Abstract
Escherichia coli strains with inactivated rluD genes were previously found to lack the conserved pseudouridines in helix 69 of 23S ribosomal RNA and to grow slowly. A suppressor mutant was isolated with a near normal growth rate that had changed the conserved Glu-172 codon to a Lys codon in prfB, encoding translation termination factor RF2. When nonsense suppression in strains with all combinations of prfB+/prfBE172K and rluD+/rluD::cat was analyzed, misreading of all three stop codons as sense codons was found to be increased by rluD inactivation: Nonsense suppression was increased 2-fold at UAG codons, 9-fold at UAA, and 14-fold at UGA. The increased read-through at UGA corresponds to reading UGA as a sense codon in 30% of the cases. In contrast, the accuracy of reading sense codons appeared unaffected by loss of rluD. When the inactivated rluD gene was combined with the altered prfB, wild-type levels of termination were restored at UAA codons and termination was more efficient than wild type at UGA. These results strongly suggest that at least one of the helix 69 pseudouridines has a function in translation termination. To our knowledge, this is the first described function for a ribosomal RNA pseudouridine modification.
Keywords: nonsense suppression, release factor 2, rluD, rRNA modification
Pseudouridine is found in many species of stable RNA. Its formation is catalyzed by several pseudouridine synthases and takes place after transcription (1). No function has so far been identified for any of the pseudouridines in ribosomal RNA (2). In Escherichia coli 7 enzymes catalyze the formation of the 11 pseudouridines in rRNA (3). The rluD gene product catalyzes the formation of the pseudouridine in helix 69 (h69) at positions 1911, 1915, and 1917 in 23S rRNA (4). In the structure of the 70S ribosome, h69 contacts both A and P site tRNA and also contributes to the universally conserved bridge B2a between the two ribosomal subunits (5). Changes in h69 can result in defects in subunit association (6, 7) and in an altered translational accuracy (8). Recently, Ali et al. (9) were able to prepare ribosomes in which h69 was deleted. These retained the ability to perform all separate ribosome functions, including proper translational accuracy and ability to translate a natural mRNA. Only peptide chain termination catalyzed by RF1 and ribosome recycling catalyzed by RRF were affected. It was therefore concluded that the main function of h69 was in translation initiation, translation termination, or ribosome recycling (9).
The presence of pseudouridines in h69 is a highly conserved feature of cytoplasmic ribosomes (4) even if the modification mechanism in eukaryotes is very different from the mechanism used by bacteria (10, 11). The question of whether all h69 pseudouridines are equally important was addressed by Hirabayashi et al. (7), who isolated strains where the ribosomal RNA did not have U1915 but retained normal cellular growth rate. They were unable to isolate variants with an altered U1917 and did not address the essentiality of U1911, but the absence of the U1911 pseudouridine modification in Haloarcula marismortui (12) would suggest that the pseudouridine U1917 has the most important function.
Despite the strong conservation of the h69 pseudouridines, the rluD gene is dispensable for the growth of E. coli. However, the inactivated rluD::cat mutant is extremely slow-growing, and faster-growing derivatives invariably arise during culture. These faster-growing derivatives still lack all three h69 pseudouridines and are presumed to contain second site mutations that partially suppress the defects of rluD inactivation (13). The slow-growing, suppressor-free rluD::cat strain and its faster-growing derivatives have been termed dust and tiny, respectively (6). The aim of this study is to characterize the rluD dust strain and to identify the function of the highly conserved, yet dispensable, pseudouridines in helix 69 of 23S ribosomal RNA.
Results
Characterization of the rluD Mutant.
In a given growth medium the cellular regulatory system aims to maintain a constant activity of ribosomes in the cell. If ribosomal activity is limited for instance by the concentration of an initiation factor, IF2 (14), or of a ribosomal protein, protein S1 (15), the cellular regulatory response is to increase the synthesis of ribosomal components, rRNA, and ribosomal proteins. Because rRNA constitutes ≈85% of total RNA, an increase in the amount of total RNA is observed under these conditions. If a ribosomal function is deficient in the rluD mutant, this is expected to increase the RNA content. The data presented in Fig. 1A show that inactivation of rluD resulted in that the dust and tiny mutants had about doubled the RNA content compared with the value expected from the growth rate. Fig. 1A also shows that each mutant had almost the same growth rate in glucose minimal medium and in the rich YT medium. This is a very unusual finding that indicated that the rluD inactivation affected growth more severely in the enriched medium.
Fig. 1.
Effect of rluD inactivation on growth. (A) RNA content of rluD wild type and mutant strains growing in different media. RNA content, in milligrams of RNA per gram of dry weight, was plotted as function of the cellular growth rate. Filled squares, wild-type MAS70 grown in glucose or YT medium; triangles, rluD:cat variant ME96 tiny grown in glucose (filled) or YT (open) medium; diamonds, ME97 rluD::cat dust grown in glucose (filled) or YT (open) medium respectively; open squares, rluD::cat, prfBE172K grown in glucose or YT medium. The experiments were performed at least three times. The standard deviations are indicated. (B) Utilization of the carbon source in wild-type and dust strains. The cultures were inoculated with the indicated amount of glucose being limiting for growth. After growth for a sufficiently long time to exhaust the carbon source, the optical density of the cultures was measured and converted to cell mass (see Materials and Methods). Squares, wild-type MAS70; open diamonds, ME97 rluD::cat dust. (C) RNA content during recovery from stationary phase. Cells were grown to stationary phase overnight in YT medium and diluted in fresh YT medium to an OD436 of ≈0.1, after which RNA contents were followed as a function of time after dilution. Squares, wild-type MAS70; diamonds, ME97 rluD::cat dust.
The growth experiments with dust were difficult because the faster-growing tiny variants appeared readily (13). Routinely, three independent cultures were inoculated from separate dust single colonies, and only cultures that had the slow dust growth rate after overnight incubation were used for experiments (see also Materials and Methods). We emphasize that the tiny strains may represent a heterogeneous group of suppressors and that the growth experiments we report for tiny have been done with the particular tiny suppressor present in the strain ME96.
To determine one of the ribosome functions directly, we measured the peptide polymerization rate in vivo by our previously developed method, which determines the time for 35S-Met pulse-labeled nascent peptides to appear in full-length protein after a chase with excess unlabeled Met. Using curve fitting that takes the localization of the Met residues in the protein into consideration, the SEM of this method is ≈1 sec (16). Table 1 summarizes these results, which indicated that the peptide chain growth rate is reduced by only15% in the dust strain, which is not enough to explain its 2-fold-lower growth rate.
Table 1.
Peptide elongation rates in the wild-type and dust strains
| Strain | EFG | DnaK | GroEL |
|---|---|---|---|
| ME3 wild type | 13.2 ± 0.7 | 13.3 ± 0.3 | 13.7 ± 1.0 |
| ME33 rluD dust | 11.4 ± 0.6 | 11.8 ± 0.6 | 11.4 ± 0.8 |
The rates are given in codons translated per second at 37°C. The rate depends on the amino acid composition and the codon usage of the protein (16), but the mRNAs used here have the codon usage typical of ribosomal proteins. The average results of three experiments and the standard errors are shown.
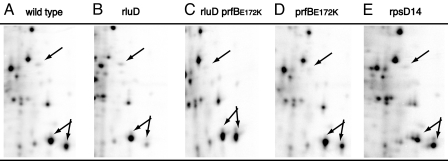
The expression level of individual proteins in the dust and wild-type strains was analyzed by two-dimensional gel electrophoresis of 35S-Met pulse-labeled extracts. Among the few and minor differences observed were an ≈20% increased synthesis of elongation factor Tu that would be compatible with an increased synthesis of ribosomal proteins and factors, and a similar increase in the synthesis of the heat-shock protein DnaK that could be caused by synthesis of aberrant and unstable proteins in the dust strain. Examination of these gels, of which sections are shown in Fig. 2, showed that all protein spots migrated at their normal positions and were without the “stuttering” spots in the isoelectric pH dimension, variants that are typically observed under conditions where misreading of sense codons takes place (17). For comparison, Fig. 2E shows a corresponding gel section from a ram strain (ribosomal ambiguity mutant, mutated in rpsD encoding ribosomal protein S4) that supports elevated levels of sense codon misreading (18) and where the characteristic stuttering spots are clearly visible. The amino acid substitutions arising from random misreading of sense codons will give a charge change of the protein in approximately one-third of the cases. Each of the proteins shown in Fig. 2 has >300 aa and therefore has >300 chances for having a codon misread during synthesis. We estimate to be able to detect whether 10% of the protein in a spot had a charge change. Fig. 2B therefore shows that the level of missense errors in the rluD inactivated strain was at most one such error per thousand codons.
Fig. 2.
Two-dimensional electrophoresis of total cell extracts from wild-type MAS70 (A), ME97 rluD::cat dust (B), ME123 rluD::cat dust, prfBE172K (C), ME125 rluD+prfBE172K (D), and the ram strain rpsD14 (E) (31). Sections of the gels showing protein with size from 55,000 to 30,000 (top to bottom) and isoelectric points from 6 to 4.5 (left to right). The position of a spot with the molecular weight and isoelectric point of RF2 is indicated with an arrow. The double arrow points to the outer membrane proteins OmpC and OmpF. Especially in this area mistranslation in the ram strain can be seen (E).
The yield of cell mass that could be generated from a given amount of glucose was reduced to ≈60% in the dust strain. This is shown in Fig. 1B, and a similar reduction was found in the YT medium (data not shown). Instead of 100 μg of glucose being used to generate ≈50 μg of cell mass found for wild-type cells, the dust cell generated only 30 μg of cell mass at the expense of oxidizing 70 μg for energy production. This indicated that extensive energy consuming, futile synthesis, and degradation processes took place as a consequence of inactivating the rluD gene.
During these studies we found that the rluD dust strain, grown to stationary phase in the rich YT medium, was able to grow with a wild-type rate for some time after being diluted into fresh medium. We measured total RNA per cell mass during such an experiment. As seen from Fig. 1C, the ratio between RNA and cell mass in both strains initially was identical and the RNA content of the dust strain initially rose steeply before reaching the higher steady-state value. During this time the cellular growth rate of the dust strain decreased from the wild-type rate to the 4-fold-lower steady-state value. We conclude from these experiments that the dust cell during steady-state growth synthesized an unstable RNA-containing growth inhibitor that in all likelihood is the incomplete ribosomal particles observed previously (6) and which had time to decay during the stationary phase.
Isolation of a Suppressor of the dust Phenotype.
During cultivation of the dust strain we observed a colony with a wild-type colony size, clearly different from that of the parental dust strain or its tiny derivatives that carry weak suppressors of rluD::cat. This mutant was still resistant to chloramphenicol, indicating that rluD was inactivated. This was verified by PCR analysis on chromosomal DNA with primers internal to rluD and flanking the chloramphenicol resistance cassette (4). The results showed that the strain contained only one copy of rluD interrupted by the cassette. The growth rate and RNA composition of the suppressor mutant were determined and are included in Fig. 1A. As shown, both growth rate and RNA content in the suppressor strain had reverted to near wild-type levels.
To characterize the suppressor, a plasmid library was made from its chromosomal DNA. We used a partial digestion with the restriction enzyme Sau3AI and inserted fragments with a length of >3,000 bp into the BamHI restriction site in the tet gene of the plasmid pBR322 (19). A “control library” was made in parallel by using DNA from the wild-type strain ME3. From the frequency of lac+ plasmids we estimate that both libraries contained sufficient plasmids to allow each gene on average to be present on six independent clones. Both libraries were transformed into an rluD::cat dust strain, and colonies with a normal colony size were isolated. In the transformation with the control library we found 40 of 33,000 clones with wild-type colony size. These were isolated, and all carried the wild-type rluD gene as judged by restriction enzyme analysis and PCR. From the library made with DNA from the rluD::cat suppressor strain, 10 of 30,000 clones (of which at least four were independent) gave rise to a normal colony size, none of which carried the rluD region. Plasmid DNA from these transformants was retransformed into the rluD::cat dust strain, and plasmid DNA that again supported normal growth was characterized by sequencing from two primers in the tet gene flanking the BamHI site. A BLAST search showed that all four plasmids carried fragments from the 65-min region of the genetic map of E. coli and that translation termination factor RF2 was the only common protein that could be expressed. The prfB gene was sequenced from one of the short clones that could encode only RF2. Only one sequence change was observed: the G in codon 172, GAA, was changed to an A, changing the wild-type Glu residue to Lys. The prfB gene from the suppressor's chromosomal DNA was then amplified by PCR and sequenced. This verified that the prfBE172K mutation was present in the original mutant. As explained in Materials and Methods we transferred the prfBE172K mutation to a new strain. In this new genetic background the suppressor still suppressed the rluD inactivation, showing that the dust phenotype can be corrected by the prfBE172K mutation alone. On the two-dimensional gels shown in Fig. 2 it can be observed that a spot with the size and isoelectric point corresponding to RF2 had changed its isoelectric point to become more basic in the rluD prfBE172K strain. We found the abundance of this protein to be approximately doubled in the rluD strain compared with the wild type.
Nonsense Codon Suppression.
Because the suppressor mutation affected RF2, we analyzed how nonsense codon suppression was affected by rluD and by the prfBE172K mutation. This was done in a strain containing the lacIZ fusion Δ14 with a stop codon in the lacI part. Here the synthesis of β-galactosidase enzyme activity is determined by the erroneous reading of the stop codon as a sense codon. We used strains with the three stop codons in two different codon contexts because nonsense suppression is influenced by the codon context. Four strains in which the lacZ gene was totally deleted and having all combinations of prfB+/prfBE172K and rluD+/rluD::cat dust were constructed. Each of seven F′lacI-lacZ translational fusions, one without a premature stop codon and six with one of the three stop codons in two different codon contexts (positions 189 and 220 in the lacI gene), was conjugated into the four strains. The β-galactosidase enzyme activities in these 28 strains were then measured, corrected for instability of the lacI-lacZ enzyme, normalized to the activity in the four strains without a stop codon, and listed in Table 2. The results were further normalized to the read-through value for the rluD+, prfB+ strain (as shown in Table 3). As seen from Tables 2 and 3, misreading of all three stop codons as sense codons was increased significantly in the rluD strain. This effect was most dramatic on codon UGA, where read-through was increased 14-fold, somewhat depending on the codon context. Misreading of UAA and UAG was also increased by 9- and 2-fold, respectively. Codon UGA is in general observed to be the least efficient of the three stop codons, and the 14-fold increase in read-through corresponds to as much as 30% nontermination on UGA (see Table 3). As seen from the absence of satellite stuttering protein spots in Fig. 2B, the accuracy of sense codon reading is not visibly affected in the rluD dust strain and any misreading of individual sense codons caused by rluD inactivation is therefore much smaller than the effect on translation termination.
Table 2.
Analysis of nonsense suppression
| Strain | No stop | UAG189 | UAA189 | UGA189 | UAG220 | UAA220 | UGA220 |
|---|---|---|---|---|---|---|---|
| prfB+, rluD+ | 100 (60.69) | 0.10 | 0.23 | 2.40 | 0.09 | 0.11 | 2.52 |
| prfB+, rluD(dust) | 100 (70.00) | 0.20 | 1.39 | 23.4 | 0.20 | 1.19 | 35.5 |
| prfBE172K, rluD+ | 100 (72.99) | 0.09 | 0.19 | 0.19 | 0.19 | 0.10 | 0.10 |
| prfBE172K, rluD(dust) | 100 (60.56) | 0.11 | 0.23 | 0.23 | 0.11 | 0.11 | 0.34 |
The specific activity of β-galactosidase in the indicated strains is shown, normalized to the activity in the corresponding strain without an internal stop codon in the lacI-lacZ gene. For these four strains the specific enzyme activities are given in parentheses in Miller units (29). All activities are corrected for instability of the lacI-lacZ fusion enzyme.
Table 3.
Analysis of nonsense suppression
| Strain | No stop | UAG189 | UAA189 | UGA189 | UAG220 | UAA220 | UGA220 |
|---|---|---|---|---|---|---|---|
| prfB+, rluD+ | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| prfB+, rluD (dust) | 1.15 | 2.2 | 7.0 | 11.2 | 2.5 | 12 | 16.3 |
| prfBE172K, rluD+ | 1.20 | 1 | 1 | 0.1 | 2.5 | 1 | 0.05 |
| prfBE172K, rluD (dust) | 1.00 | 1.1 | 1 | 0.10 | 1.25 | 1 | 0.14 |
Factor of changing the read-through. Data are the values from Table 2 normalized to the enzyme activity of the same F′ episome in the prfB+, rluD+ strain.
We speculated that tiny might have mutated either the RF2 or the RF1 gene, and we sequenced these genes from the tiny strain ME24 after amplification of the chromosomal gene by PCR. We failed to locate tiny to either prfA or prfB. Thus, rluD inactivation can be suppressed by two different mechanisms of which changing prfB suppresses the strongest.
The prfBE172K enzyme restored termination to the wild-type level on codon UAA and, independent of rluD inactivation, terminated more efficiently on codon UGA (Tables 2 and 3). We note that the prfBE172K mutation alone has no significant effect on UAG termination, confirming that RF2 does not terminate on codon UAG. This is in contrast to a previous search for omnipotent release factors (20), where a strain having the prfBE172K mutation on a high-copy-number plasmid was constructed. This strain was viable even if the prfA gene was inactivated, showing that the prfBE172K enzyme was able to read all three stop codons. We find no evidence for a reading of UAG by the prfBE172K enzyme (Tables 2 and 3), probably because in our strain the effect of prfBE172 was overruled by the presence of wild-type prfA and because prfBE172K is present in only the natural single copy.
Discussion
The presence of pseudouridine at positions 1911, 1915, and 1917 in helix 69 of 23S rRNA is a highly conserved feature of cytoplasmic ribosomes (4), yet absence of the modification can be tolerated, as shown by the existence of a slowly growing but viable rluD mutant of E. coli. The isolation of a suppressor mutation in prfB that almost completely reverted the slow growth of the rluD mutant showed that at least one of the pseudouridines in h69 of 23S rRNA is involved in translation termination by RF2. To our knowledge, this is the first time that a function for a ribosomal RNA pseudouridine has been identified. The interaction between h69 in 23S RNA and RF2 was further supported by direct measurements of the effect of rluD inactivation on translation termination at all three stop codons in two different codon contexts. The absence of the h69 pseudouridines most dramatically affected termination on stop codon UGA (being read as a sense codon in 30% of the cases in the rluD::cat strain), but also UAA (read as sense in 1.3% of the cases), and to a lesser degree UAG was affected. Importantly, missense errors were not increased to this degree by rluD inactivation (Fig. 2B).
In the three-dimensional structure of the ribosome, 23S RNA h69 is located very close to the entry site for the two translation termination factors and for all ternary complexes (5). The structure of RF2 is also known (21), and its complex with the ribosome has been solved in E. coli by cryoelectron microscopy (22) and in Thermus thermophilus by x-ray crystallography (23, 24). From these studies it can be seen that the loop residue E172 that became Lys in the rluD suppressor is located near to helix 69. We have in Fig. 3(modified from figure 3 of ref. 22) shown the approximate position of RF residue E172, and its neighborhood to h69 is clearly seen. Our genetic data therefore are in good agreement with these structural studies. Furthermore, the work of Ali et al. (9) clearly showed that only subunit association, translation initiation, termination, or ribosome recycling were affected by the absence of h69. Our results are in excellent agreement with this conclusion and suggest that an important interaction between residue 172 in RF2 (and probably also the analogous position in RF1; see below) and one of the pseudouridines in h69 takes place during the translation termination process.
Fig. 3.
Localization of the prfB mutation in RF2 in complex with the ribosome. On the model of RF2 bound to the ribosome, obtained by cryoelectron microscopy (modified from figure 3c of ref. 22, reproduced with permission), we indicated the approximate position of the loop where the suppressor had changed the Glu-172 to Lys. The basis for this localization is the linearity of the anticodon binding SPF loop, the E172 loop, and the GGQ loop that binds the peptidyl transferase center (21). Blue, the 50S ribosomal subunit; yellow, the 30S ribosomal subunit; magenta, RF2. Specific structural elements, e.g., helix 69 and the decoding center (DC), are indicated on the ribosome, and the SPF loop and GGQ loop are indicated on RF2. See ref. 22 for further details.
A negatively charged amino acid at position 172 is found in nearly all sequences of RF2 (see Materials and Methods) and is also present at the corresponding position in RF1 (prfB, SEGEVAGIK, residue 172 underlined; prfA: SEGEHGGYK). Also, both RF1 and RF2 bind the ribosome in the same manner (24). The homology between the RF1 and RF2 proteins places a negative amino acid residue close to the h69 pseudouridines also in RF1 and provides an explanation for why translation termination on codon UAG, specifically read by RF1, was also affected in the rluD mutant.
The strong conservation of the h69 pseudouridines in cytoplasmic, eukaryotic ribosomes would suggest that their function in eukaryotic translation termination is similar to the function we have identified in a bacterial ribosome. However, the sole eukaryotic translation termination factor, eRF1, has no detectable homology to the prokaryotic SEGEVAGIK sequence even if the structure of both prokaryotic and eukaryotic release factor proteins mimics that of a tRNA (25). This does not exclude a similar function for the h69 pseudouridines in eukaryotic ribosomes because the functional region in eRF1 might be too small to be noticed by a sequence homology search, as was observed in at least one other study (26).
Because the UAA stop codon is used most frequently by the highly expressed genes, the burden of making incorrectly terminated proteins in the rluD mutant strain is likely to be dominated by the effect on this codon. Tables 2 and 3 suggest that ≈1.3% of these proteins are elongated beyond their normal C terminus and are potentially unstable. This effect is probably not large enough to explain the large reduction in cellular growth rate, but the increase in the synthesis of the heat-shock proteins is compatible with an increased synthesis of aberrant proteins.
The observed slow growth rate and the doubling of the RNA content of the rluD mutant were both corrected by a mutation in RF2 (see Fig. 1A). This indicated that the primary cause of the rluD malady is the translation termination defect and that the other observed defects, e.g., in ribosome assembly (6), arise from a defective translation termination. A mechanism for this could be that a deficient translation termination process disturbed the coordinated expression of the ribosomal proteins because the synthesis of many ribosomal protein genes is translationally coupled. Such noncoordinate expression of the ribosomal proteins might affect the ribosomal assembly process. Processing of rRNA by the enzyme RNaseIII was also shown to be affected by rluD inactivation (6). This might be another secondary effect of a defect translation termination because the RNaseIII gene itself is terminated by UGA. As suggested by the data in Tables 2 and 3, one-third of the RNaseIII molecules might be elongated and unstable or inactive. Consequently, the defective ribosomal assembly process in the rluD strain has several potential sources.
Fig. 2 showed that a protein spot with the expected size and isoelectric point of RF2 was changed to become more basic in the prfBE172K strain and overproduced ≈2-fold in the rluD strain. The autoregulated expression of RF2 requires a frame-shift on an internal in-frame UGA codon (27), and the prfB gene itself is also terminated by an UGA codon. Despite this, the regulation of RF2 expression seems to be able, to some extent, to compensate for the termination defect in the rluD strain.
Tables 2 and 3 show that the prfBE172K enzyme was more efficient than the wild-type RF2 on codon UGA while retaining the wild-type efficiency on codon UAA. It is not often that a mutant enzyme functions better than the wild-type enzyme, but this increased efficiency of the prfBE172K enzyme also resulted in a 5% lower cellular doubling time (data not shown), probably because the termination process becomes slower in the mutant or because the increased termination activity of the prfBE172K enzyme led to termination at near cognate sense codons.
To conclude, we have found that the Glu residue at amino acid 172, at a critical point in the translation termination process, interacts with one or more of the pseudouridines in helix 69 of 23S ribosomal RNA. In the absence of pseudouridine in h69, a large fraction of the ribosomes continues translation past the stop codons, in particular codon UGA, and this gives abnormal and unstable proteins that induce synthesis of the heat-shock proteins. The deficient translational termination may affect the synthesis of RNaseIII and disturb the translational coupling mechanism and thereby affect the coordinated expression of the ribosomal proteins. This might influence the assembly of ribosomes, which in turn has severe consequences for the cellular growth rate, especially in the enriched medium.
Materials and Methods
Construction of Strains.
The strains used are listed in Table 4. Details in constructing the strains used here are as follows. We first made a pair of strains that were isogenic except for the rluD gene by transducing our MG1655 strain to rluD::miniTn10cat with bacteriophage P1 grown on the rluD strain supplied by J. Ofengand (University of Miami School of Medicine, Miami, FL), selecting for resistance to chloramphenicol. ME33 is thus isogenic to MG1655 except for the rluD inactivation. As observed previously (13), two variants, dust ME33 and tiny ME24, appeared after such transduction.
Table 4.
Strains of E. coli
| Strain | Relevant genotype | Construction | Source |
|---|---|---|---|
| MG1655 rluD::cat | 4 | ||
| ME3 | MG1655, rph and uracil semiauxotroph | 28 | |
| ME24 | ME3: rluD::cat tiny | P1 transduction, P1 grown on MG1655 rluD::cat | This study |
| ME33 | ME3: rluD::cat dust | P1 transduction, P1 grown on MG1655 rluD::cat | This study |
| ME41 | ME33 prfBE172K | Suppressor of ME33 | This study |
| HO847 | Δ(iciA-rpiA-PserA)101::kanaR | 29 | |
| MAS70 | Δ(lac-pro) | CSH41 (30) cured of F′lac, made gal+ by P1 transduction, P1 grown on CSH 55 (29) | This study |
| ME96 | rluD::cat tiny | MAS70 camr by P1 transduction, P1 grown on MG1655 rluD::cat | This study |
| ME 97 | rluD::cat dust | MAS70 camr by P1 transduction,P1 grown on MG1655 rluD::cat | This study |
| MC46 F′ 797 | Δ(argB-C)::Tn5, F′lacI-Z fusion Δ14,proAB+ | M. O'Connor (University of Missouri, Kansas City, MO) | |
| MC46 F′ 798 | Δ(argB-C)::Tn5 F′lacI-Z, fusion Δ14 UAG189, proAB+ | M. O'Connor | |
| MC46 F′ 799 | Δ(argB-C)::Tn5Tn5 F′lacI-Z, fusion Δ14 UAA189, proAB+ | M. O'Connor | |
| MC46 F′ 800 | Δ(argB-C)::Tn5 F′lacI-Z, fusion Δ14 UGA189, proAB | M. O'Connor | |
| MC46 F′ 801 | Δ(argB-C)::Tn5 F′lacI-Z, fusion Δ14 UAG220, proAB | M. O'Connor | |
| MC46 F′ 802 | Δ(argB-C)::Tn5 F′lacI-Z, fusion Δ14 UAA220, proAB | M. O'Connor | |
| MC46 F′ 803 | Δ(argB-C)::Tn5 F′lacI-Z, fusion Δ14 UGA220, proAB | M. O'Connor | |
| Ram D14 | rpsD14 | See ref. 31 | M. O'Connor |
To move the prfBE172K mutation from the suppressor mutant, ME41, a marker was introduced by P1 transduction. The marker was the kanamycin resistance from HO847. The rpiA gene is located at 65.8 min on the linkage map of E. coli, and prfB is located at 65.3 min. The prfB gene of ME41 Δ(iciA-rpiA-PserA)101::kanaR was sequenced to ensure the presence of the prfBE172K mutation. The kanamycin resistance from ME41 Δ(iciA-rpiA-PserA)101::kanaR was then moved to MAS70 and ME97 by P1 transduction, producing the strains ME99 and ME108, respectively. Again the prfB genes were sequenced to ensure the presence of prfBE172K. Finally, ME99 and ME108 were cured for their ribose requirement caused by the deletion of the rpiA gene (ribose phosphate isomerase A), by transduction with a P1 lysate derived from ME41, followed by selection on glucose minimal plates. This produced ME123, isogenic to MAS70 except for being prfBE172K, and ME125, isogenic to ME97 except for being prfBE172K. The different F′lac's were moved to the recipients by mating and selection for proline auxotrophy on glucose minimal media.
Growth Conditions.
Physiological experiments were performed on cultures growing exponentially at 37°C in either the rich YT medium (30) or Mops (32), supplemented with 0.2% glucose as carbon source and 10 μg/ml uracil in experiments with MG1655 derivatives. Where needed to maintain a plasmid, the appropriate antibiotic was used as suggested (30). If not otherwise stated, all cultures had grown exponentially for more than eight generations before the start of an experiment and were never allowed to exceed a cell density of 4 × 108 cells per milliliter. After each experiment with the rluD dust strain, cells were plated out to ensure that the number of tiny suppressors had not been excessive during the experiment.
Determination of the RNA and Dry Weight Content of the Cells.
Samples of 1 ml were harvested from exponentially growing cultures at 20-min intervals into 1% TCA at 0°C. The cells were collected by centrifugation, washed in 0.1% TCA, resuspended in 0.4 ml of 0.3 M NaOH, and hydrolyzed for 1 h at 42°C, after which 0.8 ml of 0.3 M HCl was added. Finally, cell debris was removed by centrifugation. The A260 values of the hydrolysates were determined and normalized to the cell density, OD436 at the time of harvest. Conversion factors were as follows: A260 = 1 equals 40 μg of hydrolyzed RNA per milliliter; OD436 = 1 equals 0.18 mg of cellular dry weight per milliliter.
Measurements of β-Galactosidase Activity.
Because of the low enzyme activity, the incubations were carried out for up to 45 h at 28°C under the conditions described previously (33) by using cell extracts prepared by sonic treatment. The enzyme activity was determined on at least six independent cell extracts that were harvested at different cellular densities from at least two independently growing cultures. The lacI-lacZ fusion enzyme was found to be unstable, with a half-life of 12.8 h, during the assay. The values in Tables 2 and 3 are corrected for this instability.
Assay for the Peptide Elongation Rate.
We examined this activity by measuring the kinetics by which the radioactivity of a 10-sec pulse of 200 μCi of carrier-free 35S-Met was chased into full-length protein. The culture was first prelabeled with 3H Lys at 120 μCi to 10 ml of culture. After the pulse, unlabeled Met was added to 1 mg/ml. Samples were then removed from the culture at 10-sec intervals into 0.5 mg/ml chloramphenicol at 0°C. Cells were collected by centrifugation, and the proteins were separated by two-dimensional gel electrophoresis (34). Finally, the incorporated radioactivity in the individual protein spots was determined. For further details and curve fitting see ref. 16. To be suitable for these measurements the protein has to be abundant and of over-average size. Here we used elongation factor EF-G and the heat-shock proteins DnaK and GroEL.
BLAST Search for Sequence Conservation Around prfB Residue 172.
We searched the database for homologies to the E. coli prfB sequence from amino acid 160 to amino acid 190. The sequence spanned the site of the suppressor mutation that changed Glu-172 to a Lys. Homology to the very conserved sequence SEGEVAGIK was found by this procedure and in all cases, except three psychrobacter species (accession nos. gi 93005664, gi 91770222, and gi 71065379) residue 172 was either Glu or Asp. The corresponding sequence in the E. coli prfA gene is SEGEHGGYK with six residues identical and two residues homologous to the prfB SEGEVAGIK sequence.
Acknowledgments
We are grateful to the late Dr. J. Ofengand for discussions and for sending us the rluD strain. We thank Dr. Stanley Brown (University of Copenhagen) for valuable discussions; Marit Warrer, Mariama Drammeh, and Jesper Tholstrup for excellent technical assistance; and Dr. Bjarne Hove-Jensen (University of Copenhagen) and Dr. Michael O'Connor for promptly supplying the requested strains. We furthermore thank Dr. Michael O'Connor for valuable comments on the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Johnson L, Söll D. Proc Natl Acad Sci USA. 1970;67:943–950. doi: 10.1073/pnas.67.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ofengand J, Malhotra A, Remme J, Gutgsell NS, Del Campo M, Jean-Charles S, Peil L, Kaya Y. Cold Spring Harbor Symp Quant Biol. 2001;66:147–159. doi: 10.1101/sqb.2001.66.147. [DOI] [PubMed] [Google Scholar]
- 3.Del Campo M, Kaya Y, Ofengand J. RNA. 2001;7:1603–1615. [PMC free article] [PubMed] [Google Scholar]
- 4.Raychaudhuri S, Conrad J, Hall BG, Ofengand J. RNA. 1998;4:1407–1417. doi: 10.1017/s1355838298981146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 6.Gutgsell NS, Deutscher MP, Ofengand J. RNA. 2005;11:1141–1152. doi: 10.1261/rna.2550105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirabayashi N, Sato NS, Suzuki T. J Biol Chem. 2006;281:17203–17211. doi: 10.1074/jbc.M511728200. [DOI] [PubMed] [Google Scholar]
- 8.O'Connor M, Dahlberg AE. J Mol Biol. 1995;254:838–847. doi: 10.1006/jmbi.1995.0659. [DOI] [PubMed] [Google Scholar]
- 9.Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF. Mol Cell. 2006;23:865–874. doi: 10.1016/j.molcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Ganot P, Bortolin ML, Kiss T. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 11.Ni J, Tien AL, Fournier MJ. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 12.Del Campo M, Recinos C, Yanez G, Pomeranz SC, Guymon R, Crain PF, McCloskey JA, Ofengand J. RNA. 2005;11:210–219. doi: 10.1261/rna.7209905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutgsell NS, Del Campo MD, Raychaudhuri S, Ofengand J. RNA. 2001;7:990–998. doi: 10.1017/s1355838201000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole JR, Olsson CL, Hershey JW, Grunberg-Manago M, Nomura M. J Mol Biol. 1987;198:383–392. doi: 10.1016/0022-2836(87)90288-9. [DOI] [PubMed] [Google Scholar]
- 15.Sørensen MA, Fricke J, Pedersen S. J Mol Biol. 1998;280:561–569. doi: 10.1006/jmbi.1998.1909. [DOI] [PubMed] [Google Scholar]
- 16.Sørensen MA, Kurland CG, Pedersen S. J Mol Biol. 1989;207:365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- 17.Parker J, Pollard JW, Friesen JD, Stanners CP. Proc Natl Acad Sci USA. 1978;75:1091–1095. doi: 10.1073/pnas.75.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjare U, Gorini L. J Mol Biol. 1971;57:423–435. doi: 10.1016/0022-2836(71)90101-x. [DOI] [PubMed] [Google Scholar]
- 19.Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 20.Uno M, Ito K, Nakamura Y. Proc Natl Acad Sci USA. 2002;99:1819–1824. doi: 10.1073/pnas.032457599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vestergaard B, Van LB, Andersen GR, Nyborg J, Buckingham RH, Kjeldgaard M. Mol Cell. 2001;8:1375–1382. doi: 10.1016/s1097-2765(01)00415-4. [DOI] [PubMed] [Google Scholar]
- 22.Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- 23.Klaholz BP, Pape T, Zavialov AV, Myasnikov AG, Orlova EV, Vestergaard B, Ehrenberg M, van Heel M. Nature. 2003;421:90–94. doi: 10.1038/nature01225. [DOI] [PubMed] [Google Scholar]
- 24.Petry S, Brodersen DE, Murphy FV, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 25.Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 26.Brown S, Thon G, Tolentino E. J Bacteriol. 1989;171:6517–6520. doi: 10.1128/jb.171.12.6517-6520.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Craigen WJ, Caskey CT. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- 28.Jensen KF. J Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hove-Jensen B, Maigaard M. J Bacteriol. 1993;175:5628–5635. doi: 10.1128/jb.175.17.5628-5635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller J. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1972. [Google Scholar]
- 31.Andersson DI, Bohman K, Isaksson LA, Kurland CG. Mol Gen Genet. 1982;187:467–472. doi: 10.1007/BF00332630. [DOI] [PubMed] [Google Scholar]
- 32.Neidhardt FC, Bloch PL, Smith DF. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ullmann A, Jacob F, Monod J. J Mol Biol. 1968;32:1–13. doi: 10.1016/0022-2836(68)90140-x. [DOI] [PubMed] [Google Scholar]
- 34.O'Farrell PH. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]