Abstract
We have developed a mathematical approach to the study of dynamical biological networks, based on combining large-scale numerical simulation with nonlinear “dimensionality reduction” methods. Our work was motivated by an interest in the complex organization of the signaling cascade centered on the neuronal phosphoprotein DARPP-32 (dopamine- and cAMP-regulated phosphoprotein of molecular weight 32,000). Our approach has allowed us to detect robust features of the system in the presence of noise. In particular, the global network topology serves to stabilize the net state of DARPP-32 phosphorylation in response to variation of the input levels of the neurotransmitters dopamine and glutamate, despite significant perturbation to the concentrations and levels of activity of a number of intermediate chemical species. Further, our results suggest that the entire topology of the network is needed to impart this stability to one portion of the network at the expense of the rest. This could have significant implications for systems biology, in that large, complex pathways may have properties that are not easily replicated with simple modules.
Keywords: dimensionality reduction, robustness, systems biology, DARPP-32
Our understanding of the signal transduction machinery used by cells to provide a coordinated set of appropriate physiological responses to multiple diverse stimuli is increasing at an extraordinary rate. A large proportion of studies in the biological sciences are now devoted to elucidating such signaling pathways, which are far more complex than had been anticipated. As a result, it is not possible intuitively to gauge the relative importance of the different components involved.
In this article, we develop a mathematical tool that can elucidate biological network properties by analyzing global features of the network dynamics. We have chosen as a model the neurotransmitter signaling cascade involving dopamine- and cAMP-regulated phosphoprotein of molecular weight 32,000 (DARPP-32) (1), because it is arguably the most thoroughly studied signal transduction cascade in the mammalian brain and plays an important role in regulating biochemical, electrophysiological, transcriptional, and behavioral responses to both physiological and pharmacological stimuli.
One of the remarkable features of molecular biological systems is their ability to respond in a controlled or coherent fashion to external stimuli, despite significant variation in their internal state. This phenomenon is often referred to as “robustness” and during the last decade, simulation has begun to play a role in elucidating its molecular basis. Most of this work has relied on using the fact that some known, simple biological function of a network remains coherent over a wide range of conditions. An important early example is the work of Barkai and Leibler (2), who showed that the topology of a small network allowed for robust adaptation of the bacterial chemotaxis system. They demonstrated that the system responded appropriately to external chemical gradients even when kinetic coefficients and chemical species concentrations were allowed to vary by orders of magnitude. Subsequent experiments (3) confirmed their hypothesis. Important progress has since been made in understanding aspects of cell cycle regulation, developmental control circuits, and signaling (4–8), nicely reviewed in ref. 9.
Broadly speaking, these earlier studies showed that a specific biological response was generated consistently from a network despite significant parameter variation. Suppose, however, that one is presented with a network but given no clear information about the desired output. How does one use the principle of robustness in a meaningful way? This is not an artificial situation; current high-throughput technology is continuing to provide vast amounts of information about network topologies with little known about the corresponding functionality. Thus, as has been noted (8–15), it will be essential to develop mathematical paradigms and computational tools that make use of high-throughput data to reveal the functional components embedded in complex networks and biochemical pathways. If successful, such tools will allow us to identify some underlying principles of biological organization.
In this study, we attempt to extend the use of modeling and robustness to the case of biochemical systems where the output has not been or may not be characterized by a simple, well defined behavior. A critical component of the study has been to use simulation to determine whether there are features whose response remains coherent over a wide range of parameter variation. In the signal transduction setting, our method simulates the response of the system to the input in the presence of noise and highlights features of the dynamics that remain well preserved. Such features are indicative of a form of robustness, and we present evidence that they are likely to correspond to biologically important functions. Before describing the method itself, we first provide a brief introduction to our target system.
The DARPP-32 Cascade
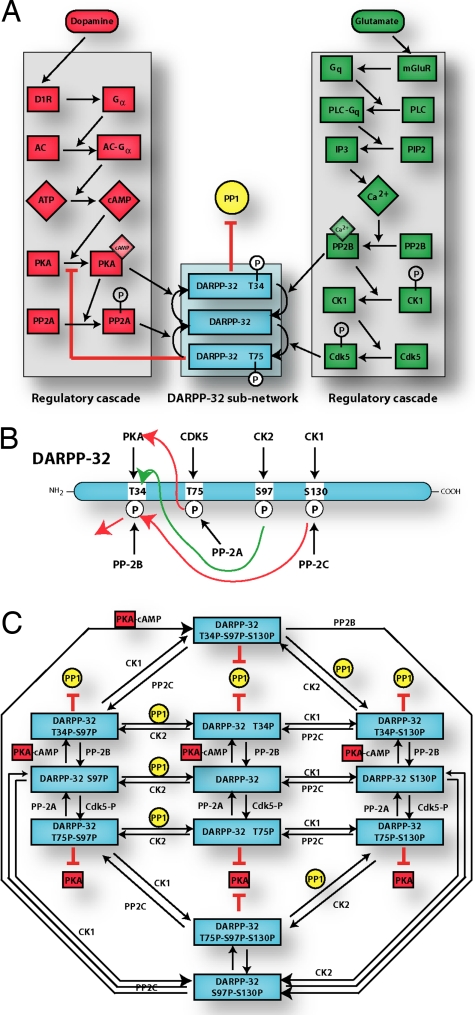
Over the past two decades, numerous studies have provided strong support for the conclusion that DARPP-32 is a key integrator of various neurotransmitter pathways. In particular, several important striatal signaling pathways responding to dopamine and glutamate stimulation have been shown to converge on DARPP-32, whose function as a regulatory protein is intimately tied to its phosphorylation state. Four phosphorylation sites are known to alter the activity of this protein (Fig. 1B): Thr-34, Thr-75, Ser-97, and Ser-130 (for mouse DARPP-32). In the striatum, increases in dopamine levels in the synaptic cleft lead to an increase in intracellular cAMP and activation of cAMP-dependent protein kinase A (PKA) by postsynaptic D1 dopamine receptors (Fig. 1A). PKA phosphorylates Thr-34 and converts DARPP-32 into a potent inhibitor of protein phosphatase-1 (PP1) (16, 17).
Fig. 1.
DARPP-32 regulatory pathways. (A) Simplified view of DARPP-32 pathway. The dopamine and glutamate cascades are shown on Left and Right (red and green), respectively. A partial view of the states of phosphorylation of DARPP-32 is shown in Center (blue). (B) Phosphorylation and dephosphorylation states of DARPP-32. DARPP-32 is phosphorylated at Thr-34 by PKA, at Thr-75 by cdk5, at Ser-97 by CK2, and at Ser-130 by CK1. Phospho-Thr-34 is preferentially dephosphorylated by PP-2B (or calcineurin); phospho-Thr-75 is preferentially dephosphorylated by PP-2A; phospho-Ser-130 is preferentially dephosphorylated by PP-2C; the phosphatase for phospho-Ser-97 is not yet characterized. Green arrow indicates positive effect; red arrows indicate negative effects. [Adapted from Nairn et al. (1).] (C) Detailed view of DARPP-32 subnetwork. Shown are the various states of phosphorylation allowed in our model.
Counteracting the effects of PKA, protein phosphatase 2B (PP2B), a Ca2+/calmodulin-activated protein phosphatase, dephosphorylates Thr-34 in response to cytosolic Ca2+ elevation induced, for example, by glutamate, another crucial neurotransmitter. Glutamate also activates cyclin-dependent protein kinase 5 (Cdk5), which phosphorylates DARPP-32 at Thr-75, converting it into an inhibitor of PKA (18).
Phosphorylation of DARPP-32 on Ser-97 converts Thr-34 into a better substrate for PKA, whereas phosphorylation of DARPP-32 on Ser-130 decreases the rate of Thr-34 dephosphorylation by PP2B. These sites are targeted by CK2 (casein kinase 2) and CK1 (casein kinase 1), respectively (19, 20). Compared with PKA, less is known about the signaling pathways involved in the regulation of these kinases.
As a consequence of phosphorylation of Thr-34 or Thr-75, DARPP-32 is a dual-function inhibitor of either PP1 or PKA, respectively. Through distinct signal transduction cascades, and differential regulation of the state of DARPP-32 phosphorylation, dopamine tends to activate PKA and inhibit PP1, whereas glutamate tends to do the reverse. Through inhibition of either PP1 or PKA, DARPP-32 regulates the state of phosphorylation and activity of key substrates, including many ion channels, pumps, neurotransmitter receptors, and transcription factors necessary for altering the physiological status of striatal neurons and, in turn, for altering the function of striatal circuits. Notably, there are “feed-forward” loops in both the dopamine and glutamate regulatory cascades. Positive feedback is also present; PKA drives DARPP-32 to the Thr-75-dephosphorylation state through activation of PP2A, thus removing its own inhibition. A more detailed picture of the allowed states of phosphorylation of DARPP-32 (the “DARPP-32 subnetwork”) is shown in Fig. 1C. In all, there are 102 chemical species in our model, including the intermediates involved in the various chemical reactions illustrated in Fig. 1 (see Appendix for a discussion of the numerical solution of the dynamical system and supporting information (SI) Text for a list of all component species).
Because much is understood about the composition of the DARPP-32 signaling cascade, it is a natural setting for computational studies. Recently, for example, two groups have simulated aspects of the network to study the effect of transient Ca2+ and dopamine (or cAMP) signals on DARPP-32 phosphorylation (21, 22). In both cases, the authors found interesting, time-dependent features of the pathway. A rather general mathematical framework for this kind of analysis has recently been developed, based on adapting dynamical systems methods (23). The authors identified phase-space domains showing high sensitivity to initial conditions that lead to qualitatively different transient activities. Our goal here is rather different (and complementary). We would like to understand why such a highly elaborate network has evolved in the first place. In particular, we would like to address two questions: (i) whether the complexity of the network plays a role in its functionality, and (ii) how robust features of an arbitrary network can be learned by simulation.
The Simulation Model
Although DARPP-32 is known to be affected by multiple neurotransmitter signals, we restrict our attention here to modeling the system's response to the competition between dopamine and glutamate. For each choice of the dopamine and glutamate signals, we allow the system to evolve from a specific initial state to a quasi-steady state by observing the chemical reactions in the network over time, until they appear to have achieved a steady state. The chemical concentrations of all component species at that “quasi-steady state” time can be viewed as a quantitative descriptor of the network response. By varying simulation conditions, we create an artificial data set that can be studied. Before discussing the data exploration step, we first describe the mathematical model in more detail.
Both the dopamine and glutamate signals are defined as functions of time according to
where p(t) is a Gaussian pulse with maximum value, Pmax, centered at t0 of variance δ:
When β = 0, the signal is entirely dopamine, and when β = 1, the signal is entirely glutamate. The full state of the network at time t is defined by the vector C(t) = (C1(t), C2(t), C3(t), …, C102(t)), where each component Ci(t) corresponds to a single chemical species (DARPP-32, PKA, cAMP, PP1, DARPP-32-Thr-34-P, etc.). The initial conditions for the ith species are defined by Ci(0). We assume each network connection describes either a binding reaction (causing an activation or deactivation) or an enzymatic reaction. The binding of dopamine to the D1 receptor (D1R), for example, which releases the active subunit denoted by Gα, is modeled as
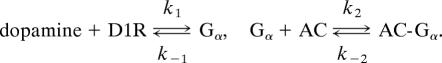 |
Thus, the differential equation for [Gα] is
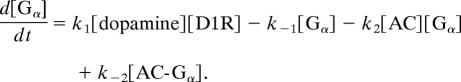 |
We do not follow strict Michaelis–Menten kinetics, because we are computing the dynamics of the enzyme concentrations. The phosphorylation of PP2A by PKA, for example, is modeled as follows:
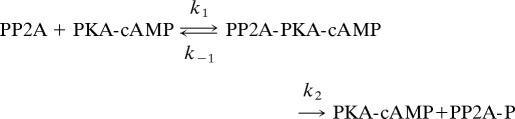 |
The first equation describes the forward enzymatic reaction and the second allows for the (nonenzymatic) dephosphorylation of PP2A-P. The ordinary differential equation for [PP2-A] takes the form
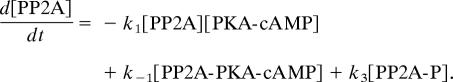 |
The complete set of components can be found in the SI Text.
In short, given initial concentrations Ci(0) for each chemical species and the kinetic coefficients ki for each reaction, the changes in concentration are determined as a function of time by solving a system of ordinary differential equations. The values of the initial concentrations and kinetic coefficients are chosen from a random distribution, with kinetic coefficients an order of magnitude larger than the concentrations. No subsequent fitting or refinement was used (see Appendix for further details). The equations are evolved until a quasi-steady-state time teq at which point the component chemical species have stopped fluctuating. The steady-state concentrations of the various species are then the components of a single point
in 102-dimensional space. Because the simulation depends on the input signals d(t), g(t), and therefore the value of the parameter β, we make this explicit and write
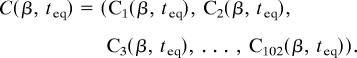 |
Suppose now that we have fixed the initial concentrations and kinetic coefficients and carried out a sequence of 1,000 simulations with β varying from 0 to 1. Assuming that the solution of the kinetic equations depends smoothly on the parameter β, the points C(β, teq) will lie on a curve in 102-dimensional space. We refer to this collection of 1,000 points as a noise-free data set. To mimic the fluctuations of a real biological system, we repeat the 1,000 simulations just discussed, but for each new value of β, we add noise. By noise, we mean that we add a random perturbation to each initial value and kinetic coefficient. We consider two cases. When the perturbations are drawn from a uniform distribution with maximum relative magnitude 30%, we refer to the perturbation as modest. When the perturbations are drawn from a uniform distribution with maximum relative magnitude 50%, we refer to the perturbation as large. The two new sets of 1,000 data points so generated are referred to as the modest perturbation and large perturbation data sets, respectively. (More thorough randomization procedures could obviously be used, but the preceding procedure is sufficient for the present study.)
In this study, we are interested in whether the modest and large perturbation data sets C(β,teq) still have any coherent behavior in response to the input signal parameter β. Because the state of phosphorylation of DARPP-32 reflects neurotransmitter signals, we are particularly interested in looking at the coherence of the DARPP-32 subnetwork and the regulatory cascades separately. For this, we want to explore the data sets visually, but unfortunately, that is difficult to do in 102 dimensions. We therefore have made use of dimensionality reduction methods to project the data onto a three-dimensional space, where direct inspection is feasible.
Dimensionality Reduction and Data Exploration
The detection of stable structures in noisy, high-dimensional data are a recurring problem in many areas of science. Fortunately, very effective approaches for this have been developed in the past few years, including locally linear embedding (LLE) (24) and Laplacian eigenmaps (LEs) (25). These have received a lot of attention in machine learning and pattern recognition, and we will not seek to review the literature here, except to note that LLE and LEs are beginning to be used in bioinformatics (26).
The intuition underlying LLE and LE is that, if the data points lie on a curve or surface (or other “low-dimensional manifold”), then a specific pattern of neighbor relations must hold. On a curve in 102-dimensional space sampled at a sequence of points, for example, each point has two nearest neighbors: the one just behind and the one just ahead. The way in which LLE and LE function is (i) to find the nearest neighbors in the original (in our case, 102-dimensional) space, and (ii) to replicate the neighbor relation in a lower dimensional “embedding” space (in our case, three-dimensional). We refer the reader to the original articles for a description of this mathematically sophisticated and highly nonlinear procedure (24, 25). Unfortunately, it should be noted that the coordinates of the points in the embedding space bear no clear relation to any of the coordinates in the original space. Nevertheless, simple visualization can be used to inspect the geometry of the data set. Curves should map to curves, two-dimensional surfaces to surfaces, etc. Details of our implementation can be found in Appendix, as can a short discussion of the underlying mathematics. Here, we illustrate the method with a simple example. For this, consider the set of points in three dimensions defined by
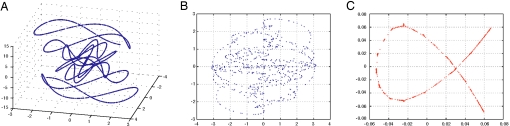
sampled at 300 points with β on the interval [−3, 3]. They clearly lie on a space curve (Fig. 2A). Imagine, however, that we were only able to visualize two-dimensional graphs. A straightforward linear projection onto a two-dimensional space is obtained by simply plotting the first two coordinates (Fig. 2B). Notably, the structure of the original data is lost. Use of LE to project onto two dimensions yields the curve shown in Fig. 2C. The fact that the data lie on a curve has been clearly and faithfully replicated.
Fig. 2.
Dimensionality reduction of a one-parameter family of data (a space curve). In A, a three-dimensional data plot reveals that the points seem to lie on a curve. In B, a two-dimensional “visualization” of the data is obtained by linear projection onto the x-y plane. The geometry of the data is difficult to see. In C, Laplacian eigenmaps are used to project the data onto a two-dimensional space in a much more revealing manner. The fact that the data lie on a curve is quite apparent. For realistic systems, like the DARPP-32 network, where the original data are high-dimensional and inaccessible to simple visualization, this is a critical tool.
Our data exploration procedure is now easy to describe. We take the data set (noise-free, modest perturbation, or large perturbation) and project it onto a three-dimensional space by using LE. If the response to variation in the parameter β is coherent and approximates a curve in 102-dimensional space, then it will be visible as a curve in three dimensions. Furthermore, we can explore the data set at will. The same method can be used to project only a subset of the components onto the three-dimensional embedding space. In the present context, for example, one can project only the DARPP-32 subnetwork or only the regulatory cascades.
It should be noted that signal transduction cascades are natural targets for this approach, because the dimensionality of the system response being sought is naturally controlled by the input parameters (here the dopamine/glutamate ratio). We briefly discuss the extension to higher-dimensional surfaces in the Appendix.
Simulation Results
Using the data generated by our simulations, we are able to identify network components that maintain coherence in the presence of noise. We are also able to identify specific topological features of the larger network that appear to be crucial for the robust behavior of those components.
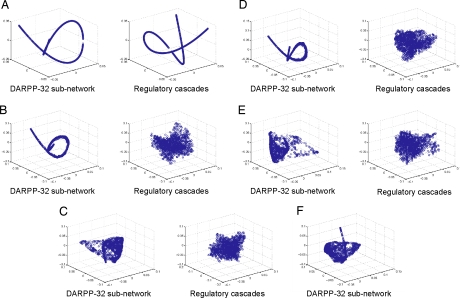
In Fig. 3 A–C, we plot the projection of the DARPP-32 subnetwork and regulatory cascades in the noise-free, modest perturbation and large perturbation cases. In the noise-free case, as expected, clean curves are traced out for both the DARPP-32 subnetwork and the regulatory cascades in response to varying the neurotransmitter signals by letting β range from 0 to 1 in increments of 0.005. In the modest perturbation case, the DARPP-32 subnetwork displays a remarkably stable response, despite the noise introduced into the system, whereas the regulatory cascades have lost coherence. In the large perturbation case, the fluctuations in initial concentrations and kinetic coefficients are sufficiently large that they overcome the inherent stability/robustness of the DARPP-32 subnetwork response.
Fig. 3.
Response manifolds for numerical simulations. (A) In the absence of perturbations, both the DARPP-32 subnetwork and the regulatory cascade follow (exactly) a simple one-parameter path, determined by the dopamine/glutamate ratio. The high-dimensional space curves are visualized in three dimensions by using Laplacian eigenmaps. (B) With modest perturbation, the response of the DARPP-32 subnetwork continues to follow a one-dimensional (but slightly noisy path). The regulatory cascades, however, no longer reflect a clear signal. (C) With sufficient noise (the large perturbation data set), the DARPP-32 response also loses its coherence, at least over some range of the dopamine/glutamate ratio. (D) Modified network 1 (see Fig. 4). cAMP is assumed to activate PP2A directly. Both PKA and PP2A are still activated in a coordinated fashion, and the DARPP-32 subnetwork clearly retains its coherence under modest perturbation. (E) Modified network 2 (see Fig. 4). All PP2A species are removed from the system (by setting their values to zero during the simulation). Clearly, the DARPP-32 subnetwork loses its coherence. (F) Modified network 3 (see Fig. 4). The regulatory cascades are eliminated by assuming that dopamine directly converts PKA to its active form (PKA-cAMP) and that glutamate directly converts PP2B to its active form (PP2B-Ca). As a result, the DARPP-32 subnetwork becomes much more sensitive to perturbations. That is, the removal of the regulatory cascades significantly affects the robustness of the signal transduction pathway.
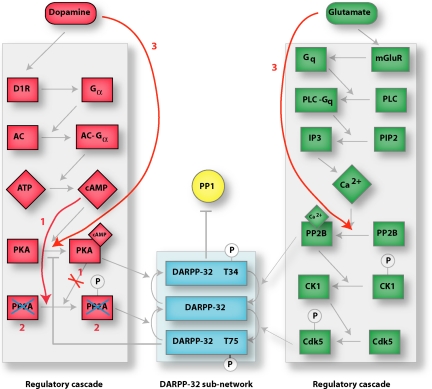
To confirm that the topology of the network plays an important role, we have broken it in three different ways (Fig. 4). In the first case, cAMP is assumed to activate PP2A directly, rather than through the intermediary PKA. Under this artificial modification, both PKA and PP2A are still activated in a coordinated fashion, and the DARPP-32 subnetwork retains its coherence under perturbations (Fig. 3D). In the second case, PP2A molecules are completely removed from the system (by setting their values to zero during the simulation) (Fig. 4); as a result, the DARPP-32 subnetwork loses its coherence (Fig. 3E).
Fig. 4.
Three simulated modified networks. In modification 1, cAMP bypasses PKA and is assumed to directly catalyze the phosphorylation of PP2A (indicated by labels 1). In modification 2, all PP2A species are deleted from the system (indicated by labels 2). In modification 3, dopamine is assumed to convert PKA to its active form (PKA-cAMP) directly and glutamate is assumed to convert PP2B to is active form (PP2B-Ca) directly (indicated by labels 3).
A final, less obvious network modification consists of trying to remove as much of the regulatory cascades as possible. For this, dopamine is assumed to convert PKA to its active form (PKA-cAMP) directly and glutamate is assumed to convert PP2B to its active form (PP2B-Ca) directly (Fig. 4). The result of this is that the DARPP-32 subnetwork becomes much more sensitive to perturbations (Fig. 3F). In other words, the regulatory cascades cannot be removed from the signal transduction pathway while retaining a robust response.
Conclusions
In the present study, we have found convincing numerical evidence that the topological structure of the DARPP-32 pathway serves a useful purpose—to faithfully translate neurotransmitter signals at the cell surface into the net state of phosphorylation of DARPP-32 in the cell's interior, despite the presence of noise. What we found particularly striking is the fact that the other components of the pathway (in the regulatory cascades) are themselves poorly controlled in response to the dopamine/glutamate signal, but that they are crucial to the overall functioning of the signaling system. This observation suggests that attempts at capturing the full network dynamics in a small module (the goal of some efforts in systems biology) may be hampered by subtle but critical topological considerations.
The tool we have developed here will allow scientists to analyze the robust response of a network to an input signal in a new way: by creating artificial data that simulate biological noise, and by exploring those data through dimensionality reduction. The most compelling feature of the method is, perhaps, that it is unsupervised—if the network is effectuating a coherent response in some subset of its components, that response should be visible to the user.
Supplementary Material
Acknowledgments
This work was supported by U.S. Army Medical Research Acquisition Activity Grant W81XWH-04-1-0307 and NYSTAR Contract C04006 (to P.B.); by Defense Advanced Research Projects Agency Defense Sciences Office Contract HR0011-04-C-0057 and Department of Energy Contract DEFG-02-00-ER25053 (to L.G.); by the National Science Foundation through the IGERT Program in Computational Biology (M.S.); acknowledge support from by U.S. Public Health Service Grants MH40899, MH074866, and DA10044 (to P.G., M.F., and A.C.N.).
Appendix
Numerical Solution of System of Differential Equations.
The dynamics of the DARPP-32 network is modeled by means of a set of enzymatic and binding reactions. Each chemical species is given an initial concentration drawn from a uniform random distribution on the interval [0, 0.1]. The kinetic coefficients (ki) in the reactions are drawn from a uniform random distribution on the interval [0, 1]. The full system of ordinary differential equations is solved for each dopamine and glutamate input signal pair (d(t), g(t)), using the CVODE package available from the Sundials site (http://acts.nersc.gov/sundials/) at Lawrence Livermore National Laboratory.
The specific integration method used is designed for stiff systems, based on BDF (backward differentiation formula) with Newton iteration.
As indicated in the text, the modest and large perturbation sets are generated as follows. As the parameter β varies from 0 to 1, a perturbation is added to each of the original initial concentrations and kinetic parameters. These perturbations are also drawn from a uniform distribution, but with maximum magnitude equal to either 30% or 50% depending on each of the original values. These define the modest and large perturbation data sets.
Implementation of Laplacian Eigenmaps.
Our visualization method relies on the method of Laplacian Eigenmaps developed by Belkin and Niyogi (25). The details of our implementation follow, where N denotes the number of high-dimensional data points.
For a fixed parameter n, add an edge between nodes i and j, if i is one of the n nearest neighbors of j, or j is one of the n nearest neighbors of i.
Let W be an N×N matrix. Set W(i, j) = 1, if there is an edge between nodes i and j and zero otherwise.
Let D be a diagonal N×N matrix with entries D(i, i) = Σj W(j,i) (column sums).
Define L = D − W, the “graph Laplacian.”
Solve the generalized eigenvalue problem Ly = λDy. Let (λ0, y0), (λ1, y1), (λ2, y2), and (λ3, y3) denote the lowest four eigenvalue/eigenvector pairs. The lowest eigenvalue λ0 is easily seen to be zero from the definitions of L, D, and W and the corresponding eigenvector is identically one. It carries no useful information.
For each high-dimensional data point Xi, compute its projection onto three-dimensional space according to the formula: Xi → Yi = (y1(i), y2(i), y3(i)).
Plot the data points Yi in three dimensions with coordinates obtained from step 6.
Plots in the article used n = 10 neighbors. The choice n = 15 produced qualitatively similar images.
Developing Quantitative Measures of Robustness.
The computational approach described in this article is based on geometric intuition. In its present form, biologists using the visualization tool are responsible for selecting the subnetworks or modules to analyze for robustness, and their own intuition in declaring that the pattern seen in the projected data is of interest. It would, therefore, be of significant interest to both quantify and automate the analytic process. For this, we need a more precise mathematical formulation of the problem.
We denote by S = {X(t)} ⊂ ℝM the full data set of trajectories, where M is the dimension of the original data vectors (M = 102 in our study) and we denote by U = {Y(t)} ⊂ ℝK the trajectories of a subset of components (K < M). We denote by P(S) or P(U) the projection of the respective trajectories onto the n-dimensional embedding space (n = 3 in our study). In essence, we are simply seeking to identify those subsets U that remain in a well defined neighborhood of the unperturbed trajectory. A measure of robustness can, therefore, be obtained by computing some statistics on P(U). In the case of a one-dimensional signal, this could simply be the radius of the tubular neighborhood around the unperturbed trajectory that contains 95% of the noisy data. Given such a measure, visual inspection would no longer be required. A more thorough development of the underlying mathematics is underway.
Automating the search process is unfortunately harder, because it is subject to “combinatorial explosion.” That is, given a system with M components, the number of potential K-dimensional modules is of the order
For large values of M, investigating all modules with more than one or two components becomes rapidly infeasible. We need to investigate hierarchical methods to overcome this barrier. For the moment, that selection process is in the hands of the user, driven by conjectures concerning the underlying biology.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709955104/DC1.
References
- 1.Nairn AC, Svenningsson P, Nishi A, Fisone G, Girault JA, Greengard P. Neuropharmacology. 2004;47(S 1):14–23. doi: 10.1016/j.neuropharm.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Barkai N, Leibler S. Nature. 1997;387:913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 3.Alon U, Surette MG, Barkai N, Leibler S. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 4.Huang CY, Ferrell JE., Jr Proc Natl Acad Sci USA. 1996;93:10078–10083. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borisuk MT, Tyson JJ. J Theor Biol. 1998;195:69–85. doi: 10.1006/jtbi.1998.0781. [DOI] [PubMed] [Google Scholar]
- 6.von Dassow G, Meir E, Munro EM, Odell GM. Nature. 2000;406:188–192. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- 7.Eldar A, Rosin D, Shilo BZ, Barkai N. Dev Cell. 2003;5:635–646. doi: 10.1016/s1534-5807(03)00292-2. [DOI] [PubMed] [Google Scholar]
- 8.Albert R, Othmer HG. J Theor Biol. 2003;223:1–18. doi: 10.1016/s0022-5193(03)00035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stelling J, Sauer U, Szallasi, Doyle FJ, III, Doyle J. Cell. 2004;118:675–685. doi: 10.1016/j.cell.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Lauffenburger DA. Proc Natl Acad Sci USA. 2000;97:5031–5033. doi: 10.1073/pnas.97.10.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 12.Strogatz SH. Nature. 2001;410:268–276. doi: 10.1038/35065725. [DOI] [PubMed] [Google Scholar]
- 13.Csete ME, Doyle JC. Science. 2002;295:1664–1669. doi: 10.1126/science.1069981. [DOI] [PubMed] [Google Scholar]
- 14.Rao CV, Wolf DM, Arkin AP. Nature. 2002;420:231–237. doi: 10.1038/nature01258. [DOI] [PubMed] [Google Scholar]
- 15.Sontag ED. Syst Biol (Stevenage) 2004;1:9–18. doi: 10.1049/sb:20045006. [DOI] [PubMed] [Google Scholar]
- 16.Walaas SI, Aswad DW, Greengard P. Nature. 1983;301:69–71. doi: 10.1038/301069a0. [DOI] [PubMed] [Google Scholar]
- 17.Hemmings HC, Jr, Greengard P, Tung HY, Cohen P. Nature. 1984;310:503–505. doi: 10.1038/310503a0. [DOI] [PubMed] [Google Scholar]
- 18.Bibb JA, Snyder GL, Nishi A, Yan Z, Meijer L. Nature. 1999;402:669–671. doi: 10.1038/45251. [DOI] [PubMed] [Google Scholar]
- 19.Girault JA, Hemmings HC, Jr, Williams KR, Nairn AC, Greengard P. J Biol Chem. 1989;264:21748–21759. [PubMed] [Google Scholar]
- 20.Desdouits F, Siciliano JC, Greengard P, Girault JA. Proc Natl Acad Sci USA. 1995;92:2682–2685. doi: 10.1073/pnas.92.7.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindskog M, Kim M, Wikstrom MA, Blackwell KT, Kotaleski JH. PLoS Comput Biol. 2006;2:e119. doi: 10.1371/journal.pcbi.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez E, Schiappa R, Girault JA, Le Novere N. PLoS Comput Biol. 2006;2:e176. doi: 10.1371/journal.pcbi.0020176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aldrich BB, Haller G, Sorger PK, Lauffenburger DA. IEEE Proc Syst Biol. 2006;153:425–432. doi: 10.1049/ip-syb:20050065. [DOI] [PubMed] [Google Scholar]
- 24.Roweis ST, Saul LK. Science. 2000;290:2323–2326. doi: 10.1126/science.290.5500.2323. [DOI] [PubMed] [Google Scholar]
- 25.Belkin M, Niyogi P. Neural Comput. 2003;15:1373–1396. [Google Scholar]
- 26.Forman JJ, Clemons PA, Schreiber SL, Haggarty SJ. BMC Bioinformatics. 2005;6:260. doi: 10.1186/1471-2105-6-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.