Abstract
Multiple genes, gene-by-gene interactions, and gene-by-environment interactions are believed to underlie most complex diseases. However, such interactions are difficult to identify. Although there have been recent successes in identifying genetic variants for complex diseases, it still remains difficult to identify gene–gene and gene–environment interactions. To overcome this difficulty, we propose a forest-based approach and a concept of variable importance. The proposed approach is demonstrated by simulation study for its validity and illustrated by a real data analysis for its use. Analyses of both real data and simulated data based on published genetic models show the effectiveness of our approach. For example, our analysis of a published data set on age-related macular degeneration (AMD) not only confirmed a known genetic variant (P value = 2E-6) for AMD, but also revealed an unreported haplotype surrounding single-nucleotide polymorphism (SNP) rs10272438 on chromosome 7 that was significantly associated with AMD (P value = 0.0024). These significance levels are obtained after the consideration for a large number of SNPs. Thus, the importance of this work is twofold: it proposes a powerful and flexible method to identify high-risk haplotypes and their interactions and reveals a potentially protective variant for AMD.
Keywords: age-related macular degeneration, genomewide association, haplotype, single-nucleotide polymorphism, tree and forest methods
It is generally accepted that the etiology of most complex diseases involves genetic and environmental factors and the interactions among them. Association study is a more powerful approach than linkage analysis when ultradense markers are genotyped. Recently, there have been landmark successes from association studies that identified genetic variants underlying a few complex traits, including age-related macular degeneration [AMD (MIM nos. 603075, 610149, 610698, and 153800)] (1–5), inflammatory bowel disease (MIM nos. 26600 and 191390) (6), cardiac repolarization (7), and Alzheimer disease (MIM no. 104300) (8). In this study, we will propose an approach for genomewide association study to identifying susceptible haplotypes and their interactions.
Epistasis is a mechanism in which the effect of the genotype in a particular locus might depend on the genotype of other loci (9). In several genetic studies, considering interactions among genes has proven to be useful in identifying susceptible loci for various scenarios (10, 11). Despite the belief that epistasis likely plays an important role in the development of complex diseases, identifying gene–gene interactions is challenging. A major cause lies in the large number of potential interactions and the resulting tests, because we generally do not know a priori which genes may be engaged in epistasis. A practical approach is to test candidate epistatic effects after a genomewide scanning reveals genes with main effects (12). However, some authors noted that if a trait is caused by several loci interacting epistatically rather than additively, then there are many situations where the main-effect-based methods may have relatively little power to detect any of those loci (13, 14). Thus, it is critical and challenging to develop powerful analytic approaches that can detect interactions and main effects.
Recently, Zhao et al. (15) introduced a test for interaction between two unlinked loci by defining the interaction as the deviance of penetrance for a haplotype at two loci from the product of the marginal penetrance of the individual alleles that span the haplotype. It is important to note, however, that haplotypes cannot be determined with certainty in the commonly available high-throughput genotype platforms such as Affymetrix GeneChip Array and Illumina BeadArray. Becker et al. (16) used maximum likelihood to estimate haplotype frequency. They then tested a global hypothesis that none of the considered single-nucleotide polymorphism (SNP) combinations showed an association with the disease. With a large number of markers such as SNPs and haplotypes, it is not an easy task to identify which haplotypes should be considered for interaction testing. Marchini et al. (17) examined the power of three strategies for analyzing gene–gene interactions in genomewide association studies: Strategy I, locus-by-locus search requiring at least one locus meeting the significant criterion; strategy II, search over all pairs of loci; and strategy III, a two-stage strategy in which all loci meeting some low threshold in a single-locus search are subsequently examined for a significant full model fit. Marchini et al. (17) suggested that strategy III is the most powerful choice in most cases. Musani et al. (18) presented a more comprehensive review of methods and issues for epistatic analysis.
Most of the efforts focused on interactions of two unlinked regions. By using the recursive partitioning technique, Zhang and Bonney (19) introduced the tree-based approach to genetic association analysis that can be used to explore gene–gene (as well as gene–environment) interactions systematically based on the available markers. Since then, the recursive partitioning technique and other machine-learning methods have been examined and applied in a number of genetic studies (20–23). All of those reports, however, evaluated the interactions among the given markers. To detect haplotypes and interactions among them, we must acknowledge the fact that haplotypes are not given and must be inferred in frequencies based on SNPs. In addition, we need to consider the uncertainties in the estimated haplotypes for association studies. To overcome this problem, we propose to use the forest-based approach to accommodate the haplotype uncertainties and variable importance to sort out significant haplotypes and their interactions in genomewide case-control association studies. In the special case when we are interested in single SNP-based analysis, our approach is similar to that of Zhang and Bonney (19) and Bureau et al. (24).
System and Methods
As described earlier, we propose a method that detects the disease-related haplotypes, some of which may act on their own, whereas others may interact with each other in manifesting their effects.
Estimated Haplotype Frequencies.
With the current high-throughput genotyping platforms, the haplotype information is not directly observed. What we can observe are the SNP genotypes. Many methods for haplotype reconstructions have been developed (25–30). We use a haplotype reconstruction program, called SNPHAP (31), to obtain haplotype frequencies for each individual. SNPHAP is a program that estimates haplotype frequencies for unrelated individuals. This program implements an expectation-maximization (EM) algorithm to calculate the maximum likelihood estimate of haplotype frequencies based on genotype data whose phase is unknown.
Recursive Partitioning.
Suppose we have n subjects with feature information (haplotypes) and their disease status or class label. Recursive partitioning is an approach to build a classifier that predicts the class membership based on the feature information. According to this information, it divides an entire study sample into smaller and smaller subgroups, called nodes, from one feature at a time in an attempt to achieve the maximum homogeneity of the subgroups in terms of the disease distribution as measured by entropy, for example. The entropy of node t is defined as it = −pt log(pt) − (1 − pt)log(1 − pt), where t is the proportion of the individuals in node t having the disease. The recursive partitioning process begins with the entire sample, namely, the root node. For clarity, let us assume for the moment that haplotypes could be observed for all individuals. We can divide the root node into two daughter nodes according to the haplotypes that one individual may or may not have. Such a split is decided after we evaluate all possible splits of the root node by using all haplotypes. We then select the one that yields the lowest weighted entropy of the two daughter nodes. The weight is proportional to the number of subjects in the daughter node, and specifically, the weighted entropy for node t with daughter nodes tL and tR is defined as i(t) = p(tL)i(tL) + p(tR)i(tR), where i(tL) and p(tL) are the entropy of node tL and the proportion of subjects in node tL, respectively. i(tR) and p(tR) are defined analogously for node tR. After the root node is divided, the same splitting process can be recursively applied to the daughters to continue the division. The root node is at the first layer of the tree, its two daughter nodes are at the second layer, the third layer consists of the daughter nodes of the nodes in the second layer, and so on. The final size and layer of the tree will be specified later. We refer to Breiman et al. (32) and Zhang and Singer (33) for details. In the description above, we assumed that haplotypes could be observed for all individuals, which is of course not the case. We resolve this problem by constructing a forest as described below.
Forest.
Because haplotypes can only be estimated by frequencies, for haplotypes in a given region, we suppose that there are K possible haplotypes H1, …, HK with frequencies q1, …, qK, respectively. The key idea is to expand our data randomly and in proportion to the frequencies q1, …, qK. For illustration, suppose we have two combinations of haplotypes, say (H1, H2) and (H3, H4), conform with the genotypes of the observed genotype for an individual and these two combinations are equally likely. We can expand one data set with the unphased genotype into two phased data sets with the first having two haploids (H1, H2) and the second having (H3, H4). Such a random expansion applies to all individuals and all regions. As a result, one unphased data set would expand to a large number of phased data sets reflecting the uncertainties of the haplotypes. Choosing an exact number of the phased data sets is usually a straightforward trial-and-error process. For example, if we begin by expanding an unphased data set into 100 phased data sets, we expect that a haplotype with the frequency of 0.1 or greater appears 10 times on average across all of the expanded data sets. Then, by doubling the number of the phased data sets a few times, we can evaluate whether the number of the phased data sets affects the result, namely, suggesting different genomic regions. For example, we used 100 data sets for the AMD study (3). For each data set after the expansion, we can construct a tree by using the recursive partitioning technique as presented above. A forest is formed after a tree is grown for every expansion data set. Because of the overfitting problem in the traditional tree framework (34), we chose to grow a maximum of seven layers in each tree. Furthermore, a tree of seven layers can accommodate many seven-way interactions and hence can represent a large and complex model. A data set with thousands of study subjects will usually not be powered to make inference on a larger model than a seven-layer tree.
The next immediate and essential question is: how do we identify genes and gene–gene interactions that contribute significantly to the disease risk? To answer this question we are required to make inference from the forest. The hypothesis is that, if certain genes and gene–gene interactions indeed underlie the disease etiology, they must manifest their contributions in the forest in patterns beyond chance. Mathematically, we use variable importance to help us identify significant patterns of genes and gene–gene interactions in the forest in association with the disease.
Importance of a Haplotype.
There are many possible ways to quantify the importance of a haplotype in its association with the disease. In the following, we propose one of such measures, which is inversely proportional to the depth of a node for which a haplotype is used to split the node. The rationale is that a variable of importance tends to appear near the top of a tree.
First, for a haplotype h in a chromosome region G, we assess its “importance” in each tree, T, of the constructed forest f. Let |T| be the number of nodes in T. Then the “importance” of the haplotype h in tree T is defined as
where Lt is the depth of node t and Gt is the χ2 independence test statistic of node t. Then, the average of “importance,”
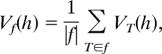 |
over all of the |f| trees in the forest f serves as the importance measure of haplotype h.
Algorithm.
We now summarize all steps in identifying haplotypes and haplotype–haplotype interactions. Visit http://c2s2.yale.edu/software/HapForest for software implementation.
Apply the recursive classification tree program (19) by using the individual SNPs as features and the disease status as the outcome.
Construct haplotype blocks containing the SNPs identified in step 1 using Hapview (35).
Use SNPHAP (31) to estimate the haplotype frequencies in the haplotype blocks identified in step 2.
A new data set is constructed from the original data set by assigning phased haplotypes in all regions (or genes) randomly according to the haplotype frequencies inferred in step 3.
Apply the recursive classification tree program to construct tree T by using the data set constructed in step 4.
Evaluate the importance, VT(h), of any haplotype h for tree T.
Repeat steps 4–6 a number of times and obtain the average importance measure.
Significance Level.
To assess the significance of a haplotype in its association with the disease, we begin with the original data set and permute the disease status among all subjects a prespecified number (e.g., 100) of times. This permutation generates the data under the null hypothesis of no association between the genotypes and the disease at genomewide level. Then, for each permuted data set, we construct a forest and then calculate the importance of a haplotype as we have done by using the original data set. This enables us to generate the distribution of the maximum importance measures for haplotypes not associated with the disease over the entire genome, which can be used to assess the significance of the importance measure of haplotypes in the original data set. It is important to note that this procedure adjusts the significance level for genomewide multiple tests, because the null distribution is derived from the genomewide data. Thus, the commonly used significance level of 0.05 is an appropriate threshold for significance. When we have multiple haplotypes, we evaluate their significance levels simultaneously through the same permutation process.
Results
Simulation Design and Power Comparison.
Even though our method does not have limitations on the number of regions or the number of SNPs in a region, to compare the power of our method with the method of Becker et al. (16), we restricted our attention to two genomic regions, each of which has three SNPs. As specified in Tables 1 and 2 we used the 12 two-locus interaction models described by Knapp et al. (36) and Becker et al. (16) and two additive models with background penetrance (Ad-1 and Ad-2). In our simulation study, we assume that each locus is diallelic and two regions are unlinked. The studied models include models of epistatis (Ep-1 to EP-6 and S-3), models of heterogeneity (Het-1 to Het-3 and S1 to S2), and models with additive effect (Add-1 to Add-2). Main effects are absent in models Ep-1 through Ep-3 and Ep-5. We refer to Knapp et al. (36) for more discussions of these models.
Table 1.
The penetrance table for the two-locus model
| Region 2 | ||||
|---|---|---|---|---|
| 0 | 1 | 2 | ||
| Region 1 | 0 | f00 | f01 | f02 |
| 1 | f10 | f11 | f12 | |
| 2 | f20 | f21 | f22 |
fij is the penetrance of the genotype carrying i and j copies of the disease haplotype at regions 1 and 2, respectively.
Table 2.
Description of two-locus segregation models
| Model | f22 | f21 | f20 | f12 | f11 | f10 | f02 | f01 | f00 | f | P1 | P2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ep-1 | f | f | 0 | f | f | 0 | 0 | 0 | 0 | 0.707 | 0.210 | 0.210 |
| Ep-2 | f | f | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.778 | 0.600 | 0.199 |
| Ep-3 | f | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.900 | 0.577 | 0.577 |
| Ep-4 | f | f | 0 | f | 0 | 0 | f | 0 | 0 | 0.911 | 0.372 | 0.243 |
| Ep-5 | f | f | 0 | f | 0 | 0 | 0 | 0 | 0 | 0.799 | 0.349 | 0.349 |
| Ep-6 | 0 | f | f | f | 0 | 0 | f | 0 | 0 | 1.000 | 0.190 | 0.190 |
| Het-1 | g | g | f | g | g | g | f | f | 0 | 0.495 | 0.053 | 0.053 |
| Het-2 | g | g | f | f | f | 0 | f | f | 0 | 0.660 | 0.279 | 0.040 |
| Het-3 | g | f | f | f | 0 | 0 | f | 0 | 0 | 1.000 | 0.194 | 0.194 |
| S-1 | f | f | f | f | f | g | f | f | 0 | 0.522 | 0.052 | 0.052 |
| S-2 | 1 | 1 | 1 | f | f | 0 | f | f | 0 | 0.574 | 0.228 | 0.045 |
| S-3 | 1 | 1 | f | 1 | f | 0 | f | 0 | 0 | 0.512 | 0.194 | 0.194 |
| Ad-1 | f | f | 0.04 | f | 0.304 | 0.02 | 0.01 | 0.01 | 0.01 | 0.799 | 0.349 | 0.349 |
| Ad-2 | f | f | 0.15 | f | 0.324 | 0.10 | 0.05 | 0.05 | 0.05 | 0.799 | 0.349 | 0.349 |
g = 2f − f 2. pi is the frequency of the disease allele at locus i. fij is defined in Table 1.
As in Becker et al. (16), we carried out our simulation studies with 300 cases and 300 controls to assess the power. Specifically, we considered two unlinked regions, each with three SNPs and all eight possible haplotypes. We considered three scenarios: (i) neither region is in linkage disequilibrium (LD) with the disease allele(s), (ii) only one region is in LD with the disease allele (D′ = 0.5), and (iii) both regions are in LD with the disease allele (D′ = 0.5). The first two are designed to examine the capability of our method to exclude false-positive SNPs or regions. We used the same code as in Becker et al. (16) to simulate the LD pattern.
The null hypothesis, as stated by Becker et al. (16), is that none of the SNPs in the two regions is associated with the disease, whether they are tested as a single SNP, in combination with other SNPs in the same region, or as interactions across the two regions.
Ideally, simulations with the whole genome data as in the AMD data would be more useful. However, we restrict our attention to two regions for two reasons. First, as stated above, we would like to compare our results with Becker et al. (16) under the same genetic models. Second, a thorough simulation with the entire genome is computationally intensive.
The power and false-positive rates are shown in Tables 3 and 4. The power is computed when at least one region is in LD with the disease allele as follows. When both regions are in LD with the disease allele (scenario iii), we considered a loose definition of power, φ1 = P (identify at least one correct haplotype), and a strict definition of power φ2 = P (identify both haplotypes correctly). When only one region is in LD with the disease allele (scenario ii), the power is defined as φ3 = P (identify the correct haplotype). The false-positive rate (FP1) is calculated as FP1 = P (identify at least one wrong haplotype).
Table 3.
Empirical power and false-positive rate when two unlinked regions are in LD with disease alleles
| Model | Forest |
FAMHAP* |
|||
|---|---|---|---|---|---|
| Power† | Power‡ | FPR§ | Power¶ | Power‖ | |
| Ep-1 | 0.995 | 0.695 | 0.035 | 1.000 | 0.010 |
| Ep-2 | 1.000 | 0.840 | 0.015 | 1.000 | 0.000 |
| Ep-3 | 1.000 | 1.000 | 0.002 | 1.000 | 0.000 |
| Ep-4 | 1.000 | 0.838 | 0.008 | 1.000 | 0.000 |
| Ep-5 | 1.000 | 0.982 | 0.017 | 1.000 | 0.000 |
| Ep-6 | 1.000 | 1.000 | 0.000 | 1.000 | 0.002 |
| Het-1 | 0.977 | 0.363 | 0.040 | 1.000 | 0.163 |
| Het-2 | 1.000 | 0.795 | 0.007 | 1.000 | 0.050 |
| Het-3 | 1.000 | 1.000 | 0.000 | 1.000 | 0.002 |
| S-1 | 0.967 | 0.347 | 0.037 | 0.998 | 0.213 |
| S-2 | 0.995 | 0.013 | 0.035 | 0.998 | 0.002 |
| S-3 | 1.000 | 0.895 | 0.013 | 1.000 | 0.003 |
| Ad-1 | 0.993 | 0.560 | 0.028 | 0.998 | 0.027 |
| Ad-2 | 0.970 | 0.187 | 0.033 | 0.995 | 0.158 |
*False-positive rate is not available.
†φ 1 = P (identify at least one correct haplotype).
‡φ 2 = P (identify both haplotypes correctly)
§False-positive rate.
¶φ 4 = P (identify at least a SNP in one region).
‖φ 5 = P (identify at least a SNP in each of the two regions).
Table 4.
Empirical power and false-positive rate when only one of the two unlinked regions is in LD with disease alleles
| Model | Forest |
FAMHAP |
||
|---|---|---|---|---|
| Power | False-positive rate | Power* | False-positive rate† | |
| Ep-1 | 0.973 | 0.032 | 0.998 | 0.088 |
| Ep-2 | 1.000 | 0.002 | 0.998 | 0.018 |
| Ep-3 | 1.000 | 0.003 | 1.000 | 0.003 |
| Ep-4 | 0.980 | 0.008 | 0.975 | 0.183 |
| Ep-5 | 1.000 | 0.005 | 0.998 | 0.015 |
| Ep-6 | 0.998 | 0.007 | 0.997 | 0.093 |
| Het-1 | 0.887 | 0.060 | 0.968 | 0.232 |
| Het-2 | 0.998 | 0.002 | 0.998 | 0.118 |
| Het-3 | 1.000 | 0.002 | 0.998 | 0.080 |
| S-1 | 0.883 | 0.047 | 0.968 | 0.250 |
| S-2 | 0.143 | 0.053 | 0.128 | 0.078 |
| S-3 | 0.992 | 0.013 | 0.997 | 0.083 |
| Ad-1 | 0.970 | 0.020 | 0.955 | 0.175 |
| Ad-2 | 0.782 | 0.058 | 0.880 | 0.270 |
*φ6 = P (identify at least a SNP in the region in LD with disease alleles).
†FP2 = P (identify at least a SNP in a neutral region).
In Table 3, the power values and false-positive rates are reported for 14 two-locus disease models under scenario iii. All of these disease models include interaction effects between the two regions, revealing the power of our method to identify the correct haplotype. Table 4 displays the power values and false-positive rates under scenario ii. Again, our approach has great power in identifying the correct high-risk haplotype (except genetic model S-2).
We should note that approaches have been proposed to test two specified regions, unlike ours that search for high-risk regions without specifying where they are a priori. For comparison purposes, we compared the power of our approach with Becker's method, called FAMHAP (16), which tests the LD of two unlinked and a priori regions with the disease allele.
Compared with the result obtained from FAMHAP, the power of our approach is compatible in identifying at least one region. In all models considered, both approaches are able to identify at least one region with power close to 1.00. However, our approach is much more powerful than the FAMHAP method in identifying the interactions of haplotypes or the combination of SNPs from two unlinked regions. For example, in the Ep-2 model, the power of our proposed approach is 0.84, whereas FAMHAP failed to identify the two regions (with the power <0.001). In most models, our proposed approach maintained its power ≈0.8 in identifying the two true haplotypes while FAMHAP often failed.
Table 4 indicates that our approach tends to have <5% false-positive rate, whereas the false-positive rate for FAMHAP is >5%. Although neither region is in LD with the disease allele(s) (scenario i), our approach had 1% false-positive rate.
Significant Genes for Age-Related Macular Degeneration
Age-related macular degeneration is the most common cause of vision loss in the elderly. Many researchers have devoted much effort to uncover the genetic mechanism of this complex disease. More information can be found in Daiger (37) and Marx (38). To test our method, we analyzed the data from a published case-control study (3). This data set contains 96 MD cases and 50 controls and has more than 100,000 SNPs for each individual.
While using more than 100,000 SNPs from each individual, we first ran the RTREE program (19) by treating each SNP as one covariate, which identified single markers potentially associated with AMD. For example, RTREE selected rs1329428 on chromosome 1 and rs10272438 on chromosome 7 as two top splits.
We then used the Hapview (35) to construct the LD blocks containing those two SNPs. We derived two haplotype blocks: one including 6 SNPs for rs1329428 (region 1) and another including 11 SNPs for rs10272438 (region 2). Region 1 consisted of rs2019727, rs10489456, rs3753396, rs380390, rs2284664, and rs1329428, and region 2 consisted of rs4723261, rs764127, rs10486519, rs964707, rs10254116, rs10486521, rs10272438, rs10486523, rs10486524, rs10486525, and rs1420150.
To assess the importance of these haplotypes, we generated 5,000 unphased data sets through permutation. Each of those data sets was randomly expanded into 100 phased data sets according to the estimated haplotype frequencies. The total running time for this data set was ≈24 h on a 3.2-GHz processor. We found that the most significant haplotype was ACTCCG in region 1 (P value = 2E-6). This finding is identical to the result reported by Klein et al. (3), which has found the same haplotype as the highest risk. These authors further validated that this polymorphism is in a region of Complement Factor H [CFH (MIM no. 134370)] and is linked to AMD. We also identified an AMD disease-related haplotpye, TCTGGACGACA, in region 2 (P value = 0.0024), which is not reported to be AMD related.
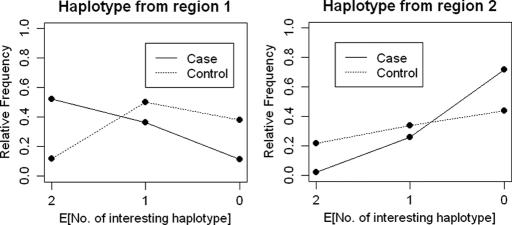
Fig. 1 represents the trends in frequency of AMD as a function of the expected number of these haplotypes. This figure confirms the role of CFH, and also suggests that the haplotype TCTGGACGACA in region 2 might be protective against the MD disease. This second region is located in the Bardet–Biedl syndrome 9 [BBS9 (MIM no. 607968)] gene, which is annotated as visual perception in gene ontology, although the function of this gene is not well understood.
Fig. 1.
The frequencies of AMD cases and controls are plotted against the expected numbers of susceptibility haplotypes. Haplotype 1 is in a region of CFH on chromosome 1, and haplotype 2 is in the BBS9 gene on chromosome 7.
Discussion
To identify haplotypes in LD with disease alleles, we proposed a forest-based method that is based on a proven statistical technique, recursive partitioning (33). This technology is known to be flexible in dealing with missing data in the predictors, to achieve variable selection and model selection simultaneously, and to avoid the colinearity problem (this could be a serious problem for genomewide data). To accommodate the uncertainties in the haplotype inference (as a result of genotyping errors and missing genotypes), we propose to randomly expand the number of data sets to reflect the haplotype distribution. To evaluate the importance of putative haplotypes, we proposed an importance measure. Our basic idea is analogous to a gold-mining process in which we shake the dirt and allow the gold to surface, and then we verify whether it is gold.
Our method can successfully identify both haplotypes with main effects and/or interactions of disease-associated haplotypes. We have demonstrated the utility of our approach through simulated data and illustrated its use by a real data study. Our approach is of particular appeal because it does not make any a priori assumption and yields a significance level that accommodates multiple tests. Although in the AMD data set, the two identified haplotypes do not appear to interact with each other, the models we used in the simulation study include interaction effects. Some of the simulated models have main effects, but in four of the simulated models, main effects are absent. This is designed to assess the specificity of our proposed method. Thus, our proposed method is designed to work when there is lack of evidence for epistasis (e.g., the AMD data set), when there is absence of main effects, or when there are presence of both haplotype heterogeneity and epistasis.
We used a permutation procedure to estimate the significance level. The computation time is reasonable for a real data set, but can be intensive for simulation studies. Methods for expediting the computation will be useful. In the simulation study, we simplified our task by focusing on the haplotypes in two different chromosomes for the comparison purpose with an existing method as well as the computational concern. Despite the restriction, our current simulation serves the purpose of evaluating the performance of our proposed method relative to an existing method. Nonetheless, it will be a worthy project to accelerate the computation and further scrutinize our proposed method.
False discovery is a major concern in disease gene identification. Through simulation studies, we demonstrated that the false-positive rate of our method is well under control and that the method can successfully distinguish disease-associated regions from neutral regions. Our reanalysis of the AMD data not only confirmed a landmark finding in genomewide association studies, but also revealed a protective variant in the BBS9 gene, for which the existing literature suggests a potential role in visual perception. We should caution that the sample size in the AMD data set is relatively small, and hence the role of the BBS9 gene warrants further investigation.
We used SNPHAP (31) to find haplotype frequencies, but other alternative methods (28, 29, 39) can also be used. Although we focused on case-control studies, our approach can be directly extended to family-based or related individuals by using programs that derive haplotype frequencies for family-based data (40) for estimating haplotype frequencies from family-based individuals.
Acknowledgments
We thank Professor Herman Chernoff and three anonymous referees for their insightful comments. This work was supported in part by National Institutes of Health Grants K02DA017713, R01DA016750, and U01HD050062.
Footnotes
The authors declare no conflict of interest.
References
- 1.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, et al. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 2.Edwards AO, Ritter R, III, Abel KJ, Manning A, Panhuysen C, Farrer LA. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 3.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dewan A, Liu M, Hartman S, Zhang SS, Liu DT, Zhao C, Tam PO, Chan WM, Lam DS, Snyder M, et al. Science. 2006;314:989–992. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, Cameron DJ, Chen H, Zhao Y, Pearson E, Li X, et al. Science. 2006;314:992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- 6.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arking DE, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, West K, Kashuk C, Akyol M, Perz S, et al. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 8.Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, et al. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlborg O, Haley CS. Nat Rev Genet. 2004;5:618–625. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- 10.Heinzen EL, Yoon W, Tate SK, Sen A, Wood NW, Sisodiya SM, Goldstein DB. Am J Hum Genet. 2007;80:876–883. doi: 10.1086/516650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kallberg H, Padyukov L, Plenge RM, Ronnelid J, Gregersen PK, van der Helm-van Mil AH, Toes RE, Huizinga TW, Klareskog L, Alfredsson L. Am J Hum Genet. 2007;80:867–875. doi: 10.1086/516736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frankel WN, Schork NJ. Nat Genet. 1996;14:371–373. doi: 10.1038/ng1296-371. [DOI] [PubMed] [Google Scholar]
- 13.Tiwari HK, Elston RC. Genet Epidemiol. 1997;14(6):1131–1136. doi: 10.1002/(SICI)1098-2272(1997)14:6<1131::AID-GEPI95>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 14.Tiwari HK, Elston RC. Theor Popul Biol. 1998;54:161–174. doi: 10.1006/tpbi.1997.1373. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, Jin L, Xiong M. Am J Hum Genet. 2006;79:831–845. doi: 10.1086/508571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker T, Schumacher J, Cichon S, Baur MP, Knapp M. Genet Epidemiol. 2005;29:313–322. doi: 10.1002/gepi.20096. [DOI] [PubMed] [Google Scholar]
- 17.Marchini J, Donnelly P, Cardon LR. Nat Genet. 2005;37:413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- 18.Musani SK, Shriner D, Liu N, Feng R, Coffey CS, Yi N, Tiwari HK, Allison DB. Hum Hered. 2007;63:67–84. doi: 10.1159/000099179. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Bonney G. Genet Epidemiol. 2000;19:323–332. doi: 10.1002/1098-2272(200012)19:4<323::AID-GEPI4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 20.Nelson MR, Kardia SL, Ferrell RE, Sing CF. Genome Res. 2001;11:458–470. doi: 10.1101/gr.172901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bastone L, Reilly M, Rader DJ, Foulkes AS. Hum Hered. 2004;58:82–92. doi: 10.1159/000083029. [DOI] [PubMed] [Google Scholar]
- 22.Cook NR, Zee RY, Ridker PM. Stat Med. 2004;23:1439–1453. doi: 10.1002/sim.1749. [DOI] [PubMed] [Google Scholar]
- 23.Foulkes AS, De Gruttola V, Hertogs K. J R Stat Soc C. 2004;53:311–323. [Google Scholar]
- 24.Bureau A, Dupuis J, Falls K, Lunetta KL, Hayward B, Keith TP, Van Eerdewegh P. Genet Epidemiol. 2005;28:171–182. doi: 10.1002/gepi.20041. [DOI] [PubMed] [Google Scholar]
- 25.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 26.Hawley ME, Kidd KK. J Hered. 1995;86:409–411. doi: 10.1093/oxfordjournals.jhered.a111613. [DOI] [PubMed] [Google Scholar]
- 27.Gusfield D. J Comput Biol. 2001;8:305–323. doi: 10.1089/10665270152530863. [DOI] [PubMed] [Google Scholar]
- 28.Qin ZS, Niu T, Liu JS. Am J Hum Genet. 2002;71:1242–1247. doi: 10.1086/344207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niu T, Qin ZS, Xu X, Liu JS. Am J Hum Genet. 2002;70:157–169. doi: 10.1086/338446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niu T. Genet Epidemiol. 2004;27:334–347. doi: 10.1002/gepi.20024. [DOI] [PubMed] [Google Scholar]
- 31.Clayton D. [Accessed November, 12 2007];SNPHAP, A Program for Estimating Frequencies of Large Haplotypes of SNPs. 2006 Available at http://www-gene.cimr.cam.ac.uk/clayton/software/snphap.txt.
- 32.Breiman L, Friedman F, Stone C, Olshen R. Classification and Regression Trees. New York: Chapman and Hall; 1984. [Google Scholar]
- 33.Zhang H, Singer B. Recursive Partitioning in the Health Sciences. New York: Springer; 1999. [Google Scholar]
- 34.Breiman L. Machine Learn. 2001;45:5–32. [Google Scholar]
- 35.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. Nat Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 36.Knapp M, Seuchter SA, Baur MP. Am J Hum Genet. 1994;55:1030–1041. [PMC free article] [PubMed] [Google Scholar]
- 37.Daiger SP. Science. 2005;308:362–364. doi: 10.1126/science.1111655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marx J. Science. 2006;314:405. doi: 10.1126/science.314.5798.405a. [DOI] [PubMed] [Google Scholar]
- 39.Lin S, Cutler DJ, Zwick ME, Chakravarti A. Am J Hum Genet. 2002;71:1129–1137. doi: 10.1086/344347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye Y, Zhong X, Zhang H. BMC Genet. 2005;6(Suppl 1):S135. doi: 10.1186/1471-2156-6-S1-S135. [DOI] [PMC free article] [PubMed] [Google Scholar]