Abstract
Isothiocyanates and phenolic antioxidants can prevent cancer through activation of Nrf2 (NF-E2 p45-related factor 2), a transcription factor that controls expression of cytoprotective genes through the antioxidant response element (ARE) enhancer. Using a human mammary MCF7-derived AREc32 reporter cell line, we now report that all-trans retinoic acid (ATRA), and other retinoic acid receptor alpha (RARα) agonists, markedly reduces the ability of Nrf2 to mediate induction of ARE-driven genes by cancer chemopreventive agents including the metabolite of butylated hydroxyanisole, tert-butylhydroquinone (tBHQ). The basal and tBHQ-inducible expression of aldo-keto reductase (AKR) AKR1C1 and AKR1C2 genes, which are regulated by Nrf2, was also repressed by ATRA in AREc32 cells. Antagonists of RARα augmented induction of ARE-driven gene expression by tBHQ, as did knockdown of RARα by using RNAi. The expression of the ARE-gene battery was increased in the small intestine of mice fed on a vitamin A-deficient diet, and this increase was repressed by administration of ATRA. By contrast, in the small intestine of Nrf2 null mice, the expression of ARE-driven genes was not affected by vitamin A status. In MCF7 cells, ATRA did not block the nuclear accumulation of Nrf2 but reduced the binding of Nrf2 to the ARE enhancer as a consequence of forming a complex with RARα. These data suggest that cross-talk between Nrf2 and RARα could markedly influence the sensitivity of cells to electrophiles and oxidative stressors and, as a consequence, to carcinogenesis.
Keywords: aldo-keto reductase, chemoprevention, Nrf2, retinoids
Chemoprevention has great potential to reduce the incidence of cancer. A major advance in this research area was the recognition that anticarcinogenic chemicals, some of which are present in edible plants, induce cytoprotective proteins through Nrf2, basic-region leucine zipper (bZIP) transcription factor (1). Proteins regulated by Nrf2 include glutathione S-transferases (GSTs), aldo-keto reductases (AKRs), and NAD(P)H:quinone oxidoreductase 1 (NQO1), as well as glutamate cysteine ligase (GCL, comprising GCLC and GCLM subunits) that catalyzes the rate-limiting step in GSH synthesis (2–6). The genes encoding these enzymes are coordinately regulated through antioxidant response elements (AREs) in their 5′-flanking promoter regions, to which Nrf2 binds as a heterodimer with small Maf proteins (7). In Nrf2-deficient mice, the basal expression of certain ARE-driven genes is reduced and induction by chemopreventive agents largely abolished. Moreover, the mutant mice are more prone to chemical carcinogenesis (8, 9).
Nrf2 is negatively regulated by Keap1, a substrate adaptor for the Cul3/Rbx1 E3 ubiquitin ligase (10). Under normal homeostatic conditions Nrf2 is rapidly ubiquitylated. However, on treatment with chemopreventive agents, Keap1 is modified and Nrf2 protein accumulates, resulting in activation of ARE-driven genes (10).
The chemopreventive agents butylated hydroxyanisole, probably through its metabolism to tert-butylhydroquinone (tBHQ), the dithiolethione oltipraz, and the isothiocyanate sulforaphane (Sul) can block carcinogenesis by activating Nrf2 and inducing its target genes (4, 8). However, increased GSH levels, coupled with overexpression of detoxification enzymes, are observed in preneoplastic nodules, tumors, and drug-resistant cancer cells (11–13), suggesting that up-regulation of the ARE-gene battery may also facilitate tumorigenesis. Indeed, it has been shown that in 19% of non-small-cell lung carcinomas Nrf2 is constitutively active through mutation of Keap1 (14).
Retinoids such as retinoic acid (RA) are chemopreventive and chemotherapeutic agents (15). One source of RA is vitamin A, derived from dietary β-carotene. RA regulates cell proliferation, differentiation, and morphogenesis. It inhibits tumorigenesis through suppression of cell growth and stimulation of cellular differentiation (16). Also, RA promotes apoptosis (17, 18), and this property may contribute to its antitumor properties.
The effects of retinoids are mediated by specific nuclear receptors, namely, retinoic acid receptors (RAR-α, -β, and -γ) and retinoid X receptors (RXR-α, -β, and -γ) (19). RXRs form heterodimers with RARs or other nuclear hormone receptors and function as transcriptional regulators. ATRA, for example, activates RAR-RXR heterodimers and exerts its biological actions by binding to retinoic acid response elements (RAREs) (20). In addition, retinoids can either activate or repress gene expression through RAR/RXR heterodimers interacting with other transcription factors, such as AP-1, estrogen receptor α, and NF-κB activities (21).
Given the potential of activators of Nrf2 and ligands of RAR in cancer chemoprevention, we investigated whether interactions exist between these pathways. Unexpectedly, we found RARα associates with Nrf2 and inhibits its activity.
Results
ATRA Can Inhibit the Induction of ARE-Driven Luciferase Activity.
We generated the stable ARE-luciferase reporter cell line called AREc32 that responds to tBHQ and Sul (22) [supporting information (SI) Fig. 7]. To determine whether retinoids influence ARE-driven transcription, we treated these cells with 1 μM ATRA for 24 h and found that this reduced basal luciferase activity between 10% and 50%. In the presence of tBHQ, Sul, or β-naphthoflavone (βNF), ATRA caused an almost complete repression of inducible luciferase activity (Fig. 1). To eliminate the possibility that inhibition of reporter activity resulted from chemical interaction between inducers and the retinoid, the effect of ATRA on ARE induction affected by overexpression of Nrf2 was studied. Twenty-four hours after transient transfection of AREc32 cells with pHyg-EF-hNrf2, luciferase activity was increased ≈4.2-fold (P < 0.001) when compared with mock-transfected cells. Inclusion of ATRA in the medium reduced the increase in reporter activity by 44% (P < 0.001). Thus, repression of luciferase activity by RA involved Nrf2 and occurred independently of the chemicals used.
Fig. 1.

All-trans-retinoic acid suppresses the induction of ARE-driven luciferase activity. AREc32 cells were incubated for 24 h with DMEM supplemented with antibiotics containing either tBHQ (10 μM), Sul (10 μM), or β-NF (10 μM). ATRA (1 μM) was added to the medium concomitantly. Luciferase activity was assayed and the activity of cells treated with DMSO (0.1% vol/vol) was set at 1. Data show mean ± SD from triplicate samples and represent the results of three separate experiments. **, P < 0.005
Time Course of all-trans-Retinoic Acid Inhibition of the ARE Response.
Inhibition of inducible ARE-driven gene expression by ATRA extended over the entire 24-h incubation period and was apparent 6 h after addition to the medium (SI Fig. 8). These data suggest that the retinoid acts directly on Nrf2 function, as there was no evidence for any morphological changes in these cells at any time point during the experiment. To establish whether short-term exposure to ATRA is sufficient to inhibit ARE activation, and also whether the effect is reversible, AREc32 cells were pretreated with 1 μM ATRA for 30 min before a 6-h incubation with tBHQ. Preexposure of AREc32 cells to ATRA, followed by transfer to medium containing tBHQ alone, reduced induction of luciferase activity from 4.6-fold to 1.6-fold (P < 0.001), indicating that repression of ARE activity by ATRA was rapid and not readily reversible.
ATRA Represses Basal and Inducible Expression of AKR1C1 and AKR1C2.
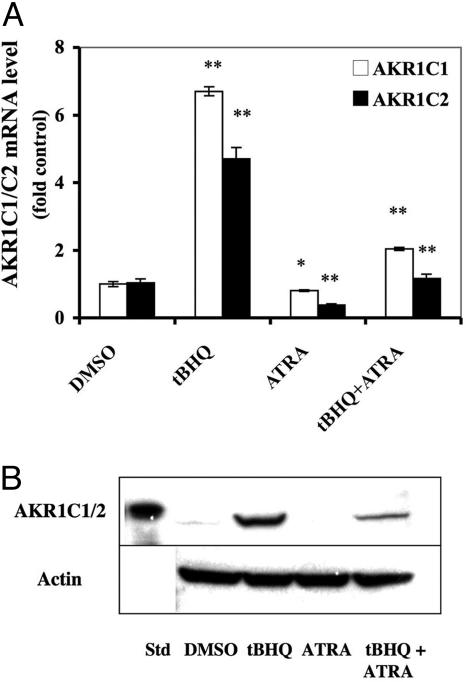
To determine whether ATRA inhibits endogenous ARE-driven gene expression, we examined AKR1C1 and AKR1C2. Treatment of AREc32 cells for 24 h with 1 μM ATRA resulted in a 20% and 40% reduction in the basal level of AKR1C1 and AKR1C2 mRNA, respectively (Fig. 2A). Also, ATRA blocked tBHQ induction of both AKR1C1 and AKR1C2 by 70% at both the mRNA and protein level (Fig. 2 A and B). These results indicate that RA represses constitutive and also inducible expression of Nrf2-regulated genes.
Fig. 2.
Inhibition of AKR1C1/2 induction by ATRA in AREc32 cells. AREc32 cells were incubated with DMEM containing either DMSO, tBHQ (10 μM), ATRA (1 μM), or tBHQ (10 μM) plus ATRA (1 μM) for 24 h. (A) AKR1C1 and C2 mRNA analysis. AKR1C1 and AKR1C2 mRNA were measured by TaqMan analysis; the level of 18S rRNA was used as an internal standard. Control cells were treated with DMSO only. The TaqMan data show mean ± SD from triplicate samples and represent the results of two separate experiments. *, P < 0.05; **, P < 0.005. (B) Repression of tBHQ-mediated induction of AKR1C1/2 protein expression by ATRA. AREc32 cell extracts were prepared and the expression of AKR1C1/2 was measured by Western blotting. The experiment was repeated three times with similar results.
Retinoic Acid Does Not Block Nuclear Accumulation of Nrf2 Affected by tBHQ.
We measured the level of Nrf2 protein in nuclear extracts from AREc32 cells treated with either tBHQ alone, or tBHQ plus ATRA, to establish how ATRA inhibits ARE-driven gene expression. Nuclear accumulation of Nrf2 by tBHQ was not inhibited by concomitant ATRA treatment (Fig. 3) suggesting that the retinoid does not affect the half-life of Nrf2 or its nuclear translocation. To further exclude the possibility that RA affects Nrf2 nuclear translocation, AREc32 cells were preincubated with tBHQ for 2 h, allowing nuclear accumulation of Nrf2 to occur, before the addition of ATRA (SI Fig. 9). The delayed addition of ATRA still inhibited induction of luciferase activity by 57% (P < 0.001) after a 6-h period.
Fig. 3.

Nrf2 nuclear translocation was not blocked by ATRA. Nuclear extracts were prepared from AREc32 cells treated with tBHQ (10 μM), ATRA (1 μM), or tBHQ (10 μM) plus ATRA (1 μM) for 24 h. Nuclear protein (20 μg) was separated on 7% SDS/PAGE and Nrf2 quantified by Western blotting. Data are representative of three separate experiments.
RAR Receptors Mediate Suppression of ARE-Driven Gene Expression by ATRA.
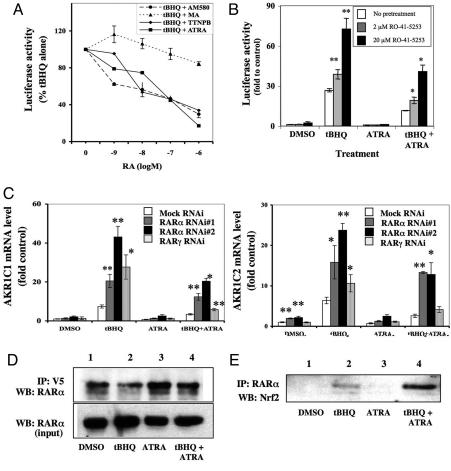
To test whether antagonism of Nrf2 by retinoids is mediated by either RAR or RXR, we treated AREc32 cells with RAR pan agonists (ATRA, TTNPB, 13-cis-RA), an RARα agonist AM580, an RXR agonist methoprene acid (MA), and a dual RAR/RXR agonist (9-cis-RA) (for reviews of ligand specificity, see refs. 23 and 24). We used MA as an RXR agonist because it has been commonly used for this purpose (25). Treatment with RAR agonists alone (ATRA, TTNPB, 13-cis-RA, or 9-cis-RA) for 24 h reduced basal luciferase activity ≈10% (data not shown). The RARα-specific agonist AM580 also suppressed basal luciferase activity to a similar degree. In contrast, the pan RXR agonist, MA, induced luciferase activity 2-fold (P < 0.05) (data not shown). Retinoids ATRA, TTNPB, 13-cis-RA or 9-cis-RA and AM580 were all potent inhibitors of tBHQ-induced ARE activity with the IC50 for individual retinoids ranging between 10 and 100 nmol/liter (Fig. 4A; SI Fig. 10). The RXR agonist, MA, had only minor effects on luciferase activity (≈20% inhibition) at the highest 1 μmol/liter dose, suggesting that the effect of ATRA was mediated by RARs rather than by RXRs. The specific RARα agonist AM580 strongly inhibited ARE-driven gene induction, suggesting that RARα was responsible for antagonizing Nrf2.
Fig. 4.
RARα, not RXR, appears to suppress ARE function. (A) Effect of RAR and RXR agonists on ARE inhibition. AREc32 cells were seeded in 96-well plates. After 24 h, tBHQ (10 μM) and the retinoids ATRA, TTNPB, AM580, or MA were added concomitantly to the medium. After a further 24 h, samples were assayed for luciferase activity. The value of luciferase activity of cells treated with 10 μM tBHQ alone (control) was taken as 100%. Values shown are mean ± SD. (B) RARα antagonist RO-41-5253 reversed RA-mediated ARE inhibition. AREc32 cells were incubated with 2 μM or 20 μM RO-41-5253 for 1 h. After pretreatment with RO-41-5253, the culture medium was replaced with fresh DMEM containing tBHQ (10 μM) in the presence or absence of ATRA (1 μM). The cells were then incubated for a further 24 h and assayed for luciferase activity, expressed relative to DMSO control. (C) Reduction of RARα expression increased induction of AKR1C1 and AKR1C2 mRNA by tBHQ and, in part, blocked the inhibitory effect of ATRA. AREc32 cells were seeded at 4 × 105 cells per well in six-well plates, containing diluted RNAi (200 pmol per well) and Lipofectamine 2000 (10 μl per well). After 24 h, the culture medium was replaced with fresh DMEM containing tBHQ (10 μM), ATRA (1 μM), or tBHQ (10 μM) plus ATRA (1 μM), and the cells were incubated for a further 24 h. The mRNA levels of AKR1C1 (Left) and AKR1C2 (Right) were measured by TaqMan. Values are expressed as the fold change relative to cells transfected with mock RNAi and treated with DMSO. The level of 18S rRNA was used as an internal standard. *, P < 0.05; **, P < 0.005. (D) V5-mNrf2 associates with endogenous RARα. AREc32 cells were seeded in a 150-mm dish at 5 × 106 cells per dish in the growth medium. After 24 h, 48 μg per dish of V5-mNrf2 plasmid DNA was transiently transfected by using Lipofectamine 2000. The culture medium was then replaced after 5 h with fresh DMEM containing 10 μM tBHQ with or without 1 μM ATRA. After 24 h Nrf2 was immunoprecipitated from the lysate by using anti-V5 antibody. The immunoprecipitated complexes were then fractionated by SDS/PAGE and immunoblotted with anti-RARα antibody. Data are representative of two independent experiments. (E) Endogenous RARα associates with Nrf2 protein. AREc32 cells were incubated with DMEM containing 10 μM tBHQ with or without 1 μM ATRA for 24 h. RARα was then immunoprecipitated from the lysate by using anti-RARα antibody. The immunoprecipitated complexes were then fractionated by SDS/PAGE and immunoblotted with anti-Nrf2 antibody. Data shown are representative of two independent experiments.
The role of RARα in inhibition was further examined by using RO-41-5253, a potent RARα antagonist. Pretreatment with RO-41-55253 for 1 h increased basal ARE activity 2-fold. Pretreatment with the antagonist also enhanced induction of luciferase activity by tBHQ (Fig. 4B); this was increased 26-fold by tBHQ alone and 38- and 72-fold by the inclusion of 2 or 20 μM RO-41-55253. In addition, the antagonist reversed inhibition of ARE-driven gene expression by ATRA in a dose-dependent fashion (Fig. 4B).
The role of RARα in Nrf2 inhibition was further investigated by knockdown experiments with two separate RNAi sequences. Twenty-four hours after transfection of AREc32 cells with RARα RNAi#1 the level of receptor protein was reduced by ≈50% (SI Fig. 11 Left); transfection with RARα RNAi#2 gave a similar reduction in receptor protein (data not shown). To investigate whether RARγ was also involved in the effect of RA on Nrf2, we performed a similar experiment to repress expression of RARγ in AREc32 cells (SI Fig. 11 Right). Knockdown of RARα by RNAi#1 increased the constitutive level of AKR1C2 mRNA ≈2-fold. RARα repression also augmented induction of AKR1C1 by tBHQ from 7-fold to >20-fold (Fig. 4C Left), and for AKR1C2 from 6-fold to >16-fold (Fig. 4C Right). The knockdown also partially abolished the inhibitory effect of ATRA on AKR1C1 and AKR1C2 induction (Fig. 4C). Although the level of RARγ protein was markedly reduced by RARγ RNAi treatment (SI Fig. 11 Right), this failed to reverse the inhibitory effect of ATRA on induction of AKR1C1 and AKR1C2 by tBHQ significantly (Fig. 4C). Knockdown of RARγ did, however, increase the induction of AKR1C1 and AKR1C2 by tBHQ, suggesting that this receptor may also antagonize Nrf2 function. Because RARβ was not detected in MCF7 cells, it was not studied.
Immunoprecipitation experiments were performed to investigate whether RARα antagonizes Nrf2 function through a physical interaction. After transfection of AREc32 cells with a plasmid encoding V5-tagged Nrf2, a V5 antibody was used to immunoprecipitate Nrf2. Immunoblots of the precipitate revealed the presence of RARα (Fig. 4D), suggesting that Nrf2 and RARα can form a complex in vivo. We next examined whether endogenous Nrf2 and RARα could associate with each other. Extracts from AREc32 cells were incubated with anti-RARα, and the immunoprecipitated proteins were analyzed by Western blotting with Nrf2 antibodies. Endogenous Nrf2 coprecipitated with RARα in the presence of tBHQ (Fig. 4E), and this association increased in AREc32 cells treated with tBHQ plus ATRA. The association between endogenous Nrf2 and RARα was barely detectable in cells treated with DMSO or ATRA alone, presumably because, in the absence of redox stress, the level of Nrf2 protein was low.
ATRA Interferes with Recruitment of Nrf2 to the ARE.
To determine whether ATRA reduced Nrf2 binding to the ARE, an electrophoretic mobility shift assay was performed. This showed three nuclear complexes (a, b, and c) could bind the enhancer specifically, all of which increased markedly in extracts prepared from tBHQ-treated cells (Fig. 5). When compared with intensities from tBHQ-treated cells, the ARE-binding complexes a and b were markedly reduced in nuclear extracts from cells treated with both tBHQ and ATRA. To confirm that ATRA can reduce DNA binding by Nrf2, nuclear extracts were incubated with a biotinylated ARE probe. After incubation, the bound protein was precipitated with streptavidin–agarose beads and immunoblotted by using anti-Nrf2 serum (SI Text). This analysis showed binding of Nrf2 to the ARE increased in response to tBHQ, but was reduced to almost control levels in the presence of ATRA (SI Fig. 12). Together these data suggest that ATRA inhibits Nrf2 function by stimulating the formation of Nrf2:RARα-containing complexes that do not bind to the ARE.
Fig. 5.

ATRA reduces binding of protein complexes to the ARE. Nuclear extracts (10 μg) from AREc32 cells treated with 10 μM tBHQ with or without 1 μM ATRA for 24 h were analyzed by EMSA. A 200-fold excess of unlabeled ARE was used to monitor specificity (lane 5). Arrows indicate the specific bands of DNA–protein complexes. The experiment was carried out three times with similar results.
Repression of Basal ARE-Gene Battery Expression by RA in Mouse Small Intestine in Vivo.
To investigate whether RA inhibits the expression of ARE-regulated genes in vivo, both Nrf2−/− and Nrf2+/+ mice were placed on a vitamin A-deficient (VAD) diet. Western blotting of proteins known to be regulated through Nrf2 revealed a profound increase in the levels of Gstm5, GCLC, NQO1, and Gsta1/2 in the small intestine of WT mice on a VAD diet (Fig. 6). No increase was observed in the levels of these proteins in Nrf2−/− mice on the VAD diet. The administration of ATRA (10 mg/kg, 2 weeks i.p.) to WT mice on the VAD diet almost completely blocked the increase in Gstm5, GCLC, NQO1, and Gsta1/2 proteins in the small intestine (Fig. 6, lane 5), demonstrating that retinoids inhibit Nrf2 function in vivo. Administration of ATRA to WT mice on a control diet did not affect the expression of Gstm5, GCLC, NQO1, or Gsta1/2 (data not shown).
Fig. 6.

Vitamin A deficiency induces ARE battery genes in the small intestine of Nrf2+/+ but not Nrf2−/− mice. Nrf2+/+ (WT) and Nrf2−/− mice were fed on a control (lanes 1, 3) or a VAD (lanes 2, 4, 5, 6) diet for 6 weeks; WT mice were given ATRA i.p. daily at a dose of 10 mg/kg for the last 2 weeks (lane 5); and corn oil (vehicle) was used as negative control (lane 6). Each lane represents a sample from a single mouse. Data represent Western blotting results from three separate experiments (n = 2–3).
We also analyzed the effect of the VAD diet on hepatic gene expression in these experiments. In one experiment involving two to three animals per group, changes similar to those observed in the gastrointestinal tract were observed (data not shown). But, in two further experiments no gene induction was observed. This finding could be due to the low abundance of RARα in hepatocytes (26).
Discussion
We provide evidence that RA antagonizes the expression of Nrf2 target genes. Using AREc32 reporter cells, we have discovered that ATRA, and other retinoids, inhibit both constitutive and inducible ARE-driven gene expression ex vivo. From placing mice on a VAD diet, we have also demonstrated that endogenous retinoids inhibit basal ARE-driven gene expression in vivo. Siddik et al. (27) reported that GST enzyme activity was increased in the liver and kidney of VAD rats. We have extended this observation considerably by showing that, in mice placed on a VAD diet, class Alpha and Mu GST subunits, as well as GCLC and NQO1, are induced substantially in the small intestine, in an Nrf2-dependent fashion.
Through serving as ligands for RARs, retinoids influence gene expression either by promoting cell growth and differentiation or by modifying individual transcription factor pathways (21). Our experiments have revealed that retinoids antagonize Nrf2 through an interaction with RARα. We found that agonists of RARα inhibit Nrf2 activity, whereas antagonists and knockdown of RARα augment Nrf2 activity. Knockdown experiments suggest that RARγ may also antagonize Nrf2, but it is not as potent as RARα in this regard. The RARα and RARγ proteins share 75% sequence identity and 82% homology. It will be informative to discover which domain of RARα is responsible for inhibiting Nrf2, because this may help explain why RARγ is a weaker inhibitor than RARα of the bZIP factor. We have not explored whether the association between Nrf2 and RARα inhibits the ability of the receptor to activate RARE-enhancer activity, but this warrants further investigation as cross-talk can occur between RARα and other transcription factors.
The finding of an interaction between Nrf2 and RARα suggests that inhibition of ARE-driven gene expression by ATRA is not due to effects on cell differentiation (19). Rather, through a direct association with RARα, Nrf2 appears to be prevented from binding the ARE. Other transcriptional repressors of ARE function have been described, such as Bach1, small Maf, and p53, all of which exert their effects by producing an inhibitory complex bound to the ARE (28–30). This mechanism of Nrf2 inhibition probably does not apply to RARα because there is no evidence that it can bind the ARE. Indeed, by using an electrophoretic mobility shift assay (EMSA), the marked increase in nuclear protein ARE-binding complexes observed after treatment of cells with tBHQ was found to be reduced substantially when cells were exposed to both tBHQ and ATRA. We found that the association of RARα with Nrf2 was increased in the presence of ATRA, suggesting that RARα may exhibit higher affinity toward Nrf2 after ligand binding. The fact that nuclear levels of Nrf2 were not affected by ATRA, but less Nrf2 was bound to the ARE, suggests that retinoids could interfere with dimerization between the bZIP factor and small Maf protein, which is required for DNA binding by Nrf2 (7). Another possibility is that RARα may cause subnuclear relocalization of Nrf2, because it has been shown that RA can affect delocalization of transcriptional intermediary factor 1β into regions of centromeric heterochromatin (31).
Besides inhibiting AP1-mediated transcription (32), RAR can antagonize several other transcription factors that contain a bZIP domain, including the mammalian C/EBPβ and NF-IL6 factors (33, 34) and viral BZLF1 (35). Our finding that RARα can inhibit Nrf2 demonstrates that a member of the cap-‘n’-collar subfamily of bZIP transcription factors are regulated in this manner. The activities of NF-E2 p45, Nrf1, and Nrf3 may also be inhibited by ATRA.
The mouse-feeding experiments involving a VAD diet show that endogenous retinoids inhibit the normal homeostatic expression of ARE-driven genes, such as Gstm5, GCLC, NQO1, and Gsta1/2, in the small intestine. We found that administration of ATRA to mice on a VAD diet restored repression of ARE-driven gene expression to the level normally observed in mice fed a control diet. However, administration of ATRA to mice on the control diet did not further repress expression of ARE-driven genes to levels below those observed under normal homeostatic conditions (data not shown). This finding could be explained by a number of factors. The constitutive expression of a number of these genes is undoubtedly not solely regulated by Nrf2 (see Fig. 6). Also, the level of constitutive expression already appears repressed by dietary retinoids; additional retinoid exposure may not add to this effect. These data suggest that constitutive retinoid levels will influence the expression of ARE-regulated genes and their induction by chemoprevention agents.
More work is required before we can understand the interplay between the impact of diet on retinoid biosynthesis, the metabolism and tissue disposition of drugs administered as RARα agonists, and the antagonism of Nrf2. This is a most important issue because in humans, dietary inducers, such as Oltipraz and broccoli sprouts, have been under clinical investigation as cancer chemopreventive agents (36). Our finding of antagonism between retinoids and the Nrf2 implies that retinoids could influence the protection conferred by inducers of the ARE-gene battery.
Retinoids and/or their precursors have been used in several chemoprevention trials, including the α-Tocopherol/β-Carotene Trial (ATBC), the β-Carotene and Retinol Efficiency Trial (CARET), and the Physicians Health Study (37). None of these investigations showed that such interventions conferred any benefit. Indeed, in the ATBC study and CARET, the incidence of lung cancer was higher in volunteers receiving β-carotene and retinol than in those given placebo (37). Various explanations have been advanced to explain these adverse effects. Our results suggest the possibility that vitamin A ± β-carotene could repress the ARE-gene battery, thereby depleting endogenous antioxidant and carcinogen detoxication systems and blocking adaptation to redox stress. The situation is difficult to assess when taking into account the complexity of carcinogenesis together with the combination of multiple mechanisms of chemoprevention (38–40). In addition, the possible in vivo antagonism of Nrf2 function, and the reports that deletion of Nrf2 increases susceptibility to carcinogens (8, 9), would have to be rationalized with the finding that Keap1, a repressor of Nrf2 activity, has been recently reported to be mutated in non-small cell lung cancer (NSCLC) (14). The complexity of this situation is also reflected in the fact that the retinal reductase AKR1B10, an Nrf2-regulated gene that would reduce retinoic acid biosynthesis, is overexpressed in NSCLC (41).
Increased intracellular levels of GSH and overexpression of the ARE-gene battery are associated with acquired resistance of tumors to chemotherapeutic agents (11–13). In such cases, it is likely that Nrf2 is constitutively activated, possibly through mutations in Keap1 or by constitutive activation of stress–response pathways. The observation that retinoids antagonize Nrf2 activation may provide an approach to treating drug-resistant tumors.
In conclusion, the inhibitory effect of retinoids on Nrf2 function could play an important role in a large variety of biological processes and should be taken into account when trying to prevent or treat cancer.
Materials and Methods
All chemicals were of analytical grade and purchased from Sigma–Aldrich, unless otherwise stated. d,l-Sulforaphane was obtained from LKT Laboratories. 4-[E2–5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl-1-propenyl] benzoic acid (TTNPB), 4-[(5, 6, 7, 8-tetrahydro-5, 5, 8, 8-tetra-methyl-2-naphthalenyl) carboxamido]-benzoic acid (AM580), methoprene acid (MA), and RO-41-5253 were purchased from Biomol International. The media supplements for cell culture were purchased from Life Technologies.
During all experimental procedures, retinoids were handled in subdued light. Retinoids used to treat mammalian cells were dissolved in DMSO. Stock solutions of retinoids were stored at −70°C in aliquots, and used only once after thawing.
Antibodies against RARα and RARγ were purchased from Santa Cruz Biotechnology. Antibody against the V5 epitope was purchased from Invitrogen. Antisera against NQO1, GCLC, GSTA1/2, GSTM5, and Nrf2 have been described (42–44), as have antisera against AKR1C1 and AKR1C2 (45).
The pHyg-EF-hNrf2 expression vector, encoding human Nrf2 tagged at its N terminus with a green fluorescent protein (GFP), was from Masayuki Yamamoto (University of Tsukuba, Japan). The V5-mNrf2 expression vector (pcDNA3.1/V5mNrf2) encodes V5-tagged mouse Nrf2 (42).
Animals.
Homozygous Nrf2 KO mice were provided by Masayuki Yamamoto (2). Two-month-old C57BL/6 Nrf2−/− and Nrf2+/+ male mice were used in this study. All animal procedures were carried out under a United Kingdom Home Office license and with local ethical approval.
Nrf2−/− and Nrf2+/+ (n = 2–3) mice were placed on a VAD (Special Diet Service) or control diet for 6 weeks and then killed. Nrf2+/+ mice were also placed on a VAD diet for 6 weeks; during the last two weeks, they received either no treatment, ATRA i.p. daily at 10 mg/kg, or the equivalent volume of corn oil. Mice were killed and the small intestine excised, washed, and frozen in liquid nitrogen.
Cell Culture and the Measurement of ARE-Driven Luciferase Activity.
The stable human mammary ARE-reporter cell line, AREc32 (22), was maintained in Dulbecco's MEM with glutamax (DMEM) supplemented with 10% FBS and penicillin-streptomycin containing 0.8 mg/ml G418, at 37°C, in 95% air/5% CO2, and was passaged every 3–4 days.
For treatment, AREc32 cells were seeded in 96-well plates at a density of 1.2 × 104 cells per well. After 24 h, culture medium was replaced with fresh DMEM supplemented with penicillin-streptomycin containing known inducers of ARE-driven gene expression or retinoids (dissolved in DMSO to give a final 0.1% vol/vol concentration of vehicle). Cells were left from 30 min to 24 h to respond to chemical agents before being harvested, and luciferase activity was measured in cell lysates as described in ref. 22. For control experiments, vehicle alone was added to the medium.
Transient Transfection.
Transfection of AREc32 cells with Nrf2 expression vectors was carried at 70–80% confluence by using Lipofectamine 2000 Reagent (Life Technologies). The culture medium was replaced 5 h after transfection with fresh DMEM containing 10 μM tBHQ in the presence or absence of 1 μM ATRA. For control experiments, mock transfections (no plasmid DNA) and vehicle alone (0.1% vol/vol DMSO) was added to the medium. Cells were left for 24 h to respond to xenobiotics before being harvested and analyzed. In control experiments, the transfection reagent alone, without DNA, was added to the cells and treated with DMSO for 2 h.
For RAR knockdown experiments in AREc32 cells, two preannealed siRNA sequences 1 (5′-GGAAUUUGUGCUGUGUAUUtt-3′) and 2 (5′-GCUCACCACAUCUUCAUCAtt-3′), which target different regions of RARα mRNA, were purchased from Ambion (Applied Biosystems). A prevalidated siRNA (5′-GGAAGCUGUGCGAAAUGACtt-′), specifically targeting human RARγ, was similarly used to transfect AREc32 cells. In these cases, the siRNA (200 pmol per well) and Lipofectamine 2000 reagent (10 μl per well) were diluted with 1 ml of Optimum (Life Technologies) in a six-well plate and incubated at 20°C for 20 min. Thereafter, 4 × 105 cells were diluted in 4 ml of growth medium without antibiotics and dispensed to each well directly. After 24 h incubation, the cells were treated for a further 24 h with 10 μM tBHQ, 1 μM ATRA, or 10 μM tBHQ plus 1 μM ATRA in fresh DMEM.
Real-Time Quantitative PCR (RT-PCR).
Taqman RT-PCR was performed as described in ref. 46. The primers were synthesized by MWG-BIOTECH AG, and the probes by Qiagen. The level of 18S rRNA was used as an internal standard. For AKR1C1 mRNA, the forward primer was 5′-CTAAAAGTAAAGCTTTAGAGGCCAC-3′; the reverse primer was 5′-ACCTGCTCCTCATTATTGTATAAATGA-3′; probe was 5′-AAATTGGCAATTGAAGCTGGCTTCCGCCATATTGA-3′. For AKR1C2 mRNA, the forward primer was 5′-GCTCTAGAGGCCGTCAAATTG-3′; the reverse primer was 5′-AACCTGCTCCTCATTATTGTAAACA-3′; the probe was 5′-AATAGAAGCCGGGTTCCACCATATTGATTCTGCA-3′.
Western Blot Analysis and Immunoprecipitation.
Whole-cell and nuclear extracts were prepared from MCF7 cells as described in ref. 22. Protein samples (5–30 μg) were separated on SDS/PAGE gels and immunoblotting was carried out as described in ref. 22. In all cases, actin was used as a loading control.
For immunoprecipitation, cells were lysed in IP buffer containing 50 mM Hepes pH 7.4, 1 mM EGTA, 1 mM EDTA, 1% Triton, 500 mM NaCl, 1 mM Na3VO4, and protease inhibitors. Aliquots (200 μg of protein) of supernatant were each incubated with 1 μg of antibody and mixed by gentle rocking at 4°C for 1 h, after which 10 μl of Protein-A Sepharose resin slurry was added to the lysate. After mixing by gentle rocking at 4°C for 1 h, the resin was collected by centrifugation and washed four times with the IP buffer. The protein immune complexes were then analyzed by Western immunoblotting.
Electrophoretic Mobility Shift Assay.
Nuclear extracts used for EMSA were prepared as described in ref. 47. A double-stranded DNA probe of the rat GSTA2 ARE [5′-GAGCTTGGAAATGGCATTGCTAATGGTGACAAAGCAACTTTG-3′ (core sequence underlined)] end-labeled with [γ-32P]ATP was used for gel shift analyses as described in ref. 48. The specificity of ARE binding was determined by competition experiments, which were carried out by adding a 200-fold molar excess of an unlabeled oligonucleotide to the reaction mixture before the labeled probe was added. Samples were separated on 4% polyacrylamide gels at 100 V and binding was analyzed by autoradiography.
Statistical Analysis
Statistical comparisons were performed by unpaired Student's t tests.
Supplementary Material
Acknowledgments
We thank Prof. Masayuki Yamamoto and Dr. Ken Itoh (University of Tsukuba, Japan) for providing the pHyg-EF-hNrf2 construct and the Nrf2−/− mice, Dr. Michael McMahon for the V5-mNrf2 construct, Olga Vassieva for discussions in the preparation of this manuscript, and Dianne Carrie for her assistance with the animal work. This work was funded by Cancer Research U.K.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709483104/DC1.
References
- 1.Talalay P, Fahey JW. J Nutr. 2001;131:3027S–3033S. doi: 10.1093/jn/131.11.3027S. [DOI] [PubMed] [Google Scholar]
- 2.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, et al. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 3.Kwak MK, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. J Biol Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 4.McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, Wolf CR, Cavin C, Hayes JD. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- 5.Devling TW, Lindsay CD, McLellan LI, McMahon M, Hayes JD. Proc Natl Acad Sci USA. 2005;102:7280–7285. doi: 10.1073/pnas.0501475102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin Y, Penning TM. Annu Rev Pharmacol Toxicol. 2007;47:263–292. doi: 10.1146/annurev.pharmtox.47.120505.105337. [DOI] [PubMed] [Google Scholar]
- 7.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Biochem J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, Kensler TW. Proc Natl Acad Sci USA. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iida K, Itoh K, Kumagai Y, Oyasu R, Hattori K, Kawai K, Shimazui T, Akaza H, Yamamoto M. Cancer Res. 2004;64:6424–6431. doi: 10.1158/0008-5472.CAN-04-1906. [DOI] [PubMed] [Google Scholar]
- 10.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. J Biol Chem. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 11.Hayes JD, Wolf CR. Biochem J. 1990;272:281–295. doi: 10.1042/bj2720281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tew KD. Cancer Res. 1994;54:4313–4320. [PubMed] [Google Scholar]
- 13.McLellan LI, Wolf CR. Drug Resist Updat. 1999;2:153–164. doi: 10.1054/drup.1999.0083. [DOI] [PubMed] [Google Scholar]
- 14.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, et al. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sporn MB, Roberts AB, Goodman DS. The Retinoids. New York: Raven Press; 1994. [Google Scholar]
- 16.Soprano DR, Qin P, Soprano KJ. Annu Rev Nutr. 2004;24:201–221. doi: 10.1146/annurev.nutr.24.012003.132407. [DOI] [PubMed] [Google Scholar]
- 17.Atencia R, Garcia-Sanz M, Unda F, Arechaga J. Exp Cell Res. 1994;214:663–667. doi: 10.1006/excr.1994.1304. [DOI] [PubMed] [Google Scholar]
- 18.Herget T, Specht H, Esdar C, Oehrlein SA, Maelicke A. J Neurochem. 1998;70:47–58. doi: 10.1046/j.1471-4159.1998.70010047.x. [DOI] [PubMed] [Google Scholar]
- 19.Rochette-Egly C, Chambon P. Histol Histopathol. 2001;16:909–922. doi: 10.14670/HH-16.909. [DOI] [PubMed] [Google Scholar]
- 20.Love JM, Gudas LJ. Curr Opin Cell Biol. 1994;6:825–831. doi: 10.1016/0955-0674(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 21.Shaulian E, Karin M. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 22.Wang XJ, Hayes JD, Wolf CR. Cancer Res. 2006;66:10983–10994. doi: 10.1158/0008-5472.CAN-06-2298. [DOI] [PubMed] [Google Scholar]
- 23.Dawson MI. Curr Med Chem Anticancer Agents. 2004;4:199–230. doi: 10.2174/1568011043352975. [DOI] [PubMed] [Google Scholar]
- 24.Liby KT, Yore MM, Sporn MB. Nat Rev Cancer. 2007;7:357–369. doi: 10.1038/nrc2129. [DOI] [PubMed] [Google Scholar]
- 25.Harmon MA, Boehm MF, Heyman RA, Mangelsdorf DJ. Proc Natl Acad Sci USA. 1995;92:6157–6160. doi: 10.1073/pnas.92.13.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhuang YH, Sainio EL, Sainio P, Vedeckis WV, Ylikomi T, Tuohimaa P. Gen Comp Endocrinol. 1995;100:170–178. doi: 10.1006/gcen.1995.1146. [DOI] [PubMed] [Google Scholar]
- 27.Siddik ZH, Mimnaugh EG, Trush MA, Gram TE. Biochem J. 1980;188:889–893. doi: 10.1042/bj1880889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen T, Huang HC, Pickett CB. J Biol Chem. 2000;275:15466–15473. doi: 10.1074/jbc.M000361200. [DOI] [PubMed] [Google Scholar]
- 29.Dhakshinamoorthy S, Jain AK, Bloom DA, Jaiswal AK. J Biol Chem. 2005;280:16891–16900. doi: 10.1074/jbc.M500166200. [DOI] [PubMed] [Google Scholar]
- 30.Faraonio R, Vergara P, Di Marzo D, Pierantoni MG, Napolitano M, Russo T, Cimino F. J Biol Chem. 2006;281:39776–39784. doi: 10.1074/jbc.M605707200. [DOI] [PubMed] [Google Scholar]
- 31.Cammas F, Oulad-Abdelghani M, Vonesch JL, Huss-Garcia Y, Chambon P, Losson R. J Cell Sci. 2002;115:3439–3448. doi: 10.1242/jcs.115.17.3439. [DOI] [PubMed] [Google Scholar]
- 32.Schule R, Rangarajan P, Yang N, Kliewer S, Ransone LJ, Bolado J, Verma IM, Evans RM. Proc Natl Acad Sci USA. 1991;88:6092–6096. doi: 10.1073/pnas.88.14.6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DiSepio D, Malhotra M, Chandraratna RA, Nagpal S. J Biol Chem. 1997;272:25555–25559. doi: 10.1074/jbc.272.41.25555. [DOI] [PubMed] [Google Scholar]
- 34.Schwarz EJ, Reginato MJ, Shao D, Krakow SL, Lazar MA. Mol Cell Biol. 1997;17:1552–1561. doi: 10.1128/mcb.17.3.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfitzner E, Becker P, Rolke A, Schule R. Proc Natl Acad Sci USA. 1995;92:12265–12269. doi: 10.1073/pnas.92.26.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang JS, Shen X, He X, Zhu YR, Zhang BC, Wang JB, Qian GS, Kuang SY, Zarba A, Egner PA, et al. J Natl Cancer Inst. 1999;91:347–354. doi: 10.1093/jnci/91.4.347. [DOI] [PubMed] [Google Scholar]
- 37.Omenn GS. Chest. 2004;125:123S–127S. doi: 10.1378/chest.125.5_suppl.123s. [DOI] [PubMed] [Google Scholar]
- 38.Kelloff GJ, Crowell JA, Boone CW, Steele VE, Lubet RA, Greenwald P, Alberts DS, Covey JM, Doody LA, Knapp GG, et al. J Cell Biochem Suppl. 1994;20:55–62. doi: 10.1002/jcb.240560906. [DOI] [PubMed] [Google Scholar]
- 39.Moon RC, Kelloff GJ, Detrisac CJ, Steele VE, Thomas CF, Sigman CC. Anticancer Res. 1994;14:5–11. [PubMed] [Google Scholar]
- 40.van Lieshout EM, Peters WH, Jansen JB. Carcinogenesis. 1996;17:1439–1445. doi: 10.1093/carcin/17.7.1439. [DOI] [PubMed] [Google Scholar]
- 41.Fukumoto S, Yamauchi N, Moriguchi H, Hippo Y, Watanabe A, Shibahara J, Taniguchi H, Ishikawa S, Ito H, Yamamoto S, et al. Clin Cancer Res. 2005;11:1776–1785. doi: 10.1158/1078-0432.CCR-04-1238. [DOI] [PubMed] [Google Scholar]
- 42.McMahon M, Itoh K, Yamamoto M, Hayes JD. J Biol Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 43.Hayes JD, Chanas SA, Henderson CJ, McMahon M, Sun C, Moffat GJ, Wolf CR, Yamamoto M. Biochem Soc Trans. 2000;28:33–41. doi: 10.1042/bst0280033. [DOI] [PubMed] [Google Scholar]
- 44.Kelly VP, Ellis EM, Manson MM, Chanas SA, Moffat GJ, McLeod R, Judah DJ, Neal GE, Hayes JD. Cancer Res. 2000;60:957–969. [PubMed] [Google Scholar]
- 45.O'Connor T, Ireland LS, Harrison DJ, Hayes JD. Biochem J. 1999;343:487–504. [PMC free article] [PubMed] [Google Scholar]
- 46.Wang XJ, Chamberlain M, Vassieva O, Henderson CJ, Wolf CR. Biochem J. 2005;388:857–867. doi: 10.1042/BJ20042087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schreiber E, Matthias P, Muller MM, Schaffner W. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moffat GJ, McLaren AW, Wolf CR. Biochem J. 1997;324:91–95. doi: 10.1042/bj3240091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.