Abstract
The Rhesus (Rh) proteins are a family of integral membrane proteins found throughout the animal kingdom that also occur in a number of lower eukaryotes. The significance of Rh proteins derives from their presence in the human red blood cell membrane, where they constitute the second most important group of antigens used in transfusion medicine after the ABO group. Rh proteins are related to the ammonium transport (Amt) protein family and there is considerable evidence that, like Amt proteins, they function as ammonia channels. We have now solved the structure of a rare bacterial homologue (from Nitrosomonas europaea) of human Rh50 proteins at a resolution of 1.3 Å. The protein is a trimer, and analysis of its subunit interface strongly argues that all Rh proteins are likely to be homotrimers and that the human erythrocyte proteins RhAG and RhCE/D are unlikely to form heterooligomers as previously proposed. When compared with structures of bacterial Amt proteins, NeRh50 shows several distinctive features of the substrate conduction pathway that support the concept that Rh proteins have much lower ammonium affinities than Amt proteins and might potentially function bidirectionally.
Keywords: ammonia channel, ammonium transport, Rhesus 50 protein, x-ray structure
The Rhesus (Rh) blood group system is the most polymorphic of the human blood groups and, next to the ABO system, is the most clinically important in transfusion medicine. The Rh antigens are carried by two red blood cell membrane proteins, RhD and RhCE (also termed Rh30; apparent molecular mass 30 kDa), which have been proposed to be associated in a complex with a related erythrocyte-specific glycoprotein RhAG (Rh50; 50 kDa) (1). The two other human nonerythroid Rh50 glycoproteins, RhBG and RhCG, are expressed in kidney, liver, brain, and skin (2). Rh50 proteins are found throughout the animal kingdom and also occur in green algae and slime molds.
Despite their clinical importance, the function of the Rh proteins remained unclear until it was recognized that they share low but significant sequence similarity with ammonium transport (Amt) proteins (3). Amt proteins are ubiquitous integral membrane channels found in bacteria, archaea, plants, and fungi (4), the best characterized being Escherichia coli AmtB (5). Structural data predict that Amt proteins function as ammonia channels and have an extracellular binding site for the ammonium ion that is deprotonated before translocation of ammonia (6–9). In vivo studies on E. coli AmtB support the model of ammonia translocation (10), but other Amt proteins have been suggested to act as NH4+ uniporters or NH3/H+ cotransporters (11, 12). Comparable studies on Rh50 proteins have mostly supported a role in ammonium transport (13), and in some cases it has been suggested that Rh50 proteins can mediate ammonia efflux (14–16). However, the Rh1 protein of Chlamydomonas reinhardtii, has been proposed to function as a CO2 channel (17, 18), and Rh30/RhAG proteins have been claimed to contribute to the high CO2 permeability of the human red cell membrane (19). Rh30 proteins appear to have evolved from Rh50 proteins early in fish speciation (20). Unlike for Rh50 proteins, there is no evidence, either from experimental data (21, 22) or from structural models (23, 24), that Rh30 proteins function as ammonium transporters, and they are currently thought to have a structural role in the erythrocyte membrane (25).
The human Rh erythrocyte complex has been proposed to be a tetramer, comprising two RhAG polypeptides and two Rh30 polypeptides (1, 26). However, x-ray crystal structures of E. coli AmtB and the related Archaeoglobus fulgidus Amt-1 protein show these proteins to be homotrimers, each subunit of which has 11 transmembrane helices (TMH) and contains a central conduction channel (6, 7, 9). Sequence alignments suggest that other Amt proteins also have 11 TMH, whereas studies on human Rh proteins suggest that they have 12 TMH, the extra helix being at the N terminus (1, 27). Both Rh50 and Rh30 proteins have been modeled on Amt proteins, and these models challenge the view of the Rh complex as a tetramer and favor a trimeric model (23, 24). They also predict that Rh proteins should lack the high-affinity ammonium binding site seen on the extracellular face of Amt proteins.
Rh and Amt proteins are clearly derived from a common ancestor (20, 28), and, although Rh and Amt genes are found together in a many lower eukaryotes, Rh genes predominate in vertebrates where Amt genes are absent (28, 29). However, recent genome sequences have revealed rare occurrences of Rh50-type genes in prokaryotes, including the chemolithoautotroph Nitrosomonas europaea (30, 31). The NeRh50 protein has been shown to function as an ammonia transporter (32, 33), and it offers an excellent opportunity to investigate the structure of a member of the Rh protein family. Here, we report the x-ray crystal structure of NeRh50 at 1.3 Å resolution and discuss its implications regarding the mechanism of ammonium translocation.
Results
Comparison of the Overall Structure of NeRh50 with Amt Proteins.
N-terminal sequence analysis revealed that NeRh50 is processed in E. coli by signal peptidase, which cleaves off the first 24 aa. The refined NeRh50 structure comprises residues 8–369; thus, residues 1–7, the most C-terminal residues 370–401, and the His6 tag are not resolved. The monomer in the asymmetric unit is part of a tightly packed trimer (Fig. 1) generated by the crystallographic threefold axis. The monomer structure is very similar to the homologous Amt structures (Fig. 2) (6, 7, 9), and major differences only occur in the loops and in the C-terminal region (CTR) after M11 [see supporting information (SI) Fig. 6]. The extra N-terminal TMH (M0) predicted to be present in human erythroid Rhesus proteins is absent in NeRh50 expressed in E. coli.
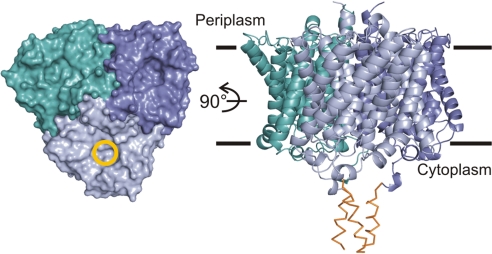
Fig. 1.
The NeRh50 trimer structure. The periplasmic face of the trimer is shown in surface representation at Left with the extracellular pore entry marked by a yellow circle in one monomer. The ribbon representation at Right shows a side view with the approximate bilayer boundaries indicated. The C-terminal segments showing only backbone density (see SI Fig. 7) are shown as orange Cα traces.
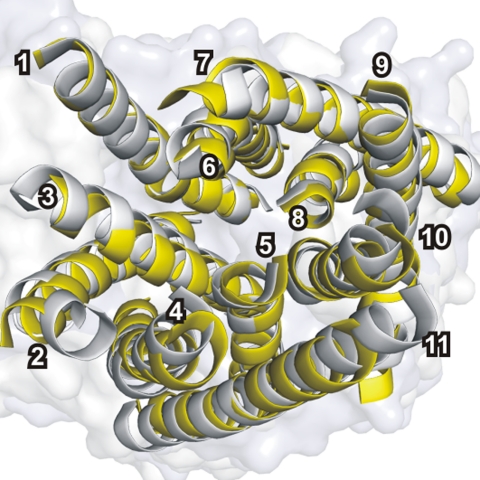
Fig. 2.
Comparison of the monomer structure of EcAmtB and NeRh50. Superposition of the transmembrane helices (numbered in white) of NeRh50 (yellow) with those of AmtB (gray, D subunit of PDB ID code 2NUU) yields an rms displacement of 1.4 Å for 268 Cα atoms.
Our structure-based alignment (Fig. 3) reveals that a bidimensional hydrophobic cluster analysis (23) correctly aligned the transmembrane helices of human (Hs) Rh proteins with EcAmtB except for M2 and M4, where their alignment is out by one and four residues, respectively. Within its structurally defined region, NeRh50 shares 21.9% (58.4%) sequence identity (similarity) with EcAmtB and 36.3% (64.9%) with HsRhAG. Furthermore, many structural differences between NeRh50 and EcAmtB are family-specific. This is particularly evident along the conduction pathway, which is primarily shaped by residues of the pseudosymmetry related M1/M3/M5 and M6/M8/M10 helices, respectively (Fig. 2). For the subset of residues of these six helices (121 residues in total), identity between NeRh50 and HsRhAG increases to 55%, whereas it remains at 22% for EcAmtB. We conclude that the NeRh50 channel architecture should be highly representative of all Rh proteins.
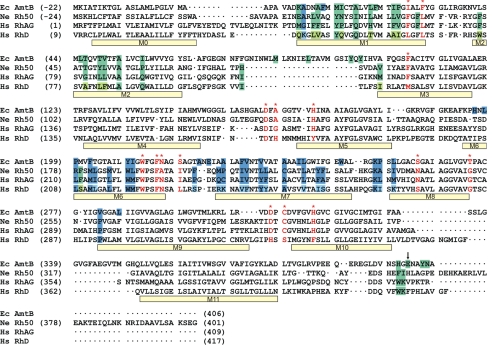
Fig. 3.
Structure-based sequence alignment of NeRh50 and EcAmtB. The program Sequoia (58) was used to carry out the structure-based alignment and combine it with a sequence-based alignment for HsRhAG and RhD. The alignment before the start of M1 was adjusted manually. Other minor adjustements were made in the structurally nonhomologous parts of loops. The vertical arrow at the CTR marks the position beyond which the alignment is not meaningful, and the sequences are merely shown for completeness. Negative sequence numbers are assigned to the processed leader peptides of EcAmtB and NeRh50, hence the mature proteins as expressed in E. coli start at residue +1. Transmembrane helices M1-M11 as observed in the NeRh50 structure and M0 as predicted (23) are indicated with bars below the aligned sequences. Residues with large interface contacts (see SI Fig. 8) are in boxes colored according to whether they belong to the green or dark blue subunit in Fig. 1. The corresponding residues in RhD are shown in lighter colors if different from those of RhAG. Selected residues along the substrate pathway as discussed in the text are marked in red.
Structural Characteristics of Channel Vestibules and Pore.
Based on the structure of Amt proteins (6, 7, 9), four regions, (i) an extracellular vestibule with an ammonium binding site, (ii) a “phenylalanine gate” at the extracellular pore entry, (iii) a central twin-histidine-containing pore, and (iv) a cytoplasmic vestibule, have been discriminated in discussing mechanistic questions related to substrate conduction (13). A primary interest is, therefore, to analyze the structural differences that discriminate Amt from Rh proteins in these functionally critical regions.
Extracellular Vestibule.
The extracellular ammonium ion binding site identified in Amt structures (6, 7, 9) is shaped by three highly conserved residues (Phe-107, Trp-148, and Ser-219 in EcAmtB) implicated in hydrogen bonding and cation-π interactions. Only the Phe residue is conserved in the Rh family, and the architecture of the solvent-accessible extracellular vestibule is not conserved between Rh and Amt proteins.
The extracellular vestibule is very differently shaped in NeRh50 (Fig. 4A), and, as predicted (7, 8, 24), no equivalent ammonium binding site exists. The NeRh50 residue that aligns with EcAmtB-Ser-219 is Ile-198 (Fig. 3), and, with few exceptions, this residue is Ile or Leu in Rh proteins. Ile-198 is quite differently positioned to AmtB-Ser-219 (Fig. 4A) and occupies space very close to the ammonium binding site in Amt protein structures. AmtB-Trp-148, located at the C terminus of M4 and implicated in a cation-π interaction with ammonium in Amt proteins, has no structurally equivalent residue in NeRh50. Five well defined water molecules occupy the corresponding free space in NeRh50.
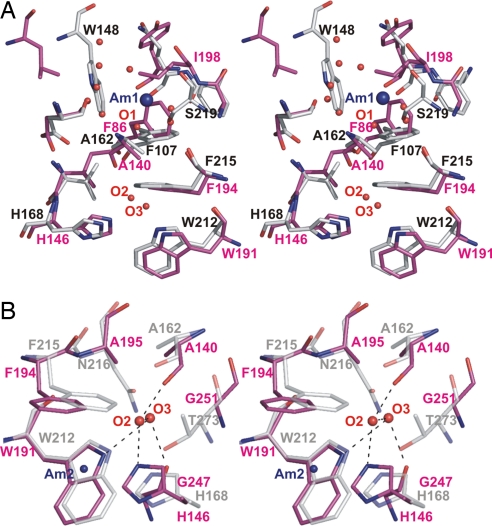
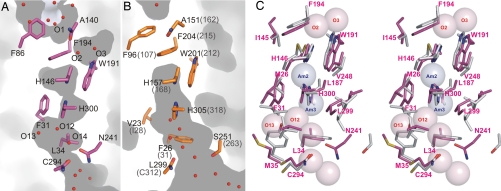
Fig. 4.
Stereo views of the periplasmic vestibule of NeRh50. NeRh50 (magenta) was superposed to EcAmtB [gray, PDB ID code 1U7G (7)], using the Cα positions of the structurally equivalent residues. Water molecules (labeled O1, O2, etc., as discussed in the text) are shown as red spheres, and the postulated ammonium/ammonia sites of EcAmtB (Am1 and Am2) are indicated with blue spheres. (A) View illustrating the absence of an ammonium ion binding site in the NeRh50 periplasmic vestibule. (B) View illustrating the significant opening in NeRh50 at the inner conserved phenylalanine (NeRh50-Phe-194). The hydrogen bonding interactions of O2 and O3 are depicted as dashed black lines.
Phenylalanine Gate.
In Amt structures, the pathway from the ammonium binding site to the central pore is severely obstructed by a “gate” formed by the stacked phenyl rings of two conserved Phe residues (Phe-107 and Phe-215 in EcAmtB). This obstruction is much reduced in NeRh50. The ring of the first Phe (Phe-86) is oriented perpendicular to that of the second (Phe-194), whereas a parallel orientation is observed in Amt protein structures. In Rh proteins, such a parallel conformation would cause a steric clash with the side chain of the well conserved Ile-145. The opening resulting from the perpendicular orientation of Phe-86 permits a water molecule (O1) to be ≈2 Å deeper along the channel entry path than the Am1 site in EcAmtB (Fig. 4A).
We also observe another opening at one edge of the phenyl ring of Phe-194. It is right above the first of the two conserved pore histidines and is occupied by two water molecules, O2 and O3 (Figs. 4 and 5C). In EcAmtB, the equivalent space is occupied by Thr-273, which structurally aligns with Gly-251 of NeRh50 (Figs. 3 and 4). This Gly is highly conserved in all known Rh proteins (29). The larger opening at the pore entrance of NeRh50 is a consequence not only of this Thr to Gly substitution but also of an increased distance between the corresponding main chains. The Cα-Cα distance between NeRh50 Phe-194 and Gly-251 is ≈1 Å larger than between EcAmtB Phe-215 and Thr-273.
Fig. 5.
Substrate conduction pathway of NeRh50 and AfAmt1/EcAmtB. (A and B) Cross-sections cutting the pores of NeRh50 (A) and AfAmt1 (B) (6). The dark areas represent cavities after removing all water molecules from the structure models. The side chains of conserved pore residues are shown as sticks, and water molecules are shown as red spheres. Af-Amt1 is shown here because it is better suited to compare hydration of the cytoplasmic vestibules (EcAmtB structures are affected by disorder, bound detergent, or bound GlnK in that region). The pore shape is very similar to that observed for EcAmtB, whose residue numbers are given in parentheses. (C) Stereoview of the superposed central pore lining residues of NeRh50 (magenta) and EcAmtB [gray, from D subunit of PDB ID code 2NUU (35)]. Selected residues of NeRh50 are labeled. Transparent spheres (1.4-Å radius) represent observed water sites in NeRh50 (O2, etc., light magenta) and the putative NH3 sites Am2 and Am3 in EcAmtB (7).
The observed water positions, O1 and O2, delineate a likely substrate path from the extracellular vestibule across the Phe gate. O1 and O2 are 5.4 Å apart and separated by a constriction formed by Phe-194, Ala-195, and Ala-140. However, the phenyl ring of Phe-194 could easily be tilted to allow substrate passage. The same rotation would also widen another constriction, between O2 and the central pore region, such that smooth substrate transfer from O1 into the central pore appears easily possible (Fig. 4).
Central “Twin-Histidine” Pore.
The central pore lumen, lined on one side by the two conserved imidazole side chains of His-146 and His-300, is similarly shaped in NeRh50 and in the known Amt structures (Fig. 5). Trp-191 is highly conserved in Amt and Rh families, whereas the pseudosymmetrically related residue Phe-31 is highly conserved in Rh proteins but is more variable in the Amt family. In AmtB, residual electron density peaks in the pore have been interpreted as partly occupied ammonia or water molecules, indicating preferred sites along the conduction pathway, and those in the central pore have been designated Am2, Am3 (7) or S2, S3, respectively (34). In the NeRh50 structure, we do not observe any such residual electron density in the lumen of the central pore.
Cytoplasmic Vestibule.
In the NeRh50 and Amt structures, the Phe gate has no comparable counterpart on the cytoplasmic side of the central pore. The residues pseudosymmetrically related to NeRh50 Phe-86 and Phe-194 are Asn-241 and Leu-34, respectively. Both are highly conserved in Rh proteins. We consider this region to be part of the cytoplasmic vestibule, because it does not severely constrict the substrate pathway as does the Phe gate on the periplasmic side. However, this asymmetry is less pronounced in NeRh50 than in Amt structures (Fig. 5).
C-Terminal Region.
The length of the CTR varies considerably within the Amt/Rh family, and the sequence of the Rh CTR differs markedly from that of Amt proteins. However, the chain traces of NeRh50 and EcAmtB superpose well over a short, type I turn segment, NeRh50 356–359 (EcAmtB 395–398) (see SI Fig. 6C). In both cases, this turn and some adjacent residues form numerous contacts to the cytoplasmic M1/M2 loop of the same subunit. In Amt proteins, the CTR reaches over into the vestibule of the adjacent subunit (6, 35, 36), which has led to speculations of cooperativity or cross-talk between the subunits (37–39), but such an interaction is not seen in NeRh50.
Electron density is well defined up to NeRh50-Glu-369, which roughly corresponds to the length of the CTR in the erythrocyte proteins RhAG and RhCE/D. Beyond this, weak density indicates that ≈12 of the remaining 32 residues may have a tendency to form a trimeric coiled coil along the threefold axis (see Fig. 1 and SI Fig. 7B). However, owing to sequence divergence, this cannot be regarded as representative of Rh proteins. An extended H-bonding network in NeRh50 involves Arg-40, Ser-44, Glu-35, and Asp-356, residues that are well conserved (R/K, S, E/D, D) in all human Rh proteins, strongly indicating a similar local structure. Contact of segment 357–361 with the M7/M8 loop of an adjacent trimer subunit is also likely to be conserved in Rh proteins (SI Fig. 7A).
NeRh50 Trimer Interface as a Model for Mammalian Rhesus Oligomers.
The erythrocyte Rh complex was originally proposed to be a heterotetramer, mainly on the basis of coimmunoprecipitation data that permit other interpretations (1). However, the homotrimeric nature of the homologous AmtB protein strongly suggested a trimeric structure for Rh proteins (24), and this is now firmly supported by the NeRh50 structure.
We have examined whether the formation of mixed oligomers between RhAG and RhCE or RhD is a plausible model. Most trimer interface residues in NeRh50 and EcAmtB (Fig. 3 and SI Fig. 8) occupy equivalent positions in the structure-based sequence alignment indicative of conservation of the quaternary structure across the whole superfamily. However, few of the interface residues are identical, so the detailed interactions are quite distinct. We selected 43 of the 76 residues involved in interface contacts in NeRh50, namely those with a contact surface >40 Å2. Most equivalent residues in AmtB (Fig. 3) are similarly involved in major interface contacts. Twenty of these 43 residues are identical in RhAG and either RhCE or RhD: a sequence identity of 46% compared with 36% overall identity. A modest increase in identity for interface residues is quite typical for homologous oligomers (40). However, here we have examined whether these proteins could form mixed oligomers with a tightly packed extended interface, which requires a very good match for both the underlying monomer structures and the packing of their interface side chains.
Our analysis suggests that the formation of mixed oligomers is unlikely. First, the overall sequence identity of 36% appears too low to be compatible with precisely matched protein backbone structures. An overall sequence identity of 75% and higher is observed in two cases of Amt family paralogs for which the formation of mixed oligomers has been postulated, namely between ScMep1 and ScMep3 (41) and between LeAMT1;1 and LeAMT1;2 (42). Second, the high number of different interface residues, 14 of 23 different side chains have very low similarity, is likely to generate severe packing mismatches in mixed interfaces. Third, analysis of only those side chains located right at the threefold axis also reveals several large discrepancies. Three such residues in NeRh50, Val-14, Ala-15, and Asn-18 are well conserved in both RhD and RhCE (Val, Ala, and Gln, respectively) but are quite different in RhAG (Phe, Glu, and Pro, respectively). Similarly, NeRh50-Phe-179 is conserved as Phe in RhAG but is Ser in RhD and RhCE. Although this does not prove that mixed trimers cannot form, it seriously questions their existence.
Discussion
The high-resolution structure of NeRh50 provides answers to several questions related to the tertiary and quaternary structure of the Rh family proteins that could not be resolved from structures of the related but distinct Amt proteins. Many of the Rh/Amt family-specific sequence differences correlate directly with specific structural differences between the channels of NeRh50 and Amt proteins. Consequently, the detailed NeRh50 channel architecture can be deduced to be highly representative of all Rh proteins.
Mechanistic Aspects of Ammonia Conduction in Amt and Rh Proteins.
A primary objective of this study was to gain insight into structural differences between the substrate conduction pores of Amt and Rh50 proteins that were predicted to differ particularly in the extracellular vestibule (7, 8, 24). The following discussion solely addresses ammonia conduction, because we see no indication from this structure that Rh proteins have a distinct substrate, namely CO2, as has been suggested (43).
As expected, the external ammonium binding site found in Amt proteins is not present, and there is no indication of a structurally well defined alternative binding site. Potentially related structural changes occur in the adjacent Phe gate region that is much less constricting than in Amt structures, although both phenylalanines are highly conserved in Amt and Rh50 proteins. The widening of this constriction is not simply the result of different side chain conformations, as observed for the first Phe, but is the combined effect of Rh/Amt family-characteristic side chain substitutions, most notably Gly for Thr at position 251 of NeRh50, and a larger separation between the corresponding main chains, namely transmembrane helices M6 and M8, in this region.
Overall, the vestibules at either end of the pore are more similar, both in their shape and their accessibility to water (or NH4+/NH3), in the NeRh50 structure than in the known Amt structures. There is a well defined path by which a substrate molecule could pass the Phe gate that separates the extracellular vestibule from the central pore lumen at the first His (Fig. 5A). This path is likely to be the same in the Amt proteins, except that it appears more difficult to generate sufficiently large transient openings for substrate passage. The main chain carbonyl group of NeRh50-Ala-140 (EcAmtB-Ala-162) appears to play an important role in providing some hydrogen bond acceptor capacity to translocating substrate molecules in this otherwise nonpolar region as suggested for AmtB (9) and observed in a molecular dynamics simulation (44).
Mechanistic aspects of ammonia transport by EcAmtB have been addressed in a number of recent molecular dynamics simulations (13, 44–51). The calculations confirm that ammonium ion transport through the central pore lumen would be energetically very unfavorable and support the current view that Amt/Rh proteins conduct uncharged NH3.
Amt proteins have to recruit ammonium at very low concentration and appear initially to bind their substrate in the more abundant ionic form (8); therefore, an efficient deprotonation mechanism is required before NH3 passage through the central pore can occur. Different mechanisms for ammonium deprotonation have been proposed, involving Ala-162:C O (44), Asp-160 (49), Ser-219 (51), or water (46) as primary proton acceptors. We consider the mechanisms where deprotonation takes place between sites Am1 and Am2 (44, 46), that is, during passage of the Phe gate, as more plausible. The proposed Ala-162:C
O (44), Asp-160 (49), Ser-219 (51), or water (46) as primary proton acceptors. We consider the mechanisms where deprotonation takes place between sites Am1 and Am2 (44, 46), that is, during passage of the Phe gate, as more plausible. The proposed Ala-162:C O deprotonation site (figure 6 of ref. 44) closely corresponds to O2 observed in the NeRh50 structure, except that the second Phe of NeRh50 is not required to rotate by 90° to provide sufficient space. This putative deprotonation site would assign a defined role to the first pore histidine, found to be essential for methylammonium transport by EcAmtB (34). In the NeRh50 structure, its Nε atom provides a hydrogen bond to water O2 located at this site. Initial proton transfer to Ala:C
O deprotonation site (figure 6 of ref. 44) closely corresponds to O2 observed in the NeRh50 structure, except that the second Phe of NeRh50 is not required to rotate by 90° to provide sufficient space. This putative deprotonation site would assign a defined role to the first pore histidine, found to be essential for methylammonium transport by EcAmtB (34). In the NeRh50 structure, its Nε atom provides a hydrogen bond to water O2 located at this site. Initial proton transfer to Ala:C O, to Nε of the first histidine, or to a water molecule accompanying NH4+ across the Phe gate, all remain viable possibilities in our opinion.
O, to Nε of the first histidine, or to a water molecule accompanying NH4+ across the Phe gate, all remain viable possibilities in our opinion.
Because Rh proteins lack a defined ammonium ion binding site (Fig. 4A), they might recognize and conduct the neutral form, in which case no deprotonation site would be needed, but a strong pH dependence of the transport rates would be expected. The mammalian Rh50 proteins fulfil their function at significantly higher substrate concentrations (52) and show apparent Km and Ki values in the millimolar range for methylammmonium transport and its inhibition by ammonium, respectively (22, 53, 54). For NH3, the respective values would be low micromolar, unreasonably low in our opinion (see SI Appendix). Furthermore, the methylamine transport rate of human Rh glycoproteins increases considerably less with pH than the concentration of the unprotonated substrate (11, 54). Therefore, despite the lack of a high-affinity ammonium binding site, Rh proteins may still rely on the recruitement and deprotonation of the charged form in the respective vestibule for optimal ammonia conduction. The structure of NeRh50 supports this view as important structural features observed in its Phe gate region are similarly observed in transient states associated with possible deprotonation states in EcAmtB MD simulations (44, 46).
Because water and ammonia are rather similar molecules, ammonia channels must discriminate against water to be effective. For Amt proteins, the ammonium ion binding site, which has micromolar affinity in EcAmtB (10), represents a highly selective filter against water, but at the price of a reduced maximum conduction rate (8). Based on simple assumptions, we estimate that Rh proteins must prefer NH3 by at least 103-fold compared with water (see SI Appendix). Discrimination at this level appears easier to achieve between NH4+, rather than NH3, and water and would have to take place in the vestibule region before deprotonation. The apparent millimolar affinity of Rh proteins for ammonium may thus reflect a weak ammonium sequestering capacity of the vestibule.
Implications for the Structures of Mammalian Rh Proteins.
The trimeric nature of NeRh50 corroborates homology modeling using Amt structures (23, 24). Hence, it seems likely that Rh proteins are trimeric rather than tetrameric, as proposed originally (1) and still widely accepted. The Rh30 proteins (RhCE/D) are expressed in red blood cells (RBC) and have been found to be in association with the RhAG glycoprotein and other erythrocyte membrane and membrane-associated proteins (55, 56). However, there is no evidence that the Rh complex is a structurally and stoichiometrically defined entity; rather, it is likely to be a complex, and probably dynamic, network of interacting molecules that appears important for the regulation of red cell shape. The trimer interface observed in NeRh50 provides a very good model for examining whether mixed trimers between Rh30s and RhAG appear plausible. Our analysis, based on examining the conservation of the most important interface residues predicted in RhAG and Rh30 proteins (Fig. 3), seriously questions the existence of mixed trimers and calls for a different interpretation of their interaction in the erythrocyte Rh complex.
Whereas Amt proteins typically have 11 TMH, human Rh proteins have been predicted to have an extra N-terminal helix (M0), and N-terminal sequencing of RhAG and RhD isolated from RBCs is consistent with this (56). However, the structural equivalent of NeRh50 M0 is a strongly predicted signal peptide that is indeed cleaved when expressed in E. coli, although we do not know whether it is naturally processed in N. europaea. A signal peptide is similarly removed during maturation of EcAmtB (57). Certainly any Rh helix M0 would have to be located on the periphery of the trimer as suggested (23, 24). In this case, the eight residues in NeRh50 connecting the predicted end of M0 and start of M1 (Fig. 3) near the threefold axis are just about sufficient to position M0 at the periphery between M2 of one monomer and M7/M9 of its neighbor.
Presence and Function of Rh50 in N. europaea.
To date, genes encoding Rh50 homologues have been identified in just four prokaryotic genomes, N. europaea (30), Nitrosospira multiformis (31), Acidobacteria bacterium, and Kuenenia stuttgartensis (32). Of these four, the two ammonia oxidizing bacteria (N. europaea and N. multiformis) lack amt genes, whereas A. bacterium and K. stuttgartensis encode both Amt and Rh proteins. When expressed in a Saccharomyces cerevisiae Δmep strain, NeRh50 can restore ammonium-dependent growth, indicating that it is competent to conduct ammonium/ammonia (32, 33). Hence, in N. europaea and N. multiformis, the Rh50 proteins may functionally replace Amt proteins to facilitate ammonium uptake, albeit with a predicted lower affinity. The supply of ammonia to ammonia monooxygenase is of fundamental importance for the metabolism of ammonia oxidizing bacteria. This process is not presently well understood, although Rh50 could potentially play a role.
Methods
Protein Preparation and Crystallization.
The Rh50 protein from N. europaea was heterologously expressed in E. coli with a hexa-histidine tag at its C terminus. The protein was solubilized from the membrane with 5% (wt/vol) n-octyl-β-d-glucopyranoside (OG) and purified by affinity chromatography, using Co2+-loaded Sepharose beads. The concentrated eluate was first desalted on a NAP-10 column and then concentrated to ≈15 mg/ml in a buffer containing 10 mM Tris·HCl (pH 7.6), 100 mM NaCl, 10% glycerol, and 0.5% OG. Crystals belonging to space group R3 (a = 100.1 Å, c = 143.8 Å) were obtained at 20°C from sitting drops set up in a 1:1 ratio from protein stock solution and reservoir solution [10 mM Tris·HCl (pH 8.5), 0.2 M NaCl, and 24% PEG 3350]. Details are given in SI Materials and Methods.
Structure Determination.
Data for crystals mounted on cryoloops were recorded at 100 K on a MarCCD detector at the protein beamline X06SA, Swiss Light Source. The structure was solved by molecular replacement, using EcAmtB structures as search models, and contains one monomer in the asymmetric unit. The final 1.3-Å resolution model has Rwork and Rfree values of 0.147 and 0.171, respectively, and excellent geometry. Details on the procedures and data collection and refinement statistics are given in SI Table 1 and SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank A. Gasperina for excellent assistance with initial protein preparation and A. Javelle for critical comments on the manuscript. This work was supported by the Swiss National Science Foundation within the framework of the National Center of Competence in Research in Structural Biology (F.K.W.) and the Biotechnology and Biological Sciences Research Council (M.M. and A.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3B9W).
This article contains supporting information online at www.pnas.org/cgi/content/full/0706563104/DC1.
References
- 1.Eyers SA, Ridgwell K, Mawby WJ, Tanner MJ. J Biol Chem. 1994;269:6417–6423. [PubMed] [Google Scholar]
- 2.Huang CH, Liu PZ. Blood Cells Mol Dis. 2001;27:90–101. doi: 10.1006/bcmd.2000.0355. [DOI] [PubMed] [Google Scholar]
- 3.Marini AM, Urrestarazu A, Beauwens R, Andre B. Trends Biochem Sci. 1997;22:460–461. doi: 10.1016/s0968-0004(97)01132-8. [DOI] [PubMed] [Google Scholar]
- 4.von Wirén N, Merrick M. Trends Curr Genet. 2004;9:95–120. [Google Scholar]
- 5.Merrick M, Javelle A, Durand A, Severi E, Thornton J, Avent ND, Conroy MJ, Bullough PA. Transfus Clin Biol. 2006;13:97–102. doi: 10.1016/j.tracli.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Andrade SL, Dickmanns A, Ficner R, Einsle O. Proc Natl Acad Sci USA. 2005;102:14994–14999. doi: 10.1073/pnas.0506254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khademi S, O'Connell J, III, Remis J, Robles-Colmenares Y, Miercke LJ, Stroud RM. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 8.Winkler FK. Pflugers Arch. 2006;451:701–707. doi: 10.1007/s00424-005-1511-6. [DOI] [PubMed] [Google Scholar]
- 9.Zheng L, Kostrewa D, Bernèche S, Winkler FK, Li X.-D. Proc Natl Acad Sci USA. 2004;101:17090–17095. doi: 10.1073/pnas.0406475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Javelle A, Thomas G, Marini AM, Kramer R, Merrick M. Biochem J. 2005;390:215–222. doi: 10.1042/BJ20042094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer M, Schaaf G, Mouro I, Lopez C, Colin Y, Neumann P, Cartron JP, Ludewig U. J Gen Physiol. 2006;127:133–144. doi: 10.1085/jgp.200509369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siewe RM, Weil B, Burkovski A, Eikmanns BJ, Eikmanns M, Kramer R. J Biol Chem. 1996;271:5398–5403. doi: 10.1074/jbc.271.10.5398. [DOI] [PubMed] [Google Scholar]
- 13.Javelle A, Lupo D, Li XD, Merrick M, Chami M, Ripoche P, Winkler FK. J Struct Biol. 2007;158:472–481. doi: 10.1016/S1047-8477(07)00165-7. [DOI] [PubMed] [Google Scholar]
- 14.Hemker MB, Cheroutre G, van ZR, Maaskant-van Wijk PA, Roos D, Loos JA, van der Schoot CE, von dem Borne AE. Br J Haematol. 2003;122:333–340. doi: 10.1046/j.1365-2141.2003.04425.x. [DOI] [PubMed] [Google Scholar]
- 15.Marini AM, Matassi G, Raynal V, Andre B, Cartron JP, Cherif-Zahar B. Nat Genet. 2000;26:341–344. doi: 10.1038/81656. [DOI] [PubMed] [Google Scholar]
- 16.Nakada T, Westhoff CM, Kato A, Hirose S. FASEB J. 2007;21:1067–1074. doi: 10.1096/fj.06-6834com. [DOI] [PubMed] [Google Scholar]
- 17.Soupene E, King N, Feild E, Liu P, Niyogi KK, Huang CH, Kustu S. Proc Natl Acad Sci USA. 2002;99:7769–7773. doi: 10.1073/pnas.112225599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soupene E, Inwood W, Kustu S. Proc Natl Acad Sci USA. 2004;101:7787–7792. doi: 10.1073/pnas.0401809101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Endeward V, Cartron JP, Ripoche P, Gros G. Transfus Clin Biol. 2006;13:123–127. doi: 10.1016/j.tracli.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Matassi G, Cherif-Zahar B, Pesole G, Raynal V, Cartron JP. J Mol Evol. 1999;48:151–159. doi: 10.1007/pl00006453. [DOI] [PubMed] [Google Scholar]
- 21.Ripoche P, Bertrand O, Gane P, Birkenmeier C, Colin Y, Cartron JP. Proc Natl Acad Sci USA. 2004;101:17222–17227. doi: 10.1073/pnas.0403704101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westhoff CM, Ferreri-Jacobia M, Mak DO, Foskett JK. J Biol Chem. 2002;277:12499–12502. doi: 10.1074/jbc.C200060200. [DOI] [PubMed] [Google Scholar]
- 23.Callebaut I, Dulin F, Bertrand O, Ripoche P, Mouro I, Colin Y, Mornon JP, Cartron JP. Transfus Clin Biol. 2006;13:70–84. doi: 10.1016/j.tracli.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Conroy MJ, Bullough PA, Merrick M, Avent ND. Br J Haematol. 2005;131:543–551. doi: 10.1111/j.1365-2141.2005.05786.x. [DOI] [PubMed] [Google Scholar]
- 25.Westhoff CM, Storry JR, Walker P, Lomas-Francis C, Reid ME. Transfusion. 2004;44:1047–1051. doi: 10.1111/j.1537-2995.2004.04003.x. [DOI] [PubMed] [Google Scholar]
- 26.Hartel-Schenk S, Agre P. J Biol Chem. 1992;267:5569–5574. [PubMed] [Google Scholar]
- 27.Avent ND, Liu W, Warner KM, Mawby WJ, Jones JW, Ridgwell K, Tanner MJ. J Biol Chem. 1996;271:14233–14239. doi: 10.1074/jbc.271.24.14233. [DOI] [PubMed] [Google Scholar]
- 28.Kitano T, Saitou N. Immunogenetics. 2000;51:856–862. doi: 10.1007/s002510000202. [DOI] [PubMed] [Google Scholar]
- 29.Huang CH, Peng J. Proc Natl Acad Sci USA. 2005;102:15512–15517. doi: 10.1073/pnas.0507886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chain P, Lamerdin J, Larimer F, Regala W, Lao V, Land M, Hauser L, Hooper A, Klotz M, Norton J, et al. J Bacteriol. 2003;185:2759–2773. doi: 10.1128/JB.185.9.2759-2773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kustu S, Inwood W. Transfus Clin Biol. 2006;13:103–110. doi: 10.1016/j.tracli.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Cherif-Zahar B, Durand A, Schmidt I, Hamdaoui N, Matic I, Merrick M, Matassi M. J Bacteriol. 2007 doi: 10.1128/JB.01089-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weidinger K, Neuhauser B, Gilch S, Ludewig U, Meyer O, Schmidt I. FEMS Microbiol Lett. 2007;273:260–267. doi: 10.1111/j.1574-6968.2007.00805.x. [DOI] [PubMed] [Google Scholar]
- 34.Javelle A, Lupo D, Zheng L, Li XD, Winkler FK, Merrick M. J Biol Chem. 2006;281:39492–39498. doi: 10.1074/jbc.M608325200. [DOI] [PubMed] [Google Scholar]
- 35.Conroy MJ, Durand A, Lupo D, Li XD, Bullough PA, Winkler FK, Merrick M. Proc Natl Acad Sci USA. 2007;104:1213–1218. doi: 10.1073/pnas.0610348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gruswitz F, O'Connell J, III, Stroud RM. Proc Natl Acad Sci USA. 2007;104:42–47. doi: 10.1073/pnas.0609796104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loque D, Lalonde S, Looger LL, von WN, Frommer WB. Nature. 2007;446:195–198. doi: 10.1038/nature05579. [DOI] [PubMed] [Google Scholar]
- 38.Neuhauser B, Dynowski M, Mayer M, Ludewig U. Plant Physiol. 2007;143:1651–1659. doi: 10.1104/pp.106.094243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Severi E, Javelle A, Merrick M. Mol Membr Biol. 2007;24:161–171. doi: 10.1080/09687860601129420. [DOI] [PubMed] [Google Scholar]
- 40.Valdar WS, Thornton JM. Proteins. 2001;42:108–124. [PubMed] [Google Scholar]
- 41.Marini AM, Springael JY, Frommer WB, Andre B. Mol Microbiol. 2000;35:378–385. doi: 10.1046/j.1365-2958.2000.01704.x. [DOI] [PubMed] [Google Scholar]
- 42.Ludewig U, Wilken S, Wu B, Jost W, Obrdlik P, El BM, Marini AM, Andre B, Hamacher T, Boles E, et al. J Biol Chem. 2003;278:45603–45610. doi: 10.1074/jbc.M307424200. [DOI] [PubMed] [Google Scholar]
- 43.Peng J, Huang CH. Transfus Clin Biol. 2006;13:85–94. doi: 10.1016/j.tracli.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 44.Nygaard TP, Rovira C, Peters GH, Jensen MO. Biophys J. 2006;91:4401–4412. doi: 10.1529/biophysj.106.089714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamoureux G, Klein ML, Berneche S. Biophys J. 2007;92:L82–L84. doi: 10.1529/biophysj.106.102756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bostick DL, Brooks CL., III PLoS Comput Biol. 2007;3:e22. doi: 10.1371/journal.pcbi.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang H, Xu Y, Zhu W, Chen K, Jiang H. Biophys J. 2007;92:877–885. doi: 10.1529/biophysj.106.090191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luzhkov VB, Almlof M, Nervall M, Aqvist J. Biochemistry. 2006;45:10807–10814. doi: 10.1021/bi0610799. [DOI] [PubMed] [Google Scholar]
- 49.Lin Y, Cao Z, Mo Y. J Am Chem Soc. 2006;128:10876–10884. doi: 10.1021/ja0631549. [DOI] [PubMed] [Google Scholar]
- 50.Bostick DL, Brooks CL., III Biophys J. 2007;92:L103–L105. doi: 10.1529/biophysj.107.109165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishikita H, Knapp EW. J Am Chem Soc. 2007;129:1210–1215. doi: 10.1021/ja066208n. [DOI] [PubMed] [Google Scholar]
- 52.Planelles G. Nephron Physiol. 2007;105:11–17. doi: 10.1159/000096979. [DOI] [PubMed] [Google Scholar]
- 53.Ludewig U. J Physiol. 2004;559:751–759. doi: 10.1113/jphysiol.2004.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mak DO, Dang B, Weiner ID, Foskett JK, Westhoff CM. Am J Physiol Renal Physiol. 2006;290:F297–F305. doi: 10.1152/ajprenal.00147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruce LJ, Beckmann R, Ribeiro ML, Peters LL, Chasis JA, Delaunay J, Mohandas N, Anstee DJ, Tanner MJ. Blood. 2003;101:4180–4188. doi: 10.1182/blood-2002-09-2824. [DOI] [PubMed] [Google Scholar]
- 56.Van Kim CL, Colin Y, Cartron JP. Blood Rev. 2006;20:93–110. doi: 10.1016/j.blre.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 57.Thornton J, Blakey D, Scanlon E, Merrick M. FEMS Microbiol Lett. 2006;258:114–120. doi: 10.1111/j.1574-6968.2006.00202.x. [DOI] [PubMed] [Google Scholar]
- 58.Bruns CM, Hubatsch I, Ridderstrom M, Mannervik B, Tainer JA. J Mol Biol. 1999;288:427–439. doi: 10.1006/jmbi.1999.2697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.