Abstract
Tumor cell proliferation requires rapid synthesis of macromolecules including lipids, proteins, and nucleotides. Many tumor cells exhibit rapid glucose consumption, with most of the glucose-derived carbon being secreted as lactate despite abundant oxygen availability (the Warburg effect). Here, we used 13C NMR spectroscopy to examine the metabolism of glioblastoma cells exhibiting aerobic glycolysis. In these cells, the tricarboxylic acid (TCA) cycle was active but was characterized by an efflux of substrates for use in biosynthetic pathways, particularly fatty acid synthesis. The success of this synthetic activity depends on activation of pathways to generate reductive power (NADPH) and to restore oxaloacetate for continued TCA cycle function (anaplerosis). Surprisingly, both these needs were met by a high rate of glutamine metabolism. First, conversion of glutamine to lactate (glutaminolysis) was rapid enough to produce sufficient NADPH to support fatty acid synthesis. Second, despite substantial mitochondrial pyruvate metabolism, pyruvate carboxylation was suppressed, and anaplerotic oxaloacetate was derived from glutamine. Glutamine catabolism was accompanied by secretion of alanine and ammonia, such that most of the amino groups from glutamine were lost from the cell rather than incorporated into other molecules. These data demonstrate that transformed cells exhibit a high rate of glutamine consumption that cannot be explained by the nitrogen demand imposed by nucleotide synthesis or maintenance of nonessential amino acid pools. Rather, glutamine metabolism provides a carbon source that facilitates the cell's ability to use glucose-derived carbon and TCA cycle intermediates as biosynthetic precursors.
Keywords: cancer, glioblastoma, Warburg effect, glutaminolysis, anaplerosis
In mammals, cell proliferation is controlled by signal transduction pathways stimulated by lineage-specific growth factors or, in tumors, constitutive activation of these pathways through oncogenic mutations. A proximal effect of signaling pathways is a robust increase in nutrient uptake (1–3). Cells must also allocate these molecules into appropriate metabolic pathways to produce and maintain pools of intermediates needed to synthesize macromolecules. Therefore, a complete understanding of the biology of cell proliferation will require a comprehensive understanding of the regulation of metabolic fluxes.
Glucose and glutamine are two of the most abundant nutrients in plasma, and together they account for most carbon and nitrogen metabolism in mammalian cells. Rapid cell proliferation has been associated with a robust but apparently wasteful metabolism of glucose. In the 1920s, Otto Warburg demonstrated that ascites tumor cells had high rates of glucose consumption and lactate production despite availability of sufficient oxygen to oxidize glucose completely (4). The “Warburg effect” is taken to be a metabolic hallmark of aggressive tumors; however, the phenotype is also observed in nontransformed cells during rapid proliferation (2, 5). Glutamine metabolism in cancer cells has also been reported to exceed the use of other nonessential amino acids (6, 7). As a result, many investigators have stressed the importance of glutamine as a source of reduced nitrogen for maintenance of nucleotide biosynthesis and nonessential amino acids.
13C NMR spectroscopy is an analytical tool that allows for noninvasive detection of selectively enriched metabolites in real time (8, 9). Multiple pathways can be probed simultaneously by using 13C-labeled nutrients, and the data can be used to measure metabolic fluxes. This approach has been used extensively for studies with intact organs (10–12). Recently, new techniques have achieved kinetic data in cultured cells (13, 14), making it possible to estimate fluxes during cell proliferation.
We anticipated that the use of 13C NMR spectroscopy to observe glucose and glutamine metabolism would yield information about how these nutrients are used in support of cancer cell growth. As a model system, we studied the human glioblastoma cell line SF188. These cells lack functional p53 but express wild-type PTEN and do not exhibit constitutive Akt activation (15, 16). When grown in serum-supplemented medium, the cells exhibit aerobic glycolysis. We perfused SF188 cultures with 13C-labeled nutrients inside an NMR spectrometer and acquired spectra continuously. The data confirmed a pronounced Warburg effect. We also detected entry of pyruvate into the tricarboxylic acid (TCA) cycle via pyruvate dehydrogenase (PDH) but not pyruvate carboxylase (PC). Exit of intermediates from the TCA cycle supplied fatty acid synthesis, and some 60% of fatty acyl carbon was glucose-derived. As expected, glutamine metabolism served to maintain pools of nonessential amino acids. Glutamine was also used to replenish TCA cycle intermediates (anaplerosis). Surprisingly, glutamine catabolism provided the cells with a robust source of NADPH production, resulting in the majority of glutamine-derived carbon being secreted as lactate. This catabolism of glutamine to lactate was also accompanied by secretion of nitrogen as ammonia and alanine. The results suggest that transformed tumor cells engage in glutamine metabolism exceeding the need for glutamine as a biosynthetic precursor.
Results
Proliferating Glioblastoma Cells Exhibit the Warburg Effect.
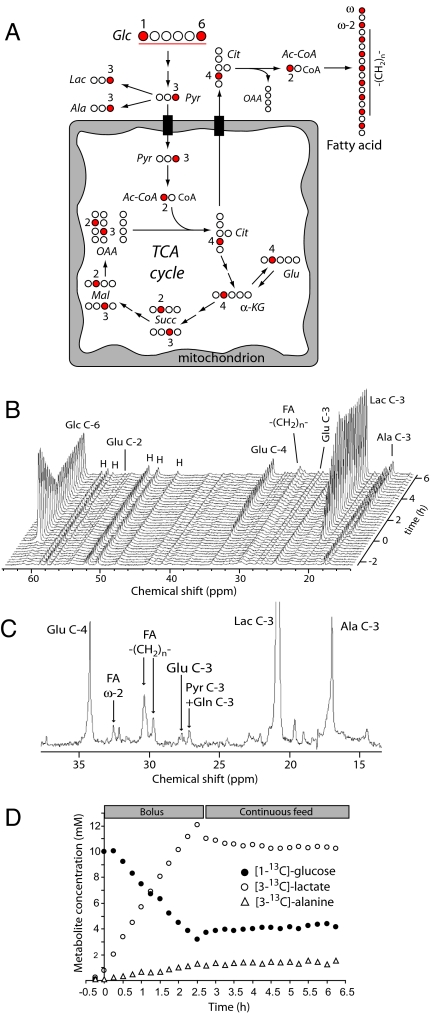
To examine glucose utilization in glioblastoma cells, we cultured SF188 cells in medium supplemented with [1,6-13C2]glucose. The pathways of intermediary metabolism through which label is transferred from glucose to other metabolites are shown in Fig. 1A. During spectroscopy, cells received a bolus of medium containing 10 mM [1,6-13C2]glucose, which recirculated for 2.5 h with constant oxygenation. Subsequently, a constant feed of fresh medium was used to maintain an extracellular glucose concentration of 4 mM. The complete series of 13C spectra is shown in Fig. 1B. A spectrum collected near the end of the perfusion (Fig. 1C) highlights labeled metabolites. We determined concentrations for individual metabolites and calculated rates from linear regressions of time courses (Fig. 1D and Table 1). This revealed a glucose consumption rate of 220 ± 7 μmol per 109 cells per hour, similar to the value obtained with off-line enzymatic measurements (207 μmol per 109 cells per hr).
Fig. 1.
Glucose metabolism in proliferating glioblastoma cells. (A) The labeling scheme highlights distribution of 13C (filled circles) from [1,6-13C2]glucose into various metabolites. Several core pathways, including glycolysis, anaerobic pyruvate metabolism, the TCA cycle, and fatty acid synthesis are shown. (B) Stacked spectra from a real-time [1,6-13C2]glucose perfusion experiment in SF188 cells. Each spectrum represents data summed over 15 min. (C) A partial spectrum from time 6 h highlights enrichment of informative metabolic intermediates. Background from the time 0-h spectrum was subtracted to emphasize the effect of [1,6-13C2]glucose metabolism. (D) Absolute concentrations of [1-13C]glucose, [3-13C]lactate, and [3-13C]alanine during the experiment.
Table 1.
Metabolic rates during culture of SF188 cells with d-[1,6-13C2]glucose
| Metabolite | Rate |
|---|---|
| [1,6-13C2]glucose consumption | 220 ± 7 |
| [3-13C]lactate production | 368 ± 6 |
| [3-13C]alanine production | 40 ± 2 |
| [4-13C]glutamate labeling (apparent) | 3.7 ± 0.2 |
| Oxygen consumption | 68 ± 2 |
The most abundant 13C-labeled metabolites produced were [3-13C]lactate and [3-13C]alanine, both of which were predominantly extracellular. The cells produced 1.8 mol of 13C-lactate plus alanine per mole of glucose consumed. Consistent with this observation, the culture used oxygen at a low rate (68 μmol per 109 cells per hr) that would support complete oxidation of only 11 μmol of glucose per 109 cells per hr, far below the measured glucose consumption rate. Therefore, glucose metabolism was largely anaerobic despite the availability of abundant oxygen, consistent with the classic pattern described by Warburg.
TCA Cycle Activity in Proliferating Glioblastoma Cells.
Cells cultured with [1,6-13C2]glucose produced [4-13C]glutamate (Fig. 1B), which resulted from conversion of glucose-derived pyruvate to acetyl-CoA by PDH and entry into the TCA cycle. The steady-state ratio of [4-13C]glutamate to [2-13C]glutamate is an indicator of the relative contributions of citrate synthase and anaplerotic reactions to TCA cycle flux (17, 18). Throughout the experiment, the level of [4-13C]glutamate was far greater than [2-13C]glutamate (Fig. 2A). Over the last 2 hours, the ratio of labeling was 13 ± 2.6:1, consistent with a large anaplerotic flux.
Fig. 2.
Tricarboxylic acid cycle activity in glioblastoma cells. (A)The intracellular concentrations of [4-13C]glutamate and [2-13C]glutamate were determined after introduction of [1,6-13C2]glucose. The [4-13C]glutamate results from PDH and the [2-13C]glutamate results from cycling of [4-13C]glutamate. (B) A 13C NMR spectrum of a cellular extract from cells cultured in [2-13C]glucose. The Inset is an expansion of the 25- to 60-ppm region highlighting glutamate carbons 2, 3, and 4. Data are from one representative experiment.
The 13C labeling of glutamate carbons is impacted by flux through the enzyme PC. To quantify PC flux, we cultured cells with [2-13C]glucose, which labels predominantly C-2 of pyruvate. PDH would transfer label to C-1 of acetyl-CoA and eventually C-5 of glutamate, whereas PC would transfer label to C-2 of OAA and eventually C-3 of glutamate. Labeling of C-2 of OAA would also result in labeling of C-3 of OAA and ultimately C-2 of glutamate if there was equilibration of OAA with malate/fumarate, but neither PC nor PDH would lead to enrichment of C-4 of glutamate. Cells were cultured with [2-13C]glucose and extracted with perchloric acid. In the resulting spectrum (Fig. 2B), the ratio of [3-13C]glutamate to [4-13C]glutamate is 1:1, demonstrating little if any PC activity. Therefore, the major route of entry of carbon from pyruvate into the TCA cycle is through PDH.
Glucose Is the Major Lipogenic Substrate in Glioblastoma Cells.
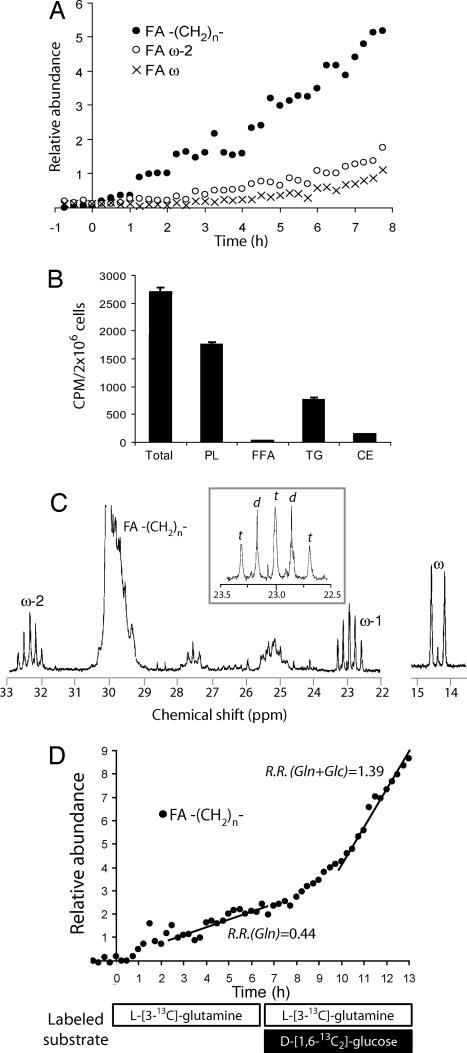
Because the TCA cycle does not function as a carbon sink, anaplerosis is coupled to the exit of anions from the TCA cycle. Fatty acid synthesis relies on citrate efflux from the mitochondria (Fig. 1A), and this pathway is required for maximal tumor cell growth (19–21). When cells were cultured with [1,6-13C2]glucose, we observed progressive labeling of fatty acids at multiple resonances (Fig. 3A). To determine the absolute rate of lipid synthesis from glucose, cells were cultured in medium containing a [U-14C6]glucose tracer, then lipids were extracted and analyzed by scintillation counting. The results indicated a rate of conversion of glucose to lipids of 2.4 ± 0.5 μmol of glucose per 109 cells per hr. Fractionation of 14C-enriched lipids revealed that phospholipids were the most abundant species synthesized (Fig. 3B).
Fig. 3.
Lipid synthesis in proliferating glioblastoma cells. (A) The labeling of three fatty acyl (FA) resonances, the ω (methyl), ω-2, and methylene [−(CH2)n−] increased throughout the experiment at a constant rate, demonstrating continuous transfer of 13C from glucose into fatty acids. (B) Cells were cultured in medium containing [U-14C6]glucose, and then lipids were extracted and analyzed by thin-layer chromatography. The average and standard deviation for three parallel cultures are shown for each lipid species. (C) Cells were cultured in medium containing 10 mM [U-13C6]glucose, and then lipids were extracted and analyzed by NMR spectroscopy. The Inset shows an expansion of the 23-ppm region, highlighting the ω-1 multiplet (d, doublet; t, triplet). (D) Cells were cultured in medium containing 4 mM [3-13C]glutamine and 10 mM unlabeled glucose (time 0–6.25 h), followed by medium containing 4 mM [3-13C]glutamine and 10 mM [1,6-13C2]glucose (time 6.25–13 h). The relative rate (R.R.) for the increase in −(13CH2)n− signal was calculated for both halves of the experiment.
The cells were also cultured with [U-13C6]glucose and extracted lipids were analyzed by NMR spectroscopy. Because fatty acid synthesis occurs via sequential addition of acetyl-CoA to an elongating chain, the methyl and ω-1 carbons are derived from one two-carbon group and the ω-2 and ω-3 carbons are derived from another. The likelihood that both the ω-1 and ω-2 carbons are 13C-labeled is thus determined by the fraction of the acetyl-CoA pool that is 13C-labeled. The lipid spectrum (Fig. 3C) contained multiplets consisting of a triplet and a doublet at ω-1 and ω-2. At ω-1 (23 ppm), the triplet corresponds to fatty acids labeled at both the methyl and ω-2 positions as well as at ω-1 (R-13CH2-13CH2-13CH3), reflecting the combination of two 13C-labeled acetyl-CoA molecules. The doublet corresponds to fatty acids labeled at the methyl and ω-1 positions, but not ω-2 (R-12CH2-13CH2-13CH3), reflecting the combination of one labeled and one unlabeled acetyl-CoA. Likewise, the ω-2 resonance (32.3 ppm) contained a triplet (R-13CH2-13CH2-13CH2-13CH3) and a doublet (R-13CH2-13CH2-12CH2-12CH3). At both ω-1 and ω-2, the triplet accounted for 60% of the total area, suggesting that 60% of the lipogenic acetyl-CoA pool is derived from glucose. We confirmed this with an alternative strategy using [1,6-13C2]glucose and [2-13C]glucose [supporting information (SI) Fig. 6].
Glutamine can also be used as a carbon source for fatty acid synthesis and can be consumed by proliferating cells more rapidly than needed to satisfy nitrogen requirements. Using real-time 13C NMR spectroscopy, we used a two-stage perfusion to compare the rate of fatty acid synthesis from glutamine to the rate from glucose plus glutamine (Fig. 3D). In the first stage, cells received complete medium containing 4 mM [3-13C]glutamine and 10 mM unlabeled glucose. In the second stage, the medium contained 4 mM [3-13C]glutamine and 10 mM [1,6-13C2]glucose. The rate of labeling of −(CH2)n− during stage 2 was triple the stage 1 rate, consistent with glucose's role as the major lipogenic precursor.
Glutamine Metabolism Supports NADPH Production and Anaplerosis in Proliferating Glioblastoma Cells.
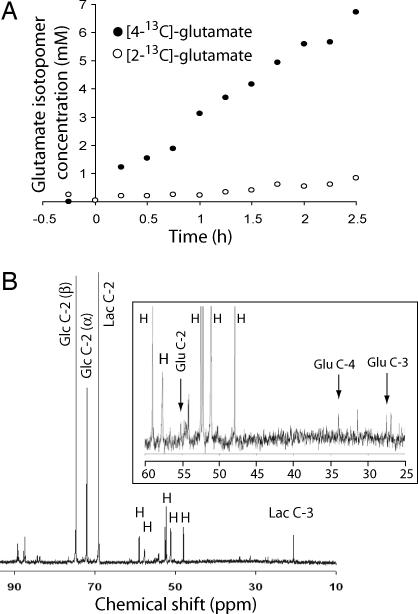
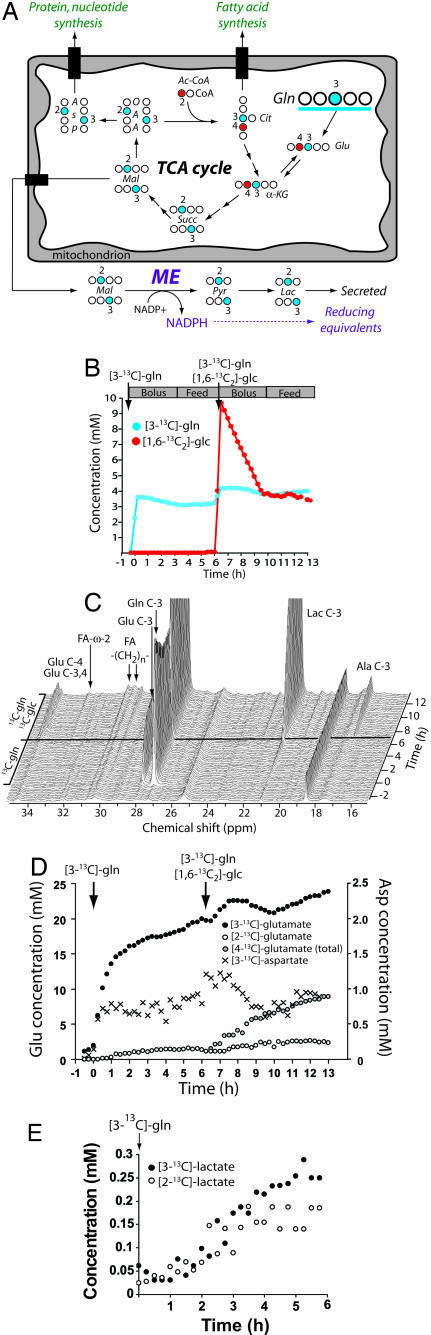
The presence of fatty acid synthesis implied the need for two supporting pathways: a source of NADPH (the electron donor for fatty acid synthesis) and an anaplerotic mechanism to replenish TCA cycle intermediates during citrate export. Because glutamine metabolism can potentially fulfill both these needs (Fig. 4A), we analyzed data from the two-stage perfusion to determine fluxes through relevant pathways (Fig. 4 B–E and Table 2). During stage 1, [3-13C]glutamate appeared rapidly and was the most abundant 13C metabolite produced (Fig. 4 C and D). Part of this pool was converted to [3-13C]α-KG and entered the TCA cycle, because soon after the appearance of [3-13C]glutamate, aspartate was labeled in C-2 and C-3. This resulted from conversion of [2-13C]OAA and [3-13C]OAA to [2-13C]aspartate and [3-13C]aspartate by aspartate aminotransferases. The intracellular [3-13C]aspartate concentration was ≈1 mM at steady state (Fig. 4D).
Fig. 4.
Glutamine metabolism in proliferating glioblastoma cells. (A) The labeling scheme begins with [3-13C]glutamine (blue) at the top right of the mitochondrion. For simplicity, extramitochondrial glutamine metabolism and aspartate production are not shown. The pathway outlines glutaminolysis, the conversion of glutamine-derived carbon to lactate by using malic enzyme (ME), which produces NADPH. The appearance of [2-13C]Ac-CoA (red) is from the addition of [1,6-13C2]glucose. (B) Design for two-stage perfusion experiment. In the first stage, cells received [3-13C]glutamine and unlabeled glucose as a bolus and then as a continuous feed. In the second stage, cells received [3-13C]glutamine and [1,6-13C2]glucose. (C) Stacked spectra acquired during the two-stage perfusion experiment. Each spectrum represents data summed over 15 min. (D) Intracellular concentrations of [3-13C]glutamate, [2-13C]glutamate, [4-13C]glutamate, and [3-13C]aspartate during both stages. (E) Concentration of [2-13C]lactate and [3-13C]lactate during perfusion with [3-13C]glutamine.
Table 2.
Metabolic rates during culture of SF188 cells with l-[3-13C]glutamine and l-[1,6-13C2]glucose
| Metabolite | Rate |
|---|---|
| [3-13C]glutamine consumption | 20 ± 1 |
| Stage 1 ([3-13C]glutamine) | |
| [3-13C]glutamate labeling (apparent) | 19 ± 2 |
| [2-13C]lactate + [3-13C]lactate production | 12 ± 1 |
| Stage 2 ([3-13C]glutamine and [1,6-13C2]glucose) | |
| [1,6-13C2]glucose consumption | 213 ± 3 |
| [3-13C]lactate production | 334 ± 3 |
| [4-13C]glutamate labeling (apparent) | 2.3 ± 0.1 |
All rates are in μmol per 109 cells per hr ± standard error. The rate of [3-13C]glutamine consumption was calculated by using data from Stage 1 and Stage 2. Cells were perfused with l-[3-13C]glutamine and d-[1,6-13C2]glucose twice. These data are from one representative experiment. Data in Figs. 3D, 4, and 5 A and B are from the same experiment.
During stage 1, glutaminolysis resulted in labeling of lactate in C-2 and C-3 (Fig. 4E). This likely reflected oxidation of glutamine-derived [2-13C]malate and [3-13C]malate by malic enzyme (ME), which produces NADPH. The cells produced [2-13C]lactate and [3-13C]lactate at a combined rate of 12 ± 1 μmol per 109 cells per hr. This is a minimum estimate for malic enzyme flux. The other major cellular source of NADPH is glucose-6 phosphate dehydrogenase (G6PDH) in the oxidative pentose phosphate shunt. To estimate this flux, we analyzed extracellular medium from the experiment in Fig. 2B, in which cells were cultured with [2-13C]glucose. The ratio of 13C label in C-2 to C-3 of lactate was 25.1:1. Based on the stoichiometry of the oxidative pentose phosphate pathway (22), the flux was calculated to be 5.8 ± 0.3% of the glycolytic flux, or 12.8 μmol per 109 cells per hour.
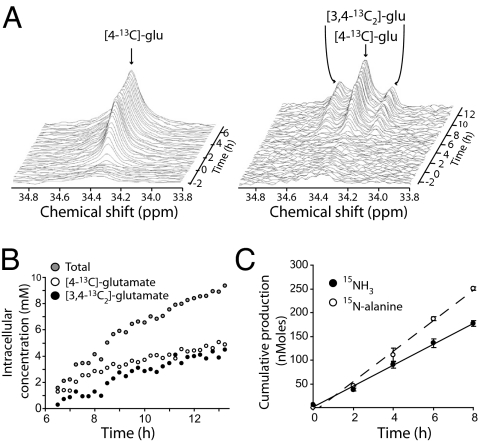
The apparent lack of PC activity implied that anaplerosis was not supplied by glucose metabolism. As shown in Fig. 4A, metabolism of [3-13C]glutamine can label C-3 of α-KG and subsequently C-2 and C-3 of OAA. Condensation of [2-13C]OAA with [2-13C]Ac-CoA derived from [1,6-13C2]glucose would result in glutamate labeled at both C-3 and C-4 ([3,4-13C2]glutamate). Addition of [1,6-13C2]glucose to the medium during steady-state perfusion with [3-13C]glutamine (stage 2) caused rapid labeling of glutamate at C-4 (Fig. 4 C and D). However, unlike the results with [1,6-13C2]glucose alone (Fig. 1B, magnified in Fig. 5A Left), the combination of substrates produced a large amount of doubly labeled glutamate ([3,4-13C2]glutamate, Fig. 5A Right). We determined that [3,4-13C2]glutamate accounted for 45 ± 3% of the total glutamate labeled at C-4 (Fig. 5B). This implies that ≈45% of the OAA pool used for citrate synthesis was labeled at C-2.
Fig. 5.
Glutamine is the major anaplerotic substrate in proliferating glioblastoma cells. (A) Stacked spectra of the 34.3-ppm region ([4-13C]glutamate) obtained during a perfusion with [1,6-13C2]glucose (Left, from Fig. 1B) and during the two-stage perfusion (Right). (B) Intracellular concentration of glutamate isotopomers labeled at C-4. “Total” concentration equals the sum of [4-13C]glutamate and [3,4-13C2]glutamate. (C) Cumulative production of extracellular 15NH3 and [15N]alanine from cells cultured with l-[α-15N]glutamine. This experiment was performed three times, and one representative time course is shown.
There are two major sources for label at OAA C-2. First, [3-13C]glutamate can enter the TCA cycle as [3-13C]α-KG and label OAA at either C-2 or C-3 (Fig. 4A). Second, [4-13C]glutamate formed after condensation of OAA with glucose-derived [2-13C]Ac-CoA can progress around the TCA cycle again, also labeling OAA at C-2 or C-3 (Fig. 1A). However, the results indicate that the contribution of “cycled” glutamate was small because the majority of label was lost with each turn. Therefore the majority of [2-13C]OAA was formed through direct anaplerotic contribution of glutamine-derived carbon. This pathway yields equal amounts of [2-13C]OAA and [3-13C]OAA, but only the former contributes to [3,4-13C2]glutamate. Consequently, the observation that 45% of the OAA pool was labeled at C-2 suggests that 90% was labeled at either C-2 or C-3, and thus that glutamine, by its conversion to α-KG and then OAA, was the major source of anaplerotic flux.
After cells convert glutamine to glutamate, α-KG can be produced by glutamate dehydrogenase (GDH) or by transamination reactions. Both mechanisms remove the α-amino group from glutamate, transferring it to either ammonia (GDH) or to nonessential amino acids (transaminases). Based on the high net rate of alanine production (Table 1), alanine aminotransferase (ALT) appeared to be the most active transaminase in these cells. We cultured cells with l-[α-15N]glutamine and analyzed the medium by mass spectrometry to follow transfer of 15N to ammonia and alanine (Fig. 5C). As calculated by this method, the cells transferred 15N to ammonia at a rate of 5.8 μmol per 109 cells per hour and to alanine at a rate of 8.2 μmol per 109 cells per hour. Thus, approximately half of the resulting amino groups were secreted from the cell as either alanine or ammonia.
Discussion
A combination of techniques has been used to study the metabolism of proliferating glioblastoma cells. Although the cells have a pronounced Warburg effect, the TCA cycle is intact. As a result of condensation of glucose-derived acetyl-CoA and glutamine-derived OAA, the cells synthesized fatty acids and lipids primarily with carbon from glucose. The cells converted ≈60% of the glutamine metabolized to lactate. This implied a malic enzyme flux high enough to supply the NADPH needed for fatty acid synthesis. A large fraction of nitrogen generated during glutamine metabolism was also released from the cell.
Like many cancer cell lines, these cells exhibited a high rate of glucose consumption and anaerobic metabolism of pyruvate. By summing the production of lactate and alanine, we accounted for >90% of total glucose metabolism. Therefore, all remaining glucose-dependent activities (glycosylation, fatty acid synthesis, glycerogenesis, nucleotide biosynthesis, pyruvate oxidation, etc.) accounted for <10% of total glucose utilization.
The function of the TCA cycle and oxidative metabolism in tumor cells has been the subject of discussion and controversy. This stemmed in part from Warburg's claim that the root cause of cancer was a permanent impairment of oxidative metabolism (23, 24). However, the data here reveal a complete TCA cycle allowing some [2-13C]glutamate to be formed during culture with [1,6-13C2]glucose (Fig. 2A). Citrate oxidation was limited by efflux from the mitochondria to supply fatty acid synthesis. Therefore, the reduction of substrate oxidation during tumor cell proliferation can be secondary to the metabolic activities needed for biosynthesis rather than to impairments in oxidative metabolism per se. Using NMR spectroscopy of total cellular lipids, we determined that glucose is the major lipogenic precursor, a finding that clarifies the association between glucose-to-lipid carbon flux and cell proliferation in other tumor cells (21).
The investigation of glutamine utilization illuminated several aspects of intermediary metabolism. Glutamine is similar to glucose in that proliferating cells metabolize it by using a variety of pathways that support bioenergetics and biosynthesis, and cultured tumor cells require at least 10 times as much glutamine as any other amino acid (25–27). Like glucose, glutamine can be degraded to lactate rather than oxidized completely (7). The data here show that lactate and alanine production accounts for 60% of total glutamine utilization. A by-product of this flux is robust NADPH production by malic enzyme. The glutaminolytic flux was at least as high as the G6PDH flux, and appeared to be higher than that needed for fatty acid synthesis. This could mean that NADPH generated during glutaminolysis also supplies other anabolic processes such as nucleotide biosynthesis.
Glutamine carbon was also used in biosynthetic pathways. First, glutamine provided a secondary source of carbon for fatty acid synthesis. Estimates using 14C labeling showed that glutamine accounted for some 25% of total fatty acyl carbon (data not shown). Second, entry of glutamine carbon into the TCA cycle resulted in rapid labeling of aspartate, a required precursor for synthesis of nucleotides, asparagine, and arginine.
Another major role for glutamine metabolism was to provide an anaplerotic source of OAA. This activity is required to maintain the TCA cycle under conditions when cells use citrate as a biosynthetic precursor. Anaplerosis is a more specific indicator of growth than glycolysis, because the latter is also stimulated by hypoxia and other stresses independently of biosynthetic activity. It was surprising that such a high fraction of the anaplerotic flux was derived from glutamine. Together, the data imply that at citrate synthase, the majority of acetyl-CoA is derived from glucose, whereas the majority of OAA is derived from glutamine. Because glucose and glutamine are rapidly consumed during proliferation of most cell types, the results may reflect a general phenomenon during rapid cell growth.
To serve as an anaplerotic precursor, the glutamate derived from glutamine must be converted to α-KG. Several enzymes can carry out this reaction, including GDH and various transaminases. In these cells, the summed activities of GDH and ALT accounted for more than half of the glutamine metabolized, and therefore these two enzymes supplied the majority of glutamine-derived α-KG. As a consequence of GDH and ALT activity, most of the α-nitrogen from glutamine degradation was secreted from the cells as ammonia and alanine. This was surprising, considering the expectation that this nitrogen would largely be used to maintain intracellular amino acid pools. The findings suggest that secretion of amino groups from glutamate is a required consequence of the use of glutamine as an anaplerotic precursor and to generate NADPH.
Materials and Methods
Cell Culture and Perfusion.
SF188 cells (University of California Brain Tumor Research Center, San Francisco, CA) were grown in T-flasks (Fisher) incubated at 37°C in a 95% air/5% CO2 incubator. The base medium was DMEM (Invitrogen) with 10% FBS (HyClone) and gentamicin sulfate (50 mg/liter, Invitrogen). Routine culture used 25 mM glucose, 25 mM Hepes, and 6 mM l-glutamine. For perfusion experiments, DMEM powder without glucose and glutamine (Sigma) was supplemented with 10 mM glucose and 4 mM l-glutamine that was either unenriched or enriched (d-[1,6-13C2]glucose, Cambridge Isotope Laboratories; l-[3-13C]glutamine, Sigma). In experiments using d-[U-13C6]glucose and l-[α-15N]-glutamine, 10% dialyzed FBS (Gemini) was used to minimize contribution of unlabeled nutrients. For perfusion inside the NMR spectrometer, cells were grown in porous microcarriers as described (13, 14). Details are in SI Materials and Methods. Oxygen utilization rates were determined by using polarographic probes.
NMR Data Analysis.
Spectra on perfused cultures were acquired with a 400 MHz Varian system (14). The 9.4 T magnet had an 89-mm vertical bore and a 72-mm i.d. room-temperature shim set. Details on spectroscopy are in SI Materials and Methods. Spectra were analyzed by using NUTS 1D software (Acorn NMR). The 31P spectra were used to determine NTP levels to estimate total cellular volume (28). Cell volume was measured with a Coulter Z2 particle analyzer to convert total cellular volume into cell number. For each experiment, an average area was determined from 5–10 spectra for the Hepes resonance at 59 ppm. This was used to convert areas for 13C nuclei into metabolite concentration, as described (13). Rates and standard errors were calculated by using Microsoft Excel. For the perchloric acid extract experiment, cells were cultured in medium containing 10 mM d-[2-13C]glucose for 6 h and then extracted with 10% perchloric acid. Soluble metabolites were analyzed by 13C NMR spectroscopy. Details are in SI Materials and Methods.
Lipid Biochemistry.
To determine the rate of lipid synthesis from glucose, 3 × 106 SF188 cells were plated into T25 flasks and cultured with DMEM supplemented with 1.5 μM d-[U-14C6] glucose per liter (0.2 mCi/ml, Sigma). After 8 h, cells were trypsinized, washed in PBS and lysed in 0.4 ml of 0.5% Triton X-100. Lipids were extracted and analyzed with a scintillation counter. To characterize lipid species, extracted lipids were analyzed by thin-layer chromatography (29). To obtain 13C-labeled lipids, 2.5 × 106 SF188 cells were cultured with DMEM containing 10 mM d-[U-13C6]-labeled glucose. The cells were expanded until total number was 24 × 106, then were trypsinized and washed in PBS. Lipids were extracted and analyzed by NMR using a Bruker Advance 400 wide-bore 9.4T instrument equipped with a 5-mm 1H/13C dual probe. Details are in SI Materials and Methods.
Gas Chromatography–Mass Spectrometry.
Cells were plated at 1.2 × 106 per 6-cm dish. At 80% confluency, they were fed with 1.5 ml of DMEM containing 4 mM l-[α-15N]glutamine (Cambridge Isotope Laboratories). Every 2 hours, medium was collected to determine the concentration of NH3 (ammonia assay kit, Sigma) and alanine (HPLC). An aliquot was used to determine isotopic enrichment in NH3 and alanine with published methods (30). Details are in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the Small Animal Imaging Facility at the University of Pennsylvania for experimental assistance and members of the C.B.T. laboratory for critical reading of the manuscript. This work was supported by National Institutes of Health Grants P01 CA104838, T32 GM008638, and K08 DK072565.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709747104/DC1.
References
- 1.Cooper EH, Barkhan P, Hale AJ. Br J Haematol. 1963;9:101–111. doi: 10.1111/j.1365-2141.1963.tb05446.x. [DOI] [PubMed] [Google Scholar]
- 2.Hedeskov CJ. Biochem J. 1968;110:373–380. doi: 10.1042/bj1100373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edinger AL, Thompson CB. Mol Biol Cell. 2002;13:2276–2288. doi: 10.1091/mbc.01-12-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warburg O. Klin Wochenschr Berl. 1925;4:534–536. [Google Scholar]
- 5.Wang T, Marquardt C, Foker J. Nature. 1976;261:702–705. doi: 10.1038/261702a0. [DOI] [PubMed] [Google Scholar]
- 6.Coles NW, Johnstone RM. Biochem J. 1962;83:284–291. doi: 10.1042/bj0830284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reitzer LJ, Wice BM, Kennell D. J Biol Chem. 1979;254:2669–2676. [PubMed] [Google Scholar]
- 8.Cohen SM, Ogawa S, Shulman RG. Proc Natl Acad Sci USA. 1979;76:1603–1609. doi: 10.1073/pnas.76.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shulman RG, Brown TR, Ugurbil K, Ogawa S, Cohen SM, den Hollander JA. Science. 1979;205:160–166. doi: 10.1126/science.36664. [DOI] [PubMed] [Google Scholar]
- 10.Shulman RG, Rothman DL. Annu Rev Physiol. 2001;63:15–48. doi: 10.1146/annurev.physiol.63.1.15. [DOI] [PubMed] [Google Scholar]
- 11.Gruetter R. Neurochem Int. 2002;41:143–154. doi: 10.1016/s0197-0186(02)00034-7. [DOI] [PubMed] [Google Scholar]
- 12.Lewandowski ED. J Nucl Cardiol. 2002;9:419–428. doi: 10.1067/mnc.2002.125811. [DOI] [PubMed] [Google Scholar]
- 13.Mancuso A, Beardsley NJ, Wehrli S, Pickup S, Matschinsky FM, Glickson JD. Biotechnol Bioeng. 2004;87:835–848. doi: 10.1002/bit.20191. [DOI] [PubMed] [Google Scholar]
- 14.Mancuso A, Zhu A, Beardsley NJ, Glickson JD, Wehrli S, Pickup S. Magn Reson Med. 2005;54:67–78. doi: 10.1002/mrm.20545. [DOI] [PubMed] [Google Scholar]
- 15.Chen P, Iavarone A, Fick J, Edwards M, Prados M, Israel MA. Cancer Genet Cytogenet. 1995;82:106–115. doi: 10.1016/0165-4608(94)00213-u. [DOI] [PubMed] [Google Scholar]
- 16.Haas-Kogan D, Shalev N, Wong M, Mills G, Yount G, Stokoe D. Curr Biol. 1998;8:1195–1198. doi: 10.1016/s0960-9822(07)00493-9. [DOI] [PubMed] [Google Scholar]
- 17.Malloy CR, Sherry AD, Jeffrey FM. FEBS Lett. 1987;212:58–62. doi: 10.1016/0014-5793(87)81556-9. [DOI] [PubMed] [Google Scholar]
- 18.Weiss RG, Chacko VP, Glickson JD, Gerstenblith G. Proc Natl Acad Sci USA. 1989;86:6426–6430. doi: 10.1073/pnas.86.16.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhajda FP, Jenner K, Wood FD, Hennigar RA, Jacobs LB, Dick JD, Pasternack GR. Proc Natl Acad Sci USA. 1994;91:6379–6383. doi: 10.1073/pnas.91.14.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pizer ES, Wood FD, Heine HS, Romantsev FE, Pasternack GR, Kuhajda FP. Cancer Res. 1996;56:1189–1193. [PubMed] [Google Scholar]
- 21.Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Mancuso A, Sharfstein ST, Tucker SN, Clark DS, Blanch HW. Biotechnol Bioeng. 1994;44:563–585. doi: 10.1002/bit.260440504. [DOI] [PubMed] [Google Scholar]
- 23.Warburg O. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 24.Warburg O. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 25.Eagle H, Oyama VI, Levy M, Horton CL, Fleischman R. J Biol Chem. 1956;218:607–616. [PubMed] [Google Scholar]
- 26.Kovacevic Z, McGivan JD. Physiol Rev. 1983;63:547–605. doi: 10.1152/physrev.1983.63.2.547. [DOI] [PubMed] [Google Scholar]
- 27.Brand K. Biochem J. 1985;228:353–361. doi: 10.1042/bj2280353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mancuso A, Fernandez EJ, Blanch HW, Clark DS. Biotechnology. 1990;8:1282–1285. doi: 10.1038/nbt1290-1282. [DOI] [PubMed] [Google Scholar]
- 29.Deberardinis RJ, Lum JJ, Thompson CB. J Biol Chem. 2006;281:37372–37380. doi: 10.1074/jbc.M608372200. [DOI] [PubMed] [Google Scholar]
- 30.Brosnan JT, Brosnan ME, Yudkoff M, Nissim I, Daikhin Y, Lazarow A, Horyn O, Nissim I. J Biol Chem. 2001;276:31876–31882. doi: 10.1074/jbc.M103890200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.