Abstract
Metabolic imaging of the relative amounts of reduced NADH and FAD and the microenvironment of these metabolic electron carriers can be used to noninvasively monitor changes in metabolism, which is one of the hallmarks of carcinogenesis. This study combines cellular redox ratio, NADH and FAD lifetime, and subcellular morphology imaging in three dimensions to identify intrinsic sources of metabolic and structural contrast in vivo at the earliest stages of cancer development. There was a significant (P < 0.05) increase in the nuclear to cytoplasmic ratio (NCR) with depth within the epithelium in normal tissues; however, there was no significant change in NCR with depth in precancerous tissues. The redox ratio significantly decreased in the less differentiated basal epithelial cells compared with the more mature cells in the superficial layer of the normal stratified squamous epithelium, indicating an increase in metabolic activity in cells with increased NCR. However, the redox ratio was not significantly different between the superficial and basal cells in precancerous tissues. A significant decrease was observed in the contribution and lifetime of protein-bound NADH (averaged over the entire epithelium) in both low- and high-grade epithelial precancers compared with normal epithelial tissues. In addition, a significant increase in the protein-bound FAD lifetime and a decrease in the contribution of protein-bound FAD are observed in high-grade precancers only. Increased intracellular variability in the redox ratio, NADH, and FAD fluorescence lifetimes were observed in precancerous cells compared with normal cells.
Keywords: diagnosis, imaging, metabolism, mitochondria, oral cancer
The electron transport chain is the most efficient means of energy production in cells. The electron transport chain produces energy in the form of ATP by transferring electrons to molecular oxygen. The metabolic coenzymes FAD and NADH are the primary electron acceptor and donor, respectively, in oxidative phosphorylation. Neoplastic cells have an increased metabolic demand relative to normal cells because of rapid cell division (1), and neoplastic metabolism is associated with changes in the relative concentrations of NADH and FAD (2–4). Many enzymes bind to NADH and FAD in the metabolic pathway (5), and, as favored metabolic pathways shift with cancer progression, the distribution of NADH and FAD binding sites also change (6). Thus, metabolic imaging of the relative amount of NADH and FAD, and their microenvironment (such as binding sites and/or the presence of local quenchers) can shed light on metabolic changes associated with carcinogenesis.
The metabolic coenzymes NADH and FAD are autofluorescent and can be monitored nondestructively and without exogenous labels, using optical techniques. The most common optical method for metabolic imaging is the “redox ratio,” which is the ratio of the fluorescence intensity of FAD and NADH (7). This optical redox ratio provides relative changes in the oxidation-reduction state in the cell. The redox ratio is sensitive to changes in the cellular metabolic rate and vascular oxygen supply (1–3, 7). A decrease in the redox ratio usually indicates increased cellular metabolic activity (8), as is typically observed in cancer cells. The redox ratio has been extensively evaluated in ex vivo normal and neoplastic tissues (2–4, 9). To date, we have found no previous studies that investigated changes in the redox ratio with precancer development in three dimensions with cellular resolution in vivo.
The fluorescence lifetime, which probes the time that a molecule remains in its excited state before decaying back to the ground state, is sensitive to fluorophore microenvironment, and provides a method for discriminating free and protein-bound components (and the relative amounts of the free and protein-bound components) of NADH and FAD. NADH has a short and long lifetime component, respectively, depending on whether it is in a free or protein-bound state (10), and FAD has a short and long lifetime component, respectively, depending on whether it is in a protein-bound or free state (11). The shorter fluorescence lifetimes of both protein-bound FAD and free NADH are due to dynamic quenching by the adenine moiety (12, 13). The fluorescence lifetime of protein-bound NADH depends on the enzyme to which it is bound (14), and this suggests that changes in NADH binding with cancer development can be probed by the lifetime of this fluorophore. The fluorescence lifetime of protein-bound FAD decreases in the presence of NAD+ because of increased intramolecular dynamic quenching in the presence of NAD+ (13). This suggests that the lifetime of protein-bound FAD is sensitive to changes in NAD+ levels. Changes in the lifetimes of free NADH and FAD could also reflect changes in quencher concentrations, such as oxygen, tyrosine, or tryptophan, or changes in local temperature and pH (12, 15, 16).
Fluorescence lifetime imaging microscopy (FLIM) of NADH and FAD remains relatively unexplored as an in vivo metabolic imaging technique. Previous studies of NADH lifetimes in cell culture (17–19) and tissue slices (20) have revealed that the fluorescence lifetime of free and protein-bound NADH, and the contribution of protein-bound NADH decrease with hypoxia. The fluorescence lifetime of protein-bound NADH and its relative contribution has also been shown to decrease with low- and high-grade precancers in the hamster cheek pouch model of oral cancer in vivo (19). However, to our knowledge, no previous studies have investigated changes in the contribution and lifetime of protein-bound and free FAD with neoplastic development.
The goal of this study was to simultaneously image and quantify the cellular redox ratio, NADH and FAD lifetimes, and subcellular morphology in an in vivo model of epithelial precancer. Although these sources of contrast have been explored individually, to our knowledge there has been no previous work that combines these four approaches to track the earliest stages of cancer development, which is clinically important. All in vivo images were collected with multiphoton microscopy (21), because the near infrared excitation used in multiphoton microscopy allows for greater penetration depths (22) and improved tissue viability (23) compared with single-photon excitation. The hamster cheek pouch model of oral cancer was used for this study, because it allowed for multiple disease states to be evaluated in vivo, and it has been shown to mimic the development of squamous epithelial cancer in the human oral cavity (24).
Results
Pathology Results.
Of the 22 cheek pouches imaged (1 per animal), 6 were diagnosed as normal, 4 with mild dysplasia, 4 with moderate dysplasia, 4 with severe dysplasia, 3 with carcinoma in situ (CIS), and 1 with squamous cell carcinoma (SCC). For statistical analyses, tissue samples were divided into normal (n = 6), low-grade (mild and moderate dysplasia, n = 8), and high-grade precancer (severe dysplasia and CIS, n = 7). The SCC sample was not included in the analysis.
In Vivo Multiphoton Microscopy Images and Statistical Analyses of Image Parameters.
Shown in Fig. 1 are in vivo three-dimensional multiphoton image stacks of the redox ratio (Fig. 1 a–c), mean NADH lifetime (the weighted average of free and protein-bound components) collected at 800-nm excitation (Fig. 1 d–f), and mean FAD lifetime collected at 890-nm excitation (Fig. 1 g–i) from three tissues diagnosed as normal (Fig. 1 a, d, and g), low-grade precancer (Fig. 1 b, e, and h), and high-grade precancer (Fig. 1 c, f, and i). The redox ratio of the normal tissue shows second harmonic generation (SHG) from fibrillar collagen (19) in the deepest image slice (Fig. 1a). The redox ratio of the high-grade precancerous cells appears to be higher in the perinuclear region than the rest of the cytoplasm (Fig. 1c, 35–55 μm). Note that the mean lifetime of FAD in the normal sample (≈0.30 ns, Fig. 1g) is much shorter than the mean lifetime of NADH (≈1.0 ns, Fig. 1d) in the same sample.
Fig. 1.
In vivo three-dimensional multiphoton images of the redox ratio (fluorescence intensity of FAD/NADH) (a–c), the mean NADH lifetime (α1 × τ1 + α2 × τ2) (d–f), and the mean FAD lifetime (g–i) from tissues diagnosed as normal (a, d, and g), low-grade precancer (b, e, and h), and high-grade precancer (c, f, and i). The numbers in the corner of each image indicate the depth below the tissue surface in μm, and each image is 100 × 100 μm. There are a different color bars for a–c (in calibrated units), d–f, and g–i. The redox images were created by merging the calibrated blue NADH image (800-nm excitation with a 490-nm short-pass filter) with the calibrated red FAD image (890-nm excitation) into an RGB image for the purposes of this figure only.
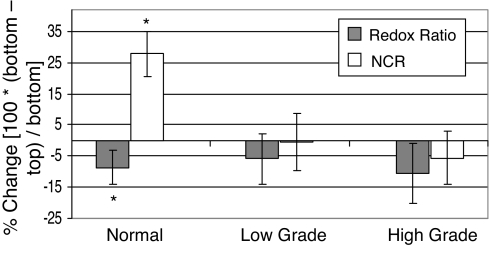
Normal stratified squamous epithelial tissues have highly proliferative basal cells located on the basement membrane [which separates the epithelium from the stroma (5)]. As the basal cells divide and mature, they move upward through the epithelium, become metabolically less active, and have a decreased nuclear-to-cytoplasmic ratio (NCR). The NCR and redox ratio quantified from in vivo multiphoton microscopy were used to determine whether the redox ratio reflects increased metabolic activity in the basal layer compared with the superficial layer within the epithelium. The percentage of change in the NCR and the redox ratio between the top (superficial) and bottom (basal) imaged plane of the epithelium within an animal is shown in Fig. 2. The NCR increases and the redox ratio decreases in the basal cell layer compared with the metabolically less active superficial cells of normal animals (P < 0.05), as expected (5). The NCR and redox ratio were unchanged with depth in the precancers (P > 0.05). Changes in the contribution and lifetimes of protein-bound and free NADH and FAD with depth within the epithelium of normal and precancerous tissues were heterogeneous and did not show the consistent trends as observed in the redox ratio (data not shown). There were also no significant changes in the cellular coefficient of variation of the redox ratio or lifetime variables between the first vs. last plane of the epithelium in either normal or precancerous tissues (P > 0.05). The thickness of the normal epithelium was 25 ± 3 μm, indicating that thickness measurements were consistent between animals and consistent with our previous studies (19, 25). The epithelial thickness in the low-grade (29 ± 9 μm) and high-grade (31 ± 9 μm) dysplastic samples were determined by using the same protocol. There were no significant differences in the epithelial thickness of normal, low-grade, or high-grade precancers in this study (P > 0.05).
Fig. 2.
Percentage of change in the nuclear-to-cytoplasmic ratio (NCR, nuclear diameter/cell diameter) and redox ratio between the top and bottom plane of the cellular epithelium within an animal, calculated from the multiphoton redox images. The mean and 95% confidence interval were calculated for animals diagnosed as normal (n = 36 bottom/top cell pairs), low-grade precancer (n = 42 bottom/top cell pairs), and high-grade precancer (n = 33 bottom/top cell pairs). Paired Wilcoxon tests revealed a significant increase in NCR and decrease in redox ratio for basal cells compared with superficial cells in normal tissue (*, P < 0.05) but no change with depth in precancerous tissues (P > 0.05).
The epithelial protein-bound NADH lifetime (volume averaged over all image planes within the epithelium) and its contribution (relative to free NADH) decreased with low- and high-grade precancers compared with normal (P < 0.05, Fig. 3a). There was also a decrease in the free NADH lifetime for high-grade precancer only (P < 0.05). The protein-bound component is also the most important component of the FAD lifetimes for precancer discrimination. The volume-averaged lifetime of protein-bound FAD increases and its contribution (relative to free FAD) decreases for high-grade precancer only (P < 0.05, Fig. 3b). Note that the short lifetime component of FAD (protein-bound FAD) dominates the fluorescence decay. There were no significant differences in the volume-averaged NADH or FAD lifetime variables between low- and high-grade precancers (P > 0.05).
Fig. 3.
Mean and 95% confidence interval of the volume-averaged NADH (a) and FAD (b) lifetime variables. All values were obtained from in vivo multiphoton FLIM images, and variables were averaged for all image planes within the epithelium of an animal for normal (n = 6), low-grade precancerous (n = 8), and high-grade precancerous animals (n = 7). Significant differences (P < 0.05) exist between normal vs. low-grade precancer and normal vs. high-grade precancer (*). There were no significant differences between low- and high-grade precancers (P > 0.05).
The error bars shown in Fig. 3 reflect the sum of the variabilities due to different animals, different scan areas within an animal, different depth planes within a scan, different pixels within a depth, and variability within a pixel due to the lifetime fits. The uncertainty in the lifetime fits per pixel due to Poisson noise was calculated by using simulated envelopes (see Materials and Methods). The contribution of the variability within a pixel to the “total” variability, represented by the error bars in Fig. 3, can be computed by using Variance component analysis (26). The standard deviation within a pixel of τ1, τ2, and α2 was computed to be ±50 ps, ±140 ps, and ±2.5%, respectively, for NADH and ±5 ps, ±40 ps, and ±0.3%, respectively, for FAD. The proportion of this “within a pixel” variability to total variability reflected by the error bars in Fig. 3 for τ1, τ2, and α2 was found to be 21 ± 3%, 34 ± 10%, and 12 ± 1%, respectively, for NADH and 6 ± 8%, 8 ± 3%, and 0.4 ± 0.2%, respectively, for FAD.
Table 1 shows the mean and standard error of the cellular coefficient of variation of the redox ratio. The cellular coefficient of variation (cell standard deviation/cell mean) is a measure of intracellular variability (27). Increased intracellular variability in the redox ratio was observed with both low- and high-grade precancers compared with normal (P < 0.05, Table 1). Precancerous protein-bound NADH lifetimes also had increased intracellular variability compared with normal (P < 0.05, Fig. 4a). Finally, the protein-bound FAD lifetime showed increased intracellular variability in high-grade precancers only (P < 0.05, Fig. 4b). There were no significant differences in intracellular variability between low- and high-grade precancers (P > 0.05).
Table 1.
Mean and standard error of the coefficient of variation (cell standard deviation/cell mean) of the redox ratio obtained from multiphoton redox images, in calibrated units
| Tissue | Redox ratio coefficient of variation |
|---|---|
| Normal | 0.92 ± 0.08 |
| Low-grade | 1.47 ± 0.09* |
| High-grade | 1.40 ± 0.04* |
Values were averaged for all cells within an animal for normal (n = 6), low-grade precancerous (n = 8), and high-grade precancerous animals (n = 7). Significant differences (P < 0.05) exist between normal vs. low-grade precancer and normal vs. high-grade precancer (*). There were no significant differences between low- and high-grade precancers (P > 0.05).
Fig. 4.
Mean and 95% confidence interval of the coefficient of variation (cell standard deviation/cell mean) of the NADH (a) and FAD (b) lifetime variables. All values were obtained from in vivo multiphoton FLIM images, and variables were averaged for all cells within an animal for normal (n = 6), low-grade precancerous (n = 8), and high-grade precancerous animals (n = 7). Significant differences (P < 0.05) exist between normal vs. low-grade precancer and normal vs. high-grade precancer (*). There were no significant differences between low- and high-grade precancers (P > 0.05). c.u., calibrated units.
Discussion
This study reports several important positive outcomes. Morphological changes consistent with standard histopathology were observed in the in vivo images. There was a significant increase in the NCR with depth within the epithelium in normal tissues, but not in precancerous tissues. The redox ratio significantly decreased in the less differentiated basal epithelial cells compared with that in the more mature cells in the superficial layer of the normal stratified squamous epithelium, indicating an increase in metabolic activity in cells with increased NCR. However, neither the redox ratio nor the NCR were significantly different between the superficial and basal cells in precancerous tissues. A significant decrease was observed in the contribution and lifetime of protein-bound NADH (averaged over the entire epithelium) in both low- and high-grade epithelial precancers compared with normal epithelial tissues. In addition, significantly increased intracellular variability in the redox ratio and NADH fluorescence lifetimes were observed in precancerous cells compared with that in normal cells.
The volume-averaged redox ratio did not show significant differences between normal and precancerous tissues (P > 0.05, data not shown), likely due to large interanimal variability. Variance component analysis, using restricted maximum likelihood estimation (SAS statistical software), showed that the main source of variability in the volume-averaged redox ratio (37–67% of the volume-averaged redox ratio) was between animals within the same tissue type, rather than between depths or between cells within an animal. The large interanimal variability in the redox ratio is likely attributed to differences in tissue absorption and scattering at the two excitation and emission wavelengths, which are difficult to account for. Changes in tissue absorption and scattering and excitation energy flux do not influence the fluorescence lifetime images as they do fluorescence intensity images. Studies have also shown inconsistent changes in the redox ratio with dysplasia (2, 3).
The lifetimes of free and protein-bound NADH and their relative contributions measured in vivo in normal tissues are consistent with previous work in cell culture (17, 18) and in vivo (19). The fluorescence lifetime of protein-bound NADH and its contribution decreased with precancer development in vivo (Fig. 3a), consistent with the results of a previous study (19). Studies have found that the lifetime of free and protein-bound NADH and the relative contribution of protein-bound NADH decrease with hypoxia in cell culture (17–19) and tissue slices (20). Under hypoxic conditions, cells favor glycolysis over oxidative phosphorylation for ATP production. Neoplastic cells also favor glycolysis over oxidative phosphorylation (1, 28), so the measured decreases in NADH lifetimes and the contribution of protein-bound NADH with precancer are consistent with the increased levels of glycolysis that are expected in neoplastic cells. Changes in the distribution of NADH enzyme binding sites associated with preferred metabolic pathways in neoplastic tissues (6) may be responsible for the change in protein-bound NADH lifetime with precancer. The decrease in the lifetime of free NADH with high-grade precancer may reflect an increase in dynamic quenching (12).
The lifetimes and relative contributions of free and protein-bound FAD measured in vivo in normal tissues are consistent with those reported in solutions (11, 29) and in the eye (30). The protein-bound FAD lifetime increased and the relative contribution of protein-bound FAD decreased with high-grade precancer only (Fig. 3b). The increase in the protein-bound FAD lifetime in high-grade precancers could indicate decreased NAD+ levels in high-grade precancers compared with normal (13).
The intracellular variability of the redox ratio increased with precancer compared with normal. Studies have reported heterogeneities in the fluorescence redox ratios and mitochondrial membrane potentials within MS57–8T carcinoma cells, and heterogeneous fluorescence redox ratios with a homogeneous distribution of mitochondrial calcium within MCF-7 carcinoma cells (31). The protein-bound NADH lifetime intracellular variability also increased with precancer compared with normal (Fig. 4a), consistent with the results of a previous study (19), and the protein-bound FAD lifetime intracellular variability increased with high-grade precancers (Fig. 4b). Heterogeneous mitochondrial function has been attributed to both functional specialization of mitochondria and diverse mitochondrial microenvironments in previous studies (31–33). The protein-bound NADH and FAD lifetimes and the redox ratio are associated with the mitochondria (34); so, together, their increased intracellular variability could indicate heterogeneous mitochondrial function and/or microenvironments with precancer development. The combination of fluorescence redox ratio and lifetime imaging at high-resolution could potentially provide a noninvasive tool for understanding the source of mitochondrial heterogeneities in cell culture and in vivo.
The contribution and lifetime of protein-bound NADH, volume averaged over the entire epithelium showed significant differences between normal tissues and precancers. A similar comparison of the redox ratio, volume-averaged over the entire epithelium did not show significant differences between normal and precancerous tissues. However, the redox ratio and the NCR were found to be significantly different between the top and bottom planes of the epithelia in all normal tissues. This phenomenon was not observed in low- and high-grade precancers. These findings indicate that the advantage of lifetime imaging is that it does not require depth-resolved imaging. In addition, lifetime measurements are considerably less sensitive to the effects of tissue absorption and scattering, and they are not susceptible to intensity calibration issues.
In this study, multiphoton redox and lifetime imaging combine structural (cellular morphology in three dimensions) and functional (metabolic mapping) information to unravel the early stages of cancer development. These findings could guide the design and development of practical time-gated and steady state fluorescence schemes for the clinical detection of human precancers and cancers. Second, this study demonstrates experimentally the attributes of multiphoton redox and lifetime imaging for high-resolution metabolic mapping in vivo. Multiphoton redox and lifetime imaging also have broad applicability in the study of diseases other than cancer where metabolism is altered, such as Alzheimer's disease and for the study of metabolism in development and aging. Moreover, these processes can be studied with multiphoton redox and lifetime imaging in vivo and do not require contrast-enhancing dyes.
Materials and Methods
DMBA-Treated Hamster Cheek Pouch Model of Oral Cancer.
A total of 16 DMBA-treated and 6 control male Golden Syrian hamsters were evaluated in this study (155 ± 15 g). This study was approved by the institutional animal use and care committee and meets National Institutes of Health guidelines for animal welfare. For each hamster, the right cheek pouch was either treated chronically with 0.5% DMBA in mineral oil (DMBA-treated animals) or mineral oil only (control animals) for 16 weeks. The treatment procedures were established from previous studies (19, 25, 35). At 16–21 weeks after the commencement of treatment, the cheek pouch of each animal was imaged by using multiphoton microscopy.
Imaging Instrumentation.
All multiphoton images were collected with the optical workstation (36) at the University of Wisconsin. The source was a titanium sapphire laser (Spectra Physics; Tsunami). The excitation and emission light were coupled through an inverted microscope (Nikon; Eclipse TE300) with a ×40 oil-immersion objective (Nikon, PlanApo, N.A. 1.3). Excitation wavelengths of 800 nm and 890 nm were used, and the average power incident on the sample was ≈15 mW. A dichromatic mirror (BG39; Schott) allowed the excitation light to reach the sample, and wavelengths between 400 and 600 nm to reach the detector. At 800-nm excitation, a 490-nm short-pass emission filter (TFI Tech) was also used to selectively collect NADH fluorescence. Fluorescence intensity and lifetime data were collected sequentially with a GaAsP PMT (Hamamatsu; H7422). Time resolved fluorescence emission was collected via time-correlated single-photon counting electronics (Becker and Hickl; SPC-730). Previous multiphoton microscopy lifetime studies (37) and our own spectroscopy results from the hamster cheek pouch (35) confirm that the main fluorophores at 800-nm (with a 490 short-pass filter) and 890-nm excitation are NADH and FAD, respectively.
In Vivo Multiphoton Redox and Fluorescence Lifetime Imaging.
Before imaging, each hamster was anesthetized with an intraperitoneal injection of a mixture of 2.5 mg/kg acepromazine, 200 mg/kg ketamine, and 5 mg/kg xylazine, and the mucosa of the cheek pouch was secured flush against the coverslip for imaging (19).
A fluorescence intensity image z-stack and then an FLIM image z-stack were collected from the same tissue volume at 800 nm, and then identical stacks were collected at 890-nm excitation. The x–y stage was then moved for another set of fluorescence intensity and FLIM image z-stacks within the biopsy site. The fluorescence intensity and FLIM image z-stacks were generated with a pixel dwell time of ≈11.5 μs, using 2-μm and 10-μm z-step sizes, respectively. The scan area at each image plane of the stack was 256 × 256 pixels (165 × 165 μm). The scan time per intensity image was ≈1 s. Single photon counting for FLIM measurements was performed at a rate of ≈5 × 104 photons per s (by adjusting the power incident on the sample). The integration time for the FLIM images was 60 to 120 s. There was no change in the photon count rates during image acquisition, ensuring that photobleaching did not occur.
Each intensity image was calibrated with Rhodamine B (Sigma; 115H3423) standard intensity images (average of three separate images) (5.8 × 10−5 g/ml Rhodamine B in distilled water) collected under conditions identical to those of the in vivo data.
The instrument response function (IRF) measured with a second harmonic signal from a β-BaB2O4 crystal (38) was used in the lifetime fit model. The IRF had a full width at half maximum of 0.313 ns. The lifetime of Fluoresbrite YG microspheres (Polysciences) were imaged at the beginning of each day of the in vivo experiments to ensure that the lifetime measurements were consistent from day to day. The microsphere lifetime decay curves were fit to a single exponential decay, resulting in a lifetime of 2.19 ± 0.02 ns (n = 12), consistent with previous studies (17, 19, 39).
Pathology.
After in vivo imaging, one biopsy was taken from the imaged sites with a 3-mm diameter dermal biopsy punch, placed in buffered formalin, and submitted for histopathology. The tissue biopsies were prepared and read by a certified pathologist (A.G.-F.). Diagnosis was based on established criteria (40). The diagnosis assigned to each image stack was based on the most severe diagnosis of the entire corresponding biopsy sample, consistent with our previous multiphoton microscopy studies in this animal model (19, 25).
Quantification of the Lifetime and Contribution of Protein-Bound and Free NADH and FAD.
SPCImage software (Becker and Hickl) was used to analyze the fluorescence lifetime decay curves, with methods identical to those reported by our group in ref. 19. The lifetime decay curve of each pixel was fit to a double-exponential decay model (19), where the free parameters are the short and long lifetime components (τ1 and τ2, respectively), and the relative contributions of the lifetime components (α1 and α2, where α1 + α2 = 100%). The presence of two distinctly different lifetimes for free and protein-bound NADH (10, 18), and for free and protein-bound FAD (11, 29) indicates that NADH and FAD fluorescence decay curves are best fit to a double-exponential decay model. When the scattering contribution was fixed at zero, the χ2 value of the fit was minimized. This offers experimental evidence that the contribution of scattered light to the decay curve was minimal. The χ2 value for all in vivo hamster cheek pouch images was 1.03 ± 0.02, indicating good fits.
The validity of using a biexponential model with three free parameters (τ1, τ2, and α1/α2) was tested with simulations. The simulations were carried out on a double-exponential decay curve [τ1 = 300 ps, τ2 = 2,000 ps, and α2 ranges from 27% to 47% in 1% increments, which are reported values for NADH (17, 20)]. These curves were fit to a double-exponential model for cases where: (i) τ1 was fixed, (ii) τ2 was fixed, (iii) τ1 and τ2 were fixed, and (iv) all were set as free parameters. Poisson noise was added to each time bin of each decay curve, using a different seed from a random number generator. The SNR was similar to that in the measured data (100 counts at peak, with Poisson noise equal to the square root of the number of counts). The χ2 value and percentage difference between actual and calculated values were comparable to within 13% for all possible combinations of free and fixed variables. The validity of biexponential fit with three free parameters were also evaluated by examining the goodness of fit (χ2 value), using measured data at 800-nm and 890-nm excitation from one normal site. Again, the measured results showed that a two-exponential fit with all parameters free gives a result that is similar to a two-exponential fit with one or two parameters fixed.
Simulated envelopes (41) were used to calculate the fitting error due to the photon counting process (Poisson noise). Poisson noise was added (as described above) to a simulated fluorescence decay curve representative of NADH (τ1 = 325 ps, τ2 = 2,356 ps, and α2 = 36%) and FAD (τ1 = 106 ps, τ2 = 2,068, and α1 = 93%) in the normal hamster cheek pouch. This process was repeated on each of the original decay curves 4,096 times. Each new decay curve was then independently analyzed to obtain a new set of fitted parameters.
Quantification of the Redox Ratio.
The redox ratio was calculated by dividing the fluorescence intensity at 890-nm excitation (FAD) by the fluorescence intensity at 800-nm excitation (NADH), pixel by pixel, using ImageJ (National Institutes of Health).
Quantification of Morphological Variables.
The epithelial layer was defined as the volume between the first layer of cells (below the acellular superficial layer) and the last layer of cells (above the stromal collagen). The thickness of the epithelial layer was determined from the total number of image planes (collected at 2-μm separations) between the bottom of the acellular surface layer (at which the epithelial cells were first visually identified) and the epithelial/stromal interface (at which collagen was first identified). The thickness measurements were averaged from each of two 150 × 150 μm sites within each animal.
The NCR was defined as the diameter of the nucleus divided by the diameter of the entire cell (along the same line, arbitrary orientation). The NCR was defined along a line rather than over an area or volume because the line-based NCR showed significant differences between categories of interest, and is the simplest metric to quantify. The NCR measurements along one arbitrary line (0.48 ± 0.13) were found to be within the standard deviation of the NCR measured along three different arbitrary lines (0.47 ± 0.11) within a cell. This was tested on three cells each from normal, low-grade, and high-grade tissues (nine cells total).
Statistical Analyses.
Wilcoxon rank-sum tests were carried out to determine whether there are significant differences in the redox ratio, the NADH and FAD lifetime variables, and the thickness of the epithelia between normal, low-grade, and high-grade tissues (unpaired comparison). Wilcoxon signed-rank tests were carried out to determine whether there were significant differences in the redox ratio, lifetime variables, and the NCR between the top and bottom epithelial planes within an image stack for all normal, low-grade, and high-grade tissues (paired comparison). Finally, Wilcoxon rank-sum tests were performed on the cellular coefficient of variation (standard deviation/mean) of the redox ratio and lifetime variables between normal, low-grade, and high-grade tissues. Nonparametric Wilcoxon tests (Matlab) were chosen for the paired (Fig. 2) and unpaired (Figs. 3 and 4 and Table 1) comparisons because of the small sample size. Exact P values were computed, all P values were two-sided, and a P value <0.05 was considered significant.
All cells in the whole field of view of each image stack were analyzed to compute the redox ratio and lifetime variables of the epithelia for each site within each animal. Three individual cells within an image plane were chosen for the redox ratio, lifetime and NCR comparison between the top and bottom planes of the epithelia, and the cellular coefficient of variation calculation, because it is labor-intensive to manually determine the NCR and the cellular coefficient of variation for all cells within each image plane. The nuclei were excluded from both the whole field of view, and individual cell analysis of the redox ratio and lifetime variables by increasing the threshold value so that the much darker nucleus was below the threshold.
Acknowledgments
We thank Axel Bergmann and Christine McMahon. This work was supported by National Institutes of Health Grant R01 EB000184 and Department of Defense Predoctoral Traineeship Grant W81XWH-04-1-0330 (to M.C.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Gulledge CJ, Dewhirst MW. Anticancer Res. 1996;16:741–749. [PubMed] [Google Scholar]
- 2.Drezek R, Brookner C, Pavlova I, Boiko I, Malpica A, Lotan R, Follen M, Richards-Kortum R. Photochem Photobiol. 2001;73:636–641. doi: 10.1562/0031-8655(2001)073<0636:AMOFCT>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 3.Ramanujam N, Kortum RR, Thomsen S, Jansen AM, Follen M, Chance B. Opt Express. 2001;8:335–343. doi: 10.1364/oe.8.000335. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z, Li H, Liu Q, Zhou L, Zhang M, Luo Q, Glickson J, Chance B, Zheng G. Biosens Bioelectron. 2004;20:643–650. doi: 10.1016/j.bios.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 5.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Mol Biol Cell. New York: Garland Science; 2002. [Google Scholar]
- 6.Banerjee S, Bhatt DK. Indian J Cancer. 1989;26:21–30. [PubMed] [Google Scholar]
- 7.Chance B, Schoener B, Oshino R, Itshak F, Nakase Y. J Biol Chem. 1979;254:4764–4771. [PubMed] [Google Scholar]
- 8.Chance B. J Appl Cardiol. 1989;4:207–221. [Google Scholar]
- 9.Zhang Z, Blessington D, Li H, Busch TM, Glickson J, Luo Q, Chance B, Zheng G. J Biomed Opt. 2004;9:772–778. doi: 10.1117/1.1760759. [DOI] [PubMed] [Google Scholar]
- 10.Lakowicz JR, Szmacinski H, Nowaczyk K, Johnson ML. Proc Natl Acad Sci USA. 1992;89:1271–1275. doi: 10.1073/pnas.89.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakashima N, Yoshihara K, Tanaka F, Yagi K. J Biol Chem. 1980;255:5261–5263. [PubMed] [Google Scholar]
- 12.Lakowicz JR. Principles of Fluorescence Spectroscopy. New York: Plenum; 1999. [Google Scholar]
- 13.Maeda-Yorita K, Aki K. J Biochem (Tokyo) 1984;96:683–690. doi: 10.1093/oxfordjournals.jbchem.a134886. [DOI] [PubMed] [Google Scholar]
- 14.Iweibo I. Biochim Biophys Acta. 1976;446:192–205. doi: 10.1016/0005-2795(76)90110-0. [DOI] [PubMed] [Google Scholar]
- 15.Muller F, Mayhew SG, Massey V. Biochemistry. 1973;12:4654–4662. doi: 10.1021/bi00747a017. [DOI] [PubMed] [Google Scholar]
- 16.Sato K, Nishina Y, Shiga K, Tanaka F. J Photochem Photobiol B. 2003;70:67–73. doi: 10.1016/s1011-1344(03)00056-3. [DOI] [PubMed] [Google Scholar]
- 17.Bird DK, Yan L, Vrotsos KM, Eliceiri KW, Vaughan EM, Keely PJ, White JG, Ramanujam N. Cancer Res. 2005;65:8766–8773. doi: 10.1158/0008-5472.CAN-04-3922. [DOI] [PubMed] [Google Scholar]
- 18.Schneckenburger H, Wagner M, Weber P, Strauss WSL, Sailer R. J Fluorescence. 2004;14:649–654. doi: 10.1023/b:jofl.0000039351.09916.cc. [DOI] [PubMed] [Google Scholar]
- 19.Skala MC, Riching KM, Bird DK, Gendron-Fitzpatrick A, Eickhoff JC, Eliceiri KW, Keely PJ, Ramanujam N. J Biomed Opt. 2007;12 doi: 10.1117/1.2717503. 024014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vishwasrao HD, Heikal AA, Kasischke KA, Webb WW. J Biol Chem. 2005;280:25119–25126. doi: 10.1074/jbc.M502475200. [DOI] [PubMed] [Google Scholar]
- 21.Denk W, Strickler JH, Webb WW. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 22.Centonze VE, White JG. Biophys J. 1998;75:2015–2024. doi: 10.1016/S0006-3495(98)77643-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Squirrell JM, Wokosin DL, White JG, Bavister BD. Nat Biotechnol. 1999;17:763–767. doi: 10.1038/11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrejevic S, Savary JF, Fontolliet C, Monnier P, van Den Bergh H. Int J Exp Pathol. 1996;77:7–14. doi: 10.1046/j.1365-2613.1996.956095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skala MC, Squirrell JM, Vrotsos KM, Eickhoff JC, Gendron-Fitzpatrick A, Eliceiri KW, Ramanujam N. Cancer Res. 2005;65:1180–1186. doi: 10.1158/0008-5472.CAN-04-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Searle S, Cassella G, McCulloch C. Variance Components. New York: Wiley; 1992. [Google Scholar]
- 27.Gibbons J, Chakraborti S. Nonparametric Statistical Inference. New York: Marcel Dekker; 1992. [Google Scholar]
- 28.Schroeder T, Yuan H, Viglianti BL, Peltz C, Asopa S, Vujaskovic Z, Dewhirst MW. Cancer Res. 2005;65:5163–5171. doi: 10.1158/0008-5472.CAN-04-3900. [DOI] [PubMed] [Google Scholar]
- 29.Konig K, Riemann I. J Biomed Opt. 2003;8:432–439. doi: 10.1117/1.1577349. [DOI] [PubMed] [Google Scholar]
- 30.Schweitzer D, Hammer M, Schweitzer F, Anders R, Doebbecke T, Schenke S, Gaillard ER, Gaillard ER. J Biomed Opt. 2004;9:1214–1222. doi: 10.1117/1.1806833. [DOI] [PubMed] [Google Scholar]
- 31.Kuznetsov AV, Usson Y, Leverve X, Margreiter R. Mol Cell Biochem. 2004;256–257:359–365. doi: 10.1023/b:mcbi.0000009881.01943.68. [DOI] [PubMed] [Google Scholar]
- 32.Saks VA, Kaambre T, Sikk P, Eimre M, Orlova E, Paju K, Piirsoo A, Appaix F, Kay L, Regitz-Zagrosek V, et al. Biochem J. 2001;356:643–657. doi: 10.1042/0264-6021:3560643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seppet EK, Kaambre T, Sikk P, Tiivel T, Vija H, Tonkonogi M, Sahlin K, Kay L, Appaix F, Braun U, et al. Biochim Biophys Acta. 2001;1504:379–395. doi: 10.1016/s0005-2728(00)00269-3. [DOI] [PubMed] [Google Scholar]
- 34.Duchen MR, Biscoe TJ. J Physiol. 1992;450:13–31. doi: 10.1113/jphysiol.1992.sp019114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skala MC, Palmer GM, Zhu C, Liu Q, Vrotsos KM, Marshek-Stone CL, Gendron-Fitzpatrick A, Ramanujam N. Laser Surg Med. 2004;34:25–38. doi: 10.1002/lsm.10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wokosin DL, Squirrell JM, Eliceiri KW, White JG. Rev Sci Instrum. 2003:74. doi: 10.1063/1.1524716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang S, Heikal AA, Webb WW. Biophys J. 2002;82:2811–2825. doi: 10.1016/S0006-3495(02)75621-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bird DK, Eliceiri KW, Fan CH, White JG. Appl Opt. 2004;43:5173–5182. doi: 10.1364/ao.43.005173. [DOI] [PubMed] [Google Scholar]
- 39.Schonle A, Glatz M, Hell SW. Applied Optics. 2000;39:6306–6311. doi: 10.1364/ao.39.006306. [DOI] [PubMed] [Google Scholar]
- 40.MacDonald DG, Saka SM. Structural Indicators of the High Risk Lesion. Cambridge, UK: Cambridge Univ Press; 1991. [Google Scholar]
- 41.Atkinson AC. Plots, Transformations and Regression: An Introduction to Graphical Methods of Diagnostic Regression Analysis. New York: Oxford Univ Press; 1985. [Google Scholar]