Abstract
Pathogens often suppress the melanization response of host insects, but the underlying molecular mechanisms are largely unknown. Here we report that Microplitis demolitor bracovirus (MdBV) carried by the wasp M. demolitor produces a protein, Egf1.0, which inhibits the phenoloxidase (PO) cascade. Egf1.0 belongs to a larger gene family that shares a cysteine-rich motif with similarities to the trypsin inhibitor-like (TIL) domains of small serine proteinase inhibitors (smapins). Gain-of-function and RNAi experiments indicated that the Egf genes are the only MdBV-encoded factors responsible for disabling the insect melanization response. Known smapins bind target proteinases in a substrate-like fashion and are cleaved at a single reactive site bond. The P1–P1′ position for Egf1.0 has the sequence Arg-Phe, which suggested that its target proteinase is a prophenoloxidase-activating proteinase (PAP). Wild-type Egf1.0 inhibited PAP-3 from Manduca sexta, whereas Egf1.0R51A, whose reactive-site arginine was replaced with an alanine, had no PAP-3 inhibitory activity. Other experiments using wild-type and mutant constructs indicated that Egf1.0 blocks activation of the PO cascade via PAP inhibition. Overall, our results identify a novel inhibitor of the PO cascade and indicate that suppression of the host melanization response is functionally important for both the virus and its associated wasp.
Keywords: melanization, virulence, immunity, parasite, antiviral
The insect immune system consists of cellular and humoral components that innately defend against a diversity of invaders (1–5). A conserved feature of the humoral immune response is the phenoloxidase (PO) cascade that regulates the formation of melanin (6, 7). The PO cascade consists of multiple serine proteinases that terminate with the zymogen pro-PO. The number of proteolytic steps in the cascade is unknown for any insect, but the serine proteinases that activate pro-PO are called prophenoloxidase-activating proteinases (PAPs; also PPAFs and PPAEs), which have been characterized from several species (8–14). Immune challenge induces upstream proteinases in the cascade to activate PAPs by cleaving them between their clip and proteinase domains. Activated PAPs then process pro-PO to PO that catalyzes the hydroxylation of monophenols to o-diphenols and oxidation of o-diphenols to quinones. Polymerized quinones form melanin that accumulates around pathogens, encapsulated parasites, and wound sites, whereas free quinones and other reactive intermediates may contribute to the killing of foreign intruders trapped by melanin (7). The main negative regulators are serine proteinase inhibitors of the serpin superfamily that maintain the PO cascade in an inactive state in the absence of immune challenge and down-regulate the cascade after infection (6–9).
Parasites and pathogens have evolved diverse strategies to evade host immune defenses (15–19). Among the most virulent immunosuppressive pathogens of insects are symbiotic polydnaviruses (PDVs) associated with thousands of parasitoid wasps (18, 20). Each PDV is genetically unique and persists as an integrated provirus in the genome of its associated wasp species. Transmission of the virus to parasitoid offspring is vertical through the germ line whereas viral replication is restricted to the reproductive tract of females. Wasps inject virions into hosts at oviposition that infect hemocytes and other tissues. PDVs do not replicate in the host, but viral gene expression suppresses several aspects of the host's immune system, including melanization. This immunosuppressive activity is essential for the survival of the wasp's offspring, but the virulence factors responsible for disabling melanization and other host defenses are largely unknown.
The braconid wasp Microplitis demolitor carries M. demolitor bracovirus (MdBV) and parasitizes the larval stage of several moth species (Lepidoptera). The 189-kb MdBV genome is divided among 15 DNA segments and encodes 61 predicted genes for products ≥100 aa (20). The majority of these genes are related variants that form four families designated as the mucin-like cell surface genes (Glc family), protein tyrosine phosphatase (PTP) genes, IκB-like genes (Vankyrins), and epidermal growth factor-like motif genes (Egf family). Here we report that the Egf family encodes proteins related to small serine proteinase inhibitors (smapins) (21) and that Egf1.0 blocks melanization by inhibiting PAPs.
Results
The Egf Gene Family Encodes Smapin-Like Proteins.
Prior studies indicated that members of the Glc, PTP, and Vankyrin families disrupt phagocytosis, encapsulation, and NF-κB signaling but are not responsible for disabling the host's melanization response (22–25). Sequence analysis, however, led us to consider the three-member Egf family located on MdBV genomic segment O (20) as melanization inhibitors (Fig. 1A). Egf1.5 and Egf1.0 encode predicted proteins of 38.8 and 26.0 kDa that possess signal peptides (SP), an identical eight-cysteine-rich domain (CD), and C-terminal repeat domains (RD) comprising six or three near-identical 35-aa repeats arranged in tandem array. Egf0.4 encodes a predicted 11.7-kDa protein with an SP, a similar but not identical CD to Egf1.5 and Egf1.0, and no RD. Database searches revealed similarities in the cysteine spacing pattern of the Egf CDs and smapins that MEROPS (http://merops.sanger.ac.uk) classifies into the I8 family on the basis of their 10 cysteine trypsin inhibitor-like (TIL) domains (26) (Fig. 1B). We also noted similarities between the Egf proteins and the I40 family, which contains a single member, the fungal serine proteinase inhibitor F (FPI-F) from the silkmoth Bombyx mori, distinguished by an eight-cysteine CD. The cysteine spacing pattern of the Egf1.5/1.0 and Egf0.4 CDs was identical to that of FPI-F, but overall sequence identity was only 27.5% and 23.5%, respectively (Fig. 1B). In contrast, the CDs of Egf1.0/1.5 and Egf0.4 shared 43.4% and 47.2% identity with the TIL domain of the I8 family member and anticoagulant AcAPc2 from the hookworm Ancylostoma caninum.
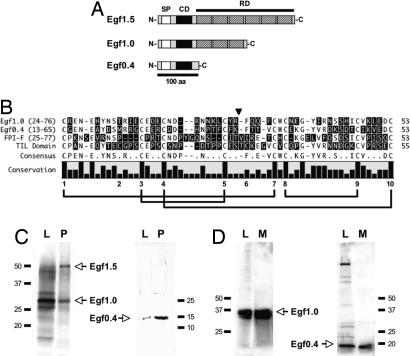
Fig. 1.
Egf family members share similar domain structures and are secreted. (A) Domain structure of Egf1.5, Egf1.0, and Egf0.4. Each contains a SP and CD, but Egf0.4 lacks a RD. (Scale bar: 100 aa.) (B) Alignment of the CDs from Egf1.0 and Egf0.4 with the CD from FPI-F (GenBank accession no. Q10731) and a prototypical TIL domain (Pfam accession no. PF01826). The consensus sequence and level of residue conservation are indicated below the alignment along with the predicted disulfide bonds for FPI-F (34) and the Egf proteins. (C) Detection of Egf1.5, Egf1.0, and Egf0.4 in hemocyte lysates (L) and cell-free plasma (P) from MdBV-infected P. includens. The immunoblot in Left was probed with anti-Egf1.0 that recognizes Egf1.5 and Egf1.0. The blot in Right was probed with anti-Egf0.4 that recognizes Egf0.4. (D) Recombinant Egf1.0 (Left) and Egf0.4 (Right) detected in High Five cell lysates (L) and conditioned medium (M) 48 h after transfection with pIZT/Egf1.0 or pIZT/Egf0.4.
MdBV preferentially infects the fat body and hemocytes of host insects as well as host-derived cell lines like High Five cells from Trichoplusia ni. Previous analyses indicated that expression of Egf family members begins within 2 h of infection and that transcript abundance for each gene remains at nearly steady-state levels over the 7 days required for M. demolitor development (22, 27–30). Egf1.5 and Egf1.0 are also the most abundant viral transcripts in infected hosts (27, 30). Antisera generated against purified Egf1.0 and Egf0.4 from Escherichia coli detected Egf1.5, Egf1.0, and Egf0.4 in hemocytes and plasma from virus-infected hosts, indicating that each protein is secreted (Fig. 1C). Anti-Egf1.0 cross-reacted with Egf1.0 and Egf1.5, as expected from their similar structures, whereas anti-Egf0.4 recognized Egf0.4 specifically. Egf1.0 and Egf0.4 were also detected in culture medium from High Five cells transfected with pIZT/Egf1.0 or Egf0.4 (Fig. 1D). No Egf proteins were detected in noninfected hosts or host cells (data not shown).
Egf1.0 Inhibits Hemolymph Melanization.
Although conserved among arthropods (6, 7), the serine proteinases and other proteins in the PO cascade are best characterized in the lepidopteran Manduca sexta (30, 31). We therefore asked whether MdBV produces factors that inhibit melanization in M. sexta using a well established assay that measures melanin formation in a pro-PO-containing fraction of plasma after induction by the bacterium Micrococcus luteus (9, 10). We first tested conditioned medium from MdBV-infected High Five cells reasoning that a viral inhibitor would most likely be secreted. Medium from virus-infected cells and cells transfected with pIZT/Egf1.0 significantly reduced PO activity, whereas medium from cells transfected with pIZT/Egf0.4 did not (Fig. 2A). Purified recombinant Egf1.0 from E. coli likewise dose-dependently reduced PO activity (Fig. 2B). These results implicated Egf1.0 as a candidate inhibitor of melanization, yet MdBV expresses many other genes that could also have anti-melanization activity. To address this possibility, we silenced egf1.0/1.5 expression in MdBV-infected High Five cells by RNAi (22). Conditioned medium from cells treated with Egf1.0/1.5 dsRNA completely lost its anti-melanization activity, whereas medium from cells treated with dsRNA to another abundantly expressed MdBV gene (glc1.8) retained its anti-melanization activity (Fig. 2C). We also adapted the M. sexta activation assay to plasma from the hosts Pseudoplusia includens and T. ni, the non-host lepidopteran B. mori, and the mosquito Aedes aegypti. Egf1.0 inhibited melanization of plasma in each (data not shown), suggesting that it interacts with a conserved feature of the insect PO cascade.
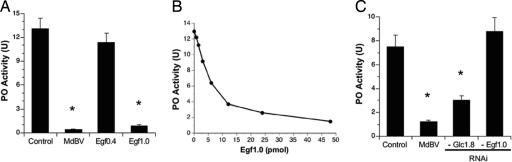
Fig. 2.
Egf1.0 inhibits melanization. (A) Inhibition experiments using conditioned medium from noninfected High Five cells (Control), MdBV-infected cells, and cells transfected with pIZT/Egf0.4 or pIZT/Egf1.0. Medium was collected 48 h after infection or transfection and concentrated, and 5 μl containing ∼50 pmol of Egf0.4 or Egf1.0 was added to the pro-PO activation fraction of M. sexta plasma. Pro-PO was then activated 10 min later by the addition of M. luteus (mean ± SD, n = 3) for 1 h. Asterisks indicate significant differences from the control (F3,8 = 244.1 and P < 0.001; followed by Dunnett's multiple-comparison procedure). (B) Dose-dependent reduction in pro-PO activation by purified recombinant Egf1.0 (1–50 pmol). (C) RNAi knockdown of egf1.0/1.5 eliminates the melanization-blocking activity of conditioned medium from MdBV-infected High Five cells. Conditioned medium from noninfected cells (Control), MdBV-infected cells, MdBV-infected cells treated with glc1.8 dsRNA (Glc1.8), and MdBV-infected cells treated with egf1.0/1.5 dsRNA (Egf1.0) was added to the pro-PO activation fraction as described in A. Asterisks indicate significant differences from the control (F3,8 = 93.6 and P < 0.001).
A Target Proteinase of Egf1.0 Is PAP-3.
Known smapins inhibit target proteinases by the Laskowski (classical) mechanism, which involves binding in a substrate-like fashion to produce an equilibrium between the uncleaved inhibitor and inhibitor cleaved at a single peptide bond in the reactive site loop (32). Comparison of the reactive site loops of FPI-F and two characterized I8 members, the cathepsin G/chymotrypsin inhibitor from the honey bee Apis mellifera (AMCI) and AcAPc2 (33–35), indicated their predicted scissile bonds reside in the same relative position (Fig. 3A). By extension, the predicted P1–P1′ position for Egf1.0 had the sequence Arg-Phe, which matched the predicted cleavage site for most insect pro-POs (Fig. 3A). This suggested that Egf1.0 blocks melanization by inhibiting a PAP. Yet Egf0.4, which did not block PO activity, had the very similar P1–P1′ sequence Lys-Phe that matches the scissile bond of endogenous PAP inhibitors like Serpin-3 from M. sexta and Spn27A from Drosophila (36–38). M. sexta produces three PAPs that localize to integument (PAP-1) and hemolymph (PAP-2 and PAP-3) (9, 10). Each hydrolyzes the synthetic substrate IEAR-p-nitroanilide and cleaves pro-PO at the correct bond, but each also requires serine proteinase homolog (SPH) cofactors to produce active PO (10, 33). To ask whether Egf1.0 and Egf0.4 are PAP inhibitors, we added recombinant Egf1.0 and Egf0.4 to purified PAP-3 plus IEAR-p-nitroanilide (9, 10). Egf1.0 strongly inhibited PAP-3 activity, but Egf0.4 did not (Fig. 3B). Reducing Egf1.0 concentration increased the amidase activity of PAP-3 (Fig. 3C), and immunoblotting experiments using antibodies to N- and C-terminal-specific epitope tags indicated that PAP-3 cleaved Egf1.0 into 20.2- and 20.9-kDa fragments corresponding to the predicted P1–P1′ site (Fig. 3D). We confirmed that the P1 arginine in Egf1.0 was required for PAP-3 inhibition by replacing the residue with an alanine to produce Egf1.0R51A. This mutant had no inhibitory activity toward PAP-3 and was not cleaved into 20.2- and 20.9-kDa fragments (Fig. 3 B and D). In contrast, replacement of the P1 arginine with a lysine, as found in Egf0.4, produced a mutant (Egf1.0R51K) that inhibited and was cleaved by PAP-3 like wild-type Egf1.0 (Fig. 3 B and D).
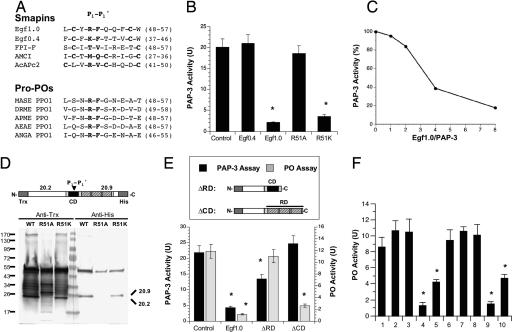
Fig. 3.
Egf1.0 inhibits PAP-3 and pro-PO activation. (A) Alignment of the reactive site loops and predicted P1–P1′ residues (bold) of Egf1.0 and Egf0.4 with FPI-F, AMCI (GenBank accession no. P56682), and AcAPc2 (GenBank accession no. AAC47080). Below shows the predicted cleavage site (bold) for pro-POs from M. sexta (MASE PP01; GenBank accession no. AAC05796), Drosophila melanogaster (DRME PP01A; GenBank accession no. BAB17671), A. mellifera (AEAE PPO; GenBank accession no. AAO72539), Ae. aegypti (AEAE PP01; GenBank accession no. AAG02219), and Anopheles gambiae (ANGA PP01; GenBank accession no. AAD01936). Positions based on the mature protein are presented in parentheses on the right. (B) PAP-3 inhibition experiments. Recombinant Egf0.4, Egf1.0, Egf1.0R51A, or Egf1.0R51K (20 pmol) was added to purified PAP-3 (2 pmol) plus substrate for 1 h. PAP-3 plus substrate alone served as the control. Asterisks indicate significant differences in amidase activity from the control (F4,14 = 110.5 and P < 0.001). (C) Concentration-dependent inhibition of PAP-3 by Egf1.0. Residual activities (percent) were plotted against the corresponding molar ratios of Egf1.0 and PAP-3. (D) Egf1.0 cleavage by PAP-3. One-tenth aliquots of the reaction mixtures from B were subjected to immunoblotting using anti-Trx (Left) and anti-His (Right) antibodies. A schematic of recombinant Egf1.0 is presented above the blot showing the predicted masses from cleavage at the predicted P1–P1′ position. Molecular mass standards are indicated to the left of the blot, and the 20.2-kDa and 20.9-kDa fragments recognized by the antibodies are noted to the right. (E) Inhibition of PAP-3 and PO activity by Egf1.0, Egf1.0ΔRD, or Egf1.0ΔCD. Egf proteins (50 pmol) were incubated with PAP-3 (2 pmol) or the pro-PO activation fraction of M. sexta plasma. Controls were PAP-3 plus substrate or pro-PO fraction plus elicitor (M. luteus) and substrate only. Schematics illustrating the structures of Egf1.0ΔRD and Egf1.0ΔCD are presented above the figure. Asterisks indicate significant differences from the controls (PAP-3 assays, F3,11 = 72.0 and P < 0.001; pro-PO assays, F3,11 = 44.9 and P < 0.001). (F) Inhibition of PO activity when Egf1.0 or Egf1.0ΔCD was added to the pro-PO activation fraction before or after the elicitor (M. luteus or PAP-3) and substrate. Treatment 1, Pro-PO fraction plus M. luteus and substrate (control); treatment 2, pro-PO plus M. luteus and substrate followed by Egf1.0 (50 pmol) 15 min later; treatment 3, pro-PO plus M. luteus and substrate followed by Egf1.0ΔCD (50 pmol) 15 min later; treatment 4, pro-PO plus Egf1.0 (50 pmol) followed by M. luteus and substrate 15 min later; treatment 5, pro-PO plus Egf1.0ΔCD (50 pmol) followed by M. luteus and substrate 15 min later. Asterisks indicate that treatments 4 and 5 differed significantly from the control (F4,14 = 33.2 and P < 0.001). Treatment 6, Pro-PO fraction plus PAP-3 (0.2 pmol) and substrate (control); treatment 7, pro-PO plus PAP-3 (0.2 pmol) and substrate followed by Egf1.0 (50 pmol) 15 min later; treatment 8, pro-PO plus PAP-3 (0.2 pmol) and substrate followed by Egf1.0ΔCD (50 pmol) 15 min later; treatment 9, pro-PO plus Egf1.0 (50 pmol) followed by PAP-3 (0.2 pmol) and substrate 15 min later; treatment 10, pro-PO plus Egf1.0ΔCD (50 pmol) followed by PAP-3 (0.2 pmol) and substrate 15 min later. Asterisks indicate that treatments 9 and 10 differed significantly from the control (F4,14 = 32.0 and P < 0.001).
FPI-F and most smapins are <100 aa with little or no flanking sequence outside of their cysteine-rich or TIL domains (21). Egf0.4 has a similar structure, but Egf1.0 and Egf1.5 differ because of their C-terminal RDs (see Fig. 1A). To determine whether the RD is functionally important for Egf1.0 activity, we produced the mutants Egf1.0ΔRD, which consisted of only the CD domain, and Egf1.0ΔCD, which consisted of only the RD domain (Fig. 3E). Egf1.0ΔRD significantly reduced the amidolytic activity of PAP-3 compared with the control, but its inhibitory activity was also weaker than wild-type Egf1.0. However, Egf1.0ΔRD did not block melanization of our plasma fraction after stimulation by M. luteus (Fig. 3E). Egf1.0ΔCD in contrast had no effect on the amidolytic activity of PAP-3 but significantly inhibited plasma melanization (Fig. 3E).
Egf1.0 Is Unable to Inhibit Already Activated PO.
The preceding results indicated that Egf1.0 is a PAP inhibitor, which in turn suggested that it suppresses the PO cascade before activation of PO itself. The data with Egf1.0ΔCD, however, raised the possibility that Egf1.0 could have other activities, including inhibition of already activated PO. If the former, we reasoned that melanin formation should be inhibited only if Egf1.0 is added to the pro-PO-containing fraction of plasma before activation by an elicitor. If Egf1.0 also inhibits activated PO, however, it should also block melanin formation when added to plasma after activation by an elicitor. Using M. luteus, we found that Egf1.0 and Egf1.0ΔCD inhibited melanization only if added to the plasma fraction before activation (Fig. 3F). We then determined that purified PAP-3 also strongly elicits melanin formation (Fig. 3F). This activity was significantly inhibited though if PAP-3 was preincubated with Egf1.0 or Egf1.0ΔCD but was not inhibited if Egf1.0 or Egf1.0ΔCD was added to plasma after activation by PAP-3 (Fig. 3F). We also tested Egf1.0 against several other serine proteinases (bovine pancreatic trypsin, human plasmin, bovine pancreatic α-chymotrypsin, and porcine pancreatic elastase) in assays conducted over a range of concentrations. No inhibitory activity was detected toward any of these targets (data not shown), further supporting the conclusion that Egf1.0 is a selective PAP inhibitor.
Egf1.0 Enhances the Survival of MdBV and M. demolitor.
MdBV enters host hemocytes within minutes of infection, but free virions remain in circulation for up to 24 h (ref. 29 and M.R.S., unpublished results). Reciprocally, M. demolitor eggs are encapsulated and melanized 18–24 h after infection if injected into hosts without MdBV (18, 19). These patterns indicate that both virus and parasitoid risk exposure to melanin or other products of the PO cascade in the absence of Egf1.0. We assessed whether suppression of the PO cascade is functionally significant by determining first whether MdBV and M. demolitor eggs elicit activation of the PO cascade. These assays revealed that both induced levels of melanin formation similar to M. luteus and that Egf1.0 blocked this response (Fig. 4A). We then assessed whether activation of the PO cascade reduced MdBV and M. demolitor survival. MdBV viability decreased more than three log units after a 6-h exposure to activated PO, but no loss of viability occurred when Egf1.0 was present (Fig. 4B). The viability of M. demolitor eggs declined 88% after only a 2-h exposure to activated PO, whereas almost no loss of viability occurred in the presence of Egf1.0 (Fig. 4C).
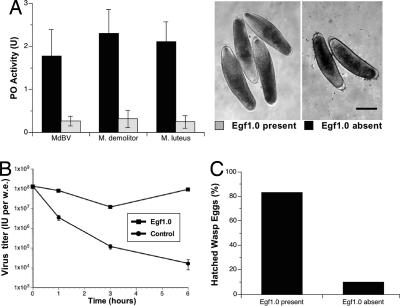
Fig. 4.
PO activity reduces the viability of MdBV and M. demolitor. (A) Activation of the pro-PO fraction of M. sexta plasma by MdBV and M. demolitor eggs. Virus and eggs similarly activated pro-PO (F2,8 = 0.4 and P = 0.71) whereas Egf1.0 inhibited activity (F2,8 = 0.1 and P = 0.94). Note that PO activity is lower for all treatments than in Figs. 2 and 3 because of buffer conditions. Micrographs show the deposition of melanin on the surface of M. demolitor eggs at 1 h in the absence of Egf1.0 and the lack of melanin on eggs when Egf1.0 is present. (B) MdBV viability after exposure to activated PO. Pro-PO was activated by MdBV as in A followed by determination of the viral titer [expressed in infectious units per wasp equivalent (w.e.)] 0–6 h after activation. MdBV viability significantly declined in the absence of Egf1.0 (F7,23 = 213.4 and P < 0.001). (C) M. demolitor viability after exposure to activated PO. Pro-PO was activated by M. demolitor eggs as in A. The percentage of eggs that hatched was significantly lower when Egf1.0 was absent (t = 18.2; df = 4; P < 0.001).
Discussion
Given the role of serpins in regulating the PO cascade, we originally speculated that PDVs inhibit the host melanization response by encoding serpins themselves. Yet sequencing of MdBV and other PDVs has thus far not identified any serpin homologues (18–20), suggesting that these immunosuppressive viruses have evolved other strategies for disabling melanization. In the case of MdBV, our biochemical data indicate that Egf1.0 suppresses activation of the PO cascade via PAP inhibition rather than by inhibiting activated PO itself. Our RNAi experiments support these results by indicating that the Egf genes are likely the only MdBV virulence factors responsible for this activity. The lack of inhibitory activity toward other serine proteinases combined with the ability of Egf1.0 to block melanization in different insects also suggests that this protein may have broad utility as a selective inhibitor of the PO cascade.
Our results with Egf1.0 indicate that both its CD and RD are important for activity and in turn suggest that Egf1.5 is also likely a PAP inhibitor. But our results do not fully reveal how the RD of Egf1.0 disables activation of pro-PO or whether the longer RD of Egf1.5 potentially confers different functional properties. The reduced inhibitory activity of Egf1.0ΔRD toward PAP-3 compared with wild-type Egf1.0 suggests that one function of the RD is in PAP recognition and positioning the reactive site loop in the CD so that it properly interacts with the catalytic domain of the enzyme. However, given that Egf1.0ΔCD does not inhibit the amidolytic activity of purified PAP-3 yet significantly inhibits melanization elicited by M. luteus or PAP-3, we suggest that RD binding must also disrupt the ability of PAP-3 to interact with the pro-PO complex and/or activate PO (6, 30, 31). The lack of PAP inhibition by Egf0.4 in contrast is likely due to the absence of an RD rather than a differing basic residue (Lys) in its P1 position because Egf1.0R51K activity was indistinguishable from wild-type Egf1.0. The target proteinase of Egf0.4 is unknown, but our results strongly indicate that it is not another PAP or proteinase in the PO cascade because Egf0.4 had no inhibitory activity in our pro-PO activation assays. In light of the broad immunosuppressive activity of MdBV (18, 19, 22–25), other serine proteinase networks of interest as targets for Egf0.4 include the Toll pathway and the pathway that regulates activation of plasmatocyte spreading peptide in Lepidoptera (39, 40).
Both melanin and other factors produced during melanin synthesis have been suggested to play a role in killing invading pathogens and parasitoids (6, 7). However, it is often unclear whether activation of the PO cascade is directly responsible for killing foreign invaders versus foreign invaders becoming melanized after dying from other causes (41, 42). Several lines of evidence also suggest that multiple melanization pathways exist in insects with some pathogens activating pro-PO by one pathway and other pathogens activating pro-PO by another (31). It was thus important in this study to determine whether MdBV and/or M. demolitor are capable of inducing pro-PO activation and, if so, whether either is adversely affected. Our results indicate that both induce activation of pro-PO although they do not reveal whether activation is through the same or different pathways. More importantly, though, our results argue that activated PO is directly responsible for reducing virus and parasitoid viability because Egf1.0, as a specific PO cascade inhibitor, protected both.
Comparative data indicate that PDVs from other Microplitis sp. encode Egf homologues (43), suggesting that other Egf variants could exist given the large number of species in this genus. In contrast, Egf genes have not been identified in PDVs from other wasp genera, indicating that they or their associated parasitoids have evolved other means for disrupting the host melanization response. One example is the identification of an SPH in the venom of a PDV-carrying wasp in the genus Cotesia that disrupts melanization by an unknown mechanism (44). Other anti-melanization strategies are also possible, as recently illustrated by an insect pathogenic bacterium (45).
Materials and Methods
Insects, Cell Cultures, and Virus.
M. demolitor, P. includens, and T. ni were reared as described (27, 28). M. sexta was reared on a diet from Carolina Supply, and Ae. aegypti was reared as outlined by Castillo et al. (46). BTI-Tn-5B1-4 (High Five) cells were maintained and MdBV was collected as previously described (22, 29). Newly laid M. demolitor eggs were collected by dissecting surface-sterilized P. includens in TC-100 medium.
Sequence Analysis and Cloning.
Sequence comparisons were made by using the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST) and MEROPS (http://merops.sanger.ac.uk) databases and conserved domain analysis (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Predicted SPs and cleavage sites were generated through the Center for Biological Sequence Analysis Servers (www.cbs.dtu.dk/services). Expression constructs for egf1.0 and egf0.4 were generated from cloned cDNAs (27, 28). For expression in High Five cells, coding sequences were PCR-amplified and cloned into pIZT/V5-His (Invitrogen) (22) using the following primer sets: 5′-AATCTAGATATGGCGAACAACATTTTC-3′ (forward) and 5′-ATACCGCGGATAATCAAGAGTT-3′ (reverse) for egf1.0; 5′-GAGGTACCAACATGGTGTCAGAAAAGTTC-3′ (forward) and 5′-GTACTAGTATACATGCCGATGATTGTTTCAGAG-3′ (reverse) for egf0.4. Egf1.0-specific primers contained XbaI and SacII sites, whereas egf0.4 primers contained Acc65I and SpeI sites (underlined). For bacterial expression, cDNAs were PCR-amplified using primers lacking SP sequences and directly cloned in frame with the C-terminal His tag into pET-32 Ek/LIC (Novagen). Egf1.0 was amplified by using the primers (82) 5′-GACGACGACAAGATGCCTACTAAAGGAAGTGAAG-3′ (103) and (699) 5′-GAGGAGAAGCCCGGTTTATCAAGAGTTTC-3′ (685). Egf0.4-specific primers were (77) 5′-GACGACGACAAGATGATCGAAACTTCGCCCATCAAC-3′ (97) and (328) 5′-GAGGAGAAGCCCGGTGCATACATGCCGATGATTGT-3′ (314). Egf1.0ΔRD was generated by using the same forward primer for Egf1.0 and the reverse primer (360) 5′-GAGGAGAAGCCCGGTGCATTGTTGGCACTGGATG-3′ (344), whereas Egf1.0ΔCD was generated by using (403) 5′-GACGACGACAAGATGGCACATGATTCGGTTTCACATAC-3′ (422) and (760) 5′-GAGGAGAAGCCCGGTCGTCGATTTAAACTTCTGCTG-3′ (740). Primer sequence extensions for LIC cloning are underlined, and primer positions corresponding to submitted cDNA sequences are indicated in parentheses. To remove the N-terminal His tag from constructs, plasmid DNA was digested with MscI and NspV, blunt-ended with T4 DNA Polymerase (Invitrogen), and religated. Egf1.0R51A and Egf1.0R51K were generated from the modified bacterial expression constructs by overlap extension PCR using the gene-specific primers 5′-GTAATAACAAACTATGCTACGCGTTCCAGCAATTCTG-3′ (forward) and 5′GCTGGAACGCGTAGCATAGTTTGTTATTAC-3′ (reverse) (Egf1.0R51A), 5′-GTAATAACAAACTATGCTACAAGTTCCAGCAATTCTG-3′ (forward) and 5′-GCTGGAACTTGTAGCATAGTTTGTTATTAC-3′ (reverse) (Egf1.0R51K) (mutated Arg underlined), and vector-specific primers 5′-AGCGGATAACAATTCCCCTCTAGAAATAAT-3′ (forward) and 5′-GCTTTGTTAGCAGCCGGATCTCAGTGGT-3′ (reverse). Full-length sequences were then recovered by PCR using vector-specific primers and overlap PCR products as templates. After subcloning and digestion with XbaI and HindIII, sequences were cloned into pET-32 Ek/LIC. Construct integrity was verified by sequencing.
Protein Expression and Immunoblotting.
Recombinant proteins were transiently expressed in High Five cells (22). Bacterial recombinant proteins were expressed in Rosetta-Gami (DE3) E. coli cells (Novagen). After induction and growth at 20°C for 4 h, cells were lysed under native conditions followed by purification using Ni-NTA-Superflow beads (Qiagen). Proteins were quantified by using the Micro BCA Protein Assay Kit (Pierce). Polyclonal antisera to Egf1.0 and Egf0.4 were produced in rabbits immunized with bacterial recombinant proteins. For immunoblots, proteins were resolved by SDS/PAGE, transferred to PVDF (Immobilon-P; Millipore), and probed with specific antisera or antibodies against thioredoxin (Sigma) and polyhistidine tags [His-probe (H-15); Santa Cruz Biotechnology]. Primary antibodies were detected with horseradish-conjugated secondary antibodies (Jackson ImmunoResearch) and visualized by using 3,3′-diaminobenzidine (Sigma).
PO Activity and RNAi Assays.
Hemolymph was collected from fifth-instar P. includens, T. ni, and M. sexta (9, 40) whereas hemolymph from Ae. aegypti was from adults (46). Hemocytes were pelleted by centrifugation, and pro-PO active fractions were prepared from plasma by adding an equal volume of saturated ammonium sulfate (pH 7.0) (9, 10). After centrifugation for 10 min at 10,000 × g, pellets were dissolved in water at one-third the initial hemolymph volume and stored at −80°C. PO activity assays (200 μl) were conducted in 96-well plates containing 20 μl of diluted pro-PO fractions followed by addition of elicitor [heat-killed M. luteus (1.5 × 104 cfu) or PAP-3 (0.2 pmol)] plus substrate [2 mM dopamine in 50 mM in sodium phosphate buffer (pH 6.5)] to activate the PO cascade. For inhibitory assays, pro-PO fractions were preincubated with medium from High Five cells or purified recombinant Egf proteins (1–50 pmol) for 10 min before addition of elicitor plus substrate. PO activity as measured by melanin production was determined at 485 nm by using a plate reader (FLUOstar Galaxy; BMG Labtech). One unit of activity was defined as ΔAbs485/min = 0.001. Treatments were replicated three times and analyzed by one-way ANOVA and Dunnett's multiple-comparison procedure using JMP (SAS Institute). RNAi assays using 10 nM dsRNA complementary to egf1.0/1.5 and glc1.8 were conducted as previously outlined (22).
Amidase Assays.
The inhibitory activity of Egf proteins was tested against the following enzymes and p-nitroanilide (pNA) substrates in 200-μl assays: 40 ng of purified PAP-3 and 50 μM N-acetyl-Ile-Glu-Ala-Arg-pNA (10); 250 ng of bovine pancreatic trypsin (Sigma) and 115 μM N-benzoyl-d,l-Arg-pNA hydrochloride (Sigma); 250 ng of human plasmin (Sigma) and 40 μM N-(p-Tosyl)-Gly-Pro-Lys-pNA acetate salt (Sigma); 125 ng of bovine pancreatic α-chymotrypsin (Sigma) and 80 μM N-succinyl-Ala-Ala-Pro-Phe-pNA (Sigma); 125 ng of porcine pancreatic elastase (Sigma) and 85 μM N-succinyl-Ala-Ala-Pro-Leu-pNA (Sigma). Enzymes were assayed in 0.1 M Tris·HCl/0.1 M NaCl (pH 8), except α-chymotrypsin, which was assayed in 0.1 M Tris·HCl/0.02 M CaCl2 (pH 8). Enzymes were preincubated with a given Egf protein at different molar ratios for 10 min before the addition of substrate-containing assay buffer. Amidase activities were measured at 405 nm and analyzed by one-way ANOVA. One unit of activity was defined as ΔAbs405/min = 0.0001.
Viability Assays.
Viability assays were conducted in 96-well plates containing a 1:20 diluted pro-PO plasma fraction from M. sexta, a physiological dose of MdBV (1.0 × 108 infectious units) (29) or 10 newly laid M. demolitor eggs, and 2 mM dopamine in TC-100 medium (1 volume) plus 50 mM sodium phosphate buffer (pH 6.5) (1 volume). Recombinant Egf1.0 was added to half of the samples 10 min before adding substrate, MdBV, or parasitoid eggs. After 2 h, PO activity was measured and compared with control samples induced by M. luteus. MdBV viability was measured 0–6 h after incubation by end-point dilution assay using High Five cells (29). M. demolitor viability was measured by collecting eggs after 2 h, rinsing in TC-100 medium, and culturing in TC-100 medium plus 30% heat-inactivated P. includens plasma (47). An egg was scored as viable if it hatched to form a first-instar parasitoid after 30 h. Data were analyzed by ANOVA as described above.
Acknowledgments
We thank R. J. Suderman and K. D. Clark for suggestions, J. Johnson and A. Robertson for assistance, and M. R. Kanost for generously providing purified PAP-3. This work was supported by the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service (Grant 2005-05382).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Gillespie JP, Kanost MR, Trenczek T. Annu Rev Entomol. 1997;42:611–643. doi: 10.1146/annurev.ento.42.1.611. [DOI] [PubMed] [Google Scholar]
- 2.Lavine MD, Strand MR. Insect Biochem Mol Biol. 2002;32:1237–1242. doi: 10.1016/s0965-1748(02)00092-9. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann JA. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 4.Bangham J, Jiggins F, Lemaitre B. Immunity. 2006;25:1–5. doi: 10.1016/j.immuni.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Strand MR. In: Insect Immunity. Beckage NE, editor. San Diego: Elsevier; 2007. in press. [Google Scholar]
- 6.Cerenius L, Soderhall K. Immunol Rev. 2004;198:116–126. doi: 10.1111/j.0105-2896.2004.00116.x. [DOI] [PubMed] [Google Scholar]
- 7.Nappi AJ, Christensen BM. Insect Biochem Mol Biol. 2005;35:443–459. doi: 10.1016/j.ibmb.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 8.Satoh D, Horii A, Ochiai M, Ashida M. J Biol Chem. 1999;274:7441–7453. doi: 10.1074/jbc.274.11.7441. [DOI] [PubMed] [Google Scholar]
- 9.Jiang H, Wang Y, Kanost MR. Proc Natl Acad Sci USA. 1998;95:12220–12225. doi: 10.1073/pnas.95.21.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang H, Wang Y, Yu X-Q, Zhu Y, Kanost MR. Insect Biochem Mol Biol. 2003;33:1049–1060. doi: 10.1016/s0965-1748(03)00123-1. [DOI] [PubMed] [Google Scholar]
- 11.Lee SY, Cho MY, Hyun JH, Lee KM, Homma KI, Natori S, Kawabata SI, Iwanaga S, Lee BL. Eur J Biochem. 1998;257:615–621. doi: 10.1046/j.1432-1327.1998.2570615.x. [DOI] [PubMed] [Google Scholar]
- 12.Tang H, Kambris Z, Lemaitre B, Hashimoto C. J Biol Chem. 2006;281:28097–28104. doi: 10.1074/jbc.M601642200. [DOI] [PubMed] [Google Scholar]
- 13.Volz J, Muller HM, Zdanowicz A, Kafatos FC, Osta MA. Cell Microbiol. 2006;8:1392–1405. doi: 10.1111/j.1462-5822.2006.00718.x. [DOI] [PubMed] [Google Scholar]
- 14.Paskewitz SM, Andreev O, Shi L. Insect Biochem Mol Biol. 2006;36:701–711. doi: 10.1016/j.ibmb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Strand MR, Pech LL. Annu Rev Entomol. 1995;40:31–56. doi: 10.1146/annurev.en.40.010195.000335. [DOI] [PubMed] [Google Scholar]
- 16.Celli J, Finlay BB. Trends Microbiol. 2002;10:232–237. doi: 10.1016/s0966-842x(02)02343-0. [DOI] [PubMed] [Google Scholar]
- 17.Mason NJ, Artis D, Hunter CA. Immunol Rev. 2004;198:48–56. doi: 10.1111/j.0105-2896.2004.00189.x. [DOI] [PubMed] [Google Scholar]
- 18.Webb BA, Strand MR. In: Comprehensive Molecular Insect Science. Gilbert LI, Iatrou K, Gill SS, editors. Vol 6. San Diego: Elsevier; 2005. pp. 323–360. [Google Scholar]
- 19.Strand MR. Encyclopedia of Virology. 2nd Ed. San Diego: Elsevier; 2007. in press. [Google Scholar]
- 20.Webb BA, Strand MR, Deborde SE, Beck M, Hilgarth RS, Kadash K, Kroemer JA, Lindstrom KG, Rattanadechakul W, Shelby KS, et al. Virology. 2006;347:160–174. doi: 10.1016/j.virol.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Zang X, Maizels RM. Trends Biochem Sci. 2001;26:191–197. doi: 10.1016/s0968-0004(00)01761-8. [DOI] [PubMed] [Google Scholar]
- 22.Beck M, Strand MR. Virology. 2003;314:521–535. doi: 10.1016/s0042-6822(03)00463-x. [DOI] [PubMed] [Google Scholar]
- 23.Beck MH, Strand MR. J Virol. 2005;79:1861–1870. doi: 10.1128/JVI.79.3.1861-1870.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thoetkiattikul L, Beck MH, Strand MR. Proc Natl Acad Sci USA. 2005;102:11426–11431. doi: 10.1073/pnas.0505240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruijssers A, Strand MR. J Virol. 2007;81:1209–1219. doi: 10.1128/JVI.02189-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rawlings ND, Morton FR, Barrett AJ. Nucleic Acids Res. 2006;34:D270–D272. doi: 10.1093/nar/gkj089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strand MR, Witherell SA, Trudeau D. J Virol. 1997;71:2146–2156. doi: 10.1128/jvi.71.3.2146-2156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trudeau D, Witherell AR, Strand MR. J Gen Virol. 2000;81:3049–3058. doi: 10.1099/0022-1317-81-12-3049. [DOI] [PubMed] [Google Scholar]
- 29.Beck MH, Inman RB, Strand MR. Virology. 2007;359:179–189. doi: 10.1016/j.virol.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Kanost MR, Jiang H, Yu X-Q. Immunol Rev. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- 31.Gorman MJ, Wang Y, Jiang H, Kanost MR. J Biol Chem. 2007;282:11742–11749. doi: 10.1074/jbc.M611243200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laskowski M, Qasim MA. Biochim Biophys Acta. 2000;1477:324–337. doi: 10.1016/s0167-4838(99)00284-8. [DOI] [PubMed] [Google Scholar]
- 33.Huang K, Strynadka NC, Bernard VD, Peanasky RJ, James MN. Structure (London) 1994;2:679–689. doi: 10.1016/s0969-2126(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 34.Pham T-N, Hayashi K, Takano R, Itoh M, Eguchi M, Shibata H, Tanaka T, Hara S. J Biochem. 1996;119:428–434. doi: 10.1093/oxfordjournals.jbchem.a021259. [DOI] [PubMed] [Google Scholar]
- 35.Murakami MT, Rios-Steiner J, Weaver SE, Tulinsky A, Geiger JH, Arni RK. J Mol Biol. 2007;366:602–610. doi: 10.1016/j.jmb.2006.11.040. [DOI] [PubMed] [Google Scholar]
- 36.De Gregorio E, Han SJ, Lee WJ, Baek MJ, Osaki T, Kawabata S, Lee BL, Iwanaga S, Lemaitre B, Brey PT. Dev Cell. 2002;3:581–592. doi: 10.1016/s1534-5807(02)00267-8. [DOI] [PubMed] [Google Scholar]
- 37.Zhu Y, Wang Y, Gorman MJ, Jiang H, Kanost MR. J Biol Chem. 2003;47:46556–46564. doi: 10.1074/jbc.M309682200. [DOI] [PubMed] [Google Scholar]
- 38.Yu X-Q, Jiang H, Wang Y, Kanost MR. Insect Biochem Mol Biol. 2003;33:197–208. doi: 10.1016/s0965-1748(02)00191-1. [DOI] [PubMed] [Google Scholar]
- 39.Kambris Z, Brun S, Jang IH, Nam HJ, Romeo Y, Takahashi K, Lee WJ, Ueda R, Lemaitre B. Curr Biol. 2006;16:808–813. doi: 10.1016/j.cub.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 40.Strand MR, Hayakawa Y, Clark KC. J Insect Physiol. 2000;46:817–824. doi: 10.1016/s0022-1910(99)00171-7. [DOI] [PubMed] [Google Scholar]
- 41.Michel K, Suwanchaichinda C, Morlais I, Lambrechts L, Cohuet A, Awono-Ambene PH, Simard F, Fontenille D, Kanost MR, Kafatos FC. Proc Natl Acad Sci USA. 2006;103:16858–16863. doi: 10.1073/pnas.0608033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schniger AK, Kafatos FC, Osta MA. J Biol Chem. 2007;282:21884–21888. doi: 10.1074/jbc.M701635200. [DOI] [PubMed] [Google Scholar]
- 43.Luo K-J, Pang Y. Acta Biochim Biophys Sin. 2006;38:577–585. doi: 10.1111/j.1745-7270.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 44.Zhang G, Lu Z-Q, Jiang H, Asgari S. Insect Biochem Mol Biol. 2004;34:477–483. doi: 10.1016/j.ibmb.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 45.Eleftherianos I, Boundy S, Joyce SA, Aslam S, Marshall JW, Cox RJ, Simpson TJ, Clarke DJ, ffrench-Constrant RH, Reynolds SE. Proc Natl Acad Sci USA. 2007;104:2419–2424. doi: 10.1073/pnas.0610525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castillo JC, Robertson AE, Strand MR. Insect Biochem Mol Biol. 2006;36:891–903. doi: 10.1016/j.ibmb.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strand MR, Wong EA. J Insect Physiol. 1991;37:503–515. [Google Scholar]