Abstract
Mutations in MECP2 cause the autism-spectrum disorder Rett syndrome. MeCP2 is predicted to bind to methylated promoters and silence transcription. However, the first large-scale mapping of neuronal MeCP2-binding sites on 26.3 Mb of imprinted and nonimprinted loci revealed that 59% of MeCP2-binding sites are outside of genes and that only 6% are in CpG islands. Integrated genome-wide promoter analysis of MeCP2 binding, CpG methylation, and gene expression revealed that 63% of MeCP2-bound promoters are actively expressed and that only 6% are highly methylated. These results indicate that the primary function of MeCP2 is not the silencing of methylated promoters.
Keywords: chromatin, DNA methylation, epigenetics, genomics, Rett syndrome
Mutations in MECP2 cause the vast majority of cases of Rett syndrome (RTT), an X-linked neurodevelopmental disease that primarily affects females (1). The symptoms of RTT include autistic features, seizures, loss of motor skills, breathing abnormalities, and sleep/wake disruption (2). MeCP2 is apparently unnecessary for prenatal brain development, as symptoms of RTT occur typically between 6 and 18 months of age (2), and the genetic reintroduction of MeCP2 in postnatal Mecp2-deficient mice can partially reverse the disease (3, 4).
MeCP2 is one member of a family of DNA-binding proteins that preferentially bind to methylated CpG dinucleotides (5, 6). Although much is known about the ability of MeCP2 to repress methylated gene transcription in vitro (5), the specific function of MeCP2 that is required in postnatal neurons in vivo is less well understood. Early studies suggest that MeCP2 bound to areas of particularly dense CpG dinucleotides or CpG islands and recruited histone HDACs and other factors involved in transcriptional silencing to nearby genes (7, 8). More recent evidence suggests that Mecp2-null mice and Rett patients do not ectopically express genes that are regulated by promoter methylation (9, 10). More diverse roles for MeCP2 have recently been shown in the direct compaction of chromatin (11, 12) and the control of alternative splicing of pre-mRNA (13). The multifunctional nature of MeCP2 is reflected by its intrinsically disordered structure of multiple autonomous domains (14), including a methyl CpG-binding domain (MBD) (15) and a transcriptional repression domain (TRD) (8). The goal of this study was to test the dominant model of MeCP2 as a proximal gene silencer by mapping MeCP2-binding sites in specific target chromosomal loci and promoters genome-wide and by correlating these with transcriptional activity of nearby genes.
Results
To identify all MeCP2-binding sites within the chromosomal loci of known or suspected target genes, a custom high-density oligonucleotide microarray was designed for ChIP–chip analysis, containing 50-mers tiled every 32 bp. Chromosomal regions included 13 Mb of 15q11–13 (SNRPN, GABRB3) (16, 17); 2.6 Mb of 11p15.5 (H19-IGF2) (18); 3.8 Mb of 7q21.3 (DLX5, DLX6) (19); 2.9 Mb of 19p13.2 (JUNB, DNAJB) (20); 0.3 Mb of 20q11.21 (ID1) (20); 0.4 Mb of 2p25.1 (ID2) (20); 0.4 Mb of 1p36.12 (ID3) (20); 2.9 Mb of 6p22.3 (ID4) (20); and 0.2 Mb of 11p14.1 (BDNF) (21, 22) for a total of 26.3 Mb with repetitive sequences removed. The human neuronal cell line SH-SY5Y has been shown to double MeCP2 expression upon differentiation (23), thus 48-h differentiated SH-SY5Y cells were used as a source of chromatin from a homogeneous neuronal cell type with a high MeCP2 expression level. A custom high-affinity IgY specific for the C terminus of MeCP2 common to both isoforms was used for ChIP in three replicate experiments. This antibody detected a single band in differentiated SH-SY5Y cells by Western blot analysis [supporting information (SI) Fig. 6A], specifically immunoprecipitated MeCP2 (SI Fig. 6 B and C), and recognized MeCP2 in ChIP analyses of chromatin from wild-type but not Mecp2-null brain (SI Fig. 6D). As a positive control, ChIP was also performed with an antibody to RNA polymerase II (Pol2). ChIP–chip data were analyzed using Nimblegen Signal Map software and a custom statistical method for identifying peaks in ChIP–chip data (24). Peaks were defined as nonrandom occurrence in at least two of three replicates and were statistically prioritized as L1–L4, with L1 being the most stringent and L4 being the least stringent (see Methods) (24).
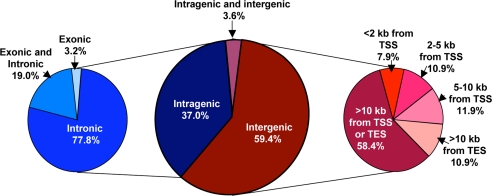
Although analyses of all four peak levels are shown in SI Table 1, Fig. 1 shows a summary of MeCP2-binding sites at the L3 level that revealed 170 MeCP2-binding sites within the 26.3 Mb assayed. Fig. 1 demonstrates that 59.4% of MeCP2-binding sites were entirely between genes (intergenic), and, of these, 58.4% were >10 kb from a transcriptional start site or transcriptional end site (TES). MeCP2 sites within 10 kb of a gene were more likely to be upstream of the gene (30.7%) than downstream (10.9%). Of the 37% of MeCP2 L3 sites that were located entirely within gene boundaries (intragenic), 77.8% were exclusively intronic, 19.0% overlapped both introns and exons, and only 3.2% were exclusively exonic. Surprisingly, the distribution of MeCP2 L3 sites relative to genes was not different from that expected of random. Of 170 total sites, only 10 or 5.9% of MeCP2 sites overlapped with CpG islands (SI Table 1). In comparison with published expression microarray analysis (20), MeCP2 only affected the expression of one of these 10 genes with an MeCP2-bound CpG island (SI Table 2). Interestingly, this affected gene, RNASEH2A, was down-regulated rather than up-regulated by MeCP2 inhibition, which is opposite to the predicted role of MeCP2 as a transcriptional silencer.
Fig. 1.
The majority of MeCP2-binding sites are intergenic or intronic. MeCP2 sites from 26.3 Mb of human genomic sequence at the moderate L3 level of statistical significance are represented. Similar analyses at all four significance levels (L1–L4) are shown in SI Table 1. (Center) A total of 170 MeCP2 sites were divided into intragenic (blue) and intergenic (red) or both (purple). (Left) Intragenic sites were further subdivided into exclusive intronic or exonic or overlapping both introns and exons (shades of blue). (Right) Intergenic sites were categorized according to distances from gene transcription start sites (43) or transcription end sites (TES) (shades of red).
Assuming that the average of one MeCP2-binding site per 209 kb holds for other chromosomal loci, the nonrepetitive genome would have ≈15,384 MeCP2 binding-sites at the L3 level of confidence. On average, L1-level peaks had a higher density of CpG sites than L3-level peaks (12.6 and 6.8 CpGs per bp, respectively), suggesting that most of the highest confidence peaks contained multiple MeCP2 molecules. Because 8.2% of L1–L3 peaks had exactly one CpG site, however, detection of one bound MeCP2 molecule is possible, similar to that observed for the transcription factor E2F1 by ChIP–chip analysis (24). Randomly selected MeCP2 L1, L3, and L4 sites were tested by standard ChIP and PCR. All 46 MeCP2 sites tested showed enrichment over total DNA and were therefore 100% validated by another method (SI Fig. 7).
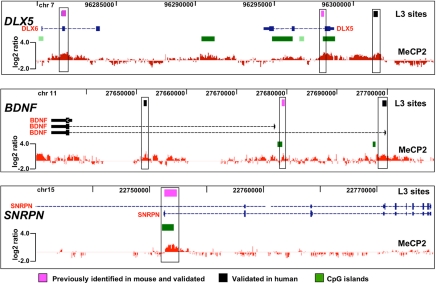
MeCP2 binding sites in target genes identified by conventional ChIP analysis in multiple previous studies were included on the custom array. These positive controls included DLX5/DLX6 in 7q21.3 (19), BDNF in 11p14.1 (21, 22), and SNRPN in 15q11–13 (20, 21). The representative results shown in Fig. 2 demonstrate the validity of our ChIP–chip approach in SH-SY5Y neurons in its remarkable consistency with previous studies using mouse brain tissue. These analyses also demonstrate conservation of MeCP2 sites in these target genes between mouse and human. Examples of additional ChIP–chip genomic tiling analyses for imprinted and nonimprinted regions are shown in SI Figs. 8 and 9. 15q11–13, containing a complex cluster of imprinted and nonimprinted genes implicated in multiple autism-spectrum disorders (25–27), contained 62 L3-level MeCP2-binding sites, including sites in UBE3A and GABRB3 affected by loss of MeCP2 in RTT and Mecp2-deficient mice (16, 28) (SI Fig. 8A). Analysis of 11p15.5, in which MeCP2 binding is correlated with significant changes in H19 and IGF2 transcription (18, 20, 29, 30), but not with changes in imprinted allelic expression (16, 31), revealed sites in the H19-imprinting control region (ICR) (32) and flanking insulin (INS) and the dopaminergic neuronal gene tyrosine hydroxylase (TH) (SI Fig. 8B). Analysis of the MeCP2 target genes ID1-4 revealed distal MeCP2-binding sites that were up to 20 kb from the target genes in addition to proximal sites (SI Fig. 9).
Fig. 2.
Validation of ChIP–chip analysis on established MeCP2-binding sites. A representative histogram of MeCP2 log2 signal ratio values is shown below the University of California, Santa Cruz, Genome Browser window containing known genes, CpG islands (green boxes), and L3 sites (black and pink boxes) for each MeCP2 target gene locus. For DLX5 and DLX6 controls, one site was previously identified in mouse (right pink box) (19) and validated (SI Fig. 7), and another site in DLX6 (left pink box) was also identified in mouse and validated by conventional ChIP analysis (19). For BDNF, an MeCP2 site in the first intron (pink box) was observed at the same position as previously shown in mouse and rat Bdnf (21, 22) and appears to control activation-induced transcription of the gene in neurons (44). Additional new MeCP2 sites were observed and validated (black boxes). A site in SNRPN that serves as the paternal imprinting control region for 15q11–13 was included (pink box) as an additional control. Analyses of this site in both mouse and human by conventional ChIP revealed binding by MeCP2 (16, 17). However, binding of MeCP2 to this site does not affect transcription of SNRPN (16, 19). The SNRPN site was validated by ChIP–chip at levels L1–L4.
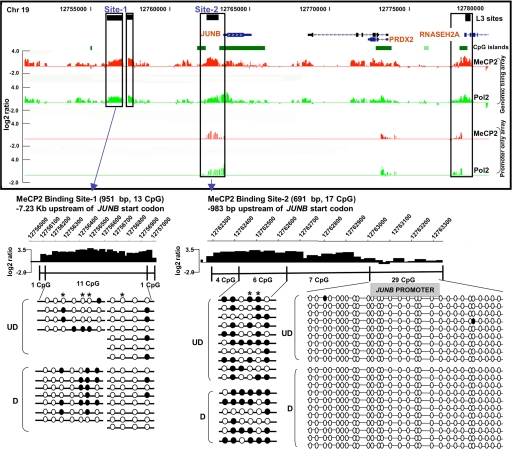
Prior expression microarray analysis of human SH-SY5Y neurons differentiated in vitro and treated with and without a CpG methylated “decoy” revealed a number of potential MeCP2 target genes located on 19p13.2 (20). One of the genes that was most altered by MeCP2 binding was the immediate-early response gene JUNB. A cluster of three MeCP2 L3 sites within 7 kb of JUNB were detected, one of which was located between two CpG islands (Fig. 3). These sites were validated by conventional ChIP analysis (data not shown). Surprisingly, MeCP2-binding levels (Fig. 3, red histogram) were frequently concordant with Pol2-binding levels (Fig. 3, green histogram) at this and other nonimprinted loci included on the custom tiling array. Higher resolution analysis of ChIP–chip histograms compared with bisulfite sequencing analysis of DNA methylation revealed that MeCP2-bound fractions were enriched in partially methylated regions −983 bp and −7.23 kb upstream of JUNB, whereas the proximal promoter CpG island was unmethylated (Fig. 3 Lower). Previous expression microarray analysis showed that JUNB expression increased after differentiation but was further increased when MeCP2 was blocked, suggesting that MeCP2 serves to down-regulate, but not silence, this active gene (20). Interestingly, these analyses also suggest that RNASEH2A is coordinately but oppositely regulated by MeCP2 binding, because expression is decreased by MeCP2 deficiency (SI Table 2). Together, these results suggest that MeCP2 binding to proximal and distal sites near active genes such as JUNB and RNASEH2A does not silence gene expression.
Fig. 3.
JUNB is a partially methylated, actively expressed gene regulated by MeCP2 binding. Three previously undescribed MeCP2-binding sites (black boxes) were found upstream of JUNB, with another in the first intron of RNASEH2A, overlapping with a CpG island (green box). (Upper) Shown in the histograms, high levels of MeCP2 binding (red histograms) were correlated with high levels of Pol2 binding (green histograms), a correlation also observed in ChIP–chip analysis of MeCP2 and Pol2 binding to genome-wide promoters. (Lower) Shown is bisulfite analysis of JUNB sites 1 and 2 compared with higher resolution of MeCP2 ChIP–chip histograms (open circles, unmethylated; filled circles, methylated CpG sites). Partial methylation of CpGs was observed in undifferentiated (UD) and differentiated (D) SH-SY5Y cells, especially methylation of CpGs next to AT runs (40), which are indicated by an asterisk. In contrast, the proximal CpG island promoter of this actively expressed gene was unmethylated and de-enriched for MeCP2 binding relative to the methylated upstream regions.
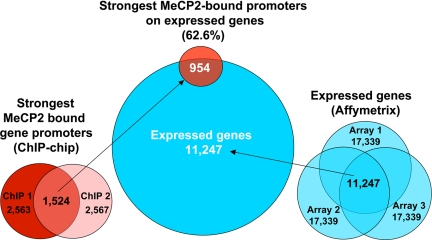
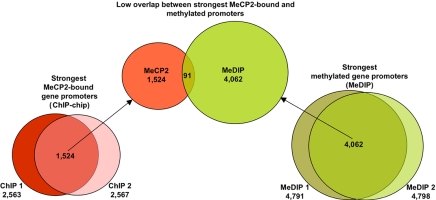
In addition to demonstrating that most of MeCP2-binding sites are distally located (>10 kb) from genes, genomic ChIP–chip analysis of selected regions such as JUNB revealed concordance of MeCP2 and Pol2 binding on promoters even in nonimprinted chromosomal loci (Fig. 3, validated in SI Fig. 10). These results suggested that MeCP2 is frequently bound to transcriptionally active promoters, a finding that directly contradicts the dominant model of MeCP2 function as that of a transcriptional silencer (33). To directly address the possibility of MeCP2 binding to actively expressed genes on a genome-wide basis, ChIP-chip analysis of MeCP2 binding to 24,275 putative human promoters (defined as 1.3 kb upstream and 0.2 kb downstream of a TSS) was performed in differentiated SH-SY5Y neurons (see Materials and Methods) and compared with expression profiling analysis of identically differentiated SH-SY5Y cells (Fig. 4). Statistical analysis of promoter ChIP–chip data indicated that 2,600–4,300 promoters were bound by MeCP2 on the basis of total signal levels and assuming a 10% false positive correction rate (SI Fig. 11). By using the most conservative threshold, comparison of the strongest hits between two replicate arrays yielded 1,524 common MeCP2-bound promoters (Fig. 4). Comparison of these promoters to the 11,247 expressed genes in differentiated SH-SY5Y cells (defined as “present” in all three arrays) revealed that 62.6% of the strongest MeCP2-bound promoters (including JUNB and BDNF) are expressed genes (Fig. 4). Furthermore, of the genes with the highest expression levels, at least twice as many displayed MeCP2 promoter binding than was expected by random chance (SI Fig. 12). As a specific example, JUNB was 51st on the list of genes ranked by expression level. Furthermore, genome-wide promoter methylation analysis by methylated DNA immunoprecipitation (MeDIP) (34, 35) from identically differentiated SH-SY5Y cells demonstrated that, of the top 4,062 promoters with the highest levels of methylation, only 2.2% were bound by MeCP2 (Fig. 5). MeDIP analysis of SH-SY5Y cells correlated well with a published study using primary human fibroblasts (35), demonstrating the general maintenance of promoter methylation in this cell line (SI Fig. 13). These combined results demonstrate that although the strongest MeCP2-binding sites are distal to promoters, promoters of many active partially unmethylated genes (such as JUNB) are bound by MeCP2.
Fig. 4.
The majority of MeCP2-bound promoters genome-wide are expressed genes. Comparison of the strongest MeCP2-bound promoters with genes expressed in differentiated SH-SY5Y neurons is shown. (Center) Shown is a Venn diagram of 1,524 MeCP2 promoter hits with 11,247 expressed genes (blue circle), which reveals that 954 or 62.6% of MeCP2-bound promoters are transcriptionally active genes. (Left) The strongest MeCP2-bound promoters were identified by comparing the top 2,600 bound promoters, removing duplicate promoters to obtain 2,563 and 2,567 hits, respectively, for two replicate arrays, ChIP-1 and ChIP-2. (Right) Expression microarray analysis of 48-h-differentiated SH-SY5Y cells in three biologic replicates (arrays 1–3) (20) was reanalyzed using a determination of expressed genes by Affymetrix mismatch analysis to identify a conservative common set of 11,247 expressed genes in SH-SY5Y neurons.
Fig. 5.
Highly methylated promoters are not bound by MeCP2. (Center) The 1,524 promoters with the highest levels of MeCP2 binding identified in Fig. 4 were compared with the 4,062 most highly methylated promoters, revealing an overlap of only 91 genes. Therefore, only 2.2% of highly methylated promoters were bound by MeCP2, and only 6.0% of MeCP2-bound promoters were highly methylated. (Left and Right) Two replicate MeDIP assays were performed on genomic DNA from SH-SY5Y cells (Right) differentiated identically to those used for MeCP2 promoter ChIP–chip (Left) and expression profiling (see Materials and Methods). The median top 5,000 log2 promoter signals were selected from each experiment, and, after the removal of duplicate promoters, the overlap produced 4,062 genes in common.
Discussion
The predominant model of MeCP2 function hypothesizes that MeCP2 binds to CpG methylated promoters and recruits histone deacetylase along with co-repressor activities, thereby silencing gene transcription (7, 8, 18, 33, 36, 37). This model of MeCP2 function is based on the behavior of artificial gene constructs transfected into cell lines. To thoroughly test the predominant model in neurons we performed a large-scale integrated analysis of endogenous MeCP2 binding with genome-wide expression and promoter methylation.
These studies reveal several important new findings of high significance to understanding MeCP2 function and Rett syndrome. First, the results show that the majority of MeCP2 bound promoters are on active genes including JUNB. Therefore, MeCP2 promoter occupancy is not consistent with gene silencing. Second, these results also show that the majority of promoters with the highest methylation levels are not bound by MeCP2. Third, ChIP-chip analysis revealed that the majority of MeCP2 binding sites were found outside of transcription units and CpG islands. Fourth, we discovered multiple intergenic MeCP2 binding sites within three imprinted loci consistent with MeCP2 organizing imprinted loci into chromatin loops (19). Finally, this study uncovered two new MeCP2 target genes of potential importance to the pathogenesis of Rett syndrome, the immediate-early response gene JUNB (38) and RNASEH2A, mutations of which cause the progressive neurological disorder Aicardi-Goutieres syndrome type 4 (AGS4) (39).
According to the dominant model of MeCP2 action, target genes such as BDNF are silenced by MeCP2 binding to the promoter (21, 22). However, combined ChIP–chip promoter and expression profiling analysis reveals that 62.6% of MeCP2-bound promoters (including JUNB and BDNF) are transcriptionally active. These studies clearly demonstrate that MeCP2 promoter occupancy does not correlate with transcriptional silencing of target genes. In a specific example in the JUNB/RNASEH2A locus, binding of MeCP2 is instead correlated with the reduction of high JUNB transcript levels and increased levels of RNASEH2A transcripts. Reintroduction of Mecp2 in a mouse model of RTT can reverse symptoms (3, 4), an outcome that would appear to be more consistent with an indirect effect or a subtle modulation of target gene expression as opposed to silencing. Recent studies demonstrating that promoter methylation is often observed on active genes (35) and MeCP2 controls alternative splicing of active gene transcripts (13) both support these findings.
The dominant model of MeCP2 function hypothesizes that promoter methylation leads to MeCP2 binding. However, the results shown here demonstrate that dense promoter methylation does not correlate with MeCP2 binding. This result may be explained by recent findings that indicate that the promoters with the highest density of potential CpG methylation sites are mostly unmethylated even when inactive (35). Therefore these promoters are probably not represented in the methylation data. These results may also be explained by previous findings demonstrating the importance of A/T sequences adjacent to methylated CpGs in determining binding of MeCP2 (40) and the methylation-independent binding and chromatin compaction by MeCP2 in vitro (11, 12).
The results have implications for genomic imprinting as well. Both 15q11–13 and 11p15.5 are complex loci containing a mixture of imprinted and nonimprinted genes. Our analysis revealed strong L1 MeCP2 binding sites in the imprinting control regions (ICR) of both 15q11–13 and 11p15.5. In addition, we have confirmed the MeCP2-binding sites in the imprinted DLX5-DLX6 locus (19). Although our studies focus on regions potentially regulated by MeCP2 binding, the data suggest that periodic binding of MeCP2 outside gene boundaries may organize chromatin into functionally important domains or loops of imprinted regions, thereby modulating gene expression in either a positive or a negative manner. Current and previous results are not consistent with the proposed role of MeCP2 as a proximal transcriptional silencer of methylated imprinted genes (7, 8, 33). In light of the findings presented here and by others, the dominant model of MeCP2 function should be reexamined, because understanding how MeCP2 regulates target genes is critical to the advancement of the field toward designing therapies for RTT patients.
Materials and Methods
ChIP–Chip.
Chromatin was prepared according to Oberly et al. (41) from three separate pools of SH-SY5Y neuroblastoma cells of 4 × 107 cells each, differentiated by 48 h of treatment with 1.6 μM Phorbol-12-Myristate-13-acetate (PMA) (EMD Biosciences). In a departure from original protocol, cells were lysed after cross-linking by dounce homogenization. Chromatin from each pool was immunoprecipitated with chicken anti-MeCP2 (custom) and anti-RNA polymerase II (Covance). To test for enrichment of MeCP2- and Pol2-bound sites, PCR amplification of SNRPN and GAPDH promoters was performed on each ChIP sample before and after ligation-mediated (LM)-PCR amplification.
A custom genomic microarray (NimbleGen Systems) covered loci on nine separate human autosomes for a total of 26.3 Mb. One microgram of amplicons from each replicate antibody ChIP was hybridized with differentially labeled total genomic DNA to each microarray. Amplicons were also applied to NimbleGen 1.5-kb human promoter arrays. The labeling and hybridization of DNA samples for ChIP–chip analysis was performed by NimbleGen Systems using established protocols. The arrays were washed using NimbleGen Wash Buffer System (NimbleGen Systems), dried by centrifugation, and scanned at 5-μm resolution by using the GenePix 4000B scanner (Axon Instruments). Fluorescence intensity raw data were obtained from scanned images of the arrays by using NIMBLESCAN 2.0 extraction software (NimbleGen Systems). For each spot on the array, log2-ratios of the Cy5-labeled test sample versus the Cy3-labeled reference sample were calculated. Then, the biweight mean of this log2 ratio was subtracted from each point; this procedure is approximately equivalent to mean-normalization of each channel.
MeCP2 peaks were assigned using an analysis method developed for identifying peaks in ChIP–chip data sets (24). The peak levels correspond to a combination of peak threshold of log2 oligomer ratios plus the P value of peak width. Thus, L1–L4 correspond to the 98th percentile threshold and P < 0.0001, the 95th percentile threshold and P < 0.0001, the 98th percentile threshold and P < 0.05, and 95th percentile and P < 0.05, respectively. Only peaks that were present on two of three replicate array hybridizations were reported.
PCR validation was performed on 100 ng of SH-SY5Y genomic DNA or ChIP DNA by using the following reaction conditions: 1× PCR buffer, 4 μM MgCl2, 1 M betaine, 400 μM dNTPs, 0.5 unit of Taq polymerase, and 0.5 μM primers for 30 cycles. Quantification of PCR products from total genomic chromatin (TR) and MeCP2 immunoprecipitated chromatin (IP) was performed using AlphaInnotech's Spot Denso analysis tool, with ratios being calculated by dividing the IP values by the TR values. Primer sequences are available upon request.
Bisulfite Treatment and Sequencing Analysis of JUNB.
Genomic DNA (gDNA) was isolated from both undifferentiated (UD) and 48-h postdifferentiated (D-48h) SH-SY5Y cells following instructions with the Puregene DNA purification kit. Bisulfite treatment of 1 μg each of UD and differentiated (D) SH-SY5Y gDNA was carried out using CpGenome Fast DNA modification kit (Chemicon). Between 1.5 and 3 μl of bisulfite-treated DNA was PCR amplified using primers directed against the promoter of JUNB, designed using MethPrimer software. PCR products were cloned into pGEM-T Easy plasmid (Promega), and ≈10 transformed white colonies under each condition and for each insert were sequenced for analyzing the percentage of methylated cytosine in every position. Primer sequences are available upon request.
Genome-Wide Promoter Methylation Analysis by MeDIP.
SH-SY5Y genomic DNA was fragmented with a Diagenode Bioruptor to an average fragment size of 200–300 bp. Immunoprecipitation of methylated DNA was performed using an anti-methylcytidine antibody (Eurogentec) as described in ref. 34, with the following modifications: a rabbit anti-mouse IgG secondary antibody (Jackson Immunoresearch) and Protein A/G beads (Pierce) were used to pull down methylated DNA–antibody complexes, and a total of six washes were performed after immunoprecipitation. A nonspecific mouse IgG was used in parallel with methyl DNA IP as a negative control. Proteinase K digestion was performed overnight and immunoprecipitated DNA was purified by phenol chloroform extraction. Immunoprecipitated and reference DNA were labeled with Cy5- and Cy3-labeled random 9-mers, respectively, and hybridized to the HG17 1.5-kb promoter arrays (NimbleGen Systems) by using the NimbleGen Array Hybridization Kit following the manufacturer's instructions. Slides were scanned with an Axon DNA Microarray Scanner and analyzed with NimbleScan 2.1 (NimbleGen Systems). DNA fragments immunoprecipitated with anti-methylcytidine antibody were hybridized along with differentially labeled genomic DNA to NimbleGen 1.5-kb promoter microarrays in two separate experiments. From each experiment, a list of mean log2 signal values was generated for all 24,275 promoters present on the array. The top 5,000 median promoter signals from the two experiments were selected on the basis of the difference between the number of promoter hits expected at random versus those observed (42). Removal of duplicate promoters yielded 4,791 and 4,798 promoter hits, respectively.
Supplementary Material
Acknowledgments
We thank Charles Nicolet, Sharon Squazzo, and Luis Acevedo for technical advice; Marc Lalande and Terumi Kohwi-Shigematsu for critical reading of the manuscript; and Prasad T. Peddada (BEA Systems, San Francisco, CA) for custom software development. This work was supported by National Institutes of Health Grants R01 HD048799, R01 HD/NS41462, and C06 RR12088; a University of California, Davis, Genome Center Expression Analysis Core Facility Pilot Grant; and an Autism Speaks/National Alliance for Autism Research Fellowship (to S.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.H.B. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE9568).
This article contains supporting information online at www.pnas.org/cgi/content/full/0707442104/DC1.
References
- 1.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 2.Shahbazian MD, Zoghbi HY. Curr Opin Neurol. 2001;14:171–176. doi: 10.1097/00019052-200104000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Giacometti E, Luikenhuis S, Beard C, Jaenisch R. Proc Natl Acad Sci USA. 2007;104:1931–1936. doi: 10.1073/pnas.0610593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meehan RR, Lewis JD, Bird AP. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 7.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 8.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 9.Tudor M, Akbarian S, Chen RZ, Jaenisch R. Proc Natl Acad Sci USA. 2002;99:15536–15541. doi: 10.1073/pnas.242566899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Traynor J, Agarwal P, Lazzeroni L, Francke U. BMC Med Genet. 2002;3:12. doi: 10.1186/1471-2350-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC. J Biol Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 12.Nikitina T, Shi X, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Woodcock CL. Mol Cell Biol. 2007;27:864–877. doi: 10.1128/MCB.01593-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, Kang D, Richman R, Johnson JM, Berget S, Zoghbi HY. Proc Natl Acad Sci USA. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams VH, McBryant SJ, Wade PA, Woodcock CL, Hansen JC. J Biol Chem. 2007;282:15057–15064. doi: 10.1074/jbc.M700855200. [DOI] [PubMed] [Google Scholar]
- 15.Nan X, Meehan RR, Bird A. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samaco RC, Hogart A, LaSalle JM. Hum Mol Genet. 2005;14:483–492. doi: 10.1093/hmg/ddi045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thatcher K, Peddada S, Yasui D, LaSalle JM. Hum Mol Genet. 2005;14:785–797. doi: 10.1093/hmg/ddi073. [DOI] [PubMed] [Google Scholar]
- 18.Drewell RA, Goddard CJ, Thomas JO, Surani MA. Nucleic Acids Res. 2002;30:1139–1144. doi: 10.1093/nar/30.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 20.Peddada S, Yasui DH, LaSalle JM. Hum Mol Genet. 2006;15:2003–2014. doi: 10.1093/hmg/ddl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 22.Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 23.Jung BP, Jugloff DG, Zhang G, Logan R, Brown S, Eubanks JH. J Neurobiol. 2003;55:86–96. doi: 10.1002/neu.10201. [DOI] [PubMed] [Google Scholar]
- 24.Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker P, Piven J, Schwartz S, Patil S. J Autism Dev Disord. 1994;24:529–535. doi: 10.1007/BF02172133. [DOI] [PubMed] [Google Scholar]
- 26.Bundey S, Hardy C, Vickers S, Kilpatrick MW, Corbett JA. Dev Med Child Neurol. 1994;36:736–742. doi: 10.1111/j.1469-8749.1994.tb11916.x. [DOI] [PubMed] [Google Scholar]
- 27.Cook EH, Jr, Courchesne RY, Cox NJ, Lord C, Gonen D, Guter SJ, Lincoln A, Nix K, Haas R, Leventhal BL, Courchesne E. Am J Hum Genet. 1998;62:1077–1083. doi: 10.1086/301832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogart A, Nagarajan RP, Patzel KA, Yasui DH, Lasalle JM. Hum Mol Genet. 2007;16:691–703. doi: 10.1093/hmg/ddm014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ballestar E, Ropero S, Alaminos M, Armstrong J, Setien F, Agrelo R, Fraga MF, Herranz M, Avila S, Pineda M, et al. Hum Genet. 2005;116:91–104. doi: 10.1007/s00439-004-1200-0. [DOI] [PubMed] [Google Scholar]
- 30.Delgado IJ, Kim DS, Thatcher KN, LaSalle JM, Van den Veyver IB. BMC Med Genet. 2006;7:61. doi: 10.1186/1471-2350-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balmer D, Arredondo J, Samaco RC, LaSalle JM. Hum Genet. 2002;110:545–552. doi: 10.1007/s00439-002-0724-4. [DOI] [PubMed] [Google Scholar]
- 32.Thorvaldsen JL, Duran KL, Bartolomei MS. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng HH, Bird A. Curr Opin Genet Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 34.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 35.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 36.Muller C, Readhead C, Diederichs S, Idos G, Yang R, Tidow N, Serve H, Berdel WE, Koeffler HP. Mol Cell Biol. 2000;20:3316–3329. doi: 10.1128/mcb.20.9.3316-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorincz MC, Schubeler D, Groudine M. Mol Cell Biol. 2001;21:7913–7922. doi: 10.1128/MCB.21.23.7913-7922.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlingensiepen KH, Wollnik F, Kunst M, Schlingensiepen R, Herdegen T, Brysch W. Cell Mol Neurobiol. 1994;14:487–505. doi: 10.1007/BF02088833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, et al. Nat Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 40.Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. Mol Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 41.Oberley MJ, Tsao J, Yau P, Farnham PJ. Methods Enzymol. 2004;376:315–334. doi: 10.1016/S0076-6879(03)76021-2. [DOI] [PubMed] [Google Scholar]
- 42.Squazzo SL, O'Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He L, Cui H, Walsh C, Mattsson R, Lin W, Anneren G, Pfeifer-Ohlsson S, Ohlsson R. Oncogene. 1998;16:113–119. doi: 10.1038/sj.onc.1201501. [DOI] [PubMed] [Google Scholar]
- 44.Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, et al. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.