Abstract
Intramolecular disulfide bonds are understood to play a role in regulating protein stability and activity. Because disulfide bonds covalently link different components of a protein, they influence protein structure. However, the effects of disulfide bonds on fast (subpicosecond to ≈100 ps) protein equilibrium structural fluctuations have not been characterized experimentally. Here, ultrafast 2D-IR vibrational echo spectroscopy is used to examine the constraints an intramolecular disulfide bond places on the structural fluctuations of the protein neuroglobin (Ngb). Ngb is a globin family protein found in vertebrate brains that binds oxygen reversibly. Like myoglobin (Mb), Ngb has the classical globin fold and key residues around the heme are conserved. Furthermore, the heme-ligated CO vibrational spectra of Mb (Mb-CO) and Ngb (Ngb-CO) are virtually identical. However, in contrast to Mb, human Ngb has an intramolecular disulfide bond that affects its oxygen affinity and protein stability. By using 2D-IR vibrational echo spectroscopy, we investigated the equilibrium protein dynamics of Ngb-CO by observing the CO spectral diffusion (time dependence of the 2D-IR line shapes) with and without the disulfide bond. Despite the similarity of the linear FTIR spectra of Ngb-CO with and without the disulfide bond, 2D-IR measurements reveal that the equilibrium sampling of different protein configurations is accelerated by disruption of the disulfide bond. The observations indicate that the intramolecular disulfide bond in Ngb acts as an inhibitor of fast protein dynamics even though eliminating it does not produce significant conformational change in the protein's structure.
Keywords: FTIR, neuroglobin, ultrafast 2D-IR
Intramolecular covalent disulfide bonds take part in the regulation of protein folding, stability, and activity (1–3). In addition, several studies have indicated that intramolecular disulfide bonds act as a regulatory switch in cells. In bacterial transcription factor OxyR, reversible disulfide bond formation induces protein conformational changes and acts as a transcriptional switch (4). Redox-dependent cysteine oxidation systems have been proposed for mammalian cells (2). Disulfide-dependent structural configurations have been investigated by NMR (5–7). Because the disulfide bonds are structural elements, local and global protein structural dynamics can be regulated by such bonds. Several molecular dynamics (MD) simulation studies have elucidated the importance of disulfide bonds on the protein motions (8, 9). However, the effects of the bonds on fast (subpicosecond to ≈100 ps) protein dynamics have not been directly characterized experimentally.
To explicate the influence of a disulfide bond on the fast protein structural dynamics, we investigated human neuroglobin (Ngb). Ngb is a recently discovered family of vertebrate globin proteins that are expressed in the brain and other nerve tissues (10). The 3D structures of Ngb from human and mouse have been published (11–13). The structure for Ngb is similar to that of myoglobin (Mb), although Ngb shows only minor correspondence at the amino acid level to Mb (10). An interesting feature for Ngb is that structural analysis has shown the presence in human Ngb of an intramolecular disulfide bond (14, 15), although the 3D structure of human Ngb was solved for the mutant protein with serines substituted for cysteines (11–13). The heme iron in both the ferrous and ferric forms of Ngb is hexacoordinated (11, 12), in contrast to mammalian Mb and Hb, which contain pentacoordinated heme iron. Although hexacoordinated heme has been reported in plant, bacteria, and invertebrate globins, its physiological significance is not yet understood (16). In Ngb, an external gaseous ligand must compete with the sixth ligand, the distal histidine (E7 in helix notation), for binding.
Eliminating the disulfide bond by substitution of the cysteine residues by serine or by reduction of the cysteines in human Ngb lowers the O2 affinity in vitro (15). Although the physiological role of the disulfide bond in human Ngb is not clear, hypoxia may induce the reduction of the disulfide bond and result in a subsequent release of O2 (15). The affinity of sixth heme ligand, the distal histidine, is also affected by the formation of the disulfide bond (15). The disruption of the disulfide bond in the ferric form of human Ngb or the reduction of the heme iron in mouse Ngb result in higher thermal stability. The increased stability is explained by a change in the affinity of the distal histidine for the heme iron (17). These results suggest that the formation of the intramolecular disulfide bond stresses the protein, and breaking the disulfide bond provides additional structural degrees of freedom of the protein, resulting in an increased affinity for O2 binding.
In this study, the effect of the intramolecular disulfide bond in human Ngb on fast protein dynamics is examined by using ultrafast 2D-IR vibrational echo spectroscopy, which has been used to measure heme protein equilibrium structural fluctuations on time scales ranging from subpicosecond to 100 ps (18–21). 2D-IR experiments have also been used to examine structure and dynamics in a variety of nonheme proteins (20–23). For heme proteins, 2D-IR vibrational echo experiments use the heme-ligated CO vibration as a direct sensor of protein dynamics by reporting on spectral diffusion (18, 24). The IR linear absorption spectrum of the CO-stretching mode of heme proteins generally displays several bands. The different bands reflect structural differences, that is, distinct structural substates, and the width of the bands reflect the range of CO vibrational energies that are associated with the distribution of protein structural configurations of each substate (18, 25). The linear IR absorption spectrum cannot provide information on a protein's structural dynamics. The dynamical information is obtained from the time evolution of the 2D-IR line shapes that are acquired with the vibrational echo experiments (24).
The FTIR spectra of CO bound to the ferrous heme iron of human Ngb were essentially unaffected by the reduction of the disulfide bond. However, 2D-IR measurements on the Ngb-CO revealed that the structural fluctuations of Ngb are constrained by the formation of the disulfide bond. When the disulfide bond is removed, the rates of fast protein structural fluctuations increase. Because the distance between the disulfide bond and heme iron is ≈20 Å, the changes in dynamics in Ngb are not induced by the direct interaction of the oxidized cysteine residues with the CO bound at the active site. Detailed analysis of the time-dependent changes in the 2D-IR vibrational echo spectra for Ngb with and without the disulfide bond demonstrate that the fast global equilibrium structural fluctuations of the protein are influenced by the presence of the disulfide bond.
Results and Discussion
Linear FTIR Spectroscopy.
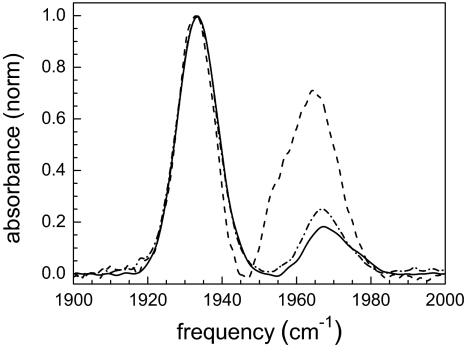
The background-subtracted linear FTIR spectra of CO bound to human Ngb with the disulfide bond (wt-Ngb), with the disulfide bond eliminated by reduction (red-Ngb), and the mutant Ngb with disulfide bond eliminated because the three cysteines are replaced by serines (CCC → SSS) (3cs-Ngb) are shown in Fig. 1. The cysteine 120 is at the surface of the protein and does not participate in the disulfide bond (15). However, we studied the 3cs-Ngb mutant because ligand binding and denaturation studies were performed on it (15, 17). In the spectrum of wt-Ngb (solid curve), there are two CO bands at 1,932 cm−1 and 1,968 cm−1. These have been called the N3 (lower frequency) and N0 (higher frequency) conformers, respectively, in analogy to the A3 and A0 bands of Mb (26–28). The significant width of the CO-stretching bands of Ngb and their Gaussian shapes implies that there is structural heterogeneity associated with each protein substate. The heterogeneity is sensed by the CO bound to the active site. Although the range of frequencies that make up a substate's absorption band is sensitive to the details of the heme pocket, it is also determined by the global structural variations of the protein (29). The frequency of N3 corresponds closely to the Mb-CO A3 conformation, which has the distal histidine localized in the heme pocket (29), whereas the frequency of N0 is almost identical to the Mb-CO A0 conformer, which has the distal histidine rotated out of the pocket (30). Recent 2D-IR vibrational echo experiments on wt-Ngb confirm that the bands are inhomogeneously broadened and report the dynamics of the N3 and N0 bands (28). The dynamics are considerably slower than those of the corresponding A3 and A0 bands of Mb-CO, but are consistent with the assignments of the N3 and N0 bands having the distal histidine in and out of the pocket, respectively. The 2D-IR spectra showed that the N0 band is actually composed of two substates, called N′0 and N0″, which overlap to such an extent that they are not resolved in the linear FTIR spectrum (28).
Fig. 1.
Normalized FTIR spectra of the CO-stretching mode bound to wt-Ngb (solid curve), red-Ngb (dot-dash curve), and 3cs-Ngb (dashed curve) in 50 mM phosphate buffer adjusted to pD 7.2.
The spectrum of red-Ngb (dot-dash curve) is almost identical to that of wt-Ngb. By using the same method that was discussed in detail for wt-Ngb (28), the 2D-IR spectrum shows that the N0 band is also composed of two substates. There may be a slight difference in the amplitudes of the N′0 and N0″ substates, but within experimental error any difference is not definitive. Experiments, theory, and MD simulations have shown that both the local and global coupling between protein structure and the CO vibrational transition frequency is mainly produced by the electric field from the protein along the CO bond (26–32). The protein residues range from charged, to polar, to nonpolar. All of these, plus to some extent the solvent, contribute to the electric field at the CO. The vibrational states couple to the electric field by the Stark effect (32). Differences in structure produce changes in the electric field at the CO and, therefore, changes in frequency. The range of frequencies that give rise to the inhomogeneous line widths of the CO bands is determined by the range of structures that produce the electric fields at the CO. The fact that there is negligible difference in the spectra of wt-Ngb and red-Ngb demonstrates that the disruption of the disulfide bond in CO adducts has no detectable influence on the distribution of protein equilibrium structures. In the case of a mutant Mb containing an artificial intramolecular disulfide bond, the formation of the disulfide bond also shows minor structural perturbation of the distribution of environments sensed by the CO ligated at the active site, but the ligand binding rate constant is changed (33). The change in the binding rate in the mutant Mb protein indicates that the protein dynamics are affected. 2D-IR vibrational echoes monitor the interconversion of the structures and provide a direct measurement of the protein structural dynamics as discussed below.
3cs-Ngb, which cannot form a disulfide bond, has significant differences in its spectrum. The N3 substate is still the major conformation, but the population of N0 substate is substantially increased. In addition, the 3cs-N0 peak is shifted somewhat to lower frequency (see Table 1), and there is a clear shoulder on the low frequency side of the line. Calling the two peaks the N′0 and N0″ substates as in the wt-Ngb and red-Ngb, the 3cs-N′0 peak is only ≈10% of the 3cs-N0″ (see Table 1). This is in contrast to the N′0 and N0″ peaks of both wt-Ngb and red-Ngb, which are approximately the same size. Because the difference between the N3 and N0 spectroscopic bands is attributed to the configuration of the distal histidine (27, 28), apparently the cysteine mutation(s) influence the heme pocket structure of Ngb and alter the distribution of the conformations, both between the main substates, the N3 and N0, and between N′0 and N0″. The exact reason for the difference in the FTIR spectrum between red-Ngb and 3cs-Ngb is not clear, but the 3D structure and MD simulation of Ngb imply that the configuration of CD-D region, including the cysteine residues in the disulfide bond, has an effect on the distal pocket (12, 34). Therefore, replacing the cysteines with serines changes the relative free energy of the N3 and N0 states (relative amplitude of the spectral peaks) and affects the pocket structure when the distal histidine is swung out (frequency of the N0 state and relative amplitudes of N′0 and N0″). The fact that the N3 frequency is not changed within experimental error suggests that the modification in structure is more pronounced with the distal histidine out of the pocket.
Table 1.
Absorption spectra parameters and lifetimes
| Protein/band | νCO, cm−1* | FWHM, cm−1 | T1, ps |
|---|---|---|---|
| wt-Ngb/N3† | 1,933.4 (84) | 12.1 | 19.3 |
| red-Ngb/N3 | 1,933.2 (80) | 12.4 | 16.0 |
| 3cs-Ngb/N3 | 1,932.7 (51) | 11.1 | 15.9 |
| wt-Ngb/N′0 and N0″† | 1,968.7 (16) | 9.8 | 18.4 |
| red-Ngb/N′0 and N0″ | 1,967.3 (20) | 12.4 | 16.1 |
| 3cs-Ngb/N′0 | 1,954.2 (5) | 6.4 | – |
| 3cs-Ngb/N0″ | 1,964.7 (44) | 14.6 | 16.0 |
wt, wild type; red, disulfide bond reduced; 3cs, disulfide bond removed by mutation.
*The values in parentheses indicate the populations of conformers.
†From ref. 28.
2D-IR Vibrational Echo Spectroscopy.
The structural evolution of a protein is reported by the effects of structural changes on the frequency of the CO vibrational mode. The experiment can be viewed qualitatively as follows. The first and second pulses, separated by a time τ that is scanned, act to label the initial frequencies of the COs. Between the second and third pulses (the time period, Tw), structural evolution of the proteins causes the initially labeled frequencies to change (spectral diffusion). The third pulse ends the evolution period, and the vibrational echo pulse, which emerges from the sample a time ≤τ after the third pulse, reads out the final frequencies. Fourier transforms of the time-dependent data produce a 2D frequency domain spectrum. Spectra are recorded as a function of Tw. As Tw increases there is more time for structural evolution, and, therefore, larger frequency changes. The frequency changes are reflected in the change in shapes of the 2D-IR vibrational echo spectra. More details are given in Materials and Methods, and full descriptions of the method have been presented (24, 35, 36).
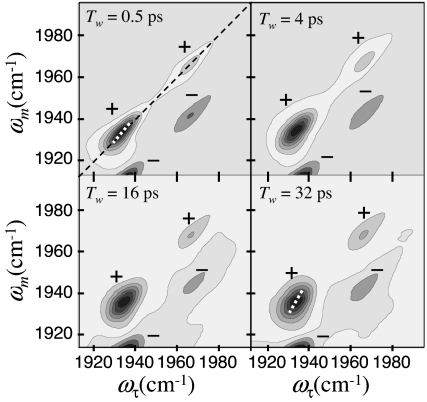
2D-IR vibrational echo spectra of red-Ngb for several increasing values of Tw are presented in Fig. 2 as contour plots. The ωτ frequency axis is associated with the first interaction with the radiation field (first IR pulse), and the ωm frequency axis is associated with the third interaction with the radiation field (third IR pulse), which is the emission frequency of the vibrational echo pulse. (ωτ and ωm are the equivalent of ω1 and ω3 in 2D NMR.) Two positive bands (labeled +) along the diagonal at (ωτ, ωm) = (1,933 cm−1, 1,933 cm−1) and (1,968 cm−1, 1,968 cm−1) arise from the 0 to 1 vibrational transitions of the CO stretch of the N3 and N0 states of wt-Ngb, respectively. (The N′0 and N0″ bands overlap to such an extent that it is not possible to distinguish their dynamics. Therefore, the N0 band is treated as a single substate.) The negative peaks (labeled −) arise from the 1 to 2 transitions and are displaced below the corresponding diagonal bands by the CO vibrational anharmonicity of ≈25 cm−1 (37). The dynamical information obtained from the 1–2 bands is the same as that obtained from the 0–1 bands; therefore, only the 0–1 bands are analyzed below.
Fig. 2.
2D-IR spectra of CO-bound red-Ngb. Each contour corresponds to a 10% signal change. The positive going peaks are labeled with + (vibrational echo emission at the 0–1 transition frequency), and the negative going peaks are labeled with − (vibrational echo emission at the 1–2 transition frequency). Conditions are 50 mM phosphate buffer adjusted to pD 7.2. The white dashed lines are the center lines.
As can be seen in Fig. 2, the shapes of the 0–1 bands (bands on the diagonal) change with increasing Tw. The change in shape of the bands as Tw is increased reflects protein structural dynamics. At short Tw the 2D line shape shows significant inhomogeneous broadening, which manifests itself as elongation along the diagonal. As Tw is increased, the experiment picks up slower protein dynamics that broaden the bands along the ωτ axis and causes them to become more symmetrical. In the long time limit, all protein structures associated with a particular substate will be sampled, and the 2D spectral bands will become round.
The changes in the shapes of the 2D bands with increasing Tw can be used to quantitatively determine the time scales and amplitudes of various contributions to the fast structural dynamics of the proteins by using methods based on diagrammatic perturbation theory (38). The frequency–frequency correlation function (FFCF) connects the experimental observables to the underlying dynamics. The FFCF is a joint probability distribution that the frequency has a certain initial value at t = 0 and another value at a later time t. As the structure evolves, the initial frequencies of the protein-bound COs change, and the FFCF decays. Once the FFCF is known, all linear and nonlinear optical experimental observables can be calculated by using time-dependent diagrammatic perturbation theory (38). Conversely, the FFCF can be extracted from 2D vibrational echo spectra with additional input from the linear absorption spectrum.
The FFCF is extracted from the Tw dependence of the 2D spectra by using the center line slope (CLS) method (39, 40). The CLS provides a useful quantity to plot for visualizing differences in the dynamics of the various proteins (39). In the CLS method, a slice through the 2D spectrum at a particular ωm is a spectrum along the ωτ axis. The peak of this spectrum is at a particular ωτ value. So the peak is a point with ωm, ωτ coordinates. Slices are taken over a range of ωm, and the resulting set of points form the center line. Two such center lines for N3 band are shown in Fig. 2 for Tw = 0.5 ps and 32 ps. In the absence of a homogeneous contribution to the spectrum (see below), at Tw = 0, the 2D line shape would be a line along the diagonal at 45° (Fig. 2 Upper Left, black dashed line). Then the slope of the center line would be 1. At very long time, the 2D line shape becomes symmetrical, and the center line is vertical (slope is infinite). The Tw-dependent part of the FFCF is equal to the Tw dependence of the inverse of the slope of the center line (39), which is referred to as the CLS. Therefore, the CLS will vary from a maximum of 1 at Tw = 0 to a minimum of 0 at sufficiently long time. In Fig. 2, it can be seen that for Tw = 32 ps the slope of the center line is steeper than at Tw = 0.5 ps. It has also been shown theoretically that combining the analysis of the CLS with the linear absorption spectrum, the full FFCF can be obtained, including the Tw-independent homogeneous component (39).
A homogeneous contribution to the 2D line shape and to the linear absorption line shape can arise from three sources: very fast structural fluctuations that produce a motionally narrowed contribution (24, 41); the vibration lifetime, T1; and orientational relaxation. Because the rotational diffusion of the protein is slow relative to the vibrational lifetime, the orientational relaxation contribution is negligible. The T1 contribution was measured independently by using IR pump–probe experiments. The T1 contribution to the homogeneous line width (1/π2T1) is quite small. It varies for the different bands, but it is ≈0.3 cm−1. The T1 values are listed in Table 1. The homogeneous contribution mainly comes from the motionally narrowed component. Motional narrowing occurs when there is some portion of the structural fluctuations that are extremely fast, such that Δτ < 1, where τ is the time scale of the fluctuations and Δ is the range (amplitude) of the frequency fluctuations. Motionally narrowed fluctuations produce a Lorentzian contribution to both the linear absorption spectrum and the 2D-IR spectrum. The Tw-independent homogeneous contribution manifests itself by broadening the 2D spectrum along the ωτ axis even at Tw = 0. This broadening reduces the initial value of the CLS to a number <1, which permits the determination of the homogeneous component (39).
A multiexponential form of the FFCF, C(t), was used to model the multi-time scale dynamics of the Ngb structural fluctuations. This form has been widely used, and it has been found applicable in studies of the structural dynamics of heme-CO proteins (18, 19, 24, 29), in particular, wt-Ngb (28). The FFCF has the form:
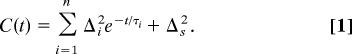 |
The Δi and τi terms are the amplitudes and correlation times, respectively, of the frequency fluctuations induced by protein structural dynamics. τi reflects the time scale of a set of structural fluctuations and the Δi is the range of CO frequencies sampled due to the structural fluctuations. The experimental time window is a few T1 values, because lifetime decay reduces the signal to zero. The 2D-IR vibrational echo experiment is sensitive to fluctuations a few times longer than this window (42), that is, ≈100 ps, because some portion of slower fluctuations will occur in the experimental window if their time scale is not too slow. Protein structural dynamics that are sufficiently slow will appear as static inhomogeneous broadening, which is reflected in C(t) by Δs, a static term. In obtaining the FFCF from the data, the Δi and the τi are determined. However, for a motionally narrow term (Δτ < 1), only the product, Δ2τ = 1/T2* can be obtained. T2* is called the pure dephasing time. The pure dephasing time is obtained by using 1/T2 = 1/T2* + 1/2T1, where T2 is determined from the CLS with use of the linear absorption spectrum (39), and T1 is obtained from IR pump–probe experiments (see Table 1).
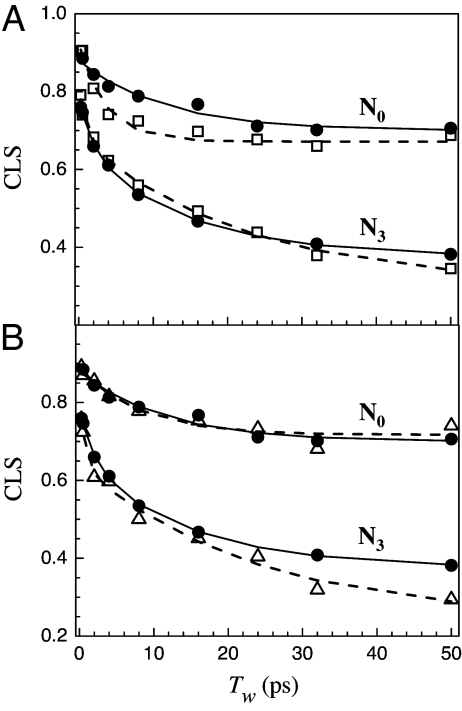
Fig. 3A shows a comparison of the 2D-IR vibrational echo CLS data for wt-Ngb (filled circles) and red-Ngb (open squares). The curves through the data points (solid, wt-Ngb; dashed, red-Ngb) are calculated from the FFCF obtained by using the CLS method. The calculated curves show the Tw-dependent portion of the FFCF. Fig. 3B again shows the wt-Ngb data and FFCF (filled circles, solid curve) and the 3cs-Ngb data and FFCF (open triangles, dashed curve). Table 2 gives all of the FFCF parameters for the N3 and N0 substates of the three proteins. Examining Fig. 3A, it is clear the reduction of the disulfide bond has increased the rate of structural fluctuations on fast time scales. Although the nature of the changes is different, both the N3 and N0 of the red-Ngb display faster decays than the wt-Ngb. For 3cs-Ngb, in which the disulfide bond is eliminated by mutation, the N3 substate shows a dramatic increase in the rate of fast structural fluctuations. The change in the N0 state is less pronounced, but there is still an increase in rate as shown in Table 2. It is important to note that the disulfide bond is ≈20 Å from the CO bound to the heme. Furthermore, there are changes in dynamics for both the N3 substate (distal histidine in the heme pocket) and for the N0 substate (distal histidine swung out of the heme pocket). Therefore, the observed changes in the dynamics with the elimination of the disulfide bond are most likely global modifications of the protein structural fluctuations rather alterations that occur very locally in the heme pocket.
Fig. 3.
Tw-dependent CLS for wt-Ngb (A and B, filled circles), red-Ngb (A, open squares), and 3cs-Ngb (B, open triangles). The solid (wt-Ngb) and dashed (A, red-Ngb; B, 3cs-Ngb) curves are calculated from the FFCFs obtained by using the CLS method.
Table 2.
FFCF parameters
| Protein/band | T2*, ps | Δ1, cm−1 | τ1, ps | Δ2, cm−1 | τ2, ps | Δs, cm−1 |
|---|---|---|---|---|---|---|
| wt-Ngb/N3* | 4.4 | 1.9 | 2.0 | 2.7 | 14 | 3.1 |
| red-Ngb/N3 | 6.0 | 3.0 | 1.3 | 3.3 | 22 | 2.9 |
| 3cs-Ngb/N3 | 6.0 | 2.0 | 0.7 | 3.0 | 23 | 2.3 |
| wt-Ngb/N0* | 9.0 | 1.8 | 11.5 | – | – | 3.5 |
| red-Ngb/N0 | 11.0 | 3.0 | 3.7 | – | – | 4.3 |
| 3cs-Ngb/N0 | 6.0 | 2.8 | 8.1 | – | – | 5.6 |
wt, wild type; red, disulfide bond reduced; 3cs, disulfide bond removed by mutation
*From ref. 28.
The FFCF parameters obtained from the 2D-IR vibrational echo experiments can be compared by examining Table 2. The homogeneous dephasing time constant, T2*, is not as useful as the other time constants because it depends on Δ2τ = 1/T2*. Therefore, the amplitude and the time constant cannot be separately determined. A change in T2* can occur because of a change in the amplitude (range) of the frequency fluctuations (Δ) or in the time constant (τ). The N3 substates of the three proteins have a homogeneously broadened component, two dynamic components that give rise to spectral diffusion within the time window of the experiment, and a static component, that is, some of the protein structures that contribute to the CO absorption line (≈25%) are sampled at rates that are too slow to measure. The time constant, τ1, becomes significantly faster when the disulfide bond is reduced and even faster when the three cysteines are replaced by serines. The τ2 component actually becomes somewhat slower when the disulfide bond is eliminated. This could occur if some portion of the very slow structural fluctuation, represented by the Δs term in the FFCF (see Eq. 1), became faster, and moved into the time window of observation. The N0 substates of all three proteins have a single dynamic component in addition to homogeneously broadened and static terms in the FFCF. For N0, in going from wt-Ngb to red-Ngb, there is a very substantial increase in the rate (1/τ1) of the fast protein structural fluctuations. The rate for 3cs-Ngb is also increased, but it is not nearly as large as that of the N3. Fig. 1 shows that the change in the N0 band in the linear spectrum for 3cs-Ngb is large in contrast to the N3 band or the bands of red-Ngb. The substitutions of cysteine residues may result in a substantial modification of the dynamics in the N0 conformer of 3cs-Ngb that is independent of the elimination of the disulfide bond. It is possible that this modification of the dynamics as sensed at the active site is in opposition to the influence of the elimination of the disulfide bond that is manifested by the change in the N0 substate dynamics in going from wt-Ngb to red-Ngb. A study has indicated that the ligand binding and distal histidine dissociation rate of Ngb are markedly changed by the disruption of the disulfide bond (15).
The differences between the results obtained for red-Ngb and 3cs-Ngb may be indicative of cysteine-mediated regulation of protein dynamics in the active site of Ngb. An MD simulation study indicated that the flexibility of the CD-D region is related to the dynamics of the heme distal site (34). The 3D structure of the CO adduct of Ngb also demonstrates that the contact between distal histidine and Tyr (CD3) is involved in the CD-D region displacement on the CO binding to the heme iron (12). The cysteine mutation(s) may change the flexibility or conformation of CD-D region and result in the changes in the FFCF parameters from those of red-Ngb, in which the cysteines are reduced by not replaced with serines and, in particular, cys120 is left unchanged. The relatively faster protein fluctuation of the N0 band for red-Ngb can be understood as occurring because of the structural restrictions of the global motion by the disulfide bond that are removed when the bond is eliminated by reduction without other changes. MD simulation of Scorpion Toxin Lqh III showed that the disulfide bond constrains the protein's fluctuations both locally and globally (9).
In a previous 2D-IR vibrational echo study, the dynamics of the N3 and N0 substates of wt-Ngb were compared with equivalent substates of Mb by using the Mb mutants, L29F (distal histidine in the active-site pocket) and H64V (no distal histidine in the pocket) as models (28). Both substates of wt-Ngb were found to have substantially slower dynamics than the corresponding Mb proteins (28). The results obtained here show that the Ngb dynamics become faster when the disulfide bond is eliminated. However, even without the disulfide bond, the dynamics of Ngb substates are still substantially slower then the dynamics of the corresponding substates of Mb. Therefore, the slower dynamics found for Ngb compared with Mb are not solely a result of the presence of the disulfide bond, which is not present in Mb. Rather, the differences between Ngb and Mb arise from the fundamental disparity in the protein structures, although they both have the classic globin fold and almost identical heme pocket structures.
NMR relaxation studies of glutaredoxin and bovine pancreatic trypsin inhibitor show that disruption of the disulfide bond, which is near the active site, increases the protein motion, primarily in the vicinity of the disulfide bond (6, 7). The 3D structure of the transcriptional factor OxyR revealed that large conformational changes induced by the disruption of the disulfide bond resulted in activation of the transcription (4). In the case of Yap1, it is thought that the elimination of the intramolecular disulfide bond produces a conformational change that masks a nuclear export sequence, preventing the insertion of the protein into the nucleus (43). 3D structure of Ngb with the disulfide bond has not been determined; only the structure of a mutant Ngb without the disulfide bond is known (11–13). However, the perturbation of the static structure produced by reduction of the disulfide bond may be small. The linear FTIR spectra of the CO adducts of wt-Ngb and red-Ngb (see Fig. 1) are very similar, indicating that changes in structure as sensed by the CO ligand bound at the active site are small. Because the CO spectrum is sensitive to both local and global changes in structure through the changes in the electric field at the CO, reduction of the disulfide bond cannot produce major structural changes. However, as can be seen in Fig. 3 and Table 2, reduction of the disulfide bond produces nonnegligible changes in the structural dynamics of Ngb.
Disruption of the disulfide bond in Ngb lowers the O2 affinity in vitro (15). The disulfide bond's influence on the protein dynamics of Ngb might play a physiological role in human Ngb. The CD-D region of Ngb, including the disulfide bond, binds the GDP-bound form of the α-subunit of heterotrimeric G protein (Gαi), which is thought to be important in the neuroprotective function of human Ngb (44). Although the disruption of the disulfide bond in Ngb does not seem to be involved directly in the Gαi binding (44), the relationship between the structural dynamics, including the active site, and the conformational freedom in the CD-D region remains to be established. Gαi binding to Ngb may restrict protein fluctuations in the CD-D region, which result in changes in the active-site dynamics.
Concluding Remarks.
The fast equilibrium structural dynamics of wild-type neuroglobin and neuroglobin with the disulfide bond eliminated were examined by using ultrafast 2D-IR vibrational echo experiments on the stretching vibration of CO bound at the active site. The experiment measures CO spectral diffusion that reports on both local and global structural fluctuations of the proteins. The disulfide bond was eliminated either by reduction of the cysteines that form it or by mutation in which three cysteines, including the two that form the disulfide bond, are replaced by serines. The experimental results show an increase in the rates of structural fluctuations when the disulfide bond is removed. The most direct comparison is between wt-Ngb and red-Ngb because the amino acid sequence is left intact. Both the N3 and N0 substates of red-Ngb show significant increases in the rates of equilibrium structural fluctuation (see Fig. 3A and Table 2), although the IR absorption spectrum of the CO bound at the active site is unchanged by reduction. The mutant, 3cs-Ngb, shows a substantial increased rate of fast dynamics for the N3 substate, but less change for the N0 substate (see Fig. 3B and Table 2). There is a significant change in the IR absorption spectrum (see Fig. 1), with the N0 band increasing in amplitude relative to the N3 band and shifting somewhat to lower frequency.
One way to view the changes of the Ngb dynamics with the reduction of the disulfide bond is in terms of the protein's energy landscape. wt-Ngb and red-Ngb have two substates, N3 and N0. A folded protein in a particular substate occupies a minimum on the free-energy landscape (45). However, the minimum is broad and rough with many local minima separated by low barriers. Transitions among these minima give rise to the structural fluctuations that cause the spectral diffusion measured by the 2D-IR vibrational echo experiment. The distribution of barrier heights gives rise to the distribution of rates of spectral diffusion that is reflected in the different terms in the FFCF. When the disulfide bond is eliminated, the rate of fast structural fluctuations is increased. However, reduction of the disulfide bond does not change the IR absorption spectrum of the CO bound at the active site, which is sensitive to both the local and the global structure of the protein. The lack of change in the CO absorption spectrum indicates that the distribution of structures in the vicinity of the folded protein free-energy minimum that are sampled under thermal equilibrium conditions are not altered significantly. These results suggest that the elimination of the disulfide bond reduces the heights of the relatively low-energy landscape barriers that determine the rate of the fast structural fluctuations.
Materials and Methods
Protein Expression and Purification.
Expression and purification of human His6-tagged Ngb and the mutant protein were performed as described in ref. 44. Purity of proteins and contamination of the disulfide-dependent formation of dimmers were checked by SDS/PAGE under reduced and nonreduced conditions. The presence of the intramolecular disulfide bond was checked by using the 4,4′-dithiodipyridine (4-PDS) method (46). The Ngb samples, containing the intramolecular disulfide bond, were reduced to the ferrous form with a small excess of sodium dithionite (Sigma Aldrich) under a CO atmosphere followed by buffer exchange to CO-saturated 50 mM phosphate D2O buffer adjusted to pD 7.2.
To break the intramolecular disulfide bond between cysteine residues in Ngb, the purified sample was incubated with 10 mM DTT (Sigma Aldrich) for 24 h. CO forms of Ngb without the disulfide bond and the mutant protein were prepared according to published protocols (29). Ngb without the disulfide bond sample contains 1 mM DTT to avoid the formation of disulfide bond during the measurements. For linear FTIR spectra and vibrational echo measurements, ≈20 μl of the sample solution was placed in a sample cell with CaF2 windows and a 50-μm Teflon spacer.
2D-IR Vibrational Echo Spectroscopy.
Three transform limited (110 fs, 150 cm−1 FWHM) mid-IR laser pulses produced with a Ti:Sapphire regenerative amplifier pumped optical parametric oscillator are sequentially time-delayed before they are brought into the sample. The vibrational echo pulse generated in the sample is emitted in the phase-matched direction, made colinear with a local oscillator pulse, and dispersed through a monochromator onto a 32-element MCT array detector. The signal is mixed with the local oscillator to obtain full time, frequency, and phase information about the vibrational echo wave packet. The laser frequency was tuned to coincide with the absorption frequencies of the proteins.
For each protein, a family of vibrational echo 2D spectra is obtained as a function of three variables: the emitted vibrational echo frequencies, ωm, and the variable time delays between the first and second pulses (τ) and the second and third pulses (Tw, “waiting” time). 2D spectra are created by numerically Fourier transforming the τ scan data as a function of the emission frequencies, ωm, at each Tw. The amplitude of the 2D-IR data at each fixed Tw is plotted as a function of the Fourier transformed τ scan interferograms, ωτ (horizontal axis in the 2D plots), and emission frequency, ωm (vertical axis of the 2D plots) (18, 24, 47, 48).
Acknowledgments
We thank Prof. R. Kopito, Stanford University, for kind permission to use his protein expression and purification equipment. We also thank Dr. J. Christianson and Dr. C. Patel, Stanford University, for invaluable assistance with sample preparation. This work was supported by the National Institutes of Health Grant 2 R01 GM-061137-05. H. I. was supported by the Human Frontier Science Program. S. K. was supported by a fellowship from the Korea Research Foundation Grant funded by the Korean Government Grant KRF-2006-214-C00038.
Footnotes
The authors declare no conflict of interest.
References
- 1.Barford D. Curr Opin Struct Biol. 2004;14:679–686. doi: 10.1016/j.sbi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 2.Liu H, Colavitti R, Rovira II, Finkel T. Circ Res. 2005;97:967–974. doi: 10.1161/01.RES.0000188210.72062.10. [DOI] [PubMed] [Google Scholar]
- 3.Hogg PJ. Trends Biochem Sci. 2003;28:210–214. doi: 10.1016/S0968-0004(03)00057-4. [DOI] [PubMed] [Google Scholar]
- 4.Kim SO, Merchant K, Nudelman R, Beyer WF, Jr, Keng T, DeAngelo J, Hausladen A, Stamler JS. Cell. 2002;109:383–396. doi: 10.1016/s0092-8674(02)00723-7. [DOI] [PubMed] [Google Scholar]
- 5.Betz SF, Marmorino JL, Saunders AJ, Doyle DF, Young GB, Pielak GJ. Biochemistry. 1996;35:7422–7428. doi: 10.1021/bi9528558. [DOI] [PubMed] [Google Scholar]
- 6.Beeser SA, Oas TG, Goldenberg DP. J Mol Biol. 1998;284:1581–1596. doi: 10.1006/jmbi.1998.2240. [DOI] [PubMed] [Google Scholar]
- 7.Kelley JJ, III, Caputo TM, Eaton SF, Laue TM, Bushweller JH. Biochemistry. 1997;36:5029–5044. doi: 10.1021/bi962181g. [DOI] [PubMed] [Google Scholar]
- 8.Tidor B, Karplus M. Proteins. 1993;15:71–79. doi: 10.1002/prot.340150109. [DOI] [PubMed] [Google Scholar]
- 9.Moghaddam ME, Naderi-Manesh H. Proteins. 2006;63:188–196. doi: 10.1002/prot.20850. [DOI] [PubMed] [Google Scholar]
- 10.Burmester T, Weich B, Reinhardt S, Hankeln T. Nature. 2000;407:520–523. doi: 10.1038/35035093. [DOI] [PubMed] [Google Scholar]
- 11.Pesce A, Dewilde S, Nardini M, Moens L, Ascenzi P, Hankeln T, Burmester T, Bolognesi M. Structure (London) 2003;11:1087–1095. doi: 10.1016/s0969-2126(03)00166-7. [DOI] [PubMed] [Google Scholar]
- 12.Vallone B, Nienhaus K, Brunori M, Nienhaus GU. Proteins. 2004;56:85–92. doi: 10.1002/prot.20113. [DOI] [PubMed] [Google Scholar]
- 13.Vallone B, Nienhaus K, Matthes A, Brunori M, Nienhaus GU. Proc Natl Acad Sci USA. 2004;101:17351–17356. doi: 10.1073/pnas.0407633101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinck E, Van Doorslaer S, Dewilde S, Moens L. J Am Chem Soc. 2004;126:4516–4517. doi: 10.1021/ja0383322. [DOI] [PubMed] [Google Scholar]
- 15.Hamdane D, Kiger L, Dewilde S, Green BN, Pesce A, Uzan J, Burmester T, Hankeln T, Bolognesi M, Moens L, et al. J Biol Chem. 2003;278:51713–51721. doi: 10.1074/jbc.M309396200. [DOI] [PubMed] [Google Scholar]
- 16.Kundu S, Trent JT, III, Hargrove MS. Trends Plants Sci. 2003;8:387–393. doi: 10.1016/S1360-1385(03)00163-8. [DOI] [PubMed] [Google Scholar]
- 17.Hamdane D, Kiger L, Dewilde S, Uzan J, Burmester T, Hankeln T, Moens L, Marden MC. FEBS J. 2005;272:2076–2084. doi: 10.1111/j.1742-4658.2005.04635.x. [DOI] [PubMed] [Google Scholar]
- 18.Finkelstein IJ, Ishikawa H, Kim S, Massari AM, Fayer MD. Proc Natl Acad Sci USA. 2007;104:2637–2642. doi: 10.1073/pnas.0610027104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massari AM, Finkelstein IJ, Fayer MD. J Am Chem Soc. 2006;128:3990–3997. doi: 10.1021/ja058745y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung HS, Khalil M, Smith AW, Ganim Z, Tokmakoff A. Proc Natl Acad Sci USA. 2005;102:612–617. doi: 10.1073/pnas.0408646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukherjee P, Kass I, Arkin IT, Zanni MT. Proc Natl Acad Sci USA. 2006;103:3528–3533. doi: 10.1073/pnas.0508833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volkov V, Hamm P. Biophys J. 2004;87:4213–4225. doi: 10.1529/biophysj.104.045435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang C, Hochstrasser RM. J Phys Chem B. 2005;109:18652–18663. doi: 10.1021/jp052525p. [DOI] [PubMed] [Google Scholar]
- 24.Finkelstein IJ, Zheng J, Ishikawa H, Kim S, Kwak K, Fayer MD. Phys Chem Chem Phys. 2007;9:1533–1549. doi: 10.1039/b618158a. [DOI] [PubMed] [Google Scholar]
- 25.Hartmann H, Parak F, Steigemann W, Petsko GA, Ponzi DR, Frauenfelder H. Proc Natl Acad Sci USA. 1982;79:4967–4971. doi: 10.1073/pnas.79.16.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morikis D, Champion PM, Springer BA, Sligar SG. Biochemistry. 1989;28:4791–4800. doi: 10.1021/bi00437a041. [DOI] [PubMed] [Google Scholar]
- 27.Sawai H, Makino M, Mizutani Y, Ohta T, Sugimoto H, Uno T, Kawada N, Yoshizato K, Kitagawa T, Shiro Y. Biochemistry. 2005;44:13257–13265. doi: 10.1021/bi050997o. [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa H, Finkelstein IJ, Kim S, Kwak K, Chung JK, Wakasugi K, Massari AM, Fayer MD. Proc Natl Acad Sci USA. 2007;104:16116–16121. doi: 10.1073/pnas.0707718104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merchant KA, Noid WG, Akiyama R, Finkelstein I, Goun A, McClain BL, Loring RF, Fayer MD. J Am Chem Soc. 2003;125:13804–13818. doi: 10.1021/ja035654x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Augspurger JD, Dykstra CE, Oldfield E. J Am Chem Soc. 1991;113:2447–2451. [Google Scholar]
- 31.Spiro TG, Wasbotten IH. J Inorg Biochem. 2005;99:34–44. doi: 10.1016/j.jinorgbio.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 32.Park ES, Boxer SG. J Phys Chem B. 2002;106:5800–5806. [Google Scholar]
- 33.Uchida T, Unno M, Ishimori K, Morishima I. Biochemistry. 1997;36:324–332. doi: 10.1021/bi960591z. [DOI] [PubMed] [Google Scholar]
- 34.Anselmi M, Brunori M, Vallone B, Di Nola A. Biophys J. 2007;93:434–441. doi: 10.1529/biophysj.106.099648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng J, Kwak K, Fayer MD. Acc Chem Res. 2007;40:75–83. doi: 10.1021/ar068010d. [DOI] [PubMed] [Google Scholar]
- 36.Park S, Kwak K, Fayer MD. Laser Phys Lett. 2007;4:704–718. [Google Scholar]
- 37.Rector KD, Kwok AS, Ferrante C, Tokmakoff A, Rella CW, Fayer MD. J Chem Phys. 1997;106:10027. [Google Scholar]
- 38.Mukamel S. Annu Rev Phys Chem. 2000;51:691–729. doi: 10.1146/annurev.physchem.51.1.691. [DOI] [PubMed] [Google Scholar]
- 39.Kwak K, Park S, Finkelstein IJ, Fayer MD. J Chem Phys. 2007;127:124503. doi: 10.1063/1.2772269. [DOI] [PubMed] [Google Scholar]
- 40.Park S, Fayer MD. Proc Natl Acad Sci USA. 2007;104:16731–16738. doi: 10.1073/pnas.0707824104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woutersen S, Pfister R, Hamm P, Mu Y, Kosov DS, Stock G. J Chem Phys. 2002;117:6833–6840. [Google Scholar]
- 42.Bai YS, Fayer MD. Phys Rev B. 1989;39:11066. doi: 10.1103/physrevb.39.11066. [DOI] [PubMed] [Google Scholar]
- 43.Wood MJ, Storz G, Tjandra N. Nature. 2004;430:917–921. doi: 10.1038/nature02790. [DOI] [PubMed] [Google Scholar]
- 44.Wakasugi K, Nakano T, Morishima I. J Biol Chem. 2003;278:36505–36512. doi: 10.1074/jbc.M305519200. [DOI] [PubMed] [Google Scholar]
- 45.Frauenfelder H. Nat Struct Biol. 1995;2:821–823. doi: 10.1038/nsb1095-821. [DOI] [PubMed] [Google Scholar]
- 46.Fago A, Hundahl C, Dewilde S, Gilany K, Moens L, Weber RE. J Biol Chem. 2004;279:44417–44426. doi: 10.1074/jbc.M407126200. [DOI] [PubMed] [Google Scholar]
- 47.Asbury JB, Steinel T, Fayer MD. J Lumin. 2004;107:271–286. doi: 10.1016/j.jlumin.2003.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zanni MT, Gnanakaran S, Stenger J, Hochstrasser RM. J Phys Chem B. 2001;105:6520–6535. [Google Scholar]