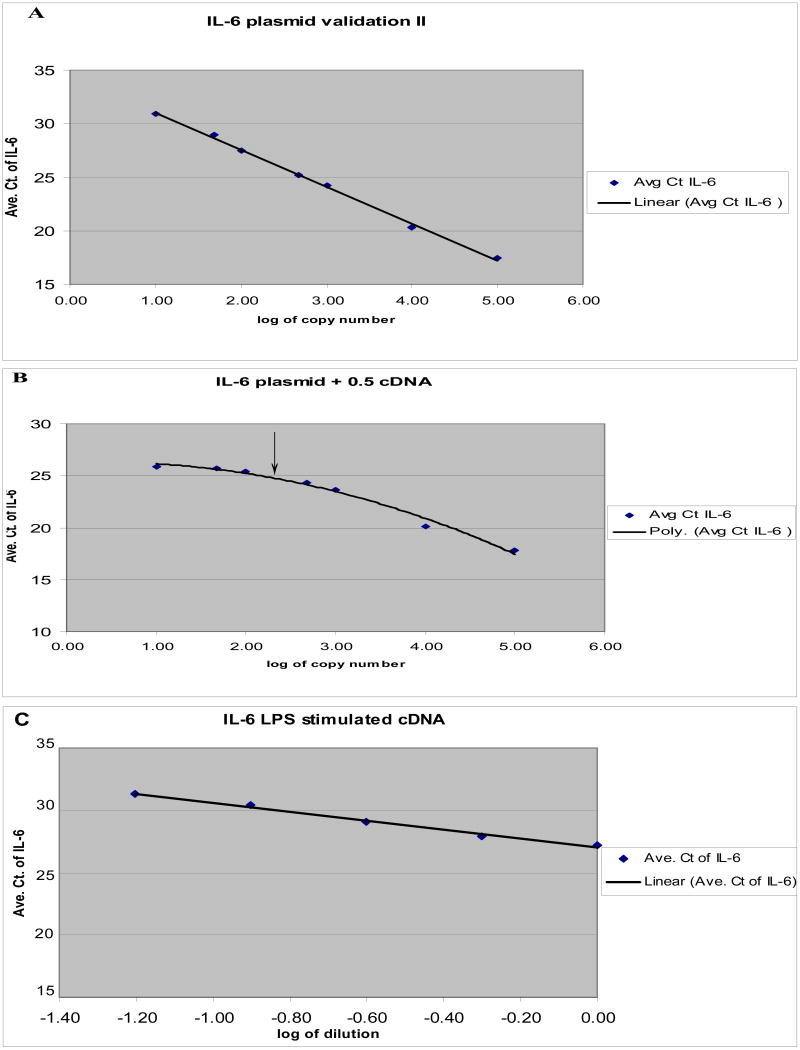
In order to determine the efficiency, repeatability, and reproducibility of the custom gene expression assays, five sets of plasmid DNA were analyzed using IL-1α, IL-1β, IL-6, IL-8, and TNF-α primers and probes. Amplification was linear over the range of 10 to 105 input copies of plasmid for all five of the equine genes. A representive curve (IL-6) is shown in Figure 1A. The correlation coefficients of the standard curves ranged from 0.9985-0.9993 (Table 2). The standard curve efficiencies of the primers and probes ranged from 99% to 101% using a plasmid DNA template (Table 2). The amplification efficiency of the target genes was approximately equal to that of 18S the housekeeping gene (data not shown).
Figure 1.
Panel A is an example of a standard curve for inter-assay validation for IL-6. On the y-axis is the average threshold cycle value for IL-6 and on the x-axis is the log of the copy number for each IL-6 plasmid dilution. Panel B is an example of a standard curve for IL-6 in the presence of background cDNA the arrow indicates the start of a plateau. Panel C is an example of a standard curve for IL-6 in the presence of LPS stimulated cDNA.
Table 2.
Average correlation coefficients and amplification efficiencies of cytokine assays with plasmid DNA templates and cDNA dilutions.
| Cytokine | R2 | Efficiency |
|---|---|---|
| IL-6 | 0.9985a | 99.0%b |
| IL-6 cDNA | 0.9918c | 91.0%d |
| IL-8 | 0.9990 | 99.0% |
| IL-8 cDNA | 0.9867 | 86.0% |
| IL-1 alpha | 0.9985 | 99.7% |
| IL-1 alpha cDNA | 0.9840 | 95.0 % |
| IL-1 beta | 0.9993 | 101.0% |
| IL-1 beta cDNA | 0.9819 | 85.0% |
| TNF-alpha | 0.9993 | 99.2% |
| TNF-alpha cDNA | 0.9959 | 83.0% |
| 18S cDNA | 0.9945 | 90.0% |
Results presented as (a) and (b) are overall averages derived from data collected to determine intra-assay and inter-assay variability Results presented as (c) and (d) are derived from dilutions of cDNA prepared from of LPS stimulated EMDM
Average correlation coefficients of combined assays
Average efficiency of combined assays
Correlation coefficient from one assay run in triplicate
Amplification efficiencies from one assay run in triplicate
Repeatability and reproducibility of the QPCR assays were determined using intra-assay and inter-assay variation respectively. Intra-assay variation was evaluated using 10 replicates containing plasmid copy numbers of approximately 105,103, 102,101. The mean CVs for each cytokine are presented in Table 3. In no case was the intra-assay CV for any cytokine, at any quantity of input plasmid, greater than 1.8%, demonstrating the repeatability of the reactions.
Table 3.
Reproducibility as measured with plasmid DNA templates
| Cytokine | Ct Rangec | Ct Ranged | Mean CV e |
|---|---|---|---|
| IL-6 inter-assaya | (17.45-17.86) | (30.51-31.07) | 0.5% |
| IL-6 intra-assayb | (17.25-17.90) | (30.39-30.76) | 0.6% |
| IL-8 inter-assay | (20.84-21.55) | (33.54-34.55) | 0.6% |
| IL-8 intra-assay | (20.66-21.11) | (33.76-35.82) | 0.8% |
| IL-1α inter-assay | (18.48-18.90) | (31.73-32.93) | 0.7% |
| IL-1α intra-assay | (19.00-19.17) | (32.00-32.74) | 0.6% |
| IL-1β inter-assay | (19.87-20.29) | (32.36-33.51) | 0.5% |
| IL-1β intra-assay | (19.44-19.70) | (32.40-33.35) | 0.6% |
| TNF-α inter-assay | (17.47-17.79) | (30.39-31.43) | 0.7% |
| TNF-α intra-assay | (17.36-17.64) | (30.38-31.18) | 0.7% |
Inter-assay variability was measured using three separate QPCR assays performed on three different days
Intra-assay variability was measured by performing a single QPCR assay
Range of cycle threshold for 10 copies of plasmid DNA
Range of cycle threshold for 105 copies of plasmid DNA
Mean of coefficient of variation
The inter-assay variation was determined by performing the assays 1-5 days apart on 3 different days using dilutions containing approximately 1× 105, 1 × 104, 1 × 103, 5 × 102, 1 ×102, 5 ×101, and 1 ×101 copies of plasmid. For equine IL-6 the mean CV for each dilution factor across reactions ranged from 0.2% to 0.9% with an overall mean CV of 0.5% (Table 3). The mean CV for equine IL-8 ranged from 0.3% to 1.1% for each dilution factor across reactions and the overall mean CV for equine IL-8 was 0.6% (Table 3). For equine IL-1α and IL-1β, the mean CV for each dilution factor across reactions ranged from 0.4% to 1.1%, and 0.2% to 1.0%, respectively. The overall mean CV for IL-1 α and IL-1 β at all copy levels was 0.7 % and 0.5%, respectively (Table 3). The mean CV for equine TNF-α ranged from 0.6% to 1.1%, with a mean overall CV of 0.7% at all copy levels (Table 3). In no case was the inter-assay CV for any cytokine, at any quantity of input plasmid greater than 1.1% demonstrating the reproducibility of the reactions. The overall mean CV across all sets of intra- and inter-assay data was 0.63% (range 0.2% to 1.8%) (Table 3). In Table 3 we also show the ranges of Ct values for lowest (10 copies) and the highest (105 copies) amounts of template. In no case was the Ct value for 10 copies of input plasmid greater than 35.82. The variability in Ct values did not increase at low template concentrations; the maximum range of Ct values was 2.06.
To test the efficiency of the primer and probe sets under more stringent conditions, two other assays were performed. First, cDNA from unstimulated EMDM was added to plasmid dilutions to test the efficiency of the primers and probes in the presence of a non-specific competitor. At input plasmid copy numbers of 5 × 102 to 105 all reactions remained linear, with amplification efficiencies similar to reactions containing only plasmid. However, at low levels of input plasmid (less than 100 copies) with the exception of IL-1α the curves were not linear, but started to plateau, as indicated by the arrow in the example for IL-6 (Figure 1B). The plateaus in the curves likely reflect the presence of cytokine mRNA in unstimulated EMDM. In the presence of equine cDNA, the linear range for IL1-β and TNF-α was from 102 copies to 105 copies and the linear range for IL-6 and IL-8 was from 5 × 102 copies to 105 copies. IL-1α remained linear to 10 copies of input plasmid (essentially identical to the curves without cDNA).
As the addition of cDNA from equine EMDM to plasmid DNA did not allow us to examine amplification efficiencies at levels below 100 copies of input plasmid, we performed dilutions of cDNA from LPS stimulated EMDM. In this instance we determined the efficiency of the equine cytokine gene expression assays and the 18S housekeeping gene using a series of 2-fold dilutions (corresponding to 0.5μl-0.03μl of cDNA) from LPS stimulated EMDM. Amplification was linear for all five cytokines and the 18S housekeeping gene and a representative curve (IL-6) is shown in Figure 1C. At these lower levels of input template the assays remained linear (correlation coefficients of the standard curves ranged from 0.984-0.9959) but amplification efficiencies dropped (Table 2). IL-1α performed the best with an efficiency of 95%; the least efficient primer and probe was TNF-α with an efficiency of 83% (Table 2).
3.2. Specificity of amplification of TaqMan assays
It is typically understood that TaqMan assays are specific, however to insure we had the desired product; gel electrophoresis was used to analyze the products generated from each cytokine assay. Bands corresponding to the expected sizes of the target genes were seen, and no nonspecific amplification products were present (data not shown). Each of the QPCR products was sequenced, and each product was found to be the sequence of interest (data not shown).
3.3. Stimulation of equine monocyte derived macrophage cells with lipopolysaccharide
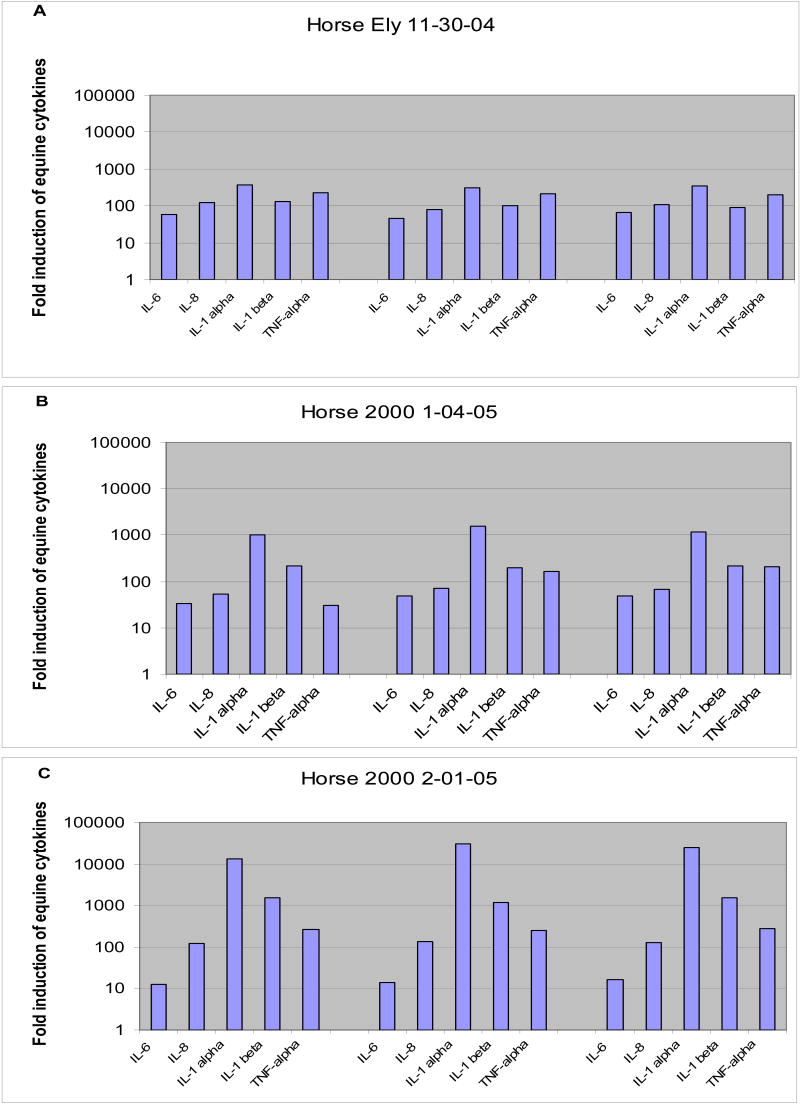
In order to provide useful comparisons for future studies three sets of cDNA from EMDM stimulated with LPS were analyzed by QPCR using IL-1α, IL-1β, IL-6, IL-8, and TNF-α primers and probes. Two EMDM cultures were from the same horse, collected one month apart, and the third set was from a different horse. RNA was isolated and cDNA was prepared as described in the materials and methods section. Cytokine expression levels varied between samples, but the general trend was the same for all three samples. IL-1α showed the greatest induction levels while IL-6 showed the lowest induction levels (Figure 2). IL-1α levels were induced approximately 300- to 30,000-fold. IL-1β levels were induced 89-fold to 1554-fold. TNF-α levels were induced from 30-fold to 280-fold. IL-8 levels were induced from 53-fold to 133-fold. IL-6 levels were induced from 12-fold to 65-fold.
Figure 2.
Induction of gene expression of equine cytokines after addition of 10ng/ml LPS to EMDM cells. Panel A horse Ely, Panel B & C horse 2000 1 month apart. For each bleed date one cDNA reaction was prepared. Three separate QPCR were performed (each assay performed in triplicate).
The constant threshold (Ct) values for the three sets of cDNA from unstimulated EMDM ranged from 26.67 for TNF-α to 37.65 for IL-α (Table 4). The three sets of cDNA from LPS stimulated EMDM had Ct values ranging from 19.22 for TNF-α to 29.35 for IL-6 (Table 4). Values for the 18S housekeeping gene ranged from 6.30 to 8.46 for cDNA from unstimulated EMDM and from 6.93 to 9.66 for cDNA from LPS stimulated EMDM (Table 4). The average Ct value using cDNA from unstimulated EMDM for 18S was 7.40, the average Ct value using cDNA from LPS stimulated EMDM for 18S was 7.95 (Table 4).
Table 4.
Cycle threshold (Ct) values for LPS stimulated and unstimulated EMDM
| Cytokinea | Mean Ctb | Ct Rangec | S.D.d | Mean CV%e |
|---|---|---|---|---|
| IL-6 unstimulated | 30.36 | (27.13-34.25) | 2.81 | 9.25 |
| IL-6 stimulated | 25.89 | (22.54-29.35) | 2.66 | 10.27 |
| IL-8 unstimulated | 27.67 | (27.15-28.15) | 0.32 | 1.16 |
| IL-8 stimulated | 21.67 | (19.67-23.79) | 1.51 | 6.97 |
| IL-1α unstimulated | 35.33 | (32.58-37.65) | 1.78 | 5.04 |
| IL-1α stimulated | 24.85 | (22.14-26.99) | 2.01 | 8.09 |
| IL-1β unstimulated | 31.41 | (30.02-32.52) | 0.98 | 3.12 |
| IL-1β stimulated | 23.73 | (20.39-26.77) | 2.38 | 10.03 |
| TNF-α unstimulated | 27.85 | (26.67-28.82) | 0.75 | 2.69 |
| TNF-α stimulated | 20.81 | (19.22-23.22) | 1.40 | 6.73 |
| 18S unstimulated | 7.40 | (6.30-8.46) | 0.58 | 7.84 |
| 18S stimulated | 7.95 | (6.93-9.66) | 0.81 | 10.19 |
Combined statistical analysis of cycle threshold values from three different samples (two samples were from same horse taken one month apart) of unstimulated and LPS stimulated EMDM measured using three separate QPCR assays performed on three different days
Mean constant threshold values for combined data from three different samples of EMDM from two different horses
Range for mean constant threshold values for combined data from three different samples of EMDM from two different horses
Standard deviation of constant threshold values for combined data from three different samples of EMDM from two different horses
Mean of coefficient of variation for combined data from three different samples of EMDM from two different horses
4. Discussion
QPCR is a rapid and reliable method for mRNA quantitation (Heid et al., 1996; Wang and Brown, 1999; Ficko and Černelč, 2005) however many QPCR primer and probe sets are poorly described and validation of many primer and probe sets is lacking. Although QPCR is a very specific and reproducible technique, in order for the results to be meaningful, validated primer and probe combinations must be available.
In this study we have described a set of QPCR primers and probes for quantifying equine IL-6, IL-8, IL-1α, IL-1β, and TNF-α, and have demonstrated that these primer and probe sets were sensitive, specific, and reproducible. Normalization to a relevant “housekeeping gene” is also important in the accuracy of QPCR. The 18S ribosomal subunit was chosen because its cellular concentration is constant within a range of 1.0-4.2 fold (Schmittgen and Zakrajsek, 2000; Aerts et al., 2004; Robinson et al., 2007) which correlates to a difference of 0.5-2.0 Ct values, and its amplification efficiency of 90% (Table 2) is similar to that of the target genes. Other housekeeping genes such as GAPDH and β-actin were considered, but were found to be inappropriate for our uses. GAPDH exhibited changes of up to 10-fold in expression in EMDM under varying conditions (J.Harrington, personal communication). Both GAPDH and β-actin are regulated within a 10-fold range and the transcription level can vary during cell proliferation, differentiation, or activation (Schmittgen and Zakrajsek, 2000; Goidin et al., 2001; Frost and Nilsen, 2003; Aerts et al., 2004; Dheda et al., 2004). 18S and β-2 microglobulin exhibit less variability and are therefore better internal controls (Goidin et al., 2001; Aerts et al., 2004).
Primer and probe sets were initially evaluated using standard curves generated from 10-fold dilutions of double stranded circular plasmids encoding the transcript of each gene of interest. The assays were linear over the range tested (10 to 105 copies of plasmid) and intra-and inter-assay variation was low. Further, there was no correlation between template copy number and assay variation, suggesting that the assays are equally robust with low template amounts. When we used more biologically complex samples (dilutions of cDNA) the primers and probes maintained acceptable, albeit lower efficiencies (Gallup and Ackermann, 2006). Valid data can be obtained from efficiencies less than 90%, if amplification is linear and the amplification efficiencies of the target and reference gene are approximately equal (http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf, 1997; Livak and Schmittogen 2001; Gallup and Ackermann, 2006; http://www.dorak.info/genetics/realtime.html, 2007).
Sensitivity of the assays was assessed using plasmid DNA dilutions. All of the assays easily detected 10 copies of input plasmid. To determine assay sensitivity for a more relevant (biologically complex) sample we added cDNA from unstimulated EMDM to plasmid dilutions. However, the addition of cDNA from EMDM to plasmid did not serve to resolve sensitivity issues because at low levels of input plasmid the curves began to plateau. It is likely that the point at which the plateau begins indicates that the number of copies of plasmid and the number of copies of cDNA are approximately the same (between 100 and 500 copies for most of the cytokines tested). In the case of IL-6, when cDNA was added to the plasmid, the curve began to plateau at Ct values between 24 and 25 (Figure 1B); this corresponds reasonably well to the observed Ct values for IL-6 in unstimulated EMDM, which were as low as 27 (Table 4). For IL-1α (data not shown) the curves were the same with (R2= 0.9925, amplification efficiency= 106%) and without (R2= 0.9985, amplification efficiency= 99.7%) cDNA. Ct values for IL-1α for unstimulated EMDM alone were between 32 and 37, similar to the Ct values obtained for 10 copies of plasmid (between 31 and 32).
We did not follow up further on the issue of absolute sensitivity as it was evident that all of the assays were capable of detecting cytokine expression in unstimulated EMDM. In these experiments we generally recovered 5 μg of total RNA from 1.5- 2 × 106 cells and 20μl of cDNA was prepared from 1μg of RNA. For all of the tested cytokine assays, amplification was positive using 0.5μl of cDNA from unstimulated EMDM. Dilutions of cDNA prepared from LPS stimulated EMDM revealed that as little as 0.03μl of cDNA was sufficient to generate a detectable signal. Therefore, while we have not absolutely determined the sensitivity of these assays, they detect as little as 10 copies of plasmid and are sufficiently sensitive to detect basal levels of cytokine expression in relevant cell types. Finally we examined product specificity by gel electrophoresis and DNA sequence analysis, to demonstrate that the assays were truly specific for the desired gene.
After extensive testing, the QPCR assays were used to determine the levels of cytokine induction in LPS-stimulated EMDM. LPS was chosen for EMDM stimulation because it is a powerful activator of the innate immune system, and it is the best known and most thoroughly characterized inducer of inflammatory cytokines (Murtaugh et al., 1996). LPS induces the synthesis of a variety of cytokines and chemokines from mononuclear phagocytes; furthermore, LPS is potent inducer of both transcription and translation; a defined set of gene products, such as IL-1α, IL-1β, and IL-8 result from the stimulation of monocytes/macrophages with LPS (Chen et al., 2003). LPS also induces the expression of TNF-α through a wide variety of agents that activate the nuclear transcription factor, NF-κ B (Myers and Murtaugh, 1995; Chen et al., 2003).
Cytokines selected for this study, IL-6, IL-8, IL-1α, IL-1β, and TNF-α, are representative of the major inflammatory cytokines produced by macrophages (Murtaugh et al., 1996). Few studies have been published that evaluate cytokine production in equine macrophages stimulated with LPS. To the authors' knowledge, only three studies have used QPCR to measure cytokine production in EMDM after LPS exposure. Sykes et al. (2005) incubated equine peripheral blood mononuclear cells (PBMC) in the presence of LPS for 0 to 48 hours and measured expression of TNF-α and IL-1β mRNA. Both TNF-α and IL-1β showed a mean increase in mRNA expression of approximately 6 fold at 6 hours; thereafter expression decreased slightly but remained significantly elevated at all time points compared with time 0 (Sykes et al., 2005).
Laan et al. (2006) exposed recurrent airway obstruction (RAO) susceptible and nonsusceptible horses to aerosolized LPS, collected bronchoalveolar lavage fluid (BALF), isolated equine alveolar macrophages at either 6 or 24 hours post-exposure and measured levels of TNF-α, IL-1β, IL-8, and IL-6 mRNA. A mean difference of approximately 7 fold was seen in TNF-α expression in RAO susceptible horses after 6 hours, but had returned to baseline after 24 hours (Laan et al., 2006). A significant increase was seen in IL-1β and IL-8 expression; a mean difference of approximately 2 fold and 3 fold was seen for IL-1β and IL-8 respectively at six hours (Laan et al., 2006). A mean difference of approximately 4 fold was seen for IL-1β in RAO susceptible horses at 24 hours (Laan et al., 2006). However, LPS stimulation did not significantly alter the expression of TNF-α, IL-1β, IL-8, or IL-6 in RAO nonsusceptible horses at either 6 or 24 hours (Laan et al., 2006).
The third study measured IL-8 using QPCR of equine alveolar macrophages exposed to LPS for 48 hours. When compared to the untreated control, alveolar macrophages stimulated with LPS showed a mean increase of approximately 14 fold in IL-8 expression (Jackson et al., 2004).
It is not possible to make a direct comparison between this study of LPS-stimulated EMDM and the aforementioned studies due to the differences in time points and study populations. In the three cytokine studies mentioned above, TNF-α, IL-1β, and IL-8 had an induction range of 2-to14-fold over a period of 6 to 48 hours. However, the induction of cytokine gene expression upon LPS exposure of EMDM seen in this study was much greater than has been previously reported. After one hour of LPS exposure, we observed increases in IL-1α gene expression ranging from approximately 300- to 30,000-fold. IL-1β mRNA levels were induced 100- to 1500-fold. TNF-α mRNA levels increased from 30-to 280-fold. IL-8 mRNA levels increased from approximately 50-to 100- fold. IL-6 mRNA levels increased from approximately 10-to 65- fold. The time points used in the other assays (earliest measurement at 6 hrs) could be an important factor in the difference between previously reported cytokine levels and our results.
Another likely explanation for the high levels of gene expression seen in this study is that the QPCR assays described herein are very sensitive and have a large linear range with baseline values easily detected in non-stimulated cells. The inability to accurately measure basal levels of gene expression can reduce the ability to determine fold changes, as can the use of assays with limited dynamic range. Another important aspect of this study was that great care was taken to use only endotoxin free media and PBS in our cell cultures, making the assay more sensitive to fold changes between the LPS stimulated and unstimulated cells. The data from this study was obtained using blood from two horses taken at three different time points which produced three independent cell cultures, each assayed in triplicate, and the general trend held true for both horses. An increase of at least 10-fold was seen for IL-6, IL-8, IL-1α, IL-1β, and TNF-α in this study for both horses. Although the absolute increases in cytokine gene expression were different for the two study animals, the relative trend in cytokine gene expression was the same. IL-1α showed the greatest induction levels while IL-6 showed the lowest induction levels for the two study animals.
In conclusion, we have validated a set of QPCR assays for the equine cytokines IL-1α, IL-1β, IL-6, IL-8 and TNF-α, and we have demonstrated these assays are sensitive, robust, and highly reproducible. We propose that these assays are a valid way to measure the effects of LPS stimulation on equine monocyte derived macrophages and that our measurements can be used as a baseline for future studies.
Acknowledgments
We would like to thank Dr. Paul Spencer for all of his advice and for introducing us to quantitative real time polymerase chain reaction (QPCR) assays. This project was supported by the National Institutes of Health CA059278 and USDA CSREES TEX09096.
Abbreviations
- ACD
anticoagulant citrate dextrose
- Ct
threshold cycle
- CV
coefficient of variation
- EMDM
equine monocyte derived macrophages
- LPS
lipopolysaccharide
- MEM α
minimum essential medium alpha
- mAb
monoclonal antibodies
- PBMC
peripheral blood mononuclear cells
- QPCR
quantitative real time polymerase chain reaction
- RAO
recurrent airway obstruction
- RT-cPCR
reverse transcription competitive chain reaction
- RT-PCR
reverse transcription polymerase chain reaction
- RPA
ribonuclease protection assay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aerts JL, Gonzales MI, Topalian SL. Selection of appropriate control genes to assess expression of tumor antigens using real-time RT-PCR. Biotechniques. 2004;36:84–91. doi: 10.2144/04361ST04. [DOI] [PubMed] [Google Scholar]
- Billinghurst RC, Fretz PB, Gordon JR. Induction of intra-articular tumor necrosis factor during acute inflammation responses in equine arthritis. Equine Vet J. 1995;27:208–216. doi: 10.1111/j.2042-3306.1995.tb03064.x. [DOI] [PubMed] [Google Scholar]
- Byrne KM, Davis WC, Holmes MA, Brassfield AL, McGuire TC. Cytokine RNA expression in an equine CD4+ subset differentiated by expression of a novel 46 kDa surface protein. Vet Immunol Immunopathol. 1997;56:191–204. doi: 10.1016/s0165-2427(96)05752-2. [DOI] [PubMed] [Google Scholar]
- Charan S, Palmer K, Chester P, Mire-Sluis AR, Meager A, Edingtion N. Transforming growth factor-β induced by live or ultraviolet-inactivated equid herpes virus type-1 mediates immunosuppression in the horse. Immunology. 1997;90:586–591. doi: 10.1046/j.1365-2567.1997.00202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Zuraw BL, Zhao M, Liu FT, Huang S, Pan ZK. Involment of protein tyrosine kinase in toll-like receptor 4-mediated NF-κB activation in human peripheral blood monocytes. Am J Physiol Lung Cell Mol Physiol. 2003;284:L607–L613. doi: 10.1152/ajplung.00116.2002. [DOI] [PubMed] [Google Scholar]
- Cherwinski HM, Schumacher JH, Brown KD, Mosmann T. Two types of mouse helper T cell clone III. Futher differences in lymphokine synthesis between Th1 and Th2 clones revealed by RNA hybridization, functionally monospecific bioassays, monoclonal antibodies. J Exp Med. 1987;166:1229–1244. doi: 10.1084/jem.166.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques. 2004;37:112–119. doi: 10.2144/04371RR03. [DOI] [PubMed] [Google Scholar]
- Ficko T, Černelč P. Real-time quantitative PCR assay for analysis of platelet gly protein IIIa gene expression. J Biochem Methods. 2005;62:241–250. doi: 10.1016/j.jbbm.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Frost P, Nilsen F. Validation of reference genes for transcription profiling in the salmon louse, Lepeophtheirus salmonis, by quantitative real-time PCR. Vet Parasitol. 2003;118:169–174. doi: 10.1016/j.vetpar.2003.09.020. [DOI] [PubMed] [Google Scholar]
- Fumaso E, Giguère S, Wade J, Rogan D, Videla-Dorna I, Bowden RA. Endometrial IL-1β, IL-6, and TNF-α, mRNA expression in mares resistant or susceptible to post-breeding endometritis. Effects of estrous cycle, artificial insemination and immunodulation. Vet Immunol Immunopathol. 2003;96:31–41. doi: 10.1016/s0165-2427(03)00137-5. [DOI] [PubMed] [Google Scholar]
- Gallup JM, Ackermann MR. Addressing fluorogenic real-time qPCR inhibition using the novel custom Excel file system ‘FocusField2-6GallupqPCRSet-upTool-001’ to attain consistently high fidelity qPCR reactions. Biol Proced Online. 2006;8:87–152. doi: 10.1251/bpo122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garton NJ, Gilleron M, Brando T, Dan HH, Giguère S, Puzo G, Prescott JF, Sutcliffe IC. A novel lipoarabinomannan from the equine pathogen Rhodococcus equi: structure and effect on macrophage cytokine production. J Biol Chem. 2002;177:31722–31733. doi: 10.1074/jbc.M203008200. [DOI] [PubMed] [Google Scholar]
- Giguère S, Prescott JF. Quantitation of equine cytokine mRNA expression by reverse transcription-competitive polymerase chain reaction. Vet Immunol Immunopathol. 1999;67:1–15. doi: 10.1016/s0165-2427(98)00212-8. [DOI] [PubMed] [Google Scholar]
- Goidin D, Mamessier A, Staquet M, Schmitt D, Berthier-Vergnes O. Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and β-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal Biochem. 2001;295:17–21. doi: 10.1006/abio.2001.5171. [DOI] [PubMed] [Google Scholar]
- Grünig G, Antczak DF. Horse trophoblasts produce tumor necrosis factor α but not interleukin 2, interleukin 4, or interferon γ. Biol Reprod. 1995;52:531–539. doi: 10.1095/biolreprod52.3.531. [DOI] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;10:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf, 1997. Applied Biosystems User Bulletin No. 2 (P/N 4303859).
- http://www.dorak.info/genetics/realtime.html, 2007. Real-Time PCR.
- Jackson KA, Stott JL, Horohov DW, Waston JL. IL-4 induced CD23 (FcεRII) up-regulation in equine peripheral blood mononuclear cells and pulmonary alveolar macrophages. Vet Immunol Immunopathol. 2004;71:197–214. doi: 10.1016/j.vetimm.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Laan TT, Bull S, Pirie R, Fink-Gremmels J. The role of alveolar macrophages in the pathogenesis of recurrent airway obstruction in horses. J Vet Intern Med. 2006;20:167–174. doi: 10.1892/0891-6640(2006)20[167:troami]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Leutenegger CM, von Rechenberg B, Huder JB, Zlinsky K, Mislin C, Akens MK, Auer J, Lutz H. Quantitative real-time PCR for equine cytokine mRNA in nondecalcified bone tissue embedded in methyl methacrylate. Calcif Tissue Int. 1999;65:378–383. doi: 10.1007/s002239900717. [DOI] [PubMed] [Google Scholar]
- Lim WS, Edwards JF, Boyd NK, Payne SL, Ball JM. Simultaneous quantitation of equine cytokine mRNAs using a multi-probe ribonuclease protection assay. Vet Immunol Immunopathol. 2003;91:45–51. doi: 10.1016/s0165-2427(02)00263-5. [DOI] [PubMed] [Google Scholar]
- Lim WS, Payne SL, Edwards JF, Kim I, Ball JM. Differential effects of virulent and avirulent equine infectious anemia virus on macrophage cytokine expression. Virology. 2005;332:295–306. doi: 10.1016/j.virol.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Murphy BA, Vick MM, Sessions DR, Cook RF, Fitzgerald BP. Acute systemic inflammation transiently synchronizes clock gene expression in equine peripheral blood. Brain Behav Immun. 2007;21:467–476. doi: 10.1016/j.bbi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Murtaugh MP, Baarsch MJ, Zhou Y, Scamurra RW, Lin G. Inflammatory cytokines in animal health and disease. Vet Immunol Immunopathol. 1996;54:45–55. doi: 10.1016/s0165-2427(96)05698-x. [DOI] [PubMed] [Google Scholar]
- Myers MJ, Murtaugh MP. Biology of Tumor Necrosis Factor. In: Myers MJ, Murtaugh MP, editors. Cytokines in Animal Health and Disease. Marcel Dekker; New York: 1995. pp. 121–151. [Google Scholar]
- Raabe MR, Issel CJ, Montelaro RC. Equine monocyte-derived macrophage cultures and their applications for infectivity and neutralization studies of equine infectious anemia virus. J Virol Meth. 1998;71:87–104. doi: 10.1016/s0166-0934(97)00204-8. [DOI] [PubMed] [Google Scholar]
- Robinson TL, Sutherland IA, Sutherland J. Validation of candidate bovine reference genes for use with real-time PCR. Vet Immunol Immunopathol. 2007;115:160–165. doi: 10.1016/j.vetimm.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Castano M, Pena L, Sanchez MA, Nieto A, Rodriguez M. Immunocytochemical detection of growth factors (PDGF and TGF β) in equine chronic pneumonia. Res Vet Sci. 1996;60:82–87. doi: 10.1016/s0034-5288(96)90137-x. [DOI] [PubMed] [Google Scholar]
- Rottman JB, Tompkins WAF, Tompkins MB. A reverse transcription-quantitative competitive polymerase chain reaction (RT-qcPCR) technique to measure cytokine gene expression in domestic mammals. Vet Pathol. 1996;33:242–248. doi: 10.1177/030098589603300217. [DOI] [PubMed] [Google Scholar]
- Sabek O, Dorak MT, Kotb M, Gaber AO, Gaber L. Quantitative detection of T-cell activation markers by real-time PCR in renal transplant rejection and correlation with histopathologic evaluation. Transplantation. 2002;74:701–707. doi: 10.1097/00007890-200209150-00019. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Method. 2000;46:69–81. doi: 10.1016/s0165-022x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- Swiderski CE, Klei TR, Horohov DW. Quantitative measurement of equine cytokine mRNA expression by polymerase chain reaction using target-specific standard curves. J Immunol Methods. 1999;222:155–169. doi: 10.1016/s0022-1759(98)00193-8. [DOI] [PubMed] [Google Scholar]
- Sykes BW, Furr M, Giguère S. In vivo pretreatment with PGG-glucan fails to alter cytokine mRNA expression of equine peripheral blood mononuclear cells exposed to endotoxin ex vivo. Vet Ther. 2005;6:67–76. [PubMed] [Google Scholar]
- Vick MM, Adams AA, Murphy BA, Sessions DR, Horohov DW, Cook RF, Shelton BJ, Fitzgerald BP. Relationships among inflammatory cytokines, obesity, and insulin sensitivity in the horse. J Anim Sci. 2007;85:1144–1155. doi: 10.2527/jas.2006-673. [DOI] [PubMed] [Google Scholar]
- Wang T, Brown MJ. mRNA quantification by real time TaqMan polymerase chain reaction: validation and comparison with RNase protection. Anal Biochem. 1999;269:198–201. doi: 10.1006/abio.1999.4022. [DOI] [PubMed] [Google Scholar]