Abstract
Contrasting taxonomic treatments of potato landraces have continued over the last century, with the recognition of anywhere from 1 to 21 distinct Linnean species, or of Cultivar Groups within the single species Solanum tuberosum. We provide one of the largest molecular marker studies of any crop landraces to date, to include an extensive study of 742 landraces of all cultivated species (or Cultivar Groups) and 8 closely related wild species progenitors, with 50 nuclear simple sequence repeat (SSR) (also known as microsatellite) primer pairs and a plastid DNA deletion marker that distinguishes most lowland Chilean from upland Andean landraces. Neighbor-joining results highlight a tendency to separate three groups: (i) putative diploids, (ii) putative tetraploids, and (iii) the hybrid cultivated species S. ajanhuiri (diploid), S. juzepczukii (triploid), and S. curtilobum (pentaploid). However, there are many exceptions to grouping by ploidy. Strong statistical support occurs only for S. ajanhuiri, S. juzepczukii, and S. curtilobum. In combination with recent morphological analyses and an examination of the identification history of these collections, we support the reclassification of the cultivated potatoes into four species: (i) S. tuberosum, with two Cultivar Groups (Andigenum Group of upland Andean genotypes containing diploids, triploids, and tetraploids, and the Chilotanum Group of lowland tetraploid Chilean landraces); (ii) S. ajanhuiri (diploid); (iii) S. juzepczukii (triploid); and (iv) S. curtilobum (pentaploid). For other classifications, consistent and stable identifications are impossible, and their classification as species is artificial and only maintains the confusion of users of the gene banks and literature.
Keywords: cultivated, microsatellites, sect. Petota, Solanum tuberosum, taxonomy
The cultivated potato represents one of the most important food plants worldwide, yet interpretation of its gene pool structure remains controversial. Contrasting taxonomic treatments of the landraces have continued over last century, with the recognition of anywhere from 1 to 21 distinct Linnean species, or of various Cultivar Groups within the single species S. tuberosum (1). For consistency in usage in our article and to maintain the names most familiar to scientists, we use the seven species terminology of Hawkes (2). Indigenous cultivated (landrace) potatoes are widely distributed in the Andes from western Venezuela, south to northern Argentina, and with another set of landraces in south-central Chile in Chiloé Island and the adjacent Chonos Archipelago. The Chilean landraces, although once proposed to have arisen independently from central Chile (3), are secondarily derived from the Andean ones (2), likely after hybridization with the Bolivian and Argentinean species Solanum berthaultii (4), a species recently combined with the formerly recognized wild species S. tarijense (5). Three of the Andean-cultivated species are hypothesized to be of hybrid origins with cultivated potatoes and wild species: S. ajanhuiri [S. stenotomum cultivated × S. megistacrolobum wild (6)], S. juzepczukii [S. stenotomum × S. acaule wild (7, 8)], and S. curtilobum [S. andigenum cultivated × S. juzepczukii (7, 8)]. The latter three “bitter potatoes” are grown in upland habitats and are not grown nearly as extensively as S. tuberosum, as outlined by Huamán and Spooner (1).
The relationships and extent of genetic differentiation between the Andean and Chilean landraces has long been controversial. Based on cytoplasmic sterility factors, geographical isolation, and ecological differences, Grun (9) suggested that Chilean landraces were distinct from Andean landraces. Hawkes (2) distinguished the tetraploid Chilean from Andean landraces by characters of the leaf and flower pedicel. Plastid restriction site data documented five genotypes (A, C, S, T, and W types) in the diploid and tetraploid Andean landraces, and the Chilean landraces had three types, A, T, and W (10, 11). The most frequently observed type in the Chilean landraces (21 of 24 or 87.5% of the accessions examined) is type T, which is characterized by a 241-bp deletion (12). Conversely, 5 of the 113 (4.4%) accessions of S. tuberosum subsp. andigenum had the T type (10–12).
Potato landraces have been classified into 21 species (13, 14), 7 species with seven subspecies (2) and 9 species with two subspecies (15, 16), or as the single species S. tuberosum with 8 user-defined Cultivar Groups (1). Cultivar Groups are taxonomic categories used by the International Code of Nomenclature of Cultivated Plants to associate cultivated plants with traits that are of use to agriculturists and are not meant to represent natural groups or species in any classification philosophy. Ploidy levels in cultivated potatoes range from diploid (2n = 2x = 24), to triploid (2n = 3x = 36), to tetraploid (2n = 4x = 48), to pentaploid (2n = 5x = 60). Huamán and Spooner (1) examined the morphological support for the various classifications of potato landraces using representatives of all seven species from the classification of Hawkes (2). The results showed some morphological support for S. ajanhuiri, S. chaucha, S. curtilobum, and S. juzepczukii, lesser support for S. tuberosum subsp. tuberosum, and no support for S. phureja and S. stenotomum. Whatever morphological support for these entities was present was only by using a suite of characters, all of which are shared with other taxa (polythetic support). These results, combined with their likely hybrid origins, multiple origins, and evolutionary dynamics of continuing hybridization, led Huamán and Spooner (1) to recognize all landrace populations of cultivated potatoes as a single species, S. tuberosum, with the eight Cultivar Groups: Ajanhuiri Group, Andigenum Group, Chaucha Group, Chilotanum Group, Curtilobum Group, Juzepczukii Group, Phureja Group, and Stenotomum Group (the latter containing all landraces of the Goniocalyx Group).
The wild relatives of these landraces (Solanum section Petota) are all tuber-bearing and include ≈190 wild species that are widely distributed in the Americas from the southwestern United States to southern Chile (17); they possess all ploidy levels of the cultivars, as well as hexaploids (2n = 6x = 72). Spooner et al. (18) studied the origin of the landraces S. tuberosum subsp. andigenum, S. tuberosum subsp. tuberosum, S. phureja, and S. stenotomum with amplified fragment length polymorphisms (AFLPs). They discovered that (i) in contrast to all prior hypotheses, these species were shown to have a monophyletic origin; (ii) the wild species progenitors were from a group of very similar wild potato species classified in the Solanum brevicaule complex; and (iii) the landraces had their origin in the highlands of southern Peru.
Although ploidy has been a major feature to define the cultivated species, many cultivated potato germplasm collections lack chromosome numbers, and many assumptions of ploidy are likely in error. For example, Ghislain et al. (19) used simple sequence repeats (SSRs) (also known as microsatellites) to assess diversity in the S. phureja collection at the International Potato Center. Solanum phureja is widely grown in the Andes from western Venezuela to central Bolivia and has been defined by short-day adaptation, diploid ploidy (2n = 2x = 24), and a lack of tuber dormancy. SSR results, in combination with chromosome counts, uncovered fully 31% (32 of 102 accessions examined) triploid and tetraploid accessions from the International Potato Center collection of S. phureja that were long assumed to be exclusively diploid.
The purpose of our study is to reexamine the support for classification categories for landrace potatoes, using nuclear SSR markers developed for optimal utility in S. tuberosum regarding polymorphism, quality scores, and genomic coverage (20), supplemented with a plastid DNA deletion marker as discussed below. Nuclear SSRs have been shown to be ideal markers for detecting phylogenetically significant diversity within cultivated potatoes (19, 21, 22). In addition, their codominant nature allows them to identify polyploids when three or four bands (alleles) are found, as was shown by Ghislain et al. (19). This is particularly important for our study, where few cultivated accessions have been characterized for chromosome number yet ploidy has been so important conceptually in defining the cultivated species. Our study also used the 241-bp plastid deletion marker distinguishing most populations of Chilean from Andean potato landraces (4, 12, 23).
Results and Discussion
SSR Neighbor-Joining (NJ) Tree.
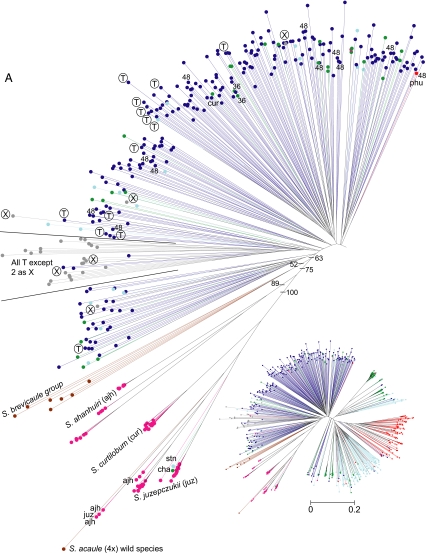
NJ results highlight a tendency to separate three broad groups: (i) putative diploids and triploids (all accessions in the diploid cluster Fig. 1), (ii) putative tetraploids and triploids (most accessions in the polyploid cluster of Fig. 1), and (iii) the hybrid cultivated species S. ajanhuiri (diploid), S. juzepczukii (triploid), and S. curtilobum (pentaploid), grouped with the wild species. Bootstrap support above 50% is common in many small groups of species in the terminal branches of the NJ tree (not shown because they greatly complicate the graphic). However, bootstrap support above 50% is present only in the lower nodes supporting S. ajanhuiri, S. juzepczukii, and S. curtilobum and the wild species.
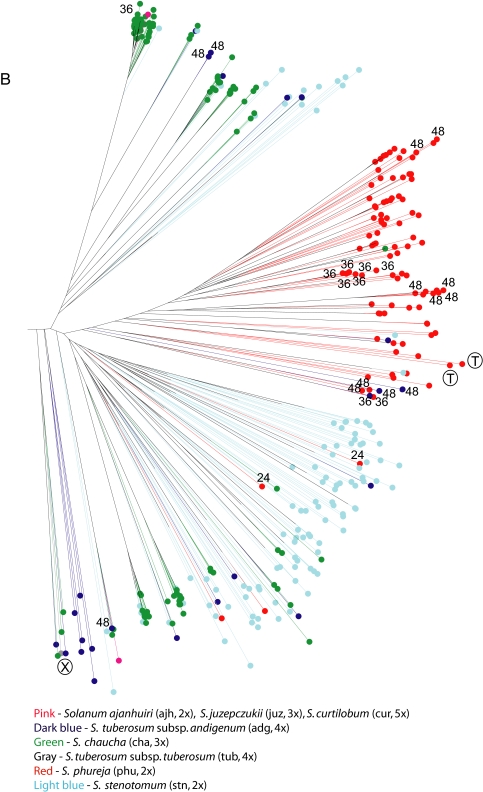
Fig. 1.
Jaccard's tree based on a dissimilarity matrix of 742 potato landraces and 8 wild species examined with 50 microsatellite primer pairs. (Inset) Overall view of the entire tree. (A) Detail of the left-hand side of this tree, which contains mostly putative polyploid accessions (the “polyploid cluster”) and accessions of S. ajanhuiri, S. curtilobum, S. juzepczukii, and wild species. (B) Mostly putative diploid accessions and some triploids (the “diploid cluster”). Chromosome numbers (36 and 48) are labeled for accessions not 24 within the main red S. phureja cluster (in red) on the left right side of the tree or previously identified as S. phureja but falling outside of the red cluster. The circled “T” (possessing a 241-bp deletion) and “X” (no deletion) designate accessions unexpected to occur in these clusters because they either lack the T cytoplasm thought be largely confined to lowland Chile (area in the wedge) or lack this deletion (area outside of the wedge containing mostly accessions from Venezuela to Argentina).
However, there are many exceptions of clustering by ploidy. Landraces of S. goniocalyx, as in the morphological study of Huamán and Spooner (1), were invariably intermixed with those of S. stenotomum and are so labeled as this species on Fig. 1. There are 28 putative triploid landraces of S. chaucha present on the main polyploid cluster of the tree and 123 on the main diploid cluster. There are 28 putative tetraploids (S. tuberosum subsp. andigenum and subsp. tuberosum) on the diploid cluster and 28 putative diploids (S. phureja and S. stenotomum) on the tetraploid cluster. In addition, S. phureja (the only cultivated species with extensive chromosome counts) shows 18 of the 89 accessions to be triploid or tetraploid. Regarding the S. phureja, Fig. 1 shows the position of all accessions formerly identified as this species in the International Potato Center collection (24) but shown to be polyploid in the study of Ghislain et al. (19). Most are present in the main “S. phureja cluster” (red dots in the diploid cluster of Fig. 1), but this cluster also contains two accessions of S. tuberosum subsp. andigenum, two of S. stenotomum, and one of S. chaucha, and with 10 accessions elsewhere on the diploid cluster and 16 on the polyploid cluster. As expected, most (22 of 27) of the S. tuberosum subsp. tuberosum accessions clustered together (the area in the wedge in the polyploid cluster in Fig. 1). However, this cluster also contained three accessions of S. tuberosum subsp. andigenum.
241-bp Plastid Deletion.
We determined the presence or absence of the 241-bp plastid deletion for all 742 cultivated accessions examined. As expected, most (22 of 23) of the S. tuberosum subsp. tuberosum accessions in the main cluster of this subspecies (designated by the wedge in the polyploid cluster of Fig. 1) possessed this deletion. Also in the area of the wedge are three tetraploid accessions from Peru (dark blue); two of these possess the deletion, and one lacks it (the two accessions lacking the deletion are marked with “X”).
All four remaining accessions from Chile falling outside of this cluster (gray dots marked with “X”) lack the 241-bp deletion characteristic of this subspecies, suggesting misidentifications of possible recent introductions of the S. tuberosum subsp. andigenum into Chile. Thirteen of the 251 S. tuberosum subsp. andigenum accessions (5.2%; marked with “T” outside of the gray S. tuberosum subsp. tuberosum cluster) possessed the deletion, similar to the 4.4% reported in prior studies (above). These 13 accessions are widely distributed throughout the Andes in Venezuela (2 accessions), Colombia (1 accession), Ecuador (13 accessions), Peru (4 accessions), Bolivia (2 accessions), and Argentina (1 accession). In addition, one of the S. stenotomum accessions (putatively diploid) and two S. phureja accessions (known as diploid) possessed this deletion, the first report of diploid potatoes possessing this deletion, because none of the accessions of S. stenotomum (215) and S. chaucha (150) previously screened was found with this marker (19, 25). Unfortunately, these three accessions do not have reliable collection information.
Reconsideration of the Classification of Cultivated Potato.
In combination with a recent morphological study (1), the SSR data support the reclassification of the cultivated potatoes into four species: (i) S. tuberosum, (ii) S. ajanhuiri (diploid), (iii) S. juzepczukii (triploid), and (iv) S. curtilobum (pentaploid). We support dividing S. tuberosum into two Cultivar Groups (Andigenum Group of upland Andean genotypes containing diploids, triploids, and tetraploids, and the Chilotanum Group of lowland tetraploid Chilean landraces). Because Cultivar Groups are taxonomic categories used to associate cultivated plants with traits that are of use to agriculturists, this classification is convenient to separate these populations that grow in different areas, are adapted to different day-length regimes, and have some degree of unilateral sexual incompatibility to the Andean populations. For the remaining “species” or Cultivar Groups, consistent and stable identifications are impossible, their classification as Linnean species is artificial, and their maintenance as either species or Cultivar Groups only serves to perpetuate confusion by breeders and gene bank managers, and the instability of names in the literature. For example, Ghislain et al. (19) showed S. phureja to be indefinable as traditionally recognized because prior authors incorrectly assumed that their assumption of diploidy was incorrect for 31% of the accessions, and our results showed many accessions of S. phureja to cluster with the polyploids. The recognition of S. phureja as either a species or Cultivar Group (Phureja Group), therefore, is no longer tenable because it is no longer diploid, does not exclusively possess low-dormancy tubers, is not short-day adapted, and is not morphologically coherent (1). The other species (or Cultivar Groups) have ploidy as a major identifying criterion. The results from S. phureja and this study indicate that chromosome counts from other accessions of cultivated potatoes will uncover a high proportion of counts not matching expectations based on their identifications.
Ploidy has been of great importance in the classification of cultivated potatoes, but our results show so many exceptions that it is a poor character to define gene pools. Cultivated potato fields contain mixtures of different ploidy levels (6, 26–32). Bukasov (33) was the first to count chromosomes of the cultivated potatoes and used ploidy variation to speculate on hybrid origins. The strong reliance on ploidy levels was clearly stated by Hawkes and Hjerting (34): “The chromosome number of 2n = 36 largely helps to identify S. chaucha, but morphological characters can also be used.”
Morphology is a poor character to define most species or Cultivar Groups except for the bitter potato species S. ajanhuiri, S. curtilobum, and S. juzepczukii. As shown by Huamán and Spooner (1), most traditionally recognized cultivated potato species have little morphological support, and then only by using a suite of characters, all of which are shared with other taxa (polythetic support).
The International Potato Center has collected cultivated potatoes for 30 years and has invested tremendous effort in their identification. An examination of identification records at the International Potato Center shows many changes over the years, further showing the lack of stability of any character set to reliably define most cultivated species.
Potato gene banks are in great need of an integrated and comprehensive program of ploidy determinations; controlled and replicated studies of tuber dormancy (which we suspect will highlight grades of dormancy, not the present/absent determinations that exist today); photographically documented determinations of tuber and flesh colors and tuber shapes; and determinations of tuber pigments, glycoalkaloid contents, carbohydrates, proteins, amino acids, minerals, and secondary metabolites, using functional genomics approaches, with all data publicly integrated into a readily searchable web-based bioinformatics database. Such a multicomponent system will serve the breeding community much better than the outdated, unstable, and phylogenetically indefensible traditional classifications that exist today.
Materials and Methods
Plant Materials.
A total of 742 potato landraces of all cultivated potato species were examined: S. tuberosum subsp. andigenum, putatively tetraploid (251 accessions); S. ajanhuiri, diploid (22); S. chaucha, triploid (151 accessions); S. tuberosum subsp. tuberosum, tetraploid (27 accessions); S. curtilobum, pentaploid (21 accessions); S. juzepczukii, triploid (35 accessions); S. phureja, diploid (104 accessions); S. stenotomum, diploid (131 accessions); 7 diploid wild species accessions in the northern S. brevicaule complex S. ambosinum Ochoa (1 accession), S. bukasovii Juz. (4 accessions), and S. multiinterruptum Bitter (2 accessions); and the wild tetraploid species S. acaule Bitter (1 accession) (750 accessions in total with the 8 wild species). Selection of these wild species is based on recent amplified fragment length polymorphism (AFLP) studies that documented the northern S. brevicaule complex wild species to be the progenitors of the cultivated potatoes and S. acaule believed to be a wild species parent in the hybrid species S. juzepczukii and S. curtilobum. We qualify landrace collection ploidy as “putative” because only S. phureja has been counted in detail (19), that showed extensive examples of incorrect assumptions of ploidy as discussed above. Data of these accessions that includes International Potato Center accession number, taxonomic identification, ploidy when known, locality of collection, and average number of SSR alleles per accession are available as a supporting information (SI) Dataset.
DNA Extraction, SSR Primers, PCR Conditions, and Electrophoresis.
Genomic DNA was obtained by using standard protocols at the International Potato Center (35). DNA concentration was calculated by using PicoGreen dsDNA quantitation reagent (Molecular Probes) and a TBS-380 Fluorometer (Turner BioSystems). DNA dilutions were performed to achieve a final concentration of 3 ng/μl, using 96-well plates. We used 50 nuclear SSRs (see SI Dataset) screened from 88 that included the 22 from the Potato Genetic Identity (PGI) kit (20), 13 from ESTs developed at the Scottish Crop Research Institute (36), 30 identified by using the potato EST database at TIGR, and 23 from the University of Idaho (37). PCR reactions were performed in a 10-μl volume containing 100 mM Tris·HCl (Sigma), 20 mM (NH4)2SO4 (Merck), 2.5 mM MgCl2 (Merck), 0.2 mM each dNTP (Amersham Biosciences), 0.3 μM labeled M13 forward primer (LI-COR IRDye 700 or 800), 0.3 μM M13-tailed SSR forward primer (Invitrogen), 0.2 μM SSR reverse primer (Invitrogen), 1 unit of Taq polymerase (GIBCO/BRL), and 15 ng of genomic DNA. PCR was carried out in a PTC-200 thermocycler (MJ Research). The program used was the following: 4 min at 94°C, followed by 33 cycles of 50 sec at 94°C, 50 sec at annealing temperature (T°a), and 1 min at 72°C, then 4 min at 72°C as a final extension step. PCR products were separated by electrophoresis on a LI-COR 4300 DNA analyzer system. The molecular weight ladder was the LI-COR IRDye 50–350 bp size standard and was loaded into gel each eight samples.
SSR Allele Scoring.
SSR alleles were detected and scored by using SAGA Generation 2 software (LI-COR). Size calibration and an SSR “smiling line” were performed by using the molecular weight ladder (LI-COR IRDye 50–350). The SSR alleles were determined for size in bp of the upper band of the allele and scored as present (1) or absent (0). Missing data were scored as “9.”
Data Analysis.
Genetic analysis was performed by using the program DARwin (38). A dissimilarity matrix was calculated by using Jaccard's coefficient, 60% of minimal proportion of valid data required for each unit pair, and 500 replicate bootstrapping. The dendrogram was built by using the NJ method, using the seven wild species accessions in the northern S. brevicaule complex as outgroup. The NJ method developed by Saitou and Nei (39) estimates phylogenetic trees. Although based on the idea of parsimony (it does yield relatively short estimated evolutionary trees), the NJ method does not attempt to obtain the shortest possible tree for a set of data. Rather, it attempts to find a tree that is usually close to the true phylogenetic tree (40). This method allows the rooting of trees on outgroups (in this case, the seven accessions of the S. brevicaule complex). The polymorphic information content (PIC) was calculated as PIC = 1 − Σ(pi2), where pi is the frequency of the ith allele detected in all accessions (41). Data of somatic chromosome counts for accessions of S. phureja were obtained from Ghislain et al. (19).
Plastid DNA Polymorphism Detection.
The 241-bp deletion was analyzed for all 742 cultivated accessions by using the primers from ref. 23. PCR amplification was performed in a volume of 10 μl consisting of 18 ng of genomic DNA, 0.4 μM each of primers (Invitrogen), 1× PCR buffer (PerkinElmer), 2.5 mM MgCl2 (PerkinElmer), 200 μM each dNTP (Amersham Biosciences), and 0.25 unit of Taq DNA polymerase (GIBCO/BRL). Thermal cycling was carried out in a PTC-200 thermocycler (MJ Research) (one cycle of 4 min at 94°C, followed by 40 cycles of 45 sec at 94°C, 45 sec at 59°C, and 45 sec at 72°C, then terminated with one cycle of 4 min at 72°C). PCR products were separated by electrophoresis in a 1% agarose gel, and lambda phage digested by PstI was used as a molecular weight marker. The 241-bp plastid polymorphism was determined for size in bp and scored as “T” (≈200 bp for deleted type) and “X” (≈440 bp for undeleted type).
Acknowledgments
We thank David Douches and Lynn Bohs for comments on an earlier draft of the manuscript. This work was supported by the International Potato Center, U.S. Department of Agriculture, Generation Challenge Program Grant SP1C2-2004-5, and National Science Foundation Grant DEB 0316614 entitled “A world-wide treatment of Solanum” (to D.M.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709796104/DC1.
References
- 1.Huamán Z, Spooner DM. Am J Bot. 2002;89:947–965. doi: 10.3732/ajb.89.6.947. [DOI] [PubMed] [Google Scholar]
- 2.Hawkes JG. The Potato: Evolution, Biodiversity, and Genetic Resources. London: Belhaven; 1990. [Google Scholar]
- 3.Ugent D, Dillehay T, Ramirez C. Econ Bot. 1987;41:17–27. [Google Scholar]
- 4.Hosaka K. Am J Potato Res. 2003;80:21–32. [Google Scholar]
- 5.Spooner DM, Fajardo D, Bryan GJ. Taxon. 2007;56:987–999. [Google Scholar]
- 6.Johns T, Huamán Z, Ochoa CM, Schmiediche PE. Syst Bot. 1987;12:541–552. [Google Scholar]
- 7.Hawkes JG. Z Pflanzenzuecht. 1962;47:1–14. [Google Scholar]
- 8.Schmiediche PE, Hawkes JG, Ochoa CM. Euphytica. 1982;31:395–707. [Google Scholar]
- 9.Grun P. Econ Bot. 1990;44(Suppl 3):39–55. [Google Scholar]
- 10.Hosaka K, Hanneman RE., Jr Theor Appl Genet. 1988;76:333–340. doi: 10.1007/BF00265332. [DOI] [PubMed] [Google Scholar]
- 11.Hosaka K, de Zoeten GA, Hanneman RE., Jr Theor Appl Genet. 1988;75:741–745. [Google Scholar]
- 12.Kawagoe Y, Kikuta Y. Theor Appl Genet. 1991;81:13–20. doi: 10.1007/BF00226106. [DOI] [PubMed] [Google Scholar]
- 13.Bukasov SM. In: Flora of Cultivated Plants. Bukasov SM, editor. Vol 9. Leningrad, Russia: Kolos; 1971. pp. 5–40. [Google Scholar]
- 14.Lechnovich VS. In: Flora of Cultivated Plants. Bukasov SM, editor. Vol 9. Leningrad, Russia: Kolos; 1971. pp. 41–302. [Google Scholar]
- 15.Ochoa CM. The Potatoes of South America: Bolivia. Cambridge, UK: Cambridge Univ Press; 1990. [Google Scholar]
- 16.Ochoa CM. Las Papas de Sudamérica: Perú, Part 1. Lima, Peru: International Potato Center; 1999. [Google Scholar]
- 17.Spooner DM, Salas A. Handbook of Potato Production, Improvement, and Post-Harvest Management. Binghamton, NY: Haworth; 2006. pp. 1–39. [Google Scholar]
- 18.Spooner DM, McLean K, Ramsay G, Waugh R, Bryan GJ. Proc Natl Acad Sci USA. 2005;120:14694–14699. doi: 10.1073/pnas.0507400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghislain M, Andrade D, Rodríguez F, Hijmans R, Spooner DM. Theor Appl Genet. 2006;113:1515–1527. doi: 10.1007/s00122-006-0399-7. [DOI] [PubMed] [Google Scholar]
- 20.Ghislain M, Spooner DM, Rodríguez F, Villamón F, Núñez J, Vásquez C, Waugh R, Bonierbale M. Theor Appl Genet. 2004;108:881–890. doi: 10.1007/s00122-003-1494-7. [DOI] [PubMed] [Google Scholar]
- 21.Raker C, Spooner DM. Crop Sci. 2002;42:1451–1458. [Google Scholar]
- 22.Ríos D, Ghislain M, Rodríguez F, Spooner DM. Crop Sci. 2007;47:1271–1280. [Google Scholar]
- 23.Hosaka K. Am J Potato Res. 2002;79:119–123. [Google Scholar]
- 24.Huamán Z, Golmirzaie A, Amoros W. In: Biodiversity in Trust: Conservation and Use of Plant Genetic Resources in CGIAR Centres. Fuccillo D, Sears L, Stapleton P, editors. Cambridge, UK: Cambridge Univ Press; 1997. pp. 21–28. [Google Scholar]
- 25.Hosaka K. Am J Potato Res. 2004;81:153–158. [Google Scholar]
- 26.Ochoa CM. Expedición Colectora de Papas Cultivadas a la Cuenca del Lago Titicaca. Vol 1. Lima, Perú: Ministerio de Agricultura; 1958. [Google Scholar]
- 27.Jackson MT, Hawkes GJ, Rowe PR. Euphytica. 1980;29:107–113. [Google Scholar]
- 28.Brush SB, Carney HJ, Huamán Z. Econ Bot. 1981;35:70–88. [Google Scholar]
- 29.Johns T, Keen SL. Econ Bot. 1986;40:409–424. [Google Scholar]
- 30.Quiros CF, Brush SB, Douches DS, Zimmerer KS, Huestis G. Econ Bot. 1990;44:254–266. [Google Scholar]
- 31.Quiros CF, Ortega R, Van Raamsdonk L, Herrera-Montoya M, Cisneros P, Schmidt E, Brush SB. Genet Res Crop Evol. 1992;39:107–113. [Google Scholar]
- 32.Zimmerer K. J Biogeogr. 1991;18:165–178. [Google Scholar]
- 33.Bukasov SM. Physis (Buenos Aires) 1939;18:41–46. [Google Scholar]
- 34.Hawkes JG, Hjerting JP. The Potatoes of Bolivia: Their Breeding Value and Evolutionary Relationships. Oxford: Oxford Univ Press; 1989. [Google Scholar]
- 35.Ghislain M, Zhang D, Herrera M, editors. Molecular Biology Laboratory Protocols: Plant Genotyping, Genetic Resources Department Training Manual. Lima, Peru: International Potato Center; 1997. [Google Scholar]
- 36.Milbourne D, Meyer R, Collins A, Ramsay L, Gebhardt C, Waugh R. Mol Gen Genet. 1998;259:233–245. doi: 10.1007/s004380050809. [DOI] [PubMed] [Google Scholar]
- 37.Feingold S, Lloyd J, Norero N, Bonierbale M, Lorenzen J. Theor Appl Genet. 2005;111:456–466. doi: 10.1007/s00122-005-2028-2. [DOI] [PubMed] [Google Scholar]
- 38.Perrier X, Jacquemoud-Collet JP. DARwin. 2006 ( http://darwin.cirad.fr/darwin), Version 5.0.
- 39.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 40.Rohlf FJ. NTSYSpc: Numerical Taxonomy and Multivariate Analysis System. Setauket, NY: Exeter Software; 1997. Version 2.0. [Google Scholar]
- 41.Nei M. Proc Natl Acad Sci USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]