Abstract
Protein synthesis in the chloroplast is carried out by chloroplast ribosomes (chloro-ribosome) and regulated in a light-dependent manner. Chloroplast or plastid ribosomal proteins (PRPs) generally are larger than their bacterial counterparts, and chloro-ribosomes contain additional plastid-specific ribosomal proteins (PSRPs); however, it is unclear to what extent these proteins play structural or regulatory roles during translation. We have obtained a three-dimensional cryo-EM map of the spinach 70S chloro-ribosome, revealing the overall structural organization to be similar to bacterial ribosomes. Fitting of the conserved portions of the x-ray crystallographic structure of the bacterial 70S ribosome into our cryo-EM map of the chloro-ribosome reveals the positions of PRP extensions and the locations of the PSRPs. Surprisingly, PSRP1 binds in the decoding region of the small (30S) ribosomal subunit, in a manner that would preclude the binding of messenger and transfer RNAs to the ribosome, suggesting that PSRP1 is a translation factor rather than a ribosomal protein. PSRP2 and PSRP3 appear to structurally compensate for missing segments of the 16S rRNA within the 30S subunit, whereas PSRP4 occupies a position buried within the head of the 30S subunit. One of the two PSRPs in the large (50S) ribosomal subunit lies near the tRNA exit site. Furthermore, we find a mass of density corresponding to chloro-ribosome recycling factor; domain II of this factor appears to interact with the flexible C-terminal domain of PSRP1. Our study provides evolutionary insights into the structural and functional roles that the PSRPs play during protein synthesis in chloroplasts.
Keywords: chloroplast protein synthesis, cryo-EM structure, ribosomal RNA, plastid ribosome recycling factor, protein synthesis
Cellular organelles, like mitochondria and chloroplasts, possess their own genome and components of gene-expression system, including ribosomes (for reviews, see refs. 1 and 2). The chloroplast (or plastid) ribosome (henceforth referred to as the “chloro-ribosome”) is specialized in that it is involved in the biosynthesis of only the small number of proteins encoded by the chloroplast genome. The complete genome sequence of the spinach (Spinacea oleracea) chloroplast identified a total of 146 genes, of which 98 encode protein products, and 45 represent structural RNAs (3). The majority of the chloroplast-encoded proteins is targeted to the thylakoid membranes and include components of the ATP synthase, cytochrome b/f, and especially photosystem I and II complexes (3). In addition, chloro-ribosomes translate NADH dehydrogenase, the large subunit of RuBisCO, RNA polymerase subunits, and a distinct subset of ribosomal proteins: 12 from the small (30S) subunit and 8 from the large (50S) subunit, and the rest are nuclear-encoded and imported into the chloroplast.
The origin of chloroplasts is understood to be through an early endosymbiotic event between a photosynthetic prokaryotic ancestor related to cyanobacteria and a eukaryotic cell (4). In fact, the overall constituents of the plastid translational machinery are similar to those of eubacteria. The ribosomal RNAs (rRNAs) are similar in length and exhibit relatively few differences [see supporting information (SI) Figs. 6–8]. The 16S rRNAs of the Escherichia coli 30S subunit has 1,542 nt, whereas the spinach chloro-30S subunit is slightly smaller, containing 1,491 nt. Although the chloro-50S subunit is composed of three rRNAs, rather than two as in E. coli, the combined size of chloro-50S subunit rRNAs is larger (3,033 nt) than the E. coli counterpart (3,024 nt) by only 9 nt.
Proteomic studies reveal that the chloro-ribosomes possess in total 58 ribosomal proteins, 25 and 33 proteins in the 30S and 50S subunits, respectively (5, 6), and have orthologs to all 52 E. coli ribosomal proteins, with the exception of L25 and L30. Plastid ribosomal proteins (PRPs) generally are larger than their E. coli counterparts, containing extensions predominantly at the N and C termini. In addition, the chloro-ribosome possesses six nonorthologous proteins, which are termed “plastid-specific ribosomal proteins” (PSRPs): four of these proteins (PSRP1–4) are associated with the 30S subunit, and two (PSRP5 and PSRP6) are associated with the 50S subunit. The larger size of the PRPs as well as the presence of six PSRPs leads to a significantly altered protein/RNA mass ratio of 2/3 in the case of chloro-ribosomes, as compared with 1/3 for E. coli (5, 6). The higher protein content of chloro-ribosomes, and especially the presence of PSRPs, has prompted the suggestion that chloroplasts have evolved specific proteins that play unique functional roles during chloroplast translation, such as the light-dependent stimulation of protein synthesis (5–7).
Here, we present a 9.4-Å 3D cryo-electron microscopic (cryo-EM) reconstruction of the spinach 70S chloro-ribosome. The chloro-ribosome is similar to bacterial 70S ribosomes, but it is larger in size and exhibits a number of specific features not observed in the structures of cytoplasmic (e.g., ref. 8) or mammalian mitochondrial (9) ribosomes. Molecular analysis of the map reveals the positions of the extensions of the orthologous PRPs and allows us to assign the locations of five of the six PSRPs. In addition, we find a mass of density corresponding to the chloroplast (plastid) ribosome recycling factor (pRRF), which is known to be tightly associated with the chloro-ribosome (5, 7). Collectively, the results suggest that PSRPs play both functional and structural roles in the chloro-ribosome.
Results and Discussion
Overall Architecture of the 70S Chloro-Ribosome.
The 3D cryo-EM map of the 70S chloro-ribosome (Fig. 1 A and B) reveals known structural features of the bacterial ribosome (Fig. 1 C and D), such as the body, head, and platform within the 30S subunit and the central protuberance (CP) and L1 and L7/L12 stalks in the 50S subunit. In addition, we find a weak mass of density corresponding to tRNA in the E-site region that copurifies with the ribosomes as observed previously (e.g., refs. 10 and 11). However, a detailed analysis reveals a number of unique structural features in the chloro-ribosome compared with that of the E. coli 70S ribosome (8): (i) the larger overall size of the chloro-ribosome (by ≈10 Å along the longest diameter); (ii) the presence of a prominent extra mass of density at the spur of the 30S subunit; and (iii) much larger densities for protein L1 protuberance and protein L7/L12 stalk base in the 50S subunit.
Fig. 1.
Comparison of the cryo-EM maps of the spinach chloroplast 70S ribosome at 9.4-Å resolution and the E. coli 70S ribosome. (A and B) The chloro-ribosome with 30S subunit (yellow) and 50S subunit (green). (C and D) The E. coli 70S ribosome (8) with 30S subunit (pale yellow) and 50S subunit (blue). In A/C and B/D, ribosomes are shown from the L7/L12 and L1 sides, respectively. Structural differences of the 30S and 50S subunits are marked by red arrows. Landmarks of the 30S subunit: h, head; pt, platform; sh, shoulder; and sp, spur. Landmarks of the 50S subunit: CP, central protuberance; H38, 23S rRNA helix 38; L1, protein L1 protuberance; and Sb, stalk base.
We have computationally separated the RNA and protein components (12) within both subunits of the chloro-ribosome (Fig. 2). Comparison of rRNA and protein maps with the x-ray structures for the E. coli (13) and Thermus thermophilus 70S (11) ribosomes, as well as a spinach 70S homology model generated from these structures (see Materials and Methods), allowed us to locate the differences in rRNA and protein distributions between the chloroplast and the bacterial ribosomes. The rRNA-only map is in agreement with the spinach homology model in terms of the locations of nucleotide insertions, or deletions, relative to bacterial rRNAs (see SI Figs. 6–8 for secondary structure differences of rRNAs), whereas the protein-only maps reveal unique protein densities that can be assigned to five PSRPs, and the extensions of the orthologous PRPs, with possible exception to regions of disordered tails or loops in these proteins. Furthermore, the protein-only map computed for the chloro-50S subunit shows a strong mass of density corresponding to pRRF domain I, located in a similar position as the bacterial RRF (14, 15). The RNA–protein separations also show that the positions and compositions of intersubunit bridges in the chloro-ribosome are mostly conserved between the bacterial (8, 11, 13, 16) and chloro-ribosomes. One possible exception is the protein–protein bridge B1b, which appears to be slightly altered because of the presence of extensions of PRPs S13 and L5 on the 30S and 50S subunits, respectively, of the chloro-ribosome.
Fig. 2.
Distribution of RNA and protein on the chloro-ribosome with positions of PSRPs and PRP extensions highlighted. (A and B) 30S subunit: pale yellow, rRNA; yellow, PRPs; green, PRP extensions; and red, PSRPs. (C and D) 50S subunit: blue, rRNA; purple, PRPs, green, PRP extensions; and red, PSRP5. In A/C and B/D, subunits are shown from the interface and solvent sides, respectively. To avoid visual complexity, only densities corresponding to PRP extensions (with suffix “ext”), conformational changes (with suffix “c”), or shifts in the position of PRPs (with suffix “s”) with respect to their bacterial orthologs, and PSRPs (1–5), are labeled. Numbers following S or L identify small and large subunit PRPs, respectively, and numbers following h or H identify 16S and 23S rRNA helices within small and large subunits, respectively. Landmarks: b, body of the 30S subunit; pRRF, domain I of pRRF; St, L7/L12 stalk of the 50S subunit. All other landmarks are the same as in Fig. 1.
Structure of the Small (30S) Ribosomal Subunit.
The striking size and shape of the foot or “spur” feature distinguishes the chloro-30S ribosomal subunit from its E. coli counterpart (Fig. 2 A and B). The secondary structure maps of the 16S rRNA (see SI Fig. 6) indicate that the major differences with E. coli are in fact the absence of nucleotides in this region, specifically, in helix 6 (h6), which forms the spur, and h10 and h17, which lie in the immediate vicinity (see ref. 17). The RNA–protein separated cryo-EM maps of the chloro-30S ribosomal subunit are in agreement with the loss of rRNA from this region with respect to E. coli but also indicate that a significant protein mass is present here. To assign the protein masses of the chloro-ribosome, a systematic assignment of all of the chloro-ribosome-specific PRP extensions and PSRP was undertaken. Most of the orthologous PRPs of the chloro-30S ribosomal subunit are larger in size when compared with their bacterial counterparts, generally possessing N-terminal extensions (NTEs) or C-terminal extensions (CTEs) (5). The homology model and the analysis of the protein-only mass of the 30S cryo-EM map allow us to assign the density located near to where the extensions of PRPs would be expected to be, for example, PRPs S5, S6, S9, S10, S13, S17, S18, S20, and S21 (Fig. 2 A and B).
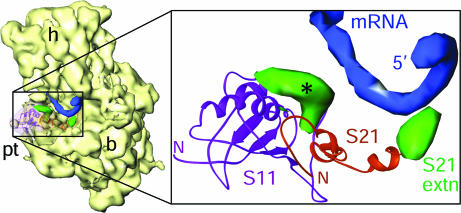
Three proteins, S3, S4 and S5, encircle the entrance of the mRNA channel in the bacterial ribosome (18). Of these, S3 and S4 are shorter in the chloro-ribosome, whereas S5 is significantly larger, with an 86-residue NTE. Other 30S subunit proteins that approach the mRNA path in bacterial ribosome are S1, S7, S11, S12, S18, and S21. PRPs S1 and S7 are smaller, and PRPs S11, S12, and S18 are larger in the chloro-30S subunit. However, none of the protein extensions, including that of PRP S5, seem to affect the overall topology of the mRNA entrance in the chloro-ribosome, except the CTE in PRP S21, which is positioned to interact directly with the 5′ untranslated region of the mRNA (19) (Fig. 3).
Fig. 3.
The mRNA exit channel. X-ray crystallographic structure of the whole 30S mRNA complex (19) was docked into the cryo-EM map of the chloro-30S subunit. Boxed region in the thumbnail (Left) is enlarged to show the 5′ terminus of mRNA (blue), PRPs S11 (magenta ribbons), and S21 (orange ribbons) with their extensions (green). The CTE of S21 is close enough to make direct contact with the mRNA, whereas the extra density associated with S11 (marked with an asterisk) is within 13 Å of the mRNA. The latter density could be partly attributable to a local conformational change.
After assignment of PRPs and the additional masses arising because of their extensions, only three unidentified protein masses remained. The lack of continuity with neighboring PRPs led to the categorization of these masses as PSRPs. The large unassigned protein mass located at the neck of the 30S subunit (Fig. 2A) has been assigned to PSRP1 on the basis that similar density arises in cryo-EM reconstructions of PSRP1 bound to the E. coli 70S ribosome (M.R.S., D.N.W., C.B., P.P.D., P.F., and R.K.A., unpublished data). This study reveals that PSRP1 is not in fact a bona fide ribosomal protein but rather a regulatory factor that has homology to the cold-shock protein pY, the binding site of which also is located in the same region on E. coli ribosomes (20). In this position, PSRP1 interacts with 16S rRNA helices 18 and 44 from the 30S body and helices 30 and 34 and PRPs S9 and S13 from the 30S head, and it is within 10 Å distance from the PRP S12.
The extra protein mass buried between the 16S rRNA helices 30, 41, 41a, 42, and 43 within the 30S head was readily assigned to PSRP4. PSRP4 is the most basic protein (pI 11.79) in the 30S subunit (5), consistent with its extensive interaction with the negatively charged rRNA. Furthermore, the NTD of PSRP4 shows homology with the Thx protein that is present in the T. thermophilus 70S ribosome (11) but absent in the E. coli 70S ribosome (13). A homology model of the NTD domain of PSRP4 based on Thx shows an excellent fit into the cryo-EM density assigned to PSRP4. Thx and PSRP4 appear to share a similar functional role in stabilizing the rRNA helices in the 30S head region.
The remaining PSRP mass is located at the bottom of the 30S subunit, where it more than compensates, in terms of its overall size, for the loss (relative to E. coli) of nucleotides in helices h6, h10, and h17 of the 16S rRNA (see SI Fig. 6). The overall size of this protein mass is too large for either PSRP2 or PSRP3 alone; however, it is consistent with the combined mass of the two PSRPs (see SI Table 1). In fact, there are two unequally sized globular masses within this PSRP mass, which are connected through a weak mass of density that disappears upon increasing the density threshold value. We tentatively assign the larger of the two masses to PSRP2 and the smaller one to PSRP3 (Fig. 2 A and B). PSRP2 and PSRP3 interact with each other (as mentioned above) and with 16S rRNA helices 6 and 10, respectively. PSRP2 shows sequence similarity to a stromal RNA-binding domain (RBD), leading to the suggestion that it binds mRNA and plays a role in translation initiation under light-synchronized protein synthesis (7). However, based on its location in our map, we believe that RBD of PSRP2 facilitates its interaction with the 16S rRNA helix 10. The combined densities of PSRP2 and PSRP3 span a large portion of the chloro-30S subunit bottom. Thus, like in the mammalian mitochondrial ribosome (mitoribosome) (9), we find that the bottom of the small subunit in the chloro-ribosome is protein-rich, as compared with its bacterial counterparts.
Structure of the Large (50S) Ribosomal Subunit.
The overall shape of the chloro-50S subunit is similar to its bacterial counterpart, except that the PRP L1 arm and L7/L12 stalk are distinguishably larger. As for the 30S subunit, the 23S rRNA secondary structure maps (see SI Fig. 7) and homology model of the chloro-ribosome are consistent with the RNA–protein separated map of the chloro-ribosome, indicating the deletion of rRNA nucleotides relative to E. coli 23S rRNA within H9, H45, H63, and H98 (see SI Fig. 7), as well as a 9-nt insertion between H1 and H2 of the 4.8S rRNA (corresponding to helices 100 and 101 in the E. coli 23S rRNA), a 20-nt loop insertion within H38 (Fig. 2D and SI Fig. 9), and 23-nt insertion in H58 (Fig. 2C). No insertions or deletions are located within domain V and VI that encompasses the peptidyl-transferase center of the ribosome, highlighting the high conservation and functional importance of this region.
Almost all of the orthologous PRPs of the 50S subunit are larger in size than the respective E. coli counterparts, because of the NTEs and CTEs, such that PRP L21 and L22 are almost twice the size (5). The extensions of most PRPs are located on the solvent side of the subunit and can be readily identified in our protein-only map (Fig. 2 C and D). Extensions of PRPs L13, L21, and L22 apparently converge into an elongated mass between PRP L11 and the polypeptide-exit tunnel. Our map does not show densities for L25 or L30, both of which are known to be absent in the chloro-ribosome, nor for deleted portions of L17 and L18 (5). L25 is a 5S rRNA binding protein, and sequence homology between PRP L22 and L25 led to the suggestion that PRP L22 may have taken over the role of L25 (5); however, the total absence of density in the L25 region and the distant localization of L22 from the 5S rRNA argue against this hypothesis. In contrast, density attributable to the 20-nt loop insertion within H38 overlaps with the L30 region, suggesting that this RNA insertion may compensate partially for the absence of L30 (SI Fig. 9).
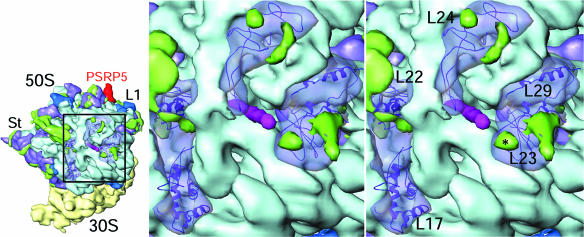
The 50S subunit possesses two small PSRPs, PSRP5 (6.6–9.2 kDa) and PSRP6 (7.4 kDa). Based on the mass of the proteins and size of unidentified density on the chloro-50S (SI Table 1), we assigned PSRP5 tentatively to a density located near the tRNA-exit, or E site, in a groove formed between 23S rRNA helices H68 and H88 (Fig. 2 C and D). This position of PSRP5 may reflect a possible role in ejection of the deacylated tRNAs from the chloro-ribosome. Because we do not observe a distinct mass of density that can be assigned to PSRP6, it is likely that PSRP6 is relatively loosely associated with the ribosome, which is consistent with the low stoichiometry of PSRP6 observed on 2D gels compared with other PRPs of similar size, e.g., L28, L32–L35 (5).
Interaction Between PSRP1 and pRRF.
Although the density for the NTD of PSRP1 is well resolved in the cryo-EM map of the chloro-ribosome, the C-terminal domain (CTD) appears to be flexible because it is partially disordered. However, the CTD shows up in a difference map calculated between the chloro- and E. coli 70S maps as a protruding mass from the NTD of PSRP1. The CTD of PSRP1 has sequence homology to a light-regulated transcript A (lrtA) present in the cyanobacterium Synechococcus sp. PCC 7002 (21), suggesting that PSRP1 may be directly involved in the light-dependent regulation of translation in chloroplasts. In our map, the density for the CTD of PSRP1 forms a connection with domain II of pRRF, sitting on the chloro-50S subunit. Like the CTD of PSRP1, domain II of pRRF appears to be flexible but is observed in the same difference map (Fig. 4; also see SI Fig. 10). When bound to the chloro-ribosome, both PSRP1 and pRRF would completely block the binding positions for A and P site tRNAs, thus supporting the role of PSRP1 as a stress response factor rather than an integral ribosomal protein. The cohabitation of pRRF with PSRP1 on the chloro-ribosome may indicate an additional role for RRF during protein synthesis. Normally, RRF binds to posttermination complexes and, in combination with elongation factor G (EF-G), splits the ribosomes, recycling the ribosomal subunits, mRNA, and tRNAs for the next round of translation (see ref. 22). A second function of pRRF may be in conjunction with PSRP1 to block translation under stress conditions by filling the tRNA binding sites. When conditions are again favorable (such as reexposure to light), pRRF may play a role in release of PSRP1 from the ribosome, analogous to its role in releasing tRNAs during bacterial ribosome recycling.
Fig. 4.
Stereoview presentation of the interaction between CTD of PSRP1 with domain II of pRRF. Difference map, showing the N-terminal domain of PSRP1 (semitransparent red), is shown with the fitted homology model (red ribbons). N-terminal (N) and C-terminal (C) ends are labeled. Difference map corresponding to pRRF (semitransparent yellow) is shown with fitted atomic structure (greenish-yellow ribbons) of Thermotoga maritima RRF (PDB ID 1DD5). Two domains (I and II) of RRF are labeled. Thumbnail to the left depicts the orientation of the chloro-ribosome with small (semitransparent pale yellow) and large (semitransparent blue) subunits identified.
The Polypeptide Tunnel Exit.
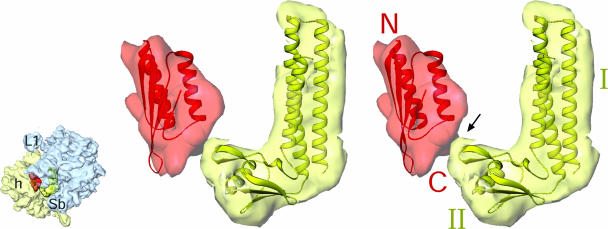
The majority of the polypeptides synthesized by the chloro-ribosome are inserted into the thylakoid membrane, therefore it might be expected that chloroplasts have evolved a more specialized polypeptide-exit tunnel for efficient posttranslational protein export. The chloro-ribosome map shows that although the overall topography of the tunnel exit is similar to that observed in the bacterial ribosomes, with ribosomal proteins L22, L23, L24, and L29 encircling the tunnel exit site (e.g., refs. 11, 13, and 23), a number of differences are evident. All these PRPs are larger in chloro-ribosome (5), and indeed we see corresponding extra densities associated with these PRPs (Fig. 5). Extensions of the PRP L23, L24, and L29 may establish additional contacts with Ffh (fifty-four homolog or SRP54) protein to compensate for the absence of the RNA moiety of the signal recognition particle (SRP) in the chloroplast. The protein-rich nature of the tunnel exit site in chloro-ribosomes also has been observed for the mitoribosome (9), suggesting that this may be conserved as an important feature of organellar ribosomes, necessary for efficient export or insertion of proteins into membranes.
Fig. 5.
Topography of the polypeptide-exit tunnel in the 50S subunit. The modeled polypeptide chain (magenta) exiting from the tunnel is shown. PRPs surrounding the tunnel exit are shown as semitransparent purple masses with homology models (shown as ribbons) of PRPs docked in. Light blue density corresponds to 23S rRNA. Thumbnail to the left shows the orientation of the ribosome with the boxed region, which has been enlarged for stereoviewing.
After the completion of this study, a cryo-EM study of chloro-ribosome from an algae Chlamydomonas reinhardtii was reported at lower resolution (24). With the exception of PSRP7, which is not present in spinach chloroplasts, this study did not assign any of the PRP extensions or additional PSRPs. Furthermore, because a cryo-EM map was not deposited in EM database, no meaningful comparison can be made between the two studies.
Materials and Methods
Preparation of Chloro-Ribosomes.
Chloro-ribosome isolation was performed according to Bartsch et al. (25). Five kilograms of spinach was deveined and washed thoroughly in precooled distilled deionized water. The leaves were homogenized (2 liters per 1 kg of leaves) using 0.7 M sorbitol in buffer A (10 mM Tris·HCl, pH 7.6, 50 mM KCl, 10 mM MgOAc, and 7 mM 2-mercaptoethanol). The homogenate was filtered through several layers of cheesecloth and one layer of Miracloth (Calbiochem) before centrifugation at 1,200 × g for 15 min. The pellet was resuspended in 0.4 M sorbitol in buffer A and recentrifuged at 1,200 × g for 15 min. The washed chloroplast pellet was resuspended in buffer A supplemented with 2% (vol/vol) Triton X-100 and incubated on ice for 30 min. The lysed suspension was clarified by centrifugation at 26,000 × g for 30 min before isolation of crude ribosomes by centrifugation at 50,000 × g for 24 h through 1 M sucrose (in buffer B: buffer A with 10% glycerol). The greenish pellet was washed and then resuspended in buffer B with gentle agitation. The crude ribosomes were clarified by centrifugation at 26,000 × g for 15 min before being snap-frozen at −80°C. Alternatively, the clarified supernatant was applied directly onto a 10–30% sucrose gradient (in buffer B) to obtain tight-couple chloroplast 70S ribosomes.
Cryo-EM and 3D Image Reconstruction.
EM data were collected on a Philips FEI (Eindhoven, The Netherlands) Tecnai F20 field emission gun electron microscope, equipped with low-dose kit and an Oxford cryo-transfer holder, at a magnification of ×50,760. Then, 164 micrographs were scanned on a Zeiss flatbed scanner (Z/I Imaging Corporation, Huntsville, AL), with a step size of 14 μm, corresponding to 2.76 Å on the object scale. The projection-matching procedure (26) within the SPIDER software was used to obtain the 3D map. We used an 11.5-Å resolution E. coli 70S ribosome map (8) as the initial reference. An 18-Å resolution map of the 70S chloro-ribosome so obtained subsequently was used as reference for iterative refinement. Initially, 192,133 images, sorted into 41 groups according to defocus value (ranging from 1.4 μm to 4.4 μm), were picked. Finally, 86,370 images were retained, after visual and cross-correlation-based screening and removal of images from overrepresented groups among the initial set of 83 representative views of the ribosome, and these were used in the final reconstructions. The resolution of the final CTF-corrected 3D maps (27), estimated by using the Fourier shell correlation with a cut-off value of 0.5 (see refs. 28 and 29), was 9.4 Å [or ≈7.5 Å by the 3σ criterion (30)].
The fall-off of the Fourier amplitudes toward higher spatial frequencies was corrected as described previously (8) by using the small-angle x-ray solution scattering intensity distribution of the E. coli 70S ribosome. RNA and protein components of the 70S chloro-ribosome map were computationally separated by using a method (12) based on differences in the density distribution of the two moieties, taking into account the molecular masses and contiguity constraints. A model for the 70S chloro-ribosome was built from homology models of the PRPs generated in HHPred (31) with E. coli (13) and T. thermophilus 70S (11) ribosomal proteins as templates. In most cases, the additional NTEs and CTEs of the PRPs were predicted to be unstructured and therefore were not included in the final model. The chloro-ribosome rRNA was modeled on the basis of the E. coli 70S structure (13) by comparison of sequence and secondary structure alignments. The rRNA and PRPs then were refined in CNS (32). Homology models of specific domains of PSRP1, PSRP2, and PSRP4 were generated with Swiss-Model (33) (http://swissmodel.expasy.org//SWISS-MODEL.html), Phyre (www.sbg.bio.ic.ac.uk/∼phyre), and HHPred (http://toolkit.tuebingen.mpg.de/hhpred). Assignments of PSRPs within the protein-only maps of both subunits are based on two main criteria: (i) the presence of protein masses in locations that are not seen in the bacterial ribosome and (ii) calculation of voxel volumes for each discrete new protein mass and a correlation between voxel volumes and molecular mass of the corresponding PSRP (see SI Table 1). Before computation of a difference map between the maps of chloro-ribosome and E. coli ribosome, the two maps were scale-matched and cross-correlated. Visualization and interpretation of the map and docking of crystallographic structures were performed by using SPIDER, IRIS Explorer (Numerical Algorithms Group, Inc., Downers Grove, IL), O (34), and Ribbons (35).
Supplementary Material
Acknowledgments
We thank Timothy Booth for assisting with cryo-EM data collection, Aruna Silasagaram for help with image processing, and Judith Wellen for help procuring copious amounts of spinach plants. We acknowledge use of the Wadsworth Center's EM infrastructure. This work was supported, in part, by National Institutes of Health Grant R01 GM61576 (to R.K.A.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The cryo-EM map of the chloroplast 70S ribosome has been deposited in the EM database, www.ebi.ac.uk (accession code EMD-1417). The homology model for the chloro-ribosome small and large subunits have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3BBN and 3BBO, respectively).
This article contains supporting information online at www.pnas.org/cgi/content/full/0709856104/DC1.
References
- 1.Harris EH, Boynton JE, Gillham NW. Microbiol Rev. 1994;58:700–754. doi: 10.1128/mr.58.4.700-754.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Brien TW. Gene. 2002;286:73–79. doi: 10.1016/s0378-1119(01)00808-3. [DOI] [PubMed] [Google Scholar]
- 3.Schmitz-Linneweber C, Maier RM, Alcaraz JP, Cottet A, Herrmann RG, Mache R. Plant Mol Biol. 2001;45:307–315. doi: 10.1023/a:1006478403810. [DOI] [PubMed] [Google Scholar]
- 4.Margulis L. Origin of Eukaryotic Cells. New Haven: Yale Univ Press; 1970. [Google Scholar]
- 5.Yamaguchi K, Subramanian AR. J Biol Chem. 2000;275:28466–28482. doi: 10.1074/jbc.M005012200. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi K, von Knoblauch K, Subramanian AR. J Biol Chem. 2000;275:28455–28465. doi: 10.1074/jbc.M004350200. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi K, Subramanian AR. Eur J Biochem. 2003;270:190–205. doi: 10.1046/j.1432-1033.2003.03359.x. [DOI] [PubMed] [Google Scholar]
- 8.Gabashvili IS, Agrawal RK, Spahn CM, Grassucci RA, Svergun DI, Frank J, Penczek P. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- 9.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 10.Agrawal RK, Spahn CM, Penczek P, Grassucci RA, Nierhaus KH, Frank J. J Cell Biol. 2000;150:447–460. doi: 10.1083/jcb.150.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 12.Spahn CMT, Penczek PA, Leith A, Frank J. Structure (London) 2000;8:937–948. doi: 10.1016/s0969-2126(00)00185-4. [DOI] [PubMed] [Google Scholar]
- 13.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 14.Agrawal RK, Sharma MR, Kiel MC, Hirokawa G, Booth TM, Spahn CM, Grassucci RA, Kaji A, Frank J. Proc Natl Acad Sci USA. 2004;101:8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson DN, Schluenzen F, Harms JM, Yoshida T, Ohkubo T, Albrecht R, Buerger J, Kobayashi Y, Fucini P. EMBO J. 2005;24:251–260. doi: 10.1038/sj.emboj.7600525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 17.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 18.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 19.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 20.Vila-Sanjurjo A, Schuwirth BS, Hau CW, Cate JHD. Nat Struct Mol Biol. 2004;11:1054–1059. doi: 10.1038/nsmb850. [DOI] [PubMed] [Google Scholar]
- 21.Samartzidou H, Widger WR. Plant Physiol. 1998;117:225–234. doi: 10.1104/pp.117.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barat C, Datta PP, Raj VS, Sharma MR, Kaji H, Kaji A, Agrawal RK. Mol Cell. 2007;27:250–261. doi: 10.1016/j.molcel.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Schlunzen F, Wilson DN, Tian P, Harms JM, McInnes SJ, Hansen HA, Albrecht R, Buerger J, Wilbanks SM, Fucini P. Structure (London) 2005;13:1685–1694. doi: 10.1016/j.str.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Manuell AL, Quispe J, Mayfield SP. PLoS Biol. 2007;5:e209. doi: 10.1371/journal.pbio.0050209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartsch M, Kimura M, Subramanian AR. Proc Natl Acad Sci USA. 1982;79:6871–6875. doi: 10.1073/pnas.79.22.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penczek PA, Grassucci RA, Frank J. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 27.Penczek PA, Zhu J, Schroder R, Frank J. Scanning Microscopy. 1997;11:147–154. [Google Scholar]
- 28.Bottcher B, Wynne SA, Crowther RA. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 29.Malhotra A, Penczek P, Agrawal RK, Gabashvili IS, Grassucci RA, Junemann R, Burkhardt N, Nierhaus KH, Frank J. J Mol Biol. 1998;280:103–116. doi: 10.1006/jmbi.1998.1859. [DOI] [PubMed] [Google Scholar]
- 30.Orlova EV, Dube P, Harris JR, Beckman E, Zemlin F, Markl J, van Heel M. J Mol Biol. 1997;271:417–437. doi: 10.1006/jmbi.1997.1182. [DOI] [PubMed] [Google Scholar]
- 31.Söding J, Biegert A, Lupas AN. Nucl Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunger A, Adams P, Clore G, DeLano W, Gros P, Grosse-Kunstleve R, Jiang J, Kuszewski J, Nilges M, Pannu N, et al. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Schwede T, Kopp J, Guex N, Peitsch MC. Nucl Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones TA, Zhou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr a A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 35.Carson M. Appl Crystallogr. 1991;24:103–106. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.