Abstract
The life cycle of human papillomaviruses (HPVs) is linked to epithelial differentiation, with late viral events restricted to the uppermost stratified layers. Our studies indicated that HPV activates capases-3, -7, and -9 upon differentiation, whereas minimal activation was observed in differentiating normal keratinocytes. Activation occurred in the absence of significant levels of apoptosis, suggesting a potential role for caspases in the viral life cycle. In support of this, the addition of caspase inhibitors significantly impaired differentiation-dependent viral genome amplification. A conserved caspase cleavage motif was identified in the replication protein E1 (46DxxD49) that was targeted in vitro by both recombinant caspase-3 and caspase-7. Mutation of this site inhibited amplification of viral genomes, indicating that caspase cleavage is necessary for the productive viral life cycle. Our study demonstrates that HPV activates caspases upon differentiation to facilitate productive viral replication and represents a way by which HPV controls viral gene function in differentiating cells.
Keywords: replication, pathogenesis, apoptosis
Human papillomaviruses (HPVs) are small, double-stranded DNA viruses that exhibit tropism for epithelial cells (1). Approximately one-third of the >100 HPV types identified infect epithelial cells of the genital tract, and a subset are the etiological agents of cervical cancers. HPVs infect cells in the basal layer of the epithelium, which become exposed through microlesions (2). After infection, viral genomes are established as extrachromosomal elements in the nucleus and are maintained at ≈50–100 copies per cell. Epithelial differentiation triggers the productive phase of the HPV life cycle, which includes genome amplification, activation of late gene expression, and assembly of mature virions. HPV-positive cells reenter into the S phase upon differentiation because of the actions of the E6 and E7 proteins (3, 4). E6 binds a number of factors, including the ubiquitin ligase E6-AP, directing it to the tumor suppressor p53, resulting in its accelerated turnover (5–7). E7 binds and targets members of the retinoblastoma family of proteins (pRb, p107, p105) for degradation, resulting in the activation of E2F transcription factors (8, 9), which leads to accelerated cell cycle progression and increased proliferation (10). E6 and E7 can abrogate normal cellular checkpoints and interfere with processes intended to eliminate aberrant cells, including apoptosis and senescence.
Apoptosis, a form of programmed cell death, is a tightly regulated process that plays important roles in development and homeostasis, as well as functioning as an antiviral defense mechanism (11, 12). The primary effectors of the apoptotic response are a group of cysteine proteases termed caspases that are synthesized as inactive zymogens and become activated by oligomerization or processing (13–15). Two pathways can be stimulated to initiate the caspase cascade. The extrinsic pathway is triggered by the binding of ligands to their cognate death receptors, resulting in the activation of caspase-8. The intrinsic, or mitochondrial-mediated, pathway is initiated in response to DNA damage, growth factor withdrawal, or viral infection, resulting in the activation of the initiator caspase, caspase-9. Both pathways converge to activate the same executioner caspases, including caspase-3 and -7, which then cleave important cellular proteins, resulting in cell death (16).
Many viruses have developed strategies to block host-mediated apoptosis, to preserve the cellular machinery necessary for viral gene expression, as well as replication (17). In this study, we investigated the effect of HPV-31 on the expression profile of apoptotic caspases and determined that HPV proteins activate caspases upon differentiation, which in turn is necessary for productive replication of viral genomes.
Results
Differentiation of HPV-31-Positive Keratinocytes Leads to Caspase Activation in an E6- and E7-Dependent Manner.
To determine whether caspase activation was altered by HPV proteins upon differentiation, we first performed immunohistochemical analysis to examine whether the active form of caspase-3 was present in organotypic raft cultures generated from normal human foreskin keratinocytes (HFKs), as well as from human keratinocytes that stably maintain HPV-31 episomes (HFK-31). In raft cultures of normal HFKs, a small number of caspase-3-positive cells were observed that were generally restricted to the basal layer (Fig. 1A). Surprisingly, the number of caspase-3-positive cells was significantly increased in rafts of HPV-31-containing cells, and these were distributed throughout all layers of the stratified epithelia (Fig. 1A), suggesting that HPV proteins induce rather than block caspase activation upon differentiation.
Fig. 1.
HPV induces caspase activation in cells induced to differentiate in organotypic raft cultures and by suspension in methylcellulose. (A) Sections of organotypic raft cultures generated from untransfected HFKs (Upper) and HFKs stably transfected with HPV-31 (HFK-31) (Lower) were stained with H&E (I and IV), an antibody specific for active caspase-3 (II and V), as well as DAPI to visualize nuclei (III and VI). Arrows indicate the basal layer of the epithelium. (B) Western blot analysis of cell lysates from untransfected HFKs (HFK) and HPV-31-positive HKFs (HFK-31) by using antibodies specific for the active and pro forms of capsase-9, -3, and -7. Lysates were harvested from undifferentiated (T0) monolayer cultures or after suspension in methylcellulose to induce differentiation for 12 and 24 h. GAPDH served as a loading control. The results shown are representative of three independent experiments by using two different HFK backgrounds. Hr, hours; MC, methylcellulose.
To ensure that the activation of caspases by HPV proteins was not specific for the organotypic raft culture system, we examined caspase levels in keratinocytes induced to differentiate by two alternate methods: suspension in methylcellulose and growth in high-calcium-containing medium. Suspension of HPV-31-positive cells in methylcellulose for 24 h is sufficient to activate late events in the viral life cycle, including capsid gene expression and genome amplification (18). Normal HFKs, as well as HPV-31-positive keratinocytes (HFK-31), were suspended in methylcellulose, protein extracts were harvested, and caspase-3 activation was examined by Western blot analysis (Fig. 1B). Similar to the observations in raft cultures, HPV-31-positive cells exhibited activation of caspase-3 upon differentiation in methylcellulose (Fig. 1B), with the cleaved (active) form of caspase-3 detected as early as 2 h after suspension [supporting information (SI) Fig. 7A]. Increased activation of the upstream initiator caspase, caspase-9, as well as another downstream executioner caspase, caspase-7, was also detected (Fig. 1B). Unfortunately, we were unable to detect the active forms of caspase-7 and caspase-9 in organotypic raft cultures by immunohistochemistry because of high background. Similar results for caspase activation were observed for HPV-31 cell lines generated from multiple HFK backgrounds, as well as CIN612 cells that are derived from a HPV-31-positive cervical biopsy (19) (data not shown). Consistent with observations in raft cultures, little to no activation of any of these caspases was detected in Western blot analysis of normal HFKs (Fig. 1B). HPV-31-positive cells also exhibited increased cleavage of the caspase-3 and caspase-7 substrate poly(ADP-ribosyl) polymerase upon differentiation (SI Fig. 7B), which was indicative of caspase activation. A similar pattern of caspase activation was observed in cells that were induced to differentiate in high-calcium medium (SI Fig. 8A). Whereas increased activation of caspase-3 and caspase-7 was observed in differentiating HPV-31-positive HFKs, as well as the CIN612 cell line, little to no activation was observed in normal HFKs (SI Fig. 8A). We were unable to detect any of the active forms of caspase-8 by Western blot analysis in either HPV-positive HFKs or normal HFKs induced to differentiate in methylcellulose or high calcium (data not shown), indicating that the intrinsic rather than the extrinsic pathway was being activated in differentiating cells. Based on these results, obtained by using three different methods for differentiation, we conclude that caspase activation occurs during the differentiation-dependent phase of the viral life cycle.
Because the E6 and E7 oncoproteins have been shown to modulate apoptotic pathways in response to various stimuli (20, 21), we next investigated whether either of these proteins could be responsible for caspase activation observed in differentiating cells. HFKs stably expressing the retroviral vector pLXSN alone, pLXSN-E6 or -E7 alone, or pLXSN–E6/E7 were suspended in methylcellulose, and cell extracts were harvested as a function of time. Western blot analysis was then performed to examine the state of caspase activation (Fig. 2). Consistent with previous observations, normal HFKs demonstrated little to no activation of caspase-7 and caspase-3 after differentiation in methylcellulose (Fig. 2). In contrast, cells stably expressing E6 alone, E7 alone, or the combination of E6/E7 exhibited increased caspase activation. Cells expressing E7 alone appeared to induce the greatest amount of the active forms of these caspases. Similar results were observed upon calcium-induced differentiation of HFKs stably expressing E6, E7, or E6/E7 (data not shown). Our results indicate that E7, as well as E6, can independently induce activation of caspases upon differentiation.
Fig. 2.
High-risk E6 and E7 can independently activate caspases upon differentiation. Western blot analysis was performed on cell lysates harvested from HFKs retrovirally transduced with pLXSN alone or pLXSN-HPV-31 E6, E7, or E6/E7. Lysates were harvested from undifferentiated cells (T0) and cells induced to differentiate in methylcellulose for 24 and 48 h. Primary antibodies used were specific for the active form of caspase-3 and caspase-7 and for procaspase-3 and -7. GAPDH was used as a loading control. The results are representative of three experiments by using two different HFK backgrounds.
HPV Mediates an Antiapoptotic Response Despite Caspase Activation upon Differentiation.
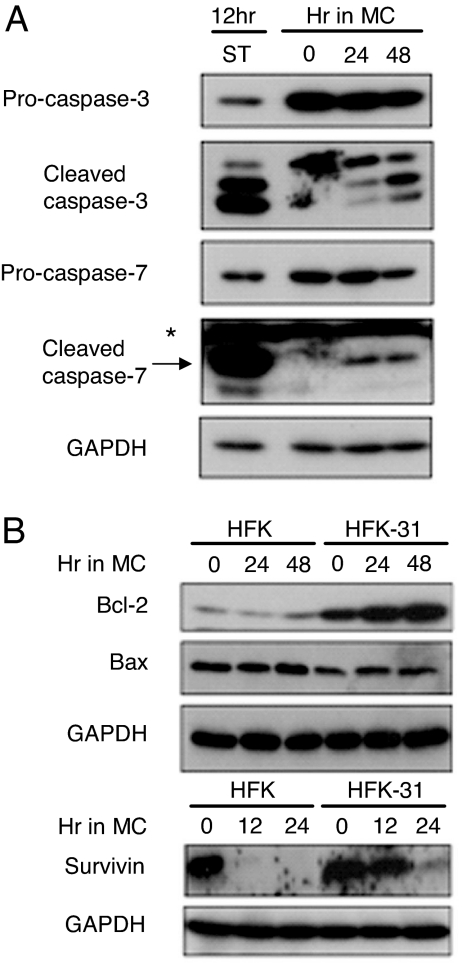
The observation that HPV proteins activate caspases upon differentiation was surprising given that productive viral replication depends on epithelial differentiation and requires cells to be active during the S phase (4, 19). However, several studies have indicated that caspases can regulate a number of nonapoptotic cellular processes (22, 23). Microscopic examination of cross-sections of organotypic raft cultures of HPV-31-positive cells failed to identify a large number of cells exhibiting characteristics of apoptosis, such as membrane blebbing or loss of intact nuclei. To determine whether the level of caspase activation observed in differentiating HPV-positive cells was similar to that of cells undergoing apoptosis induced by chemical agents, HPV-31-positive HFKs were treated with the cell death stimulus staurosporine or were induced to differentiate in methylcellulose. As shown in Fig. 3A, treatment with staurosporine resulted in high levels of the active forms of caspase-3 and caspase-7. In contrast, the level of caspase activation observed in an equal number of differentiating HPV-31-positive cells after suspension in methylcellulose was reduced (Fig. 3A), which may have contributed to the limited apoptosis seen in differentiating HPV-positive cells. By using an enzymatic assay to measure caspase-3/caspase-7 activity, we found that HPV-positive cells exhibited a 2-fold lower induction of caspase activity upon differentiation compared with cells undergoing staurosporine-induced apoptosis (SI Fig. 9). We also observed that levels of antiapoptotic Bcl-2 were increased in HPV-positive cells, whereas those of proapoptotic Bax were not significantly altered between HPV-31 HFKs and normal HFKs (Fig. 3B). Because Bcl-2 is important in regulating antiapoptotic responses through the mitochondrial pathway, it is possible that increased levels of this protein may be necessary to block or limit apoptosis in differentiating HPV-positive cells. We also observed that survivin, a member of the inhibitor of apoptosis family, was maintained in differentiating HPV-31-positive HFKs but not in normal HFKs, suggesting that its activity may be important for viral functions. Similar levels of survivin were also observed in HPV-positive cells after calcium-induced differentiation (SI Fig. 8B). These results indicate that HPV-mediated caspase activation coincides with increased levels of antiapoptotic factors, such as survivin and Bcl-2, and this may be important for maintaining the viability of HPV-positive cells upon differentiation.
Fig. 3.
HPV proteins activate low levels of caspases upon differentiation while simultaneously increasing levels of antiapoptotic proteins. (A) Analysis of caspase activation in apoptotic vs. differentiating HFK-31 cells. HFK-31 cells were treated with 0.5 μM staurosporine (ST) for 12 h or were suspended in methylcellulose to induce differentiation. Whole cell lysates were harvested from undifferentiated monolayer cultures (T0) or from cells suspended in methylcellulose for 24 and 48 h. Immunoblots were probed by using primary antibodies that recognize either the active form of caspase-3 and -7 or the procaspase forms. GAPDH was used as a loading control. *, nonspecific band. (B) Analysis of antiapoptotic proteins in HPV-31-positive cells induced to differentiate in methylcellulose. Whole cell lysates were harvested from HPV-31-positive HFKs (HFK-31) and untransfected HFKs at time 0 (undifferentiated) and 12, 24, and 48 h after suspension in methylcellulose to induce differentiation. Blots were probed with antibodies to Bcl-2, Bax, and survivin. GAPDH served as a loading control. The data shown are representative of three independent experiments.
HPV-Mediated Caspase Activation Is Necessary for Viral Genome Amplification upon Differentiation.
To determine whether caspase activation contributed to some aspect of the productive life cycle of HPV, the effect of peptide caspase inhibitors on the differentiation-dependent event of viral genome amplification was examined. The presence of caspase inhibitors in methylcellulose failed to block caspase activation in suspended cells (data not shown), indicating that cellular uptake of the inhibitors did not occur; therefore, for these studies, we focused on differentiation induced by high calcium. Growth of HPV-31-positive cells in high-calcium medium induces viral DNA amplification after 48 to 72 h (C.A.M., unpublished data). To examine the effect of inhibiting caspases on the amplification of viral genomes, HPV-31-positive cells were grown in high-calcium medium for up to 96 h in the presence of DMSO alone as a vehicle control or 20 μM the general caspase inhibitor Z-VAD-FMK (Fig. 4). At this concentration, Z-VAD-FMK inhibited activation of caspase-3/caspase-7 upon differentiation of HPV-positive cells (SI Fig. 9). Total DNA was isolated, and Southern blot analysis was performed to screen for amplification of viral genomes (Fig. 4A). Whereas the DMSO-treated cells exhibited no inhibitory effect on the amplification of HPV episomes, the Z-VAD-FMK-treated cells were impaired in this ability (Fig. 4A Upper). Similar results were observed with a peptide inhibitor specific for caspase-3 (Z-DMQD-FMK) (Fig. 4A Lower), as well as for caspase-3 and caspase-7, but not with an inhibitor to caspase-8 (data not shown). These results indicate that activation of caspase-3 and/or caspase-7 may be necessary for the productive replication of HPV DNA upon differentiation.
Fig. 4.
Differentiation-dependent viral genome amplification requires caspase activation. (A) HPV-31-positive CIN612 cells were treated with DMSO alone as a vehicle control, 20 μM the general caspase inhibitor Z-VAD-FMK (Upper), or 20 μM the caspase-3 inhibitor Z-DMQD-FMK (Lower) and grown in high-calcium medium for the indicated times. DNA was harvested from both control and treated cells and examined by Southern blot analysis by using the HPV genome as a probe. (B) Expression of keratin 10 (K10) in differentiating CIN612 cells. CIN612 cells were treated with DMSO as a vehicle control or 15 or 20 μM the general caspase inhibitor Z-VAD-FMK and were grown in high-calcium medium for 48 and 96 h. Lysates were analyzed by immunoblotting by using an antibody specific for K10. Experiments were repeated a minimum of three times with similar results.
To ensure that the reduction in viral genome amplification observed in the presence of caspase inhibitors was not attributable to inhibition of differentiation, we examined the expression of cytokeratin 10 (K10), which is a marker of differentiation and is expressed concurrently with genome amplification (24). Expression of K10 increased upon calcium-induced differentiation of CIN612 cells and was not diminished by treatment with Z-VAD-FMK (Fig. 4B) or the caspase-3 inhibitor Z-DMQD-FMK (data not shown). Similar results were observed for another marker of differentiation, cytokeratin 1 (data not shown). Overall, these results suggest that the ability of HPV-31-positive cells to efficiently amplify viral genomes upon differentiation is directly dependent on caspase activation.
HPV-31 E1 Is a Substrate for Caspase-3 and -7 Processing.
We next screened HPV genome sequences for putative caspase cleavage sites and identified a potential caspase-3/7 cleavage motif in the HPV-31 replication protein E1 at position 46–49, corresponding to the sequence DMVD (Fig. 5A). Importantly, this cleavage site is conserved at similar locations in E1 proteins from both high-risk (types 16 and 18) and low-risk (types 6 and 11) viruses. To determine whether HPV-31 E1 is a substrate for caspase-3 and/or -7, we first used GST fusion proteins encoding amino acids 1–170 of E1 together with recombinant caspase-3 and caspase-7 in in vitro caspase cleavage assays (Fig. 5B and SI Fig. 10A). The products of the cleavage reactions were examined by Western blot analysis by using an antibody specific for GST to detect the cleaved forms of E1. As shown in Fig. 5B, the HPV-31 GST–E1 fusion protein was cleaved upon incubation with caspase-3 (lanes 2 and 3), whereas pretreatment with a caspase-3-specific inhibitor significantly impaired cleavage (lane 4). Recombinant caspase-7 was also able to cleave GST–E1, although less efficiently than caspase-3 (SI Fig. 10A). Mutation of the aspartic acid residue at position 49 of the cleavage site to an alanine (E1D49A) abrogated cleavage by both caspase-3 and caspase-7, confirming that the 46DMVD49 motif serves as a caspase-3/7 recognition and cleavage site in vitro and possibly in vivo (Fig. 5B, lanes 6 and 7, and SI Fig. 10A). The E1 protein is expressed at low levels from the viral genome and can be detected only by using overexpression systems. To determine whether E1 could be cleaved in vivo in the presence of activated caspases, C33A cells expressing YFP-tagged HPV-31 E1 or a YFP-E1 D49A mutant were stimulated to undergo apoptosis by treatment with staurosporine. Western blot analysis was then performed to screen for cleavage products (SI Fig. 10B). Whereas treatment with staurosporine resulted in cleavage of WT E1, mutation of the caspase cleavage motif abrogated E1 processing, indicating that E1 can serve as a caspase substrate in vivo. In addition, these results indicate that the conserved caspase cleavage motif (46DMVD49) is the only site recognized by activated caspases.
Fig. 5.
Recombinant caspase-3 cleaves GST-31 E1 in vitro. (A) Schematic of E1 showing the location of the caspase cleavage site at amino acids 46–49. (B) Purified GST-31 E1 WT (amino acids 1–170) and the GST-31 E1D49A cleavage mutant were incubated with increasing concentrations of recombinant caspase-3 (C3) [2.5 ng (lanes 2 and 6), 25 ng (lanes 3 and 7)] for 1 h at 37°C. Indicated samples were also incubated with the caspase-3 inhibitor Z-DMQD-FMK (C3i) (50 μM). Cleavage reactions were analyzed by Western blot analysis by using an antibody that recognizes GST. Full-length and cleaved forms of E1 are indicated. (C) buffer control.
Mutation of the Caspase Cleavage Site in E1 Reduces Viral Genome Amplification upon Differentiation.
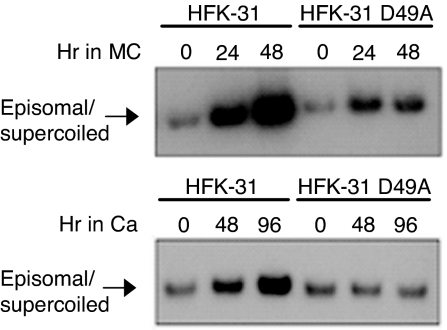
To examine the effects of blocking caspase cleavage of E1 on activation of differentiation-dependent viral functions, we constructed genomes in which amino acid 49 of the caspase cleavage site was mutated from an aspartic acid to an alanine (E1D49A). HFKs were transfected with WT and E1D49A mutant genomes, and stable cell lines were generated. Southern blot analysis performed on DNA harvested from monolayer cultures demonstrated that the E1D49A mutant genomes were stably maintained at a copy number similar to cells containing WT genomes (Fig. 6), suggesting that caspase-mediated cleavage of E1 is not necessary for genome maintenance in undifferentiated cells. In contrast, after suspension of these cells in methylcellulose or exposure to high calcium, the E1D49A mutant genomes were found to be greatly impaired in the ability to amplify compared with WT HPV-31 (Fig. 6). Similar results were observed in two additional cell lines containing mutant genomes, indicating that caspase-mediated cleavage of E1 upon differentiation plays an important role in the viral life cycle.
Fig. 6.
Mutation of the E1 caspase cleavage site reduces differentiation-dependent viral genome amplification. HFKs that stably transfected with WT 31 genomes or genomes containing the E1 D49A caspase cleavage mutant were suspended in methylcellulose for 24 and 48 h (Upper) or grown in high calcium for 48 and 96 h (Lower) to induce differentiation. DNA was harvested from undifferentiated, monolayer cultures (T0) and differentiated cells at the indicated times. Southern blot hybridization was performed by using the HPV genome as a probe. A minimum of three independent experiments were performed by using three different HFK backgrounds.
Discussion
Viruses often inactivate apoptotic pathways to ensure completion of their life cycle. Our studies indicate that HPV proteins activate rather than suppress caspases of the intrinsic apoptotic pathway and that this activation is necessary for the productive HPV life cycle. We observed that HPV proteins activate caspases-9, -3, and -7 upon differentiation, with little to no activation observed in undifferentiated or differentiated normal keratinocytes (Fig. 1B and SI Fig. 8A). The level of caspase activation induced by HPV proteins was reduced from that seen in cells undergoing staurosporine-induced apoptosis (Fig. 3A) and was accompanied by activation of antiapoptotic proteins, including Bcl-2 and survivin (Fig. 3B). Bcl-2 works at the mitochondrial membrane to interfere with cytochrome c release, and E6 has been shown to increase levels of this protein by as yet undefined mechanisms (25, 26). The mechanism by which survivin mediates its antiapoptotic effects is not well understood but may involve inhibition of both initiator (caspase-9) as well as effector caspase (caspase-3 and -7) activity (27, 28). Interestingly, p53 down-regulates survivin expression, and E6, which targets p53 for degradation, has been implicated in survivin activation (29). In our studies, we failed to observe high levels of apoptosis in differentiating HPV-positive cells. It is possible that the expression of antiapoptotic proteins, coupled with a low level of caspase activation, may be important in providing a balance between cell viability and cell death upon differentiation. It is also possible that some degree of apoptosis occurs after productive replication to facilitate postassembly events, such as virion release.
The E6 and E7 proteins were found to independently activate caspases upon differentiation (Fig. 2). E7 promotes reentry of infected cells into the S phase upon differentiation through destabilization of Rb family members, which leads to the release of E2F factors, including E2F1 and E2F3, resulting in transcription of genes involved in the apoptotic response (8, 9, 30). However, because expression of the procaspases was not increased in HPV-positive cells, it is likely that other mechanisms are responsible for the caspase activation we observed. Another consequence of E7-mediated Rb degradation is up-regulation of p53 (31, 32), which in normal cells can lead to cell cycle arrest or apoptosis. In HPV infections, the presence of E6 counteracts this increase by causing a rapid turnover of p53 (5–7). It is therefore possible that E7-mediated activation of caspases is not a physiologically relevant phenomenon because it may occur only in the absence of E6. In addition to p53, E6 binds a number of cellular factors, and one or more of these interactions may be responsible for its role in caspase activation upon differentiation (2). Most of these interactions have been shown to interfere with apoptosis, but the consequences of only a limited number of these factors have been examined upon differentiation (20, 21). It is also not clear at what stage HPV proteins target the intrinsic pathway to activate the caspase cascade, although it is most likely upstream of caspase-9 activation. We have also observed that low risk HPV 11 E6 and, to a lesser degree, E7 activate caspases upon differentiation in methylcellulose, indicating that this property is shared among genital HPV types (M. Beglin, C.A.M., and L.A.L., unpublished data).
The activation of caspases was found to be necessary for high levels of differentiation-dependent amplification of HPV-31 genomes. Treatment of HPV-31-positive cells with caspase inhibitors significantly reduced viral genome amplification (Fig. 4A), suggesting that caspase activation is important for the productive replication of the virus. High-level expression of the HPV E1∧E4 protein occurs in the uppermost layers of the epithelium and can be used as a marker for cells that are positive for viral genome amplification (33). When raft cultures were examined by immunohistochemistry, we were unable to correlate E1∧E4 expression with high levels of caspase-3 activation (C.A.M. and L.A.L., unpublished data). However, because amplification can be activated by low levels of active caspases, we suspect that these activated enzymes are present in low amounts throughout the differentiated layers and coincide with cells amplifying viral DNA.
The identification of a caspase-3/7 cleavage site (46DxxD49) in the viral replication protein E1, which is conserved in all genital HPVs, suggests this motif provides an important function in the differentiation-dependent life cycle of papillomaviruses. Consistent with this hypothesis, mutation of this sequence in the context of the complete HPV-31 genome significantly impaired the ability of the viral DNAs to amplify upon differentiation (Fig. 6). Processing at this cleavage site yields two E1 peptides, and it is unclear whether one or both of these peptides provide replication functions. In preliminary studies using transient assays, we have observed that the C-terminal cleavage product, designated E1-Casp, significantly enhances the ability of E1 by itself to mediate transient replication of origin-containing plasmids (C.A.M. and L.A.L., unpublished data). The N terminus of E1 mediates binding to the cellular factor p80, which negatively regulates the stability of E1 and limits viral copy number in undifferentiated cells (A. Côté-Martin, C.A.M., A. Fradet-Turcotte, C. D'Abramo, S. Joubert, et al., unpublished data). Cleavage and removal of the N-terminal portion of E1 may abrogate the ability of p80 to control E1 levels and activity. Alternatively the cleaved E1 products could also affect multimer formation or stabilize the binding of the full length protein to the origin. In addition to E1, the E2 and E7 proteins also contain conserved caspase cleavage sites, and the cleaved products may provide important functions during productive replication. Examination of these factors and their roles in mediating productive viral replication will be important in understanding how activation of caspases by HPV proteins regulates productive viral replication.
Experimental Procedures
Cell Culture.
Human foreskin keratinocytes (HFKs) were derived from neonatal human foreskin epithelia and grown, transfected, or infected with retroviruses as described (34, 35). Raft cultures were grown and analyzed as described (36). Differentiation in methylcellulose was induced as described (36). To induce differentiation in high calcium, cells were cultured in keratinocyte basal medium (KBM) with growth supplements for at least 24 h and then switched to KBM (without supplements) containing 1.5 mM CaCl2.
Plasmids and Chemicals.
The pBR322min plasmid, pSG5–E1 and pSG5–E2, and HPV-31 Luc have been described (37). The E1D49A mutant was constructed in the pSG5–E1 background by using QuikChange XL site-directed mutagenesis according to the instructions of the manufacturer (Stratagene). The GST–E1 fusion construct contains amino acids 1–170 of E1 inserted into the pGEX-4T-1 expression vector. The E1-Casp expression vector was generated by inserting the PCR-amplified DNA sequence encoding amino acids 50–580 containing an initiator methionine at the N terminus into the pSG5 vector. The EYFP–HPV-31 E1 construct was generated by inserting the HPV-31 E1 ORF into the pEYFP–C1 vector (Clontech). Staurosporine was purchased from Sigma–Aldrich. Purified recombinant caspase-3 and caspase-7 were purchased from Biomol International. A peptide inhibitor of caspase-3 (Z-DMQD-FMK) and a nonpeptide inhibitor of caspase-3/7 were purchased from Calbiochem. The general caspase inhibitor, Z-VAD-FMK and the caspase-8 inhibitor (Z-IETD-FMK) were purchased from R&D Systems.
Western and Southern Blot Analyses.
Lysates and Western blot analyses were performed as described (35). Primary antibodies were as follows: anticleaved caspase-3, anticaspase-3, anticleaved caspase-7, anticaspase-7, anticleaved caspase-9, anticaspase-9, anti-Bcl-2, anti-Bax (Cell Signaling Technology), antisurvivin and anti-K10 (Santa Cruz Biotechnology), anti-GFP (JL-8) (Clontech), and anti-GAPDH (Abcam). DNA isolation and Southern blot analysis were performed as described (36).
In Vitro Cleavage Assay of E1 Fusion Proteins by Caspases.
The GST–E1 and –E1D49A fusion proteins were expressed in Escherichia coli BL21 cells and purified according to the instructions of the manufacturer (Sigma). Proteins were quantified by using the Bio-Rad protein assay. Two hundred nanograms of GST–E1 and GST–E1D49A were incubated alone or with 2.5 or 25 ng of the purified recombinant caspases for 1 h in a buffer containing 20 mM Hepes (pH 7.4), 0.1 M NaCl, and 1 mM DTT in the presence or absence of 50 μM Z-DMQD-FMK or Z-VAD-FMK. Reactions were terminated by the addition of SDS/PAGE loading buffer and analyzed on 10% denaturing polyacrylamide gels.
In Vivo Cleavage of Full-Length E1.
HPV-negative C33A cells were transfected with YFP-tagged E1 or YFP–E1D49A mutant. Two days after transfection, cells were treated with either DMSO or 0.5 or 0.8 μM staurosporine for 4 h. Lysates were then harvested and analyzed by Western blot analysis for cleavage products by using an antibody to GFP.
Analysis of Enzymatic Caspase-3/7 Activity.
Enzymatic caspase-3/-7 activity was determined by using the Apo 3/7 HTS fluorometric assay according to the instructions of the manufacturer (Cell Signaling Technology). For this assay, HPV-31 HFKs were treated with either DMSO as a vehicle control or 20 μM the caspase inhibitor ZVAD-FMK and grown in high calcium for 96 h to induce differentiation. As a positive control, HPV-31-positive HFKs were treated with 0.5 μM staurosporine for 12 h. Cells (106) from each culture were incubated in the presence of the caspase-3/caspase-7 substrate (z-DEVD)2-R110. Free R110, generated because of cleavage of the DEVD motif, was measured after 30 min by using a fluorometer at excitation and emission wavelengths of 488 and 515, respectively. The assay was repeated three times with similar results.
Supplementary Material
Acknowledgments
We thank members of the laboratory of L.A.L., as well as Kathy Rundell and John Sixbey, for thoughtful discussions. C.A.M. was supported by American Cancer Society Fellowship PF-06-177-01-MBC and National Institutes of Health Postdoctoral Training Grant 5T32 AR007593. This work was supported by National Cancer Institute Grants RO1CA CA59655 and RO1CA74202 (to L.A.L.) and the Canadian Institutes for Health Research (J.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707947104/DC1.
References
- 1.Howley PM. In: Fundamental Virology. Fields BN, Knipe DM, Howley PM, editors. Philadelphia: Lippincott–Raven; 1996. pp. 947–978. [Google Scholar]
- 2.Hebner CM, Laimins LA. Rev Med Virol. 2006;16:83–97. doi: 10.1002/rmv.488. [DOI] [PubMed] [Google Scholar]
- 3.Cheng S, Schmidt-Grimminger DC, Murant T, Broker TR, Chow LT. Genes Dev. 1995;9:2335–2349. doi: 10.1101/gad.9.19.2335. [DOI] [PubMed] [Google Scholar]
- 4.Hummel M, Hudson JB, Laimins LA. J Virol. 1992;66:6070–6080. doi: 10.1128/jvi.66.10.6070-6080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 6.Werness BA, Levine AJ, Howley PM. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 7.Huibregtse JM, Scheffner M, Howley PM. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chellappan S, Kraus VB, Kroger B, Munger K, Howley PM, Phelps WC, Nevins JR. Proc Natl Acad Sci USA. 1992;89:4549–4553. doi: 10.1073/pnas.89.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyson N, Howley PM, Munger K, Harlow E. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 10.Weintraub SJ, Chow KN, Luo RX, Zhang SH, He S, Dean DC. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 11.Hay S, Kannourakis G. J Gen Virol. 2002;83:1547–1564. doi: 10.1099/0022-1317-83-7-1547. [DOI] [PubMed] [Google Scholar]
- 12.Steller H. Science. 1995;267:1445–1449. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 13.Cohen GM. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y. Mol Cell. 2002;9:459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- 15.Thornberry NA, Lazebnik Y. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 16.Boatright KM, Salvesen GS. Curr Opin Cell Biol. 2003;15:725–731. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Benedict CA, Norris PS, Ware CF. Nat Immunol. 2002;3:1013–1018. doi: 10.1038/ni1102-1013. [DOI] [PubMed] [Google Scholar]
- 18.Ruesch MN, Stubenrauch F, Laimins LA. J Virol. 1998;72:5016–5024. doi: 10.1128/jvi.72.6.5016-5024.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bedell MA, Hudson JB, Golub TR, Turyk ME, Hosken M, Wilbanks GD, Laimins LA. J Virol. 1991;65:2254–2260. doi: 10.1128/jvi.65.5.2254-2260.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finzer P, Aguilar-Lemarroy A, Rosl F. Cancer Lett. 2002;188:15–24. doi: 10.1016/s0304-3835(02)00431-7. [DOI] [PubMed] [Google Scholar]
- 21.Garnett TO, Duerksen-Hughes PJ. Arch Virol. 2006;151:2321–2335. doi: 10.1007/s00705-006-0821-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Algeciras-Schimnich A, Barnhart BC, Peter ME. Curr Opin Cell Biol. 2002;14:721–726. doi: 10.1016/s0955-0674(02)00384-8. [DOI] [PubMed] [Google Scholar]
- 23.Launay S, Hermine O, Fontenay M, Kroemer G, Solary E, Garrido C. Oncogene. 2005;24:5137–5148. doi: 10.1038/sj.onc.1208524. [DOI] [PubMed] [Google Scholar]
- 24.Carson A, Khan SA. J Virol. 2006;80:4356–4362. doi: 10.1128/JVI.80.9.4356-4362.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du J, Chen GG, Vlantis AC, Chan PK, Tsang RK, van Hasselt CA. Cancer Lett. 2004;205:81–88. doi: 10.1016/j.canlet.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 26.Cory S, Adams JM. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 27.Shin S, Sung BJ, Cho YS, Kim HJ, Ha NC, Hwang JI, Chung CW, Jung YK, Oh BH. Biochemistry. 2001;40:1117–1123. doi: 10.1021/bi001603q. [DOI] [PubMed] [Google Scholar]
- 28.Altieri DC. Oncogene. 2003;22:8581–8589. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
- 29.Borbely AA, Murvai M, Konya J, Beck Z, Gergely L, Li F, Veress G. J Gen Virol. 2006;87:287–294. doi: 10.1099/vir.0.81067-0. [DOI] [PubMed] [Google Scholar]
- 30.Nahle Z, Polakoff J, Davuluri RV, McCurrach ME, Jacobson MD, Narita M, Zhang MQ, Lazebnik Y, Bar-Sagi D, Lowe SW. Nat Cell Biol. 2002;4:859–864. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- 31.Demers GW, Halbert CL, Galloway DA. Virology. 1994;198:169–174. doi: 10.1006/viro.1994.1019. [DOI] [PubMed] [Google Scholar]
- 32.Jones DL, Munger K. J Virol. 1997;71:2905–2912. doi: 10.1128/jvi.71.4.2905-2912.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doorbar J, Foo C, Coleman N, Medcalf L, Hartley O, Prospero T, Napthine S, Sterling J, Winter G, Griffin H. Virology. 1997;238:40–52. doi: 10.1006/viro.1997.8768. [DOI] [PubMed] [Google Scholar]
- 34.Fehrmann F, Laimins LA. Methods Mol Biol. 2005;292:317–330. doi: 10.1385/1-59259-848-x:317. [DOI] [PubMed] [Google Scholar]
- 35.Hebner CM, Wilson R, Rader J, Bidder M, Laimins LA. J Gen Virol. 2006;87:3183–3193. doi: 10.1099/vir.0.82098-0. [DOI] [PubMed] [Google Scholar]
- 36.Fehrmann F, Klumpp DJ, Laimins LA. J Virol. 2003;77:2819–2831. doi: 10.1128/JVI.77.5.2819-2831.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hubert WG, Laimins LA. J Virol. 2002;76:2263–2273. doi: 10.1128/jvi.76.5.2263-2273.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.