Abstract
Lafora disease is a progressive myoclonus epilepsy with onset typically in the second decade of life and death within 10 years. Lafora bodies, deposits of abnormally branched, insoluble glycogen-like polymers, form in neurons, muscle, liver, and other tissues. Approximately half of the cases of Lafora disease result from mutations in the EPM2A gene, which encodes laforin, a member of the dual-specificity protein phosphatase family that additionally contains a glycogen binding domain. The molecular basis for the formation of Lafora bodies is completely unknown. Glycogen, a branched polymer of glucose, contains a small amount of covalently linked phosphate whose origin and function are obscure. We report here that recombinant laforin is able to release this phosphate in vitro, in a time-dependent reaction with an apparent Km for glycogen of 4.5 mg/ml. Mutations of laforin that disable the glycogen binding domain also eliminate its ability to dephosphorylate glycogen. We have also analyzed glycogen from a mouse model of Lafora disease, Epm2a−/− mice, which develop Lafora bodies in several tissues. Glycogen isolated from these mice had a 40% increase in the covalent phosphate content in liver and a 4-fold elevation in muscle. We propose that excessive phosphorylation of glycogen leads to aberrant branching and Lafora body formation. This study provides a molecular link between an observed biochemical property of laforin and the phenotype of a mouse model of Lafora disease. The results also have important implications for glycogen metabolism generally.
Keywords: polyglucosan, Lafora bodies, Epm2a, phosphate
Glycogen is a branched polymer of glucose that serves as an energy reserve in many cell types (1). The primary polymerization in glycogen is via α-1,4-glycosidic linkages, formed by the action of glycogen synthase, with branchpoints created by α-1,6-glycosidic linkages introduced by the branching enzyme (1). Mutations in these and other glycogen metabolizing enzymes result in a series of glycogen storage diseases characterized by aberrant glycogen deposits (2–4). Lafora disease (OMIM254780) is an autosomal recessive juvenile-onset myoclonus epilepsy that, although not normally viewed as a classic example of a glycogenosis, nonetheless is characterized by abnormal glycogen accumulation. The hallmark of the disease is the formation of Lafora bodies, which contain insoluble, poorly branched glycogen-like polysaccharide or “polyglucosan” (5–7). These bodies develop in liver, muscle, heart, skin, and neurons that normally accumulate little glycogen (8).
There have been major advances in understanding the genetics of Lafora disease, with the identification of two genes, EPM2A and EPM2B (NHLRC1), whose mutation accounts for ≈90% of the cases (5). However, analysis of the corresponding proteins, laforin and malin, has thus far revealed little about the molecular basis for the disease or the formation of Lafora bodies. Laforin belongs to the dual-specificity protein phosphatase family and additionally contains a highly conserved polysaccharide binding domain. Laforin binds to glycogen (9, 10), and virtually all of the laforin in a mouse skeletal muscle extract is recovered in the high-speed glycogen pellet (data not shown). Laforin is very sensitive to inhibition by polysaccharides, when assayed by using the generic phosphatase substrate p-nitrophenylphosphate (pNPP) (11). One disease mutation, W32G (6), is located in the conserved carbohydrate binding domain (12) and, when transferred to recombinant laforin, eliminates glycogen binding by recombinant laforin without completely abolishing pNPPase activity (10, 11). Thus, impaired glycogen binding by laforin may be sufficient to cause disease. Another link between laforin and glycogen comes from the observation that the laforin protein level correlates with the amount of glycogen in a series of mouse models in which the muscle glycogen content was genetically manipulated (13). Malin is an E3 ubiquitin ligase and has been reported to act on laforin (14). In a cell model, expression of malin results in the degradation of laforin. However, because Lafora disease is recessive, it is hard to envisage how loss-of-function mutations to either malin or laforin cause the same phenotype. If the function of malin is to promote laforin degradation, mutation of malin should increase laforin level and presumably function.
A prominent hypothesis in Lafora research has been that the polyglucosan might result from an imbalance between the activities of glycogen synthase and branching enzyme. There is precedent for this proposal. Glycogenosis type IV, Andersen disease, is caused by mutations in the branching enzyme gene and leads to the accumulation of polyglucosan (15). A somewhat similar situation exists in glycogenosis type VII, Tarui disease, in which mutation of the phosphofructokinase gene causes a build-up of glycolytic intermediates that are thought to drive excessive glycogen synthesis through elevation of the allosteric activator of glycogen synthase, glucose-6-P (16). In addition, we observed that a mouse in which hyperactive glycogen synthase was overexpressed in muscle both overaccumulated glycogen and developed structures reminiscent of Lafora bodies (17, 18). Therefore, several groups have looked for ways by which laforin could affect glycogen-synthesizing enzymes. Glycogen synthase is inactivated through phosphorylation by the protein kinase GSK-3, which itself contains an inhibitory phosphorylation site (19). Two groups (20, 21) have proposed that the inhibitory phosphate of GSK-3 can be removed by laforin, thus potentially leading to activation of glycogen synthase to cause the biosynthetic imbalance. However, Worby et al. (22) reported no evidence for laforin action on GSK-3, and we saw no significant change in glycogen synthase activation or GSK-3 phosphorylation in a mouse model of Lafora disease in which an inactive mutant of laforin, C266S, was overexpressed transgenically (23). We also found no evidence that laforin can dephosphorylate GSK-3 in vitro under conditions where the inhibitory phosphate can be removed by the λ-phosphatase [supporting information (SI) Fig. 5]. Notwithstanding the controversy over whether laforin acts on GSK-3, identification of physiological laforin substrates remains critical to understanding the molecular basis for Lafora disease. A potentially important advance came with the observation by Worby et al. (22) that laforin could release phosphate from amylopectin, a plant polysaccharide structurally related to glycogen, which is known to contain covalent phosphate attached as monoesters to the C3 and C6 positions of the glucose units. The level of phosphorylation is low, 0.25% by weight or 0.57% mol phosphate per mol glucose in our hands, but mutations in plants that affect amylopectin phosphorylation cause profound effects on starch metabolism (24). We report here that laforin is able to release phosphate also from mammalian glycogen and, furthermore, that disruption of the laforin gene in a mouse results in increased glycogen phosphorylation in vivo.
Results and Discussion
Dephosphorylation of Glycogen by Laforin.
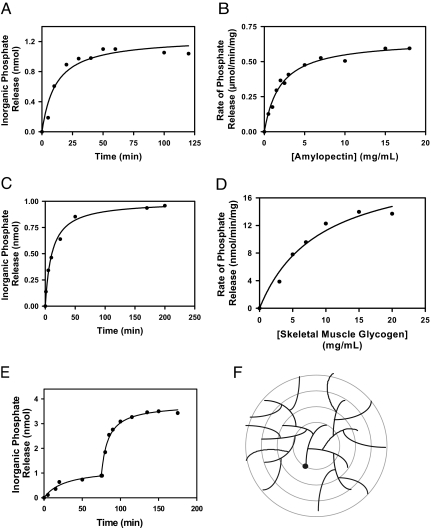
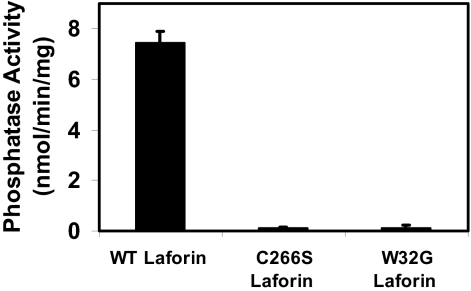
Mammalian glycogen has been reported to contain small amounts of covalent phosphate (25, 26). Worby et al. (22) attempted to measure dephosphorylation of commercially available rabbit liver glycogen by laforin but were unsuccessful, possibly because of the lack of assay sensitivity and/or the low phosphate content of the glycogen. Rabbit liver glycogen has been reported to contain significantly less phosphate than rabbit muscle glycogen (26). In the present study, we first confirmed the observation that amylopectin is a substrate for purified recombinant laforin (Fig. 1 A and B). Some 80–90% of the phosphate was released in a time-dependent manner. Varying the amylopectin concentration indicated a saturable process that followed a hyperbolic curve. We then analyzed glycogen that was isolated from rabbit skeletal muscle and showed that it contained 0.17% mol phosphate per mol glucose. Laforin released 20–25% of the total phosphate present, and the glycogen concentration dependence of the reaction was close to hyperbolic (Fig. 1 C and D). Unlike amylopectin, glycogen is believed to exist as a series of concentric shells of glucose residues (Fig. 1F) (1), and phosphate in the interior layers may not be accessible to laforin in the intact glycogen molecule. Therefore, after phosphate release had reached a maximum, we added the glycogen-degrading enzymes, α-amylase and amyloglucosidase, to disrupt the glycogen structure. Most of the remaining phosphate was now released and was by inference present in the inner tiers of the molecule (Fig. 1E). Exposure of glycogen to the glucosidases in the absence of laforin did not release inorganic phosphate, and likewise treatment of laforin with the glucosidases led to no phosphate release (data not shown). The laforin mutant C266S, in which a critical active-site residue is altered, did not hydrolyze phosphate from glycogen, confirming that phosphate release was not an artifact and was an enzyme-catalyzed reaction (Fig. 2). Some Lafora disease mutants are located in the glycogen binding domain, and, with one of these, W32G, previous studies have shown that the enzyme retains significant activity toward p-nitrophenolphosphate (10, 11). Recombinant W32G laforin was unable to dephosphorylate glycogen (Fig. 2), demonstrating that laforin action requires binding to glycogen by this noncatalytic domain. This finding is consistent with the idea that laforin is associated with glycogen during its metabolism, monitoring, and opposing the excessive introduction of phosphate.
Fig. 1.
Dephosphorylation of amylopectin and glycogen by laforin. The data shown are representative of at least five independent experiments. (A) Time-dependent release of phosphate from potato amylopectin (0.5 mg/ml) by purified, recombinant laforin (2.5 μg/ml). At the indicated times, the reaction was terminated by the addition of N-ethylmaleimide, and the inorganic phosphate was determined. (B) Concentration dependence of the rate of dephosphorylation of amylopectin by laforin. The reaction was for 15 min in the presence of 2.5 μg/ml laforin, at which time the reaction was close to linear. The apparent Km for amylopectin was 1.5 mg/ml. (C) Time-dependent release of phosphate from rabbit skeletal muscle glycogen (5 mg/ml). Laforin was present at 25 μg/ml. (D) Concentration dependence of glycogen dephosphorylation by laforin. Laforin was present at 25 μg/ml, and the reaction time was 40 min. The apparent Km for glycogen was 4.5 mg/ml. (E) Dephosphorylation of rabbit skeletal muscle glycogen in the presence of α-amylase and amyloglucosidase. Glycogen (5 mg/ml) dephosphorylation by laforin (25 μg/ml) was allowed to proceed until the reaction was essentially complete, at which time α-amylase (0.3 mg/ml) and amyloglucosidase (0.3 mg/ml) were added. Subsequent phosphate release was monitored. (F) Model for glycogen structure. Glycogen is believed to exist as a series of concentric shells of glucose residues, so that inner tiers would not be on the surface of the molecule. A full-size molecule would consist of 12 tiers.
Fig. 2.
Glycogen dephosphorylation requires the polysaccharide binding domain of laforin. WT laforin releases phosphate from glycogen (7.5 mg/ml). Mutation at the active site (C266S) essentially eliminated activity. Mutation in the carbohydrate binding domain (W32G), which is known to eliminate binding to polysaccharides, also abolished laforin's ability to dephosphorylate glycogen. The laforin proteins were present at 25 μg/ml, and the assay duration was 60 min.
Analysis of Epm2a−/− Mice.
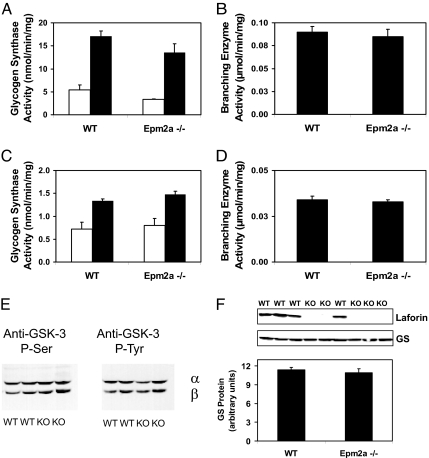
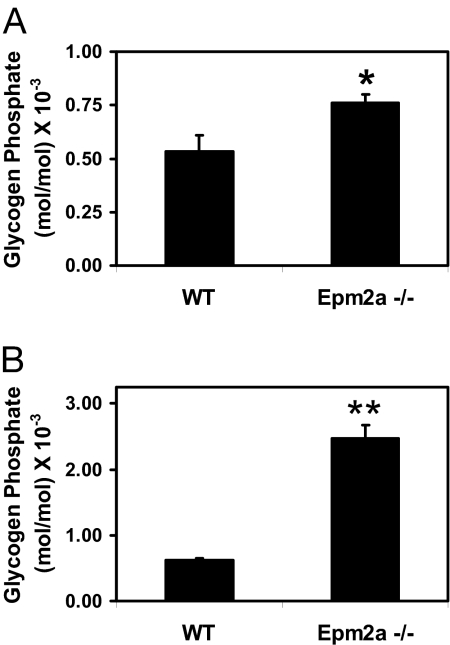
Mice with the Epm2a gene disrupted develop many of the characteristics of Lafora disease (27). The animals had Lafora bodies in liver, muscle, and brain, impaired behavioral responses, and ataxia, and they ultimately underwent spontaneous myoclonic seizures. We determined the activities of the glycogen synthetic enzymes, glycogen synthase, and branching enzyme in WT and Epm2a−/− mice. Glycogen synthase is typically measured in the absence and presence of the allosteric activator glucose-6-P, which overcomes the inactivation by phosphorylation of the Gys1 isoform of the enzyme, which is expressed in muscle and brain. The −/+ glucose-6-P ratio is a kinetic index of phosphorylation state, and the plus glucose-6-P activity should mirror enzyme level. Glycogen synthase activity was not altered in muscle or brain of Epm2a−/− mice whether measured in the absence or presence of glucose-6-P (Fig. 3 A and C). Consistent with this observation, analysis of muscle extracts by Western blotting for glycogen synthase also revealed no difference in protein levels (Fig. 3F). Also, GSK-3 phosphorylation state, an index of activity, was unchanged in Epm2a−/− mice (Fig. 3E), again consistent with the lack of effect on glycogen synthase activity. Disruption of the laforin gene had no effect on branching enzyme activity in muscle or brain (Fig. 3 B and D). All of these findings with Epm2a−/− mice were essentially consistent with our results with C266S overexpressing mice, which are another model of Lafora disease (23). Because of our finding that laforin could release covalent phosphate from glycogen in vitro, we sought to analyze the phosphate content of glycogen from Epm2a knockout animals. The glycogen isolated for our standard enzymatic determination of glucose equivalents is unsuitable for measurement of covalent phosphate because it contains too much contaminating inorganic phosphate. Therefore, we developed a protocol for the isolation of highly purified glycogen lacking this contamination (see Materials and Methods). From analysis of this glycogen, we determined an ≈4-fold increase in the phosphate content of muscle glycogen from Epm2a−/− mice compared with WT controls (Fig. 4B). In liver glycogen there was a smaller, 40%, increase that was statistically significant (Fig. 4A).
Fig. 3.
Glycogen synthase and branching enzyme activity in tissues from WT and Epm2a−/− mice. (A) Glycogen synthase activity in skeletal muscle of Epm2a−/− mice (n = 4) and WT controls (n = 4). Empty bars indicate activity in the absence of the activator glucose-6-P, and filled bars indicate activity in the presence of glucose-6-P. (B) Branching enzyme activity in skeletal muscle Epm2a−/− (n = 4) and WT (n = 4) mice. (C) Glycogen synthase activity in brain of Epm2a−/− (n = 5) and WT (n = 4) mice. (D) Branching enzyme activity in brain of Epm2a−/− (n = 5) and WT (n = 4) controls. No difference between genotypes was statistically significant. (E) Phosphorylation of GSK-3 in brain of WT and Epm2a−/− mice (KO) analyzed by using phospho-specific antibodies. (F) (Upper) Glycogen synthase in muscle of WT and Epm2a−/− mice (KO) analyzed with antiglycogen synthase antibodies. (Lower) Quantitation is shown. Also shown is the detection of laforin with an antilaforin antibody.
Fig. 4.
Phosphate content of glycogen from tissues of Epm2a−/− and WT mice. Glycogen was isolated from liver or muscle of individual mice, and covalent phosphate content, expressed as mol phosphate/mol glucose, was determined. (A) Liver glycogen phosphate content of Epm2a−/− (n = 4) and WT (n = 3) mice. The 40% increase in phosphate content in the Epm2a−/− mice is significant with P = 0.03 (∗). (B) Skeletal muscle glycogen phosphate content of Epm2a−/− (n = 8) and WT (n = 8) mice. Epm2a−/− mice have 4-fold greater phosphate content, significant at P = 0.0001 (∗∗).
Laforin as a Polysaccharide Phosphatase.
The present study extends the work of Worby et al. (22) to demonstrate that glycogen, as well as amylopectin, is a substrate for laforin. There is precedent for members of the dual-specificity phosphatase family hydrolyzing phosphate from nonprotein substrates because both PTEN and myotubularin dephosphorylate phospholipids (28, 29). Although glycogen and amylopectin share the chemistry of their polymerization and branching linkages, their structures are thought to be quite different (30). Amylopectin has a generally higher molecular weight and is significantly less branched (1 in ≈30 residues) than glycogen (1 in ≈12 residues). Whereas glycogen is believed to be formed of concentric tiers of glucose residues (Fig. 1F), amylopectin has a more extended structure that includes semicrystalline regions of unbranched glucose chains lined up in parallel (31). This difference likely accounts for the fact that most of the phosphate in amylopectin is readily released by laforin and is presumably superficial and available, whereas only a fraction of the phosphate is accessible on the surface of the glycogen molecule. Phosphate from the inner tiers cannot be accessed until the structure is disrupted by hydrolyzing enzymes.
It is interesting that the rate of phosphate release from both amylopectin and glycogen is saturable by substrate and follows approximately hyperbolic kinetics. The apparent Kms for amylopectin and glycogen were 1.5 and 4.5 mg/ml, respectively. For glycogen with Mr 107, the Km would be 0.45 μM. Expressed in terms of covalent phosphate concentrations, the values would be 47.3 and 42.5 μM for amylopectin and glycogen, respectively. However, if only 25% of the glycogen phosphate is assumed to be accessible, the Km would be reduced to ≈10 μM. Nonetheless, these Km values are quite close to each other and would be consistent with some commonality in the reaction mechanism for phosphate removal from both polysaccharides. However, interpreting the kinetics of polysaccharide dephosphorylation by laforin is complex. First, both glycogen and amylopectin are polydisperse. Second, the analysis is complicated by the fact that laforin contains a carbohydrate binding domain separate from the catalytic site. In fact, if the inhibition of laforin pNPPase activity by polysaccharide (11) reflects binding, then, even at the lowest substrate concentrations of Fig. 1, virtually all of the laforin would be bound. Thus, simple enzyme kinetic formulations may not be appropriate.
Glycogen Phosphorylation and Lafora Disease.
This study links a defect in a Lafora disease gene with a biochemical change in vivo compatible with an observed property of the corresponding protein. How malin deficiency would relate to Lafora body formation is unclear. The role of laforin as a physiological GSK-3 phosphatase is controversial and, in any event, Lafora bodies form in Epm2a−/− or the C266S (27, 32) overexpressing mice in tissues in which GSK-3 phosphorylation is not altered (23) (Fig. 3E). The elevated phosphate content in glycogen from Epm2a−/− mice is consistent with laforin acting as a glycogen phosphatase in vivo and additionally implicates glycogen phosphorylation as a determinant of its branching structure. One earlier report (33) had also described increased phosphate associated with Lafora bodies in neural tissue of patients with Lafora disease. We propose that excessive phosphorylation of glycogen can impair branching, by a yet-to-be-determined mechanism, and that laforin, which binds tightly to the polymer, is normally present during glycogen metabolism to limit this phosphorylation. Because phosphate is a relatively scarse constituent of glycogen, such a role would also explain why defective laforin does not immediately result in grossly abnormal glycogen structure and associated pathology. Only with aging would glycogen particles gradually overaccumulate phosphate past a threshold for the formation of aberrant structure and ultimately Lafora bodies. The results of this study provide insight into the mechanistic basis for Lafora disease and have important implications for glycogen metabolism generally.
Materials and Methods
Reagents.
Potato amylopectin, pNPP, and malachite green oxalate were from Sigma–Aldrich. Recombinant His-tagged mouse laforin, or mutants thereof, were prepared as described (11). Amyloglucosidase from Aspergillus niger was from Fluka. α-Amylase from Bacillus species was from Sigma. Anti-phospho-GSK-3α/β (Ser-21/Ser-9) antibody was from Cell Signaling Technology, and anti-phospho-GSK-3α/β (Tyr-279/Tyr-216) antibody was from Upstate Biotechnology. Antilaforin antibodies were from Abnova. Antiglycogen synthase antibodies raised in chicken was obtained from the late J. C. Lawrence, Jr. (University of Virginia, Charlottesville).
Mice and Tissue Harvesting.
The Epm2a−/− mice have been described (27). Animals, males 3–4 months old, were killed by cervical dislocation, and tissues were rapidly excised, immersed in liquid nitrogen, and stored at −80°C. Muscle or liver was pulverized in liquid nitrogen and processed for measurement of enzyme activities or glycogen as described (23).
Glycogen Purification.
Glycogen for use as a laforin substrate was isolated from rabbit skeletal muscle by a variant of previously described procedures (26, 34). Briefly, a muscle extract was treated with 10% trichloroacetic acid (TCA), the glycogen was precipitated from the supernatant with ethanol, and the solubilized pellet was shaken with chloroform/octanol. After another ethanol precipitation, the glycogen was incubated with SDS, harvested by ultracentrifugation, and subjected to further ethanol precipitation. The glycogen contained 0.17% mol/mol inorganic phosphate. Glycogen for covalent phosphate determination was prepared from WT or Epm2a−/− mouse skeletal muscle or liver as follows. The tissue was boiled in 30% KOH and then filtered to remove floating fat. The glycogen was precipitated with ethanol and redissolved in water. Ten volumes of 4:1 methanol/chloroform was added, and the solution was mixed and heated at 80°C for 5 min. The glycogen was recovered, ethanol-precipitated, and redissolved in 10% TCA. The resulting solution was centrifuged, and the glycogen was recovered from the supernatant by ethanol precipitation. After extensive dialysis, the glycogen was precipitated again with ethanol. By this procedure, lipids were removed by the methanol/chloroform extraction. Proteins and nucleic acids were absent as determined by the Bradford protein determination assay (35) and A260 absorbance, respectively.
Inorganic Phosphate and Glycogen Determination.
Measurement of inorganic phosphate, whether in laforin enzyme assays or hydrolysates of glycogen, used the highly sensitive malachite green method (22, 36). Glycogen was hydrolyzed in 10 μl of 5 M H2SO4 and 30 μl of 60% HClO4 according to the protocol of Hess and Derr (37) before phosphate measurement. The glycogen was quantitated by measuring glucose equivalents after digestion by α-amylase and amyloglucosidase (38).
Enzyme Assays.
Glycogen synthase and branching enzyme assays were as described (23). Laforin phosphatase activity toward pNPP used a colorimetric assay (22).
Western Blotting.
Analysis of GSK-3 phosphorylation by using phospho-specific antibodies was as described (23). Similar procedures were used for laforin and glycogen synthase.
Statistics.
Data from animal groups are displayed as mean ± SEM. Statistical significance was evaluated by using an unpaired Student t test.
Supplementary Material
Acknowledgments
We thank Ms. Dyann Segvich for expert technical assistance. This research was supported in part by National Institutes of Health Grant DK27221 (to P.J.R.), the Canadian Institutes for Health Research and Canada Research Chair in Pediatric Neurogenetics (B.A.M.), and the Indiana University Diabetes and Obesity Training Program (V.S.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707952104/DC1.
References
- 1.Roach PJ. Curr Mol Med. 2002;2:101–120. doi: 10.2174/1566524024605761. [DOI] [PubMed] [Google Scholar]
- 2.Cavanagh JB. Brain Res. 1999;29:265–295. doi: 10.1016/s0165-0173(99)00003-x. [DOI] [PubMed] [Google Scholar]
- 3.Wolfsdorf JI, Weinstein DA. Rev Endocrine Metabolic Disorders. 2003;4:95–102. doi: 10.1023/a:1021831621210. [DOI] [PubMed] [Google Scholar]
- 4.DiMauro S, Lamperti C. Muscle Nerve. 2001;24:984–999. doi: 10.1002/mus.1103. [DOI] [PubMed] [Google Scholar]
- 5.Chan EM, Andrade DM, Franceschetti S, Minassian B. Adv Neurol. 2005;95:47–57. [PubMed] [Google Scholar]
- 6.Ganesh S, Puri R, Singh S, Mittal S, Dubey D. J Hum Genet. 2006;51:1–8. doi: 10.1007/s10038-005-0321-1. [DOI] [PubMed] [Google Scholar]
- 7.Delgado-Escueta AV, Ganesh S, Yamakawa K. Am J Med Genet. 2001;106:129–138. doi: 10.1002/ajmg.1575. [DOI] [PubMed] [Google Scholar]
- 8.Gruetter R. J Neurosci Res. 2003;74:179–183. doi: 10.1002/jnr.10785. [DOI] [PubMed] [Google Scholar]
- 9.Ganesh S, Tsurutani N, Suzuki T, Hoshii Y, Ishihara T, Delgado-Escueta AV, Yamakawa K. Biochem Biophys Res Commun. 2004;313:1101–1109. doi: 10.1016/j.bbrc.2003.12.043. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Stuckey JA, Wishart MJ, Dixon JE. J Biol Chem. 2002;277:2377–2380. doi: 10.1074/jbc.C100686200. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Roach PJ. Biochem Biophys Res Commun. 2004;325:726–730. doi: 10.1016/j.bbrc.2004.10.083. [DOI] [PubMed] [Google Scholar]
- 12.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Biochem J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, Parker GE, Skurat AV, Raben N, DePaoli-Roach AA, Roach PJ. Biochem Biophys Res Commun. 2006;350:588–592. doi: 10.1016/j.bbrc.2006.09.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentry MS, Worby CA, Dixon JE. Proc Natl Acad Sci USA. 2005;102:8501–8506. doi: 10.1073/pnas.0503285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moses SW, Parvari R. Curr Mol Med. 2002;2:177–188. doi: 10.2174/1566524024605815. [DOI] [PubMed] [Google Scholar]
- 16.Nakajima H, Raben N, Hamaguchi T, Yamasaki T. Curr Mol Med. 2002;2:197–212. doi: 10.2174/1566524024605734. [DOI] [PubMed] [Google Scholar]
- 17.Raben N, Danon M, Lu N, Lee E, Shliselfeld L, Skurat AV, Roach PJ, Lawrence JC, Jr, Musumeci O., Shanske S, et al. Neurology. 2001;56:1739–1745. doi: 10.1212/wnl.56.12.1739. [DOI] [PubMed] [Google Scholar]
- 18.Pederson BA, Csitkovits AG, Simon R, Schroeder JM, Wang W, Skurat AV, Roach PJ. Biochem Biophys Res Commun. 2003;305:826–830. doi: 10.1016/s0006-291x(03)00862-3. [DOI] [PubMed] [Google Scholar]
- 19.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 20.Lohi H, Ianzano L, Zhao XC, Chan EM, Turnbull J, Scherer SW, Ackerley CA, Minassian BA. Hum Mol Genet. 2005;14:2727–2736. doi: 10.1093/hmg/ddi306. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Liu Y, Wu C, Zhang H, Zheng X, Zheng Z, Geiger TL, Nuovo GJ, Liu Y, Zheng P. Cancer Cell. 2006;10:179–190. doi: 10.1016/j.ccr.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Worby CA, Gentry MS, Dixon JE. J Biol Chem. 2006;281:30412–30418. doi: 10.1074/jbc.M606117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, Lohi H, Skurat AV, DePaoli-Roach AA, Minassian BA, Roach PJ. Arch Biochem Biophys. 2007;457:264–269. doi: 10.1016/j.abb.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blennow A, Nielsen TH, Baunsgaard L, Mikkelsen R, Engelsen SB. Trends Plant Sci. 2002;7:445–450. doi: 10.1016/s1360-1385(02)02332-4. [DOI] [PubMed] [Google Scholar]
- 25.Lomako J, Lomako WM, Whelan WJ, Marchase RB. FEBS Lett. 1993;329:263–267. doi: 10.1016/0014-5793(93)80234-l. [DOI] [PubMed] [Google Scholar]
- 26.Lomako J, Lomako WM, Kirkman BR, Whelan WJ. Biofactors. 1994;4:167–171. [PubMed] [Google Scholar]
- 27.Ganesh S, Delgado-Escueta AV, Sakamoto T, Avila MR, Machado-Salas J, Hoshii Y, Akagi T, Gomi H, Suzuki T, Amano K, et al. Hum Mol Genet. 2002;11:1251–1262. doi: 10.1093/hmg/11.11.1251. [DOI] [PubMed] [Google Scholar]
- 28.Maehama T, Dixon JE. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 29.Taylor GS, Maehama T, Dixon JE. Proc Natl Acad Sci USA. 2000;97:8910–8915. doi: 10.1073/pnas.160255697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ball S, Guan HP, James M, Myers A, Keeling P, Mouille G, Buleon A, Colonna P, Preiss J. Cell. 1996;86:349–352. doi: 10.1016/s0092-8674(00)80107-5. [DOI] [PubMed] [Google Scholar]
- 31.Zeeman SC, Smith SM, Smith AM. Biochem J. 2007;401:13–28. doi: 10.1042/BJ20061393. [DOI] [PubMed] [Google Scholar]
- 32.Chan EM, Ackerley CA, Lohi H, Ianzano L, Cortez MA, Shannon P, Scherer SW, Minassian BA. Hum Mol Genet. 2004;13:1117–1129. doi: 10.1093/hmg/ddh130. [DOI] [PubMed] [Google Scholar]
- 33.Sakai M, Austin J, Witmer F, Trueb L. Neurology. 1970;20:160–176. doi: 10.1212/wnl.20.2.160. [DOI] [PubMed] [Google Scholar]
- 34.Hubbard MJ, Cohen P. Eur J Biochem. 1989;180:457–465. doi: 10.1111/j.1432-1033.1989.tb14668.x. [DOI] [PubMed] [Google Scholar]
- 35.Bradford MM. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 36.Maehama T, Taylor GS, Slama JT, Dixon JE. Anal Biochem. 2000;279:248–250. doi: 10.1006/abio.2000.4497. [DOI] [PubMed] [Google Scholar]
- 37.Hess HH, Derr JE. Anal Biochem. 1975;63:607–613. doi: 10.1016/0003-2697(75)90388-7. [DOI] [PubMed] [Google Scholar]
- 38.Bergmeyer HU. Methods of Enzymatic Analysis. New York: Academic; 1974. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.