Abstract
IGFBP-1 is elevated in fetuses with long-term, chronic hypoxia and intrauterine growth restriction. We investigated the hypothesis that hypoxia regulates IGFBP-1 in the human fetus in vivo and IGFBP-1 gene expression and protein in vitro. Umbilical artery IGFBP-1 levels (mean ± SEM) from term babies with respiratory acidosis (acute hypoxia), normal babies, and those with mixed respiratory/metabolic acidosis (more profound and prolonged hypoxia) were measured using an immunoradiometric assay. IGFBP-1 levels were similar in normal (n = 12) and acutely hypoxic (n = 6) babies (189.1 ± 71.8 vs. 175.8 ± 45.9 ng /ml, respectively, P = 0.789). However, with more profound and prolonged hypoxia (n = 19), IGFBP-1 levels were markedly elevated (470.6 ± 80.0 ng /ml, P = 0.044). To investigate IGFBP-1 regulation by hypoxia in vitro, HepG2 cells were incubated under hypoxia (pO2 = 2%) and normoxia (pO2 = 20%). IGFBP-1 protein and mRNA increased 8- and 12-fold, respectively, under hypoxic conditions. Hypoxia did not affect protein or mRNA levels of IGFBP-2 or -4. IGFBP-5 and -6 mRNAs, undetectable in control cells, were not induced by hypoxia, whereas minimally expressed IGFBP-3 mRNA increased twofold. Investigation into IGFBP-1 gene structure revealed three potential consensus sequences for the hypoxia response element (HRE) in the first intron. To investigate functionality, a 372-bp fragment of IGFBP-1 intron 1, containing putative HREs, was placed 5′ to a heterologous hsp70 promoter in a plasmid using luciferase as a reporter gene. Under hypoxia, reporter gene activity increased up to 30-fold. Mutations in the middle HRE abolished reporter activity in response to hypoxia, suggesting that this HRE is functional in the IGFBP-1 hypoxia response. Cotransfection of HRE reporter genes with a constitutively expressing hypoxia-inducible factor 1 plasmid in HepG2 cells resulted in a fourfold induction of reporter activity, suggesting a role for hypoxia-inducible factor 1 in hypoxia induction of IGFBP-1 gene expression. These data support the hypothesis that hypoxia regulation of IGFBP-1 may be a mechanism operating in the human fetus to restrict insulin-like growth factor-mediated growth in utero under conditions of chronic hypoxia and limited substrate availability.
Keywords: growth factor/gene expression/fetus/newborn
Fetal growth is a complex process influenced by genetic, nutritional, environmental, placental, and maternal factors, each of which can have profound effects on fetal outcome (1). In humans intrauterine growth restriction (IUGR, or birthweight <10th percentile for gestational age) is a leading cause of severe fetal and neonatal morbidity and mortality. Decreased placental perfusion and placental and fetal hypoxia are leading causes of IUGR (1). The role of oxygen in fetal growth is also underscored by observations that IUGR accompanies pregnancies complicated by decreased maternal oxygen-carrying capacity and fetal anemia and hypoxia. In addition to oxygen, growth factors and particularly the fetal and maternal insulin-like growth factor (IGF) systems are important regulators of fetal growth (2, 3). IGF-I and IGF-II gene knockout studies have demonstrated that homozygous mouse pups are born with marked IUGR (4). A naturally occurring deletion of exons 4 and 5 of the human IGF-I gene results in severe intrauterine and postnatal growth restriction (5). In humans there is also a direct correlation between IGF-I concentrations in cord blood and fetal size and birthweight (6).
IGFBP-1 regulates minute-to-minute bioavailability of IGFs in the circulation and inhibits IGF actions (7). In humans and animals there is a striking inverse correlation between maternal and fetal circulating levels of IGFBP-1 and fetal size, and IGFBP-1 levels are elevated up to 20-fold in fetuses with marked IUGR (6, 8–17). Recent evidence suggests that hypoxia and the IGF system are interrelated. Animal models of uteroplacental insufficiency (caused by uterine artery ligation or maternal hypoxia) result in fetal hypoxia and IUGR, with markedly elevated circulating levels of IGFBP-1 and overexpression of IGFBP-1 mRNA in the fetal liver, the primary source of circulating IGFBP-1 (11, 12, 18–21). These studies suggest that IGFBP-1 may be an important regulator of fetal growth by regulating IGF bioavailability.
IGFBP-1 gene regulation is complex, with multiple regulatory elements in its promoter that act independently or coordinately to regulate expression (7, 22). The specific stimuli for elevated IGFBP-1 in the fetal circulation in pregnancies complicated by IUGR and hypoxia have not been determined, although stress (glucocorticoids) and hypoinsulinemia have been implicated (2). Hypoxia may itself be a stimulus for elevated IGFBP-1 levels observed in these babies. Hypoxic regulation of gene expression involves binding of a nuclear transcription factor, hypoxia-inducible factor 1 (HIF-1), to a hypoxia response element (HRE) (23, 24). In the current study, we report markedly elevated levels of IGFBP-1 in babies with intrauterine hypoxia, the presence of a functional HRE in intron 1 of the IGFBP-1 gene, evidence in vitro that IGFBP-1 mRNA and protein are induced in HepG2 cells by hypoxia, that this increase is mediated directly through an HRE located in the first intron of the hIGFBP-1 gene, and that HIF-1 is likely involved in hypoxia regulation of IGFBP-1 gene expression. We propose that transcriptional regulation of IGF-BP-1 by hypoxia is a mechanism to explain the observed increased levels of IGFBP-1 found in babies with intrauterine hypoxia and IUGR.
MATERIALS AND METHODS
Fetal Cord Samples.
Umbilical cord arterial samples, containing blood returning from the fetus to the placenta, were obtained after delivery of 37 term newborns from uncomplicated pregnancies at Stanford Hospital Labor and Delivery Suite. Umbilical cords were clamped and cut between 30 and 60 s of delivery. After double clamping the cord and isolating a small segment, blood was obtained from one umbilical artery using heparinized syringes which were immediately evacuated of air, capped, and analyzed for pH, pO2, pCO2, and HCO3− (25). Hypoxia in the newborn can be defined by assessing its acid base balance. Acute respiratory acidosis occurs with acute hypoxemia and mixed respiratory/metabolic acidosis occurs with more prolonged or profound hypoxia, resulting in accumulation of CO2 and fixed organic acids (25, 26). The following criteria (26) for umbilical cord arterial blood were used to define acute respiratory acidosis: pH < 7.2, elevated pCO2 (>2 SD above the mean), and normal plasma HC03−; and mixed respiratory/metabolic acidosis: pH < 7.2, normal pCO2, decreased plasma HC03−, and elevated base deficit (amount of buffer below the normal level). Six babies had acute terminal bradycardia just before delivery, with accompanying acute hypoxia and respiratory acidosis. Nineteen babies had mixed respiratory/metabolic acidosis and meconium aspiration, and 12 had uneventful intrapartum courses and normal cord gases. All babies had normal birthweights. Samples (plasma) were also assayed for IGFBP-1 (vide infra). The protocol was approved by the Stanford University Human Subjects Committee and was in accordance with the Declaration of Helsinki.
IGFBP-1 Assays.
IGFBP-1 concentrations in fetal cord samples and in media conditioned by HepG2 cells (see below) were measured using a two-site coated tube IRMA (Active; DSL, Webster, TX), performed in duplicate according to the manufacturer’s instructions. The sensitivity was 330 pg/ml. The intraassay coefficients of variation were 5.2%, 4.6%, and 2.7% for IGFBP-1 concentrations of 5.2, 50.2, and 144.6 ng/ml, respectively. The interassay coefficients of variation were 3.5%, 6.0%, and 3.6% for IGFBP-1 concentrations of 5.1, 47.1, and 142.0 ng/ ml, respectively.
Cell Cultures.
A human hepatocellular carcinoma cell line, HepG2, obtained from the American Type Culture Collection (HB 8065) was used to investigate the effects of hypoxia on IGFBP-1 expression and in transfection experiments. Cells were plated in duplicate and were maintained in MEM (GIBCO) with 10% fetal calf serum, 1 mM sodium pyruvate, 2 mM glutamate, nonessential amino acids, and 10 μg/ml gentamicin. Cells were plated in custom-made 60-mm glass plates free of O2, with notched sides to allow gas exchange, at 2–3 × 106 cells/plate. To determine the effect of oxygen on IGFBP-1 expression, cells were cultured in serum-free media (RPMI 1640; GIBCO) in a specially designed hypoxia chamber (27, 28) which was sealed and subjected to successive rounds of evacuation, followed by flushing with 95% N20/5% CO2 mix to produce a specific pO2 as determined by an oxygen electrode (27, 28). Conditioned medium was collected at various times and was assayed for IGFBP-1 by immunoradiometric assay as described above. Levels were normalized to total protein concentration in the media using the Bradford technique (Bio-Rad).
Northern Blot Analyses.
Total RNA was isolated from HepG2 cells (29), using Trizol (Gibco/BRL), according to the manufacturer’s instructions. Total RNA (10 μg) was denatured in formamide/formaldehyde and electrophoresis of total RNA samples was carried out on a 1.2% agarose-formaldehyde gel, as described in ref. 30. RNA was transferred to nitrocellulose by capillary transfer. cDNA probes for human IGFBP-1 through IGFBP-6 were as follows: a 938-bp EcoRI fragment for IGFBP-1 (31); a 1435-bp EcoRI fragment of the IGFBP-2 cDNA (from J. Schwander, Basel, Switzerland; ref. 32); a 1950-kb EcoRI fragment of the human IGFBP-3 cDNA (from P. Cohen, University of Pennsylvania, Philadelphia, PA ref. 33); a 503-bp EcoRI-HindIII fragment for IGFBP-4 (from S. Shimasaki, University of California, San Diego, CA; ref. 34); a 317-bp SacI-SacII fragment for IGFBP-5 and a PstI 267-bp cDNA for IGFBP-6 (both from S. Shimasaki; refs. 35 and 36); and a 1200-bp DNA fragment from the 18S rRNA cDNA, using a reverse transcription–PCR kit (CLONTECH). Probes were labeled with [α-32P]dCTP using a random primer kit (Ready-to-Go; Pharmacia Biotech) and purified on nick spin columns (Pharmacia Biotech). They were hybridized to nitrocellulose filters overnight at 42°C. Filters were washed twice at room temperature, twice at 42°C, and twice at high stringency at 65°C and air dried. Autoradiography was performed at −80°C for 2 hr to 4 days. Blots were stripped and reprobed with each cDNA probe. Gels were stained with ethidium bromide to assess RNA integrity and uniformity of loading.
Western Ligand Blots.
Samples of conditioned medium from HepG2 cells (75 μl) were run overnight on a 12% acrylamide gel and electrotransferred to nitrocellulose membranes as described (37). Blocking was with 1% BSA. Membranes were then incubated with 1.5 × 106 cpm/ml 125I-labeled IGF-I and -II overnight at 4°C, washed, air dried, and bands visualized by autoradiography. Controls included nonpregnant human serum (10 μl/well, control for IGFBP-3), seminal plasma (10 μl/well, control for IGFBP-2 and -4), and human midgestation amniotic fluid (2 μl, control for IGFBP-1).
Transient Transfection Assays.
All presented sequence coordinates of the hIGFBP-1 gene are relative to the transcription start site (ref. 38; GenBank accession no. M59316). A 372-bp fragment of intron 1, spanning from bp 614–985 of the hIGFBP-1 gene and containing three putative HREs (656-TACGTGCT-663, 714-GACGTGCT-721, and 915-CACGTGCT-922) and CA repeat (751-CACACA-756) was produced by PCR with the following synthetic oligonucleotide primers: 5′-GCAGGGGCTTGTCCACAACT-3′ and 5′-ACTCCTGGGGAGGCCAGC-3′. PCR fragment sequence was confirmed by DNA sequence analysis. The fragment was then ligated into a SmaI site of the pGUP.8 vector which contains a weak hsp70 promoter with TATA box, luciferase reporter, and an ampicillin resistance gene (39). Additionally, a fragment with two copies of the 372-bp fragment (in tandem) was generated. A mutant fragment was produced by PCR with a mutation containing the forward primer in the HRE at 5′ bp 705–724-3′ (5′-TCTTGGCAGGAAAAGCTCTG-3′) with the sequences at 716–718 (TACGTGCT → TAAAAGCT) along with the same 3′ primer as the wild-type fragment. Both wild-type and mutated fragments were inserted into the same SmaI site of the plasmid. HepG2 cells were transiently transfected with plasmids using Superfect (Qiagen, Chatsworth, CA). Transfected cells were maintained in serum-containing medium overnight, washed with PBS, and then switched to serum-free medium (RPMI 1640) at the onset of the experimental protocol. After treatment (24 hr), cells were washed and harvested in cold PBS, then lysed with luciferase lysis buffer (Promega), and luciferase activity was measured using a luminometer.
To determine whether HIF-1 mediates hypoxia induction of IGFBP-1, transient cotransfection studies were conducted. A plasmid constitutively expressing HIF-1α (40) was kindly provided by G. Semenza (Johns Hopkins University, Baltimore, MD). Plasmids containing the vascular endothelial growth factor (VEGF) promoter-reporter construct containing one HRE or four HREs, previously shown to be functional (27), were utilized as positive controls. The VEGF reporter genes were generated by PCR amplification from the VEGF promoter using forward primers, HIF-1 wild-type 5′-CCACAGTGCATACGTGGCTCC-3′ and HIF-1 mutant 5′-CCACAGTGCATAAAAGGCTCC-3′ and a reverse primer at positions −782 5′-CTGGCCTGCAGACATC-3′. The amplified sequence corresponds to position −982 to −782 in the VEGF promoter. Five micrograms of the HIF-1α plasmid or Bluescript plasmid and 5 μg of the IGFBP-1 HRE reporter gene or VEGF reporter gene were electroporated (Gene Pulser; Bio-Rad) into 3 × 106 HepG2 cells in 200 μl at 240 V, with a capacitance of 960 μF. Cells were plated on glass plates and allowed to recover for 17 hr in serum-containing medium prior to further incubation in serum-free media. Cells were lysed and analyzed for luciferase activity as described above.
Densitometry.
Autoradiographs of Northern blots and Western ligand blots were analyzed on a PDI Desk Top Scanner (Protein DNA ImageWare Systems, Huntington, NY). The integrated areas under the absorbance curves were measured for each band and were used to determine the relative amounts of mRNAs and IGFBPs.
Statistical Analysis.
Experimental variables were tested in duplicate or triplicate cultures. One-way ANOVA was used for statistical evaluation of data and significance of the differences of mean ± SD of IGFBP-1 levels in HepG2 cell conditioned media in the hypoxic and normoxic treatment groups, with P < 0.05 considered significant. For Northern blot analyses, the mean ± SD of the absorption measurements of the 1.5-kb IGFBP-1 mRNA transcript was calculated for three separate experiments. For Western ligand blot analyses, the mean ± SD of the absorption measurements for bands corresponding to the individual IGFBPs were calculated for three separate experiments. One-way ANOVA was used for statistical evaluation of data and significance of the differences between groups was determined by Dunnet’s or Scheffe’s test, as appropriate, with P < 0.05 considered significant. For the fetal data, the mean ± SEM were calculated for the umbilical cord samples and comparisons between groups were made with an unpaired t test. P < 0.05 was considered significant.
RESULTS
IGFBP-1 in Fetal Cord Samples.
Mean IGFBP-1 levels in babies with normal intrapartum courses and normal umbilical cord arterial blood gases were calculated, and the results are shown in Table 1. In addition, levels were determined in babies with meconium aspiration and mixed respiratory and metabolic acidosis and in babies with terminal bradycardia and acute respiratory acidosis (Table 1). The mean ± SEM of umbilical artery IGFBP-1 levels in normal babies (n = 12) was 189.0 ± 71.8 ng /ml. Six babies who had terminal bradycardia and respiratory acidosis had a mean IGFBP-1 of 175.8 ± 45.9 ng /ml. This was not significantly different from the normal babies, by unpaired t test (P = 0.789). In contrast, babies with mixed respiratory and metabolic acidosis (n = 19) had significantly higher levels of IGFBP-1 in their circulation (470.6 ± 80.0 ng/ml) compared with the normal babies, which was significant at P = 0.044.
Table 1.
Newborn umbilical artery IGFBP-1 levels
| Condition | IGFBP-1, ng/ml | P* |
|---|---|---|
| Normal | 189.0 ± 71.82 | na† |
| Respiratory acidosis | 175.8 ± 45.93 | 0.789 |
| Mixed metabolic/respiratory acidosis | 470.6 ± 80.02 | 0.044* |
P, compared to normal, by unpaired t test. P < 0.05 = significant.
na, not applicable.
Hypoxia Regulation of IGFBP-1 in HepG2 Cells.
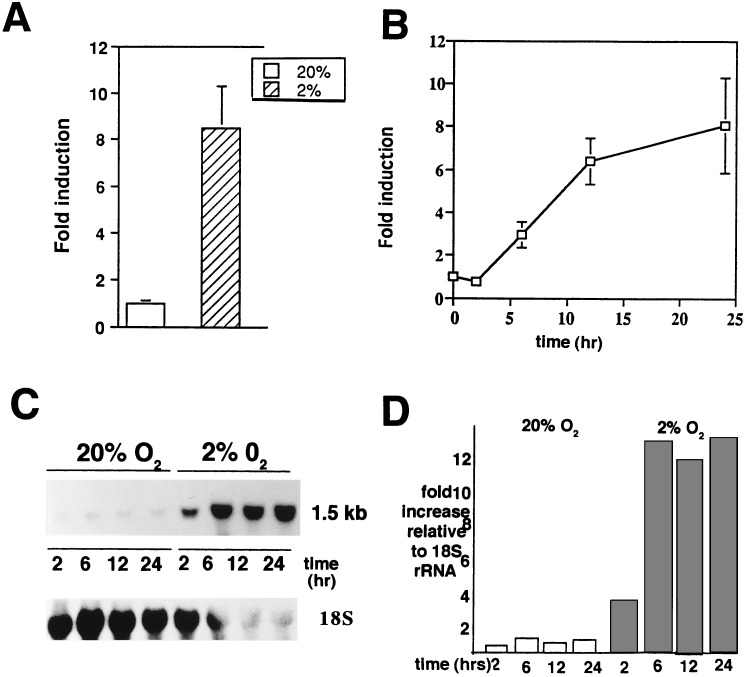
To determine whether hypoxia has an effect on IGFBP-1 expression, a human hepatocellular carcinoma cell line (HepG2), known to produce IGFBP-1 (review in ref. 7), was used as a model system to study this relationship in vitro. The effects of hypoxia on the induction of IGFBP-1 secretion into conditioned medium and on steady-state levels of IGFBP-1 mRNA expression in HepG2 cells were investigated, and the results are shown in Fig. 1. Under hypoxic conditions (2% O2) for 24 hr, IGFBP-1 levels in conditioned medium (Fig. 1A) were ≈8-fold higher than with normoxia (20% O2). A time course (Fig. 1B) revealed elevated IGFBP-1 levels in the conditioned medium at 6 hr with a maximum (≈8-fold) achieved at 24 hr compared with normoxic controls. The effects of hypoxia on steady-state levels of IGFBP-1 were analyzed by Northern blot analysis. Kinetic analysis (Fig. 1C) revealed induction of the 1.5-kb IGFBP-1 mRNA by 2 hr of exposure to hypoxia, with a maximum expression (≈12-fold) by 6 hr and maintained through 24 hr of culture. Quantitation of steady-state IGFBP-1 mRNA levels (normalized to 18S rRNA) is shown in Fig. 1D. Similar time courses for hypoxia induction of erythropoietin and VEGF mRNA and protein were also observed (data not shown).
Figure 1.
Hypoxia induction of IGFBP-1 in HepG2 cells. (A) Cells were grown in serum-containing medium in the presence of 20% and 2% O2, respectively. After 24 hr, the conditioned medium was collected and assayed in triplicate for IGFBP-1 using an immunoradiometric assay. Values shown are the mean ± SEM of six different experiments. (B) Time course of hypoxia experiments, with media being harvested after 2, 6, 12, and 24 hr of exposure of HepG2 cells at 2% and 20% pO2. Data represent the mean of four experiments run in triplicate and are reported as “fold induction.” (C) At the end of each time period, total RNA was collected from the cells and a 1.5-kb mRNA transcript was detected by Northern blot analysis using an EcoRI fragment (938 bp) of the IGFBP-1 cDNA. 18S rRNA shown at the bottom. (D) Densitometry of Northern blot shown in C, with fold increase normalized to 18S rRNA.
Specificity of IGFBP-1 Response to Hypoxia.
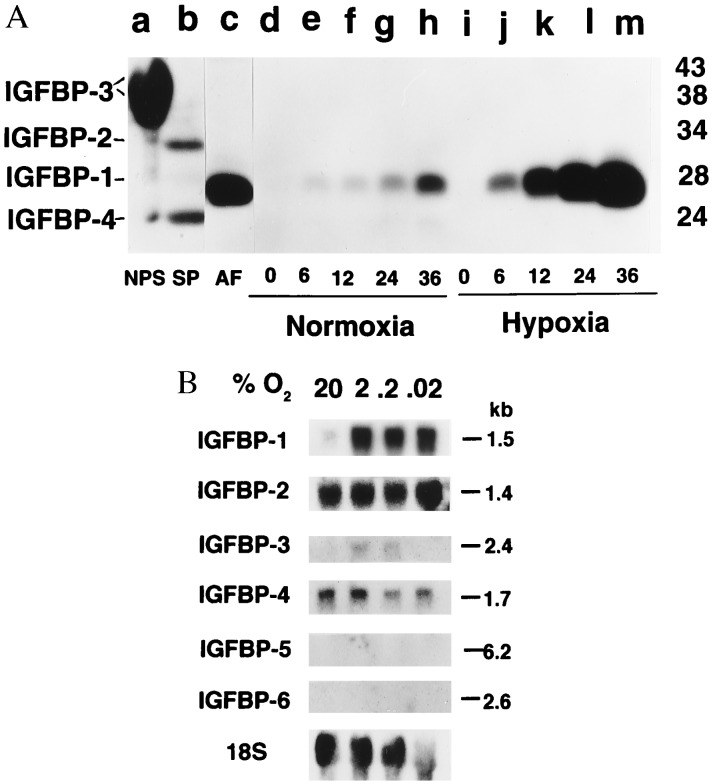
At present, six IGFBPs have been identified (41). Whether hypoxia specifically induces IGFBP-1, compared with the other five high-affinity IGFBPs, in HepG2 cells was investigated. Fig. 2A shows the complement of IGFBPs detectable by Western ligand blot analysis (see controls, lanes a–c). The 28-kDa IGFBP-1 was the primary IGFBP detected in media conditioned by HepG2 cells under normoxic conditions. Very small amounts of IGFBP-2 and IGFBP-4 are detectable only after prolonged exposure of the autoradiograms. IGFBP-1 was markedly augmented (≈6-fold) by hypoxia (2% O2), without a concomitant increase in IGFBP-2 or IGFBP-4. Fig. 2B shows a Northern blot analysis of total RNA from HepG2 cells under hypoxic and normoxic conditions with 32P-labeled cDNA probes for IGFBP-1 through -6. IGFBP-1 mRNA increased nearly 6-fold under hypoxic, compared with normoxic, conditions. Maximal stimulation was observed at 2% O2 and did not increase significantly with decreasing pO2. IGF-BP-2 mRNA and IGFBP-4 mRNA were constitutively expressed and were not affected by hypoxia. IGFBP-3 mRNA was minimally expressed by HepG2 cells, and its expression increased about 2-fold under hypoxic, compared with normoxic, conditions. IGFBP-5 and IGFBP-6 were not expressed by HepG2 cells under normoxia and were not induced under hypoxia. These results are consistent with the Western ligand blots of IGFBPs in media conditioned by these cells.
Figure 2.
Specificity of IGF-binding protein response to hypoxia. (A) Autoradiogram of representative Western ligand blot of media conditioned by HepG2 cells cultured under normoxia (20% O2, lanes d–h) and hypoxia (2% O2, lanes i–m) for time (in hr) shown. Controls are in lanes a–c. Nonpregnant human serum (NPS, lane a) demonstrates IGFBP-3, a prominent doublet with relative molecular mass between 38 and 43 kDa. Lane b shows IGFBP-2 (34 kDa) and IGFBP-4 (24 kDa) in human seminal plasma (SP). Human midgestation amniotic fluid (AF, lane c) shows IGFBP-1 (28 kDa). Molecular mass markers are shown on the right in kDa and migration of IGFBPs is shown on the left. (B) Autoradiogram of representative Northern blot of total RNA isolated from HepG2 cells cultured 24 hr in normoxia (20%) or hypoxia at % O2 shown. Membrane was probed sequentially with 32P-labeled cDNA probes (see text and Materials and Methods). Transcript sizes are shown on the right in kb. Exposure times were: for IGFBP-1, 2 hr; IGFBP-2, 3 days; IGFBP-3, 30 days; IGFBP-4, 3 days; IGFBP-5 and IGFBP-6, 7 days; 18S, 16 hr.
HRE in the IGFBP-1 Gene.
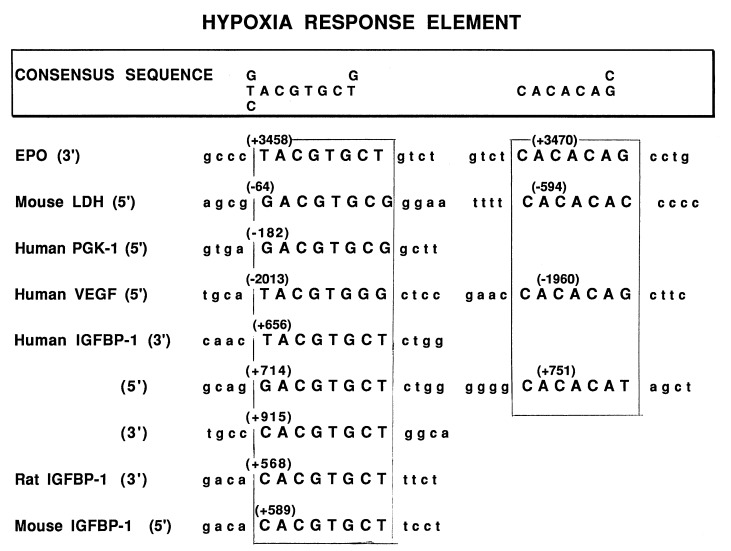
Since IGFBP-1 protein and mRNA levels are induced by hypoxia, we analyzed the structure of the hIGFBP-1 gene (38) to determine whether any consensus HREs were present that might account for our observations in the hypoxic human fetus and in HepG2 cells. As shown in Fig. 3, three potential HREs were identified in intron 1 of the hIGFBP-1 gene. The consensus HRE sequence was based on sequences of elements shown to confer the stimulatory effect of hypoxia on expression of other genes which are also presented in Fig. 3. In addition to the 8-bp HRE present in these other genes, most also have a CACACA/G repeat which may be necessary for full inducibility of these genes by hypoxia (42). One of the HREs in intron 1 of the hIGFBP-1 gene (714-GACGTGCT-721) has a nearby CA repeat (751-CACACA-756), but the other two HREs do not. Of interest, a single HRE is present in the proximal intron 1 region of the mouse and rat IGFBP-1 genes (Fig. 3), but neither is associated with a CA repeat (43, 44).
Figure 3.
Comparison of the HRE consensus sequences and CACACAG/C elements in proximal intron 1 of human, rat, and mouse IGFBP-1 genes and select hypoxia-regulated genes. Nucleotide numbers are those used in GenBank sequences [human (37), rat (42), mouse (43)]. The putative HREs are shown in uppercase letters and are encased by a box. LDH, lactate dehydrogenase; PGK, phosphoglycerokinase.
Hypoxia Induction of IGFBP-1 Is Mediated via the HRE.
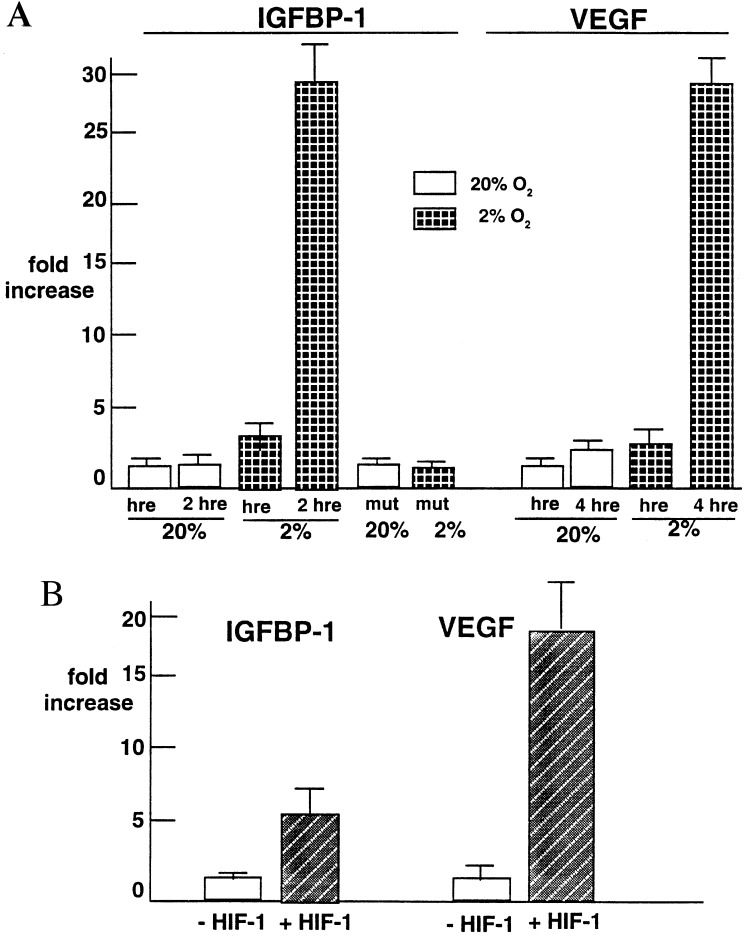
To determine whether the observed induction of IGFBP-1 by hypoxia is mediated by the HREs in intron 1 of the IGFBP-1 gene, a 372-bp fragment of IGFBP-1 that contains the putative HREs was ligated 5′ to a basal hsp70 promoter (39) that drives luciferase expression. Fig. 4A shows the effect of hypoxia on this HRE-luciferase reporter gene activity. A 2-fold induction of luciferase activity was observed under hypoxic versus normoxic conditions with one copy of the 372-bp fragment, whereas approximately a 30-fold induction was observed with two copies compared with control. Similar results were obtained when the vector was transfected into NIH 3T3 Ras-transformed cells (data not shown). A VEGF reporter gene (27) was also transfected with one or four copies of its HRE for comparison. Reporter constructs of VEGF containing a single copy of its HRE resulted in ≈2-fold induction of reporter gene activity, which increased ≈30-fold, when a 4x polymer of the HRE was used. No increase in luciferase reporter activity above baseline was observed with the pGUP.8 vector alone (data not shown). Importantly, when cells were transiently transfected with the mutant IGFBP-1 HRE (716-AAA-718) reporter construct, hypoxia failed to induce reporter gene activity (Fig. 4A). This observation suggests that this consensus sequence is a functional HRE in the hypoxia response of IGFBP-1 regulation by hypoxia.
Figure 4.
HRE and HIF-1 involvement in IGFBP-1 response to hypoxia. (A) Induction of reporter gene activity in HepG2 cells by hypoxia (expressed as fold increase above control, see text). Shown are the mean ± SD for four different experiments for IGFBP-1 HRE constructs and three different experiments with VEGF HRE constructs. Two-way analysis of variance revealed significantly increased reporter gene activity with a single copy of the IGFBP-1 HRE in the construct (P = 0.02) and with two copies of it (P = 0.0001). In normoxia, two copies of the IGFBP-1 HRE construct did not result in an increase in luciferase activity. hre, hypoxia response element; mut, mutant. (B) Transient transfection studies with the HRE in intron 1 of the IGFBP-1 gene attached to a luciferase reporter gene in a reporter construct cotransfected with a constitutively expressing HIF-1α plasmid (+HIF-1) or without (−HIF-1). Fold increase in luciferase activity is shown. VEGF served as a control (see text). Results show the mean ± SD of three different experiments. Two-way ANOVA revealed significantly increased reporter gene activity with cotransfection with the HIF-1α plasmid compared with controls (P = 0.001) and with two copies (P = 0.0001).
HIF-1 and Induction of IGFBP-1.
HIF-1 is a basic helix–loop–helix protein that activates transcription of hypoxia-inducible genes. It consists of an α subunit (HIF-1α) that is unique to HIF-1 and a β subunit (HIF-1β), which is the same as the arylhydrocarbon nuclear transcription subunit ARNT (40, 45) and is constitutively expressed in most cells. To determine whether HIF-1 mediates hypoxia induction of IGFBP-1, we conducted transient cotransfection studies. Cotransfection of the 372-bp fragment containing the transcriptionally responsive HRE and the other two HREs in IGFBP-1 intron 1, with a constitutively expressing HIF-1α plasmid under normoxic conditions, resulted in a 4-fold induction in reporter activity (Fig. 4B). Cotransfection of the 4x HRE-VEGF luciferase reporter gene with the constitutively expressed HIF-1α plasmid showed nearly a 20-fold induction of reporter activity (Fig. 4B; refs. 27 and 46). Thus, these experiments identify a transcriptionally active HRE in the IGF-BP-1 intron 1 responsive to both hypoxia and overexpression of the HIF-1α gene.
DISCUSSION
Acid-base analysis of umbilical artery cord blood is a reliable indicator of fetal oxygenation in utero and at birth. Acute respiratory acidosis and hypoxemia is a common intrapartum event usually associated with umbilical cord compression in which the hypoxic insult is intermittent and occurs within minutes. It is unlikely that the hypoxia is of sufficient duration or magnitude to result in significant tissue hypoxia (25, 26). We speculate that this is why the IGFBP-1 levels are similar in the babies with acute respiratory acidosis and hypoxemia compared with the normal babies. However, with mixed respiratory/metabolic acidosis, tissue hypoxia and acidosis are more likely to occur and are less easily reversible due to more profound hypoxia or longer duration of hypoxia (usually hours). Our findings of increased IGFBP-1 levels in this group of babies suggest that fetal tissue(s), yet to be identified although likely to include the liver (47), may respond to the hypoxic insult by the induction of IGFBP-1. Animal models of chronic intrauterine hypoxia [e.g., uterine artery ligation (18, 20) and maintaining pregnant rodents for several days in hypoxia chambers (21)] demonstrate elevation of IGFBP-1 mRNA overexpression in fetal liver, elevated IGFBP-1 in the fetal circulation, and restricted fetal growth in utero. In humans, we and others have reported elevated levels of IGFBP-1 in babies with long-term, chronic in utero hypoxia due to uteroplacental insufficiency and resultant intrauterine growth restriction (6, 8–17). The process of restricted growth likely occurs over long periods of time (weeks-months), and chronically elevated levels of IGFBP-1 are believed to decrease IGF-I available for somatic growth in utero (below).
There are several reports noting a striking inverse relationship among fetal size, hypoxia, and circulating fetal IGFBP-1 concentrations in humans (11, 12, 17). It is noteworthy that hypoxia impacts on common pathways of glucose utilization. Glucose transporter 1, for example, is induced by hypoxia, as are the glycolytic enzymes, aldolase, triose phosphate isomerase, glyceraldehyde-3-phosphate dehydrogenase, pyruvate kinase, and lactate dehydrogenase (42)—presumably to increase substrate availability. This is in contrast to inhibition of enzymes of gluconeogenesis, e.g., phosphoenolpyruvate carboxykinase. Thus, in a major effort to conserve fuel utilization in the human fetus, mechanisms likely have evolved to limit use of substrates for growth and to direct their utilization for organism homeostasis. Induction of IGFBP-1 with its inhibitory effects on IGF action may be part of this regulatory mechanism during fetal development.
Since most of the six high-affinity IGFBPs inhibit IGF action, the question arises as to why IGFBP-1 selectively is regulated by hypoxia. Of all of the IGFBPs, IGFBP-1 regulates acute bioavailability of the IGFs. In physiologic conditions where conservation of substrate is imperative for survival, limiting bioavailability of a major fetal growth factor, e.g., IGF-I, is important teleologically to the organism. We have analyzed the genomic sequences of the other five high-affinity IGFBPs and have found HREs in IGF-BP-2 and IGFBP-3. There is an HRE in exon 1 of the IGFBP-3 gene and the modest increase in IGFBP-3 mRNA observed in HepG2 cells under hypoxic conditions is intriguing and may reflect responsiveness of this HRE to hypoxia, although at a low level of expression in these cells. With regard to IGFBP-2, the entire gene for this binding protein has not yet been sequenced, mainly because of a large intron 1. Therefore, there may be HREs in this region which are, as yet, unrecognized. At present, there is controversy about hypoxia regulation of IGFBP-2 in animal models of intrauterine fetal hypoxia, and our data are consistent with either the HREs in the IGFBP-2 gene not being responsive to hypoxia in HepG2 cells or that the hypoxia response of different IGFBPs is cell or tissue specific. In support of the latter are observations that IGFBP-2 is induced in some transformed leukemic cells under hypoxic conditions (N. Denko, A. J. Fornace, A.J.G., unpublished data). The tissue-specific regulation of IGFBP expression under low oxygen conditions is unclear, but may involve the requirement for essential transcriptional coactivators whose activation is also induced by hypoxia. Future studies will be directed at understanding the differences in IGFBP expression in different tissues, especially in the developing fetus.
The hypoxia response of IGFBP-1 mRNA and protein in HepG2 cells is time dependent and likely involves activation of IGFBP-1 gene transcription. The transient transfection studies reported herein support direct activation of IGFBP-1 gene transcription involving intron 1. It is also possible that hypoxia may enhance IGFBP-1 mRNA stabilization and studies are underway to determine whether this is an additional mechanism involved in the IGFBP-1 response to hypoxia. Mutations in the HRE +716 to +718 that eliminated the hypoxic response strongly suggest that this HRE is functionally important in hypoxia regulation of IGFBP-1 gene expression. The other two HREs as well as the CACACA sequence in intron 1 may also be important in the hypoxic response of this gene. The CA repeat seems to enhance hypoxia inducibility in other systems (42), and studies are currently underway to investigate the importance of the HRE consensus sequences in intron 1, the CA repeat, and other sequences in hypoxia regulation of IGFBP-1 gene expression.
The observed differences in response between IGFBP-1 and VEGF reporter genes may be due to the number of HRE copies present, differences in the HRE sequences, and/or important sequences surrounding the HREs. In our studies the IGFBP-1 reporter gene had two copies of the HRE, whereas the VEGF reporter had four copies. This difference may account for the different responses observed. However, as shown in Fig. 3, the VEGF and IGFBP-1 HREs are not identical. It is possible that HIF-1 binds with higher affinity to the VEGF HRE than the IGFBP-1 HRE(s), which could lead to increased promoter activity. Also, surrounding sequences are important in transcription factor binding/activation, suggesting that those surrounding the VEGF HRE may allow that HRE to bind/activate putatively HIF-1 to a greater extent than those surrounding the IGFBP-1 HREs. These sequences are currently under investigation.
IGFBP-1 intron 1 HREs cotransfected with a constitutively expressing HIF-1α plasmid resulting in reporter gene activity under normoxic conditions is consistent with HIF-1 acting as a transactivating factor in the hypoxic response of IGFBP-1 gene expression. HepG2 cells contain hypoxia-inducible nuclear proteins that complex with the HRE-containing IGFBP-1 intron 1 probe (unpublished data), although supershift assays to identify HIF-1 have not been possible due to lack of available antiserum. Taken together these studies suggest, but do not prove, that HIF-1 is the responsible transcription factor conferring the hypoxia effects to the IGFBP-1 gene, although additional or different transcription factors may be involved.
The process of oxygen sensing in mammals is incompletely understood, although a heme protein is believed to be involved which has two domains: a heme (redox) domain and a kinase domain (48). The kinase is believed to phosphorylate and activate HIF-1. Others have shown that when HepG2 cells are incubated with CoCl2, there is a dose-dependent increase in erythropoietin mRNA and protein (40). Cobalt as well as iron chelators have been hypothesized to bind to the heme portion of the hypoxia sensor and induce HIF-1, as does hypoxia (42). Increased IGF-BP-1 expression in HepG2 cells in response to CoCl2 (S.I.T., N.M.M., A.J.G., and L.C.G., unpublished data) supports involvement of a heme protein or a similar oxygen sensor in hypoxia regulation of IGFBP-1 gene expression. Identification of the hypoxia sensors and transcription factors involved in inducing IGFBP-1 in response to hypoxia in the human fetus remain challenges for the future.
In pregnancies complicated by uteroplacental insufficiency, decreased placental perfusion results in poor oxygen transfer with resulting fetal hypoxia and also poor nutrient transfer to the fetus. In addition, the fetus is chronically stressed in this environment. The IGFBP-1 promoter contains a cAMP response element and two glucocorticoid response elements that regulate induction of IGFBP-1 gene transcription and an insulin-response element that regulates inhibition of IGFBP-1 gene expression (7, 22). Glucocorticoids associated with fetal stress may contribute to elevated levels of IGFBP-1 in the setting of intrauterine hypoxia and IUGR due to uteroplacental insufficiency (14, 49). Alternatively, low circulating levels of insulin, due to poor nutrient transfer, may contribute to elevated IGFBP-1 levels (50–52). How these cis-regulatory elements interact with hypoxia in controlling IGFBP-1 gene expression is not well understood, but may also be important in understanding the role of IGFBP-1 in regulation of fetal growth in utero.
Acknowledgments
We thank Dr. Ruth Ann Crystal and the staff of Stanford Labor and Delivery Suite and the Lucile Packard Children’s Hospital at Stanford Blood Gas Analysis Laboratory for their help in accessing samples and blood gas analysis, respectively. We also thank Drs. Y. El-Sayed and P. Meyer of the Stanford Gynecology/Obstetrics Department for helpful discussions. This work was supported in part by the Walter and Idun Berry Fellowship (to S.I.T.), National Institutes of Health Grants HD 31398 (to L.C.G.) and CA 73832, and the Naomi Vanden Horn Cancer Research Fund (to A.J.G.).
ABBREVIATIONS
- IUGR
intrauterine growth restriction
- IGF
insulin-like growth factor
- HRE
hypoxia response element
- HIF-1
hypoxia-inducible factor 1
- VEGF
vascular endothelial growth factor
References
- 1.Creasy R K, Resnik R. In: Maternal-Fetal Medicine: Principles and Practice. Creasy R K, Resnik R, editors. Philadelphia: Saunders; 1993. pp. 547–563. [Google Scholar]
- 2.Bang P, Giudice L C, Rosenfeld R G. Front Endocrinol. 1994;6:197–212. [Google Scholar]
- 3.Chard T. Growth Regul. 1994;4:91–100. [PubMed] [Google Scholar]
- 4.Baker J, Liu J P, Robertson E J, Efstratiadis A. Cell. 1993;75:73–82. [PubMed] [Google Scholar]
- 5.Woods K A, Camacho-Hubner C, Savage M O, Clark A J L. N Engl J Med. 1996;335:1363–67. doi: 10.1056/NEJM199610313351805. [DOI] [PubMed] [Google Scholar]
- 6.Fant M, Salafia C, Baxter R C, Schwander J, Vogel C. Regul Pept. 1993;48:29–39. doi: 10.1016/0167-0115(93)90333-4. [DOI] [PubMed] [Google Scholar]
- 7.Lee P D K, Giudice L C, Conover C A, Powell D R. Trans Am Soc Exp Med Biol. 1997;216:319–357. doi: 10.3181/00379727-216-44182. [DOI] [PubMed] [Google Scholar]
- 8.Howell R J S, Perry L A, Choglay N S, Bohn H, Chard T. Br J Obstet Gynaecol. 1985;92:1141–1144. doi: 10.1111/j.1471-0528.1985.tb03026.x. [DOI] [PubMed] [Google Scholar]
- 9.Hall K, Hasson U, Lundin G, Sara V. J Clin Endocrinol Metab. 1986;63:1300–1306. doi: 10.1210/jcem-63-6-1300. [DOI] [PubMed] [Google Scholar]
- 10.Crystal R A, Giudice L C. In: Modern Concepts of Insulin-like Growth Factors. Spencer E M, editor. New York: Elsevier; 1991. pp. 395–408. [Google Scholar]
- 11.Langford, K. S., Blum, W. F., Nicolaides, K., McGregor, A. M. & Miell, J. P. (1993) J. Endocrinol. 139, Suppl., 96–101.
- 12.Unterman T G, Simmons R A, Glick R P, Ogata E S. Endocrinology. 1993;132:327–336. doi: 10.1210/endo.132.1.7678218. [DOI] [PubMed] [Google Scholar]
- 13.Verhaeghe J, Van Bree R, Van Herck E, Laureys J, Bouillon R, van Assche F A. Am J Obstet Gynecol. 1993;169:89–97. doi: 10.1016/0002-9378(93)90137-8. [DOI] [PubMed] [Google Scholar]
- 14.Hills F, Crawford R, Harding S, Farkas A, Chard T. Am J Obstet Gynecol. 1994;171:1292–1295. doi: 10.1016/0002-9378(94)90150-3. [DOI] [PubMed] [Google Scholar]
- 15.Giudice L C, De Zegher F, Gargosky S E, Nonoshita L, Rosenfeld R G, Hintz R L. J Clin Endocrinol Metab. 1995;80:1548–1555. doi: 10.1210/jcem.80.5.7538146. [DOI] [PubMed] [Google Scholar]
- 16.Ostlund E, Bang P, Hagenas L, Gried G. Hum Reprod. 1997;12:840–844. doi: 10.1093/humrep/12.4.840. [DOI] [PubMed] [Google Scholar]
- 17.Larsen T, Main K, Andersson A M, Juul A, Greisen G, Skakkebaek N E. Clin Endocrinol. 1996;45:315–319. doi: 10.1046/j.1365-2265.1996.553812.x. [DOI] [PubMed] [Google Scholar]
- 18.Unterman T, Lascon R, Gotway M B, Oehler D, Gounis A, Simmons R A, Ogata E S. Endocrinology. 1990;127:2035–2037. doi: 10.1210/endo-127-4-2035. [DOI] [PubMed] [Google Scholar]
- 19.Price W A, Rong L, Stiles A D, D’Ercole A J. Endocrinology. 1991;32:291–295. [Google Scholar]
- 20.McLellan K C, Hooper S B, Bocking A D, Delhanty P J D, Phillips I D, Hill D J, Han V K M. Endocrinology. 1992;131:1619–1628. doi: 10.1210/endo.131.4.1382958. [DOI] [PubMed] [Google Scholar]
- 21.Tapanainen P J, Bang P, Wilson K, Unterman T G, Vreman H J, Rosenfeld R G. Pediatr Res. 1994;36:152–158. doi: 10.1203/00006450-199408000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Powell D R, Allander S V, Scheimann A O, Wasserman R M, Durham S K, Suwanichkul A. Prog Growth Factor Res. 1995;6:93–101. doi: 10.1016/0955-2235(95)00034-8. [DOI] [PubMed] [Google Scholar]
- 23.Wang G L, Semenza G L. J Biol Chem. 1993;268:21513–2158. [PubMed] [Google Scholar]
- 24.Wang G L, Semenza G L. Proc Natl Acad Sci USA. 1993;90:4304–4308. doi: 10.1073/pnas.90.9.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorp J A, Dildy G A, Yeomans E R, Meyer B A, Parisi V M. Am J Obstet Gynecol. 1996;175:517–521. doi: 10.1053/ob.1996.v175.a74401. [DOI] [PubMed] [Google Scholar]
- 26.American College of Obstetricians and Gynecologists. ACOG Tech Bull. 1995;216:1–5. [PubMed] [Google Scholar]
- 27.Mazure N M, Chen E Y, Yeh P, Laderoute K R, Giaccia A J. Cancer Res. 1996;15:3436–3440. [PubMed] [Google Scholar]
- 28.Mazure N M, Chen E Y, Laderoute K R, Giaccia A J. Blood. 1997;90:3322–3331. [PubMed] [Google Scholar]
- 29.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 30.Giudice L C, Milkowski D A, Lamson G, Rosenfeld R G, Irwin J C. J Clin Endocrinol Metab. 1991;72:779–787. doi: 10.1210/jcem-72-4-779. [DOI] [PubMed] [Google Scholar]
- 31.Lee Y-L, Hintz R L, James P M, Lee P D K, Shively J E, Powell D R. Mol Endocrinol. 1988;2:404–411. doi: 10.1210/mend-2-5-404. [DOI] [PubMed] [Google Scholar]
- 32.Binkert C, Landwehr J, Mar J-L, Schwander J, Heinrich G. EMBO J. 1989;8:2497–2502. doi: 10.1002/j.1460-2075.1989.tb08386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood W I, Cachianes G, Henzel W J. Mol Encdocrinol. 1989;2:1176–1185. doi: 10.1210/mend-2-12-1176. [DOI] [PubMed] [Google Scholar]
- 34.Shimasaki S, Uchiyama F, Shimonaka M, Ling N. Mol Endocrinol. 1990;4:1451–1458. doi: 10.1210/mend-4-10-1451. [DOI] [PubMed] [Google Scholar]
- 35.Shimasaki S, Shimonaka M, Zhang H-P, Ling N. J Biol Chem. 1991;266:10646–10653. [PubMed] [Google Scholar]
- 36.Shimasaki S, Gao L, Shimonaka M, Ling N. Mol Endocrinol. 1991;5:938–948. doi: 10.1210/mend-5-7-938. [DOI] [PubMed] [Google Scholar]
- 37.Hossenlopp P, Seurin D, Segovia-Quinson B, Hardouin S, Binoux M. Anal Biochem. 1986;154:138–143. doi: 10.1016/0003-2697(86)90507-5. [DOI] [PubMed] [Google Scholar]
- 38.Cubbage M L, Suwanichkul A, Powell D R. Mol Endocrinol. 1989;3:846–851. doi: 10.1210/mend-3-5-846. [DOI] [PubMed] [Google Scholar]
- 39.Funk W D, Pak D T, Karas R H, Wright W E, Shay J W. Mol Cell Biol. 1992;6:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang G L, Jiang B-H, Rue E A, Semenza G L. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones J I, Clemmons D R. Endocr Rev. 1995;16:3–34. doi: 10.1210/edrv-16-1-3. [DOI] [PubMed] [Google Scholar]
- 42.Bunn H F, Poyton R O. Physiol Rev. 1996;76:839–885. doi: 10.1152/physrev.1996.76.3.839. [DOI] [PubMed] [Google Scholar]
- 43.Lee J, Greenbaum L, Haber B A, Nagle D, Lee V, Miles V, Mohn K L, Bucan M, Taub R. Hepatology. 1994;19:656–665. doi: 10.1002/hep.1840190317. [DOI] [PubMed] [Google Scholar]
- 44.Lacson R, Oehler D, Yang E, Goswami R, Unterman T G. Biochim Biophys Acta. 1994;1218:95–98. doi: 10.1016/0167-4781(94)90106-6. [DOI] [PubMed] [Google Scholar]
- 45.Wang G L, Semenza G L. J Biol Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- 46.Semenza G L, Agani F, Booth G. Kidney Int. 1997;51:553–555. doi: 10.1038/ki.1997.77. [DOI] [PubMed] [Google Scholar]
- 47.Pannier E M, Irwin J C, Giudice L C. Am J Obstet Gynecol. 1994;171:746–752. doi: 10.1016/0002-9378(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 48.Goldberg M A, Donning S P, Bunn H F. Science. 1988;242:1412–1415. doi: 10.1126/science.2849206. [DOI] [PubMed] [Google Scholar]
- 49.Crawford R A F, Hills F A, Farkas A, Chard T. Br J Obstet Gynaecol. 1995;102:538–540. doi: 10.1111/j.1471-0528.1995.tb11356.x. [DOI] [PubMed] [Google Scholar]
- 50.Shambarugh G, Cglick R, Radosevich J, Unterman T. Ann NY Acad Sci. 1993;692:270–272. doi: 10.1111/j.1749-6632.1993.tb26231.x. [DOI] [PubMed] [Google Scholar]
- 51.Langford K, Blum W, Nicolaides K, Jones J, McGregor A, Miell J. Eur J Clin Invest. 1994;24:851–856. doi: 10.1111/j.1365-2362.1994.tb02030.x. [DOI] [PubMed] [Google Scholar]
- 52.Gallagher B W, Oliver M H, Elchhorn K, Kessler U, Kiess W. Eur J Endocrinol. 1994;131:398–404. doi: 10.1530/eje.0.1310398. [DOI] [PubMed] [Google Scholar]