Abstract
Myosin-motors are conserved from yeast to human and transport a great variety of cargoes. Most plus-end directed myosins, which constitute the vast majority of all myosin motors, form stable dimers and interact constitutively with their cargo complexes. To date, little is known about regulatory mechanisms for cargo-complex assembly. In this study, we show that the type V myosin Myo4p binds to its cargo via two distinct binding regions, the C-terminal tail and a coiled-coil domain-containing fragment. Furthermore, we find that Myo4p is strictly monomeric at physiologic concentrations. Because type V myosins are thought to require dimerization for processive movement, a mechanism must be in place to ensure that oligomeric Myo4p is incorporated into cargo-translocation complexes. Indeed, we find that artificial dimerization of the Myo4p C-terminal tail promotes stabilization of myosin-cargo complexes, suggesting that full-length Myo4p dimerizes in the cocomplex as well. We also combined the Myo4p C-terminal tail with the coiled-coil region, lever arm, and motor domain from a different myosin to form constitutively dimeric motor proteins. This heterologous motor successfully translocates its cargo in vivo, suggesting that wild-type Myo4p may also function as a dimer during cargo-complex transport.
Keywords: cell asymmetry, Myo4p, RNA localization, She3p, motor protein
Directional transport of cytoplasmic cargoes like mRNA, proteins, vesicles, and organelles is indispensable for basic cellular functions, the establishment of cell asymmetry, and the coordination of cell differentiation (1–3). Such cellular cargo is usually transported as part of large, motor protein-containing translocation complexes to its site of destination. In recent years, great progress has been made in understanding the motile activities of motor proteins like kinesin and myosin at the molecular level. Most myosins either require dimerization through their coiled-coil domain or clustering of multiple monomeric motors onto the cargo complex for processive movement along actin filaments (3–5). In contrast to their motile activity, little is known about how myosins mediate the assembly of translocation complexes to fulfill their cellular function, cargo translocation.
Recently, a first model for translocation-complex formation has been derived from an unusual subclass of myosins that move toward the minus-end of actin. These type VI myosins (MyoVI) are fully functional only within translocation particles (5). However, these studies provide little direct information on the interaction of MyoVI with its binding partner of the cargo complex. For plus-end-directed myosins, which constitute the vast majority of all myosin motors, only few cargo-complexes have been studied in detail (3, 4).
Among plus-end-directed myosins, type V myosin (MyoV) is one of the most conserved and best studied motor proteins (3, 4, 6). Recent cryo-EM studies indicate that vertebrate MyoV forms stable dimers through its coiled-coil domain (7–9). In a variety of organisms, MyoV translocates mRNAs, vesicles, and organelles (3, 10–12). In humans, MyoV-dependent transport of melanosomes has been linked to the rare autosomal Griscelli syndrome (13). In Xenopus, this process is regulated by phosphorylation of MyoV (14).
To date, one of the most comprehensively studied MyoV-driven translocation events is the localization of the so-called She complex in yeast (11). In this translocation complex, the myosin motor, termed Myo4p, interacts with the N-terminal half of its adapter protein She3p to translocate at least 26 different types of mRNA as well as endoplasmic reticulum (ER) (15–24). For mRNA transport, the adapter protein She3p binds with its C-terminal half to the RNA-binding protein She2p (16, 19, 22, 23). She2p itself interacts with target mRNAs, allowing for MyoV-dependent mRNA translocation to the tip of the bud cell (16, 19, 21, 23, 25–27). In contrast to mRNA transport, the mode of interaction between She3p and ER remains elusive (17, 20). In summary, the interaction between Myo4p and She3p is the central step toward translocation-complex assembly for the transport of both types of cargo (16, 17, 19, 20, 22, 23).
Here, we provide insights into how a motor-translocation complex is assembled by a type V myosin motor. For complex assembly, two binding regions of Myo4p, a coiled-coil region-containing fragment and the C-terminal tail, interact with its cargo adapter She3p. To our surprise, we find that Myo4p alone is monomeric even far above physiological concentrations. In addition, we provide evidence that dimerization of Myo4p may occur within the cocomplex with She3p. Based on our in vitro and in vivo results, we propose a multistep assembly of Myo4p-motor complexes.
Results
Myo4p Binds She3p with High Affinity.
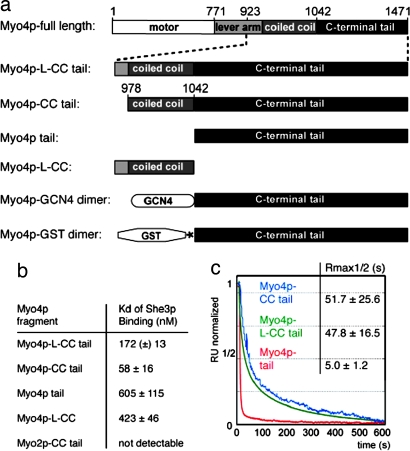
In two hybrid and in vitro studies, a Myo4p fragment consisting of the C-terminal tail, the coiled-coil region, and part of the lever arm (Myo4p-L-CC tail; Fig. 1a) was sufficient for binding to the N-terminal domain of its adapter protein She3p (She3p-N) (16). To characterize this interaction in more detail, we performed surface plasmon resonance (SPR) experiments with surface-coupled She3p-N. Steady-state binding studies revealed that the Myo4p-L-CC tail binds to She3p-N with an equilibrium dissociation constant (Kd) of 172 nM (Fig. 1b). Because lever arms of type V myosins do not participate in myosin dimerization (3, 4), we also analyzed a shorter fragment lacking the lever arm. This Myo4p fragment (Myo4p-CC tail; Fig. 1a) bound She3p-N with a Kd of 58 nM (Fig. 1b), indicating that it contains all regions necessary for efficient She3p binding. Specificity of the She3p-Myo4p interaction was confirmed by experiments with the tail region of the yeast paralog Myo2p [Myo2p-CC tail; supporting information (SI) Fig. 8].
Fig. 1.
Binding of different Myo4p fragments to the cargo adapter She3p. (a) Schematic drawing of Myo4p fragments used in this study. Asterisk indicates site-specific protease cleavage site. (b) Steady-state binding analyses of She3p-N and different myosin fragments by using surface plasmon resonance (SPR). (c) Normalized SPR plot from dissociation reactions of different Myo4p fragments from surface-coupled She3p-N. Table quotes the time-point at which 50% of the complexes have dissociated (Rmax1/2). ±, standard deviation; (±), deviation between two independent experiments.
Myo4p Requires Regions Outside the C-Terminal Tail for Binding to She3p.
In the prevailing model, plus-end-directed type V myosins interact through their C-terminal tail domain with cargo-adapter proteins (3). When we characterized a Myo4p fragment consisting only of the C-terminal tail (Myo4p tail; Fig. 1a), She3p-N was bound with an affinity that is 10-fold lower (Kd = 605 nM; Fig. 1b) than a fragment that also contains the coiled-coil region (Myo4p-CC tail). A Myo4p fragment consisting only of part of the lever arm and the coiled-coil region (Myo4p-L-CC; Fig. 1a) bound She3p-N with a Kd of 423 nM (Fig. 1b). These results demonstrate that the C-terminal tail of Myo4p alone is insufficient and that a second, additional binding region is required to achieve full binding to She3p-N.
The Coiled-Coil Region Stabilizes Complexes with She3p.
For cellular transport, Myo4p should form stable cargo complexes that do not dissociate during translocation. To find out how stable different Myo4p fragments bind to She3p, we analyzed their respective kinetics, by using again SPR. To compare the stabilities of complexes with Myo4p and She3p-N, we determined the time-point at which half of the complexes have disassembled. Our studies revealed that the half-life of complexes consisting of She3p-N and the Myo4p-L-CC tail is ≈48 s (Fig. 1c). She3p-N was bound by Myo4p-CC tail with similar stability (half-life = 52 s), whereas half of the complexes with Myo4p tail dissociated already after 5 s (Fig. 1c). Thus, the She3p-N cocomplexes with Myo4p-L-CC tail and Myo4p-CC tail are ≈10-fold more stable than with Myo4p tail alone.
Myo4p Is Monomeric at Physiological Concentrations.
Because the coiled-coil region-containing fragment of Myo4p participates in She3p binding, we analyzed this region by performing coiled-coil predictions with the paircoil algorithm (28). The prediction indicates an unusually weak and short coiled-coil region of only 31 aa (SI Fig. 9), raising the question whether Myo4p forms stable dimers.
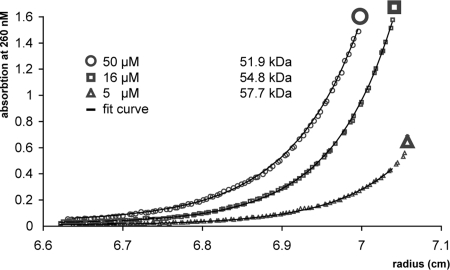
To characterize the dimerization state of Myo4p, we performed analytical ultracentrifugation (AUC). In sedimentation velocity experiments with the Myo4p-CC tail at concentrations of 5, 16, and 50 μM, consistently only a single oligomerization species with a sedimentation coefficient of 3.7 S was observed (data not shown). In subsequent sedimentation equilibrium experiments at identical Myo4p concentrations, the molecular mass of this species was determined to range between 51.9 and 57.7 kDa (Fig. 2). Because the theoretical molecular mass of a single Myo4p-CC tail heavy chain is 56.3 kDa, we conclude that Myo4p-CC tail does not dimerize even at 50 μM concentration.
Fig. 2.
Determination of the oligomerization state of Myo4p-CC tail. Sedimentation equilibrium experiments were performed with different concentrations. Resulting fit curves indicate that the Myo4p-CC tail has a molecular mass between 51.9 and 57.7 kDa. Because the theoretical molecular mass of Myo4p-CC tail is 56.3 kDa, we conclude that Myo4p is monomeric.
When assuming an average yeast cell volume of 30 μm3, Myo4p is expressed at a cellular concentration of ≈120 nM (29, 30). This in vivo concentration is >400-times lower than the highest measured concentration at which Myo4p-CC tail is still entirely monomeric (50 μM). These data imply that unbound Myo4p is present as monomer in the cell, even if it reaches significantly higher local concentrations. Type V myosins are thought to be processive only in their dimeric state (4). Thus, the monomeric state of Myo4p represents a potential problem for the assembly of functional translocation particles.
Complex Affinities upon Myo4p Dimerization.
Because all type V myosins studied so far form stable dimers via their coiled-coil region (7–9), we speculated that coiled-coil-dependent Myo4p oligomerization might occur with the help of She3p within the cocomplex.
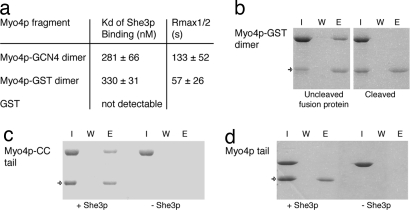
However, if Myo4p would bind cargo complexes as monomers, artificial dimerization of Myo4p is likely to result in sterical hindrance and thus interference with complex formation. To find out whether Myo4p dimerization indeed hinders complex formation, we substituted the coiled-coil region of Myo4p by the 32-aa-long coiled-coil region of GCN4 (Myo4p-GCN4 dimer; Fig. 1a), preserving the heptad register. The GCN4 coiled-coil region forms stable dimers (31) and dimerizes a fused Myo4p fragment (SI Fig. 10). SPR-binding experiments with Myo4p-GCN4 dimer and surface-coupled She3p-N showed a Kd of 281 nM (Fig. 3a), indicating that artificial Myo4p dimerization by an unrelated coiled-coil region allows for complex formation and modest increase of affinity. We also substituted the Myo4p coiled-coil region by the structure-and sequence-wise unrelated glutathion-S transferase (GST). This fusion fragment (Myo4p-GST dimer; Fig. 1a) forms dimers (SI Fig. 10) and yields a Kd of 330 nM for She3p-N binding (Fig. 3a). Together, these results indicate that dimerization of Myo4p does not interfere with complex formation, suggesting that Myo4p dimerization could, in principle, occur within the cocomplex.
Fig. 3.
Requirement of Myo4p dimerization for complex stability. (a) SPR binding studies with surface-coupled She3p-N and Myo4p fragments that contain heterologous dimerization domains. ±, standard deviation. (b) Immobilized She3p-N was used for pulldown-interaction studies with Myo4p-GST dimer. Efficient binding (Left) was abolished after the GST-dimerization domain was cleaved from the Myo4p tail (Right). This result indicates the requirement of dimeric Myo4p within the She3p cocomplex. (c and d) In control experiments, the Myo4p-CC tail (c) interacted efficiently with She3p, whereas the Myo4p tail (d) did not. Control experiments without She3p-ND, showed no Myo4p binding (c and d Right). Cropped images of Coomassie blue-stained polyacryl amid gels show input (I), final wash (W), and elution (E) fractions, arrow indicates the position of She3p-N.
Complex Stabilization upon Myo4p Dimerization.
To test how dimerization of Myo4p influences complex stability, we assessed the dissociation of She3p-N complexes with the Myo4p-GCN4 dimer and the Myo4p-GST dimer by SPR. Both dimerized Myo4p fragments showed considerably prolonged complex stabilities (Fig. 3a; 133 s and 57 s, respectively), when compared with the Myo4p tail alone (5 s; Fig. 1c). This effect is comparable with cocomplex stabilities of Myo4p fragments that contain the native coiled-coil region in addition to the C-terminal tail (Fig. 1 a and c). In summary, these results suggest a functional relevance of Myo4p dimerization for complex formation.
Disruption of Myo4p Dimerization Results in Disassembly of Cocomplexes with She3p.
Because Myo4p is monomeric in the absence of She3p (Fig. 2), the complex-stabilizing effect of dimerization is likely to be required only after the initial binding to She3p. To test this assumption, we coupled the His-tagged She3p-N to Ni-Sepharose beads, added the constitutively dimeric Myo4p-GST dimer, and performed pulldown experiments. These experiments were performed at high protein concentrations that allowed for full binding saturation. Thus, detection of bound protein mainly reflects the stabilities of complexes. As expected, the Myo4p-GST dimer (half-life = 57 s) reacted efficiently with She3p-N (Fig. 3b Left). After this initial binding reaction, we added a site-specific protease that cleaves between the GST and the Myo4p-tail fragment (Fig. 1a, asterisk), generating monomeric Myo4p tails and free GST. We reasoned that if Myo4p dimerization is required within the cocomplex, Myo4p monomerization after complex formation should result in the disassembly of these cocomplexes. Indeed, we observed that protease treatment resulted in a loss of stable cocomplexes (Fig. 3b Right). As control, we performed pulldown experiments with the Myo4p-CC tail (half-life = 52 s), which reacted efficiently with She3p-N (Fig. 3c) and with the Myo4p tail (half-life = 5 s), which did not (Fig. 3d). GST alone could also not be pulled down by She3p-N (SI Fig. 11).
In summary, the protease-cleavage experiment suggests that dimerization of Myo4p by GST within the complex is necessary to increase complex stability. These results further suggest that Myo4p-CC tail might also dimerize within the cocomplex, resulting in stable cocomplexes.
Myo4p-L-CC Tail Outcompetes Endogenous Myo4p from She3p Binding in Vivo.
To test whether C-terminal Myo4p fragments are able to compete with endogenous full-length Myo4p in vivo, we performed in vivo interference experiments. Yeast cells were transformed with a construct that ectopically expresses the motor-lacking Myo4p-L-CC tail fragment. Interaction of She3p with endogenous Myo4p leads to normal cargo-complex assembly, whereas She3p-binding to ectopically expressed Myo4p-L-CC tail should result in immobile complexes. The more successful that ectopic Myo4p-L-CC tail competes for She3p binding, the fewer cocomplexes with endogenous full-length Myo4p should form.
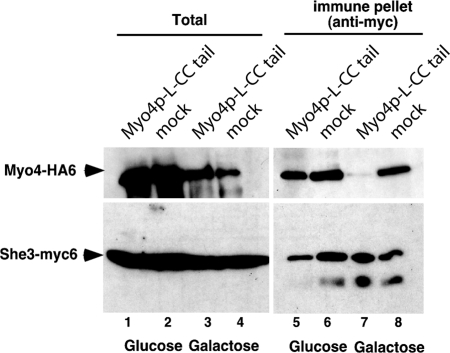
To detect potential Myo4p competition, we performed immunoprecipitation experiments with Myc-tagged She3p. Coimmunoprecipitation of endogenous Myo4p was specifically abolished upon induction of Myo4p-L-CC tail expression (Fig. 4; compare lanes 5–8). These results suggest efficient out-competition of She3p binding by the Myo4p-L-CC tail and indicate that our quantitative in vitro studies correctly reflect the binding level of full-length Myo4p.
Fig. 4.
Overexpression of the Myo4p-L-CC tail results in reduction of the She3p interaction with endogenous Myo4p. After overexpression of the Myo4p-L-CC tail in Myc-She3p- and HA-Myo4p-expressing cells, immunoprecipitation with anti-Myc antibody and Western-blot experiments against Myc- and HA-tags were performed. In glucose-containing medium, no Myo4p-L-CC tail-specific effect on the interaction between HA-Myo4p and Myc-She3p was observed (compare lane 1 with lane 2 and lane 5 with lane 6). Upon galactose-induction of Myo4p-L-CC tail expression, complex formation between HA-Myo4p and Myc-She3p was significantly reduced (compare lane 3 with lane 4 and lane 7 with lane 8).
Myo4p-L-CC Tail Efficiently Interferes with Cargo Translocation in Vivo.
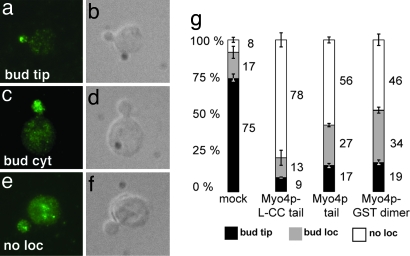
An efficient interference of cargo-complex assembly should also result in reduced localization of cargo complexes to the bud. We used immunofluorescence staining to detect whether interference of cargo-complex assembly affects She3p localization. For a comparison, we distinguished between translocation-complex localization (i) to the bud tip (Fig. 5 a and b and SI Fig. 12 a–d, (ii) to the bud cytoplasm (Fig. 5 c and d and SI Fig. 12 e–h), or (iii) no distinct localization (Fig. 5 e and f and SI Fig. 12i–l), by using antibody staining against HA-She3p.
Fig. 5.
In vivo interference assay with overexpressed Myo4p fragments. (a–f) Representative images of cells displaying bud-tip localization (a and b), bud localization (c and d), or no localization (e and f) of HA-tagged She3p. a, c, and e show immunofluorescence stainings of HA-She3p; b, d, and f are corresponding Nomarski optics images. (g) Graph summarizing interference experiments. Numbers beneath graph indicate percentage of a particular effect; ±, standard deviation. For more details, refer to SI Fig. 12.
When the Myo4p-L-CC tail was used for interference, She3p translocation was disrupted in 78% of the cells (Fig. 5g and SI Fig. 12m; mock control: 8%). Compared with the Myo4p-L-CC tail, interference by the Myo4p tail was weaker, and localization of She3p failed in 56% of the cells (Fig. 5g and SI Fig. 12m). Cells expressing the Myo4p-GST dimer failed to localize She3p in a similar range (46%; Fig. 5g and SI Fig. 12m). Western blot analysis revealed that the Myo4p tail fragment is expressed at levels higher than the other fragments (SI Fig. 13).
In summary, we observed strong interference of cargo translocation with the Myo4p-L-CC tail and a weaker interference with the Myo4p tail and the Myo4p-GST dimer. These findings confirm that regions outside the C-terminal tail are required for the efficient formation of translocation complexes (see also Fig. 1 b and c). The fact that we observed interference by the Myo4p-GST dimer indicates that enforced Myo4p dimerization allows for complex formation in vivo.
She3p Is Transported in Vivo by a Dimeric Hybrid MyoV Motor.
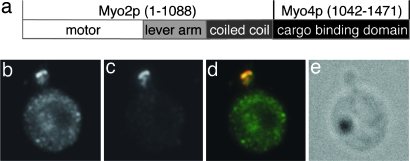
The observed cocomplex formation with artificially dimerized Myo4p tail fragments (Figs. 3a and 5g) suggests that a Myo4p tail fused to a heterologous, dimerizing motor protein would perhaps even allow for cargo translocation. To test this possibility, we first fused the Myo4p tail domain to the coiled-coil region of its ortholog Myo2p (Myo2p4p dimer). Compared with Myo4p, the coiled-coil region of Myo2p is considerably longer and is predicted with full probability to form homodimers (SI Fig. 9). We confirmed that the fusion protein indeed forms dimers by size-exclusion chromatography (SI Fig. 10). Next we fused the motor domain, lever arm, and coiled-coil domain of Myo2p with the Myo4p tail domain (Fig. 6a; Myo2p4p hybrid). We used this construct to perform rescue experiments of She3p localization in myo4Δ cells. In these experiments, we detected bud or bud tip localized She3p in 49% of all Myo2p4p-hybrid-expressing cells (Fig. 6 b–e; mock control: 0%, data not shown; wild-type localization: 92%, see Fig. 5g, mock). Because the Myo2p4p-hybrid lacks one of the two binding regions for high-affinity complex formation (Myo4p-L-CC), only a partial rescue of localization is observed. In summary, artificial dimerization of the Myo4p tail is not only compatible with complex formation and stabilization (Figs. 3a and 5 a–g) but also with cargo transport in vivo.
Fig. 6.
In vivo translocation of She3p by an artificial hybrid motor protein. (a) Schematic drawing of the Myo2p4p-hybrid motor. (b–e) Representative images of myo4Δ-rescue experiments with the Myo2p4p-hybrid showing immunofluorescence staining of HA-She3p (b), Myc-tagged Myo2p4p-hybrid (c), overlay of b and c (d), and the corresponding Normarski image (e).
Discussion
Two Binding Regions of Myo4p Interact with She3p.
The C-terminal tail and the coiled-coil-containing region of Myo4p bind to She3p with Kd of 605 nM and 423 nM, respectively (Fig. 1b). Fragments containing both regions (Myo4p-L-CC tail and Myo4p-CC tail) bind with considerably higher affinity (Fig. 1b), suggesting that they act synergistically for full binding. In vivo interference studies support this conclusion by showing a strong interference effect only with the Myo4p-L-CC tail (Fig. 5). Together, these finding shows that cargo binding by type V myosins may involve regions outside the C-terminal tail.
Myo4p Is Monomeric at Physiologic Concentrations.
We observed that Myo4p does not dimerize at concentrations up to 50 μM. When considering a cellular Myo4p concentration of 120 nM (see above), this motor protein should be monomeric in vivo. This rather surprising result demonstrates that not all type V myosins are constitutively dimeric. Vertebrate type V myosins require dimerization to move efficiently along actin filaments (4). Therefore it is likely that assembly of translocation complexes might also require Myo4p dimerization.
Myo4p Dimerization and Cocomplex Formation.
She3p binding of the Myo4p tail alone is transient, whereas longer fragments that include the coiled-coil region (Myo4p-L-CC tail and Myo4p-CC tail) or heterologous dimerization domains form more stable cocomplexes (Figs. 1c and 3a).
We were unable to obtain reliable information on the half-life of the short Myo4p-L-CC fragment in complex with She3p by using SPR. Instead, we resorted to pulldown experiments in which observed complex formation mainly reflect complex stabilities (compare Figs. 1 b and c and 3a with Fig. 3 b–d). We repeated this pull-down experiment with the Myo4p-L-CC fragment and detected no interaction (SI Fig. 14). This observation indicates a rather short half-life for the She3p complex with Myo4p-L-CC. Although indirect, this result suggests stabilization of cocomplexes by cooperativity of both binding regions and not by intrinsically high binding stability of the coiled-coil region alone. This conclusion is also consistent with the observation that artificial dimerization of the C-terminal tail is sufficient to induce complex stabilization (Fig. 3a).
Because the coiled-coil region of Myo4p has no detectable dimerization capability on its own, Myo4p dimerization before cocomplex formation is not crucial. Consistently, we found that stable complexes can form between She3p-N and Myo4p-CC tail even at nanomolar concentrations (Fig. 1 b and c). However, once the Myo4p-She3p cocomplex has formed, Myo4p dimerization within the complex may be required for complex stabilization. This conclusion is based on our observation that proteolytic cleavage of the Myo4p-GST dimer after the initial binding reaction results in the disassembly of the cocomplex (Fig. 3 b–d).
We should note that we cannot formally exclude the possibility that Myo4p is assembled in the cocomplexes as oligomers higher than dimers. However, type V myosins have thus far been described only as dimers. In addition, artificial dimerization of the Myo4p tail via the GCN4 coiled-coil and GST is compatible with complex formation and stabilizes complexes considerably (Fig. 3a). Consistent with this finding is the observation that also a Myo4p-GST dimer is able to interfere with She3p localization in vivo (Fig. 5). Furthermore, the observed She3p localization by the Myo2p4p-hybrid motor (Fig. 6) indicates that dimerization is also compatible with cargo translocation. In addition, in in vitro motility assays Trybus and colleagues (32) showed that a hybrid motor containing the Myo4p motor domain fused to the lever arm, and the stable dimer-forming coiled-coil domain of murine MyoV (SI Fig. 9) is processive. This finding indicates that Myo4p may move processively once it is dimerized.
Two Regulatory Mechanisms for Myo4p Motility.
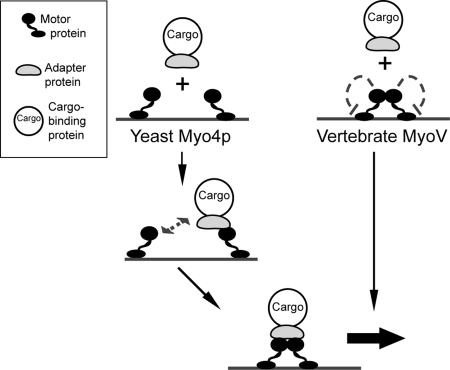
Vertebrate MyoV dimers are autoinhibited (7–9) unless they are bound by their cargo-interaction partners. According to this model, dimeric Myo4p in the cell would be autoinhibited until She3p-binding activates its motor function (Fig. 7, right path). Because Myo4p is monomeric under physiological conditions, autoinhibition of Myo4p is likely not required. Instead, we suggest that binding of the cargo adapter She3p to monomeric Myo4p induces the assembly of translocation complexes (Fig. 7, left path), which may contain dimeric Myo4p. In both scenarios, the cargo adapter controls the motile activity of the motor protein.
Fig. 7.
Model for the assembly of Myo4p-containing translocation complexes. At physiological concentrations, Myo4p is monomeric. In an initial priming step, monomeric Myo4p binds to the adapter protein She3p (left path). Then Myo4p may dimerize in complex with She3p, resulting in the formation of a stable translocation complex. Because Myo4p itself does not dimerize, autoinhibition as observed in vertebrate MyoV (right path) (7–9) is unlikely to be required.
Lacking Processivity of Myo4p in Vitro.
In vitro-motility assays suggested that Myo4p is not a processive motor (33). In vivo, however, Myo4p has been shown to move cargo complexes in a way that is consistent with processive motor activity (34). In addition, the observed motility of the previously mentioned Myo4p motor-containing hybrid myosin (32) implies that full-length Myo4p may also moves as dimers.
Because in motility assays, wild-type Myo4p was analyzed at low concentrations and in the absence of stoichiometric amounts of its cargo complex (33), our results suggest that Myo4p might have been monomeric and thus nonprocessive (3–5). On the other hand, in the in vivo study the motility of entire translocation complexes was characterized (34). Here, She3p might have helped Myo4p to dimerize and to become processive. Based on our results, we propose that the recapitulation of in vitro-motility studies in presence of stoichiometric amounts of She3p increases the processivity of Myo4p.
Importance of Complex Stability for Directional Transport.
Time-lapse microscopy revealed that the Myo4p complex translocates one of its cargo mRNAs, termed ASH1 mRNA, in ≈2 min to the bud tip (34). Thus, our measured complex half-life of ≈50 s between Myo4p and She3p is likely to be sufficient for complete cargo-complex translocation in vivo. Interestingly, ASH1 mRNA contains four localization elements (35). Each localization element is sufficient for bud localization, but full translocation efficiency requires the presence of all four elements (27). It is tempting to speculate that multiple localization elements recruit multiple motors, resulting in increased translocation efficiency. IST2 mRNA, which is also translocated by Myo4p, contains only one localization element (25). Consistent with our interpretation, IST2 mRNA is less efficiently transported and is less stringently localized to the bud tip (36).
Regulation of Motor-Complex Assembly in Higher Eukaryotes.
Plant myosin type XI is closely related to MyoV (3, 6). It is interesting to note that two recent studies on myosin XI function show a requirement of their C-terminal tail domain together with the coiled-coil domain for correct localization of myosin fragments (37, 38). These results suggest that myosin XI-dependent cargo-complex assembly may uses a mechanism related to the Myo4p and She3p interaction.
A recent study showed that mouse MyoV transports specific mRNA in hippocampal neurons (12). In this study, overexpression of various MyoV-tail fragments suppressed translocation in a manner consistent with our interpretation. Only MyoV tail fragments containing the coiled-coil domain affected the translocation of mRNPs in neuronal cell culture. Although the authors do not follow up on this initial observation, our study provide a mechanistic explanation for such transport processes in vertebrates and yeast. In light of these data, our study suggests that binding regions outside of the C-terminal globular tail might be a universal feature for translocation-complex assembly at least of MyoV-dependent mRNA-transport.
Materials and Methods
Detailed information on plasmids and yeast strains are available in SI Methods.
Protein Purification.
All myosin fragments were GST-tagged, and She3p-N was His-tagged. All fragments were expressed in Escherichia coli and isolated to a purity of ≥95% by using standard chromatographic techniques (40). GST-tags were removed by protease cleavage, unless stated otherwise.
Analytical Ultracentrifugation.
Experiments were performed with an XL-I analytical ultracentrifuge (Beckman Coulter) and data processed by using the software Ulatrascan 9.0 (B. Demeler, University of Texas Health Sciences Center, San Antonio, TX) and Origin. Rotation speed for sedimentation velocity experiments was 55,000 rpm at 20° and for sedimentation equilibrium experiments 18,000 rpm at 4° in a buffer containing 10 mM Tris·HCl (pH 8.25), and 200 mM NaCl. To allow for measurements at high protein concentrations, data collections in these samples were performed at 260 nm, otherwise at 280 nm.
Surface Plasmon Resonance.
All experiments were performed by using a Biacore 3000 system with a CM-5 chip (Biacore). Experiments were performed as triplicates. For further details, see SI Methods.
In Vitro Pulldown.
Fifty micrograms of His-tagged She3p-N and 50 μg of different Myo4p fragments were incubated with 50 μl of Ni-Sepharose (Amersham Biosciences) for 1 h in 20 mM Tris·HCl (pH 7.5), 200 mM NaCl, and 15 mM Imidazole. Binding reactions were centrifuged and pellets washed five times with 200 μl of reaction buffer, followed by a final wash step with 50 μl. Bound proteins were eluted with 50 μl of elution buffer [20 mM Tris·HCl (pH 7.5), 200 mM NaCl, and 750 mM Imidazole]. One-tenth of the input, 1/5 of the last wash, and 1/5 of the elution fraction was analyzed by SDS/PAGE and Coomassie blue staining.
Yeast Growth Conditions.
Cells were grown in SC–trp medium containing 2% raffinose. Ectopic overexpression of proteins was induced by the addition of galactose (control glucose) to a concentration of 2%. At 90 min after galactose induction, cells were prepared for immunoprecipitation and immunofluorescence microscopy.
Immunoprecipitation of Epitope-Tagged She3p.
Immunoprecipitation of Myc-She3p from cell lysates was performed with monoclonal anti-Myc antibody (9E10, Roche) and Protein G-Sepharose (Amersham Biosciences) at 4°C. Beads were eluted with hot Laemmli buffer. Proteins were separated by SDS/PAGE, blotted, and detected by monoclonal anti-HA and anti-Myc antibodies. For further details, see SI Methods.
Western blots were performed by using standard procedures. Myo4p fragments were detected with a 1:1,000 dilution of mouse monoclonal anti-myc antibody (9E10; Roche) and actin with a 1:1,000 dilution of mouse anti actin antibody (Millipore). As secondary antibody, we used a 1:5,000 dilution of HRP-anti mouse IgG antibody (Millipore). Visualization was performed with an ECL kit (Applichem).
Fluorescence Microscopy.
Indirect double immunofluorescence against Myc- or HA-tagged proteins was essentially performed as described (16, 18). For further details, see SI Methods.
Interference Assays.
Quantification of HA3-She3p localization was obtained by evaluating three times 150 cells with medium sized buds (diameter ≈2 μm).
Myo2p4p-Hybrid Translocation in Vivo.
Quantification of HA3-She3p localization was performed three times with 50 cells of medium sized buds each (diameter ≈2 μm).
Supplementary Material
Acknowledgments
We thank Michela Bertero, Christopher Hare, Jerzy Adamski, and Gabriele Möller for their contribution. This work was supported by German Research Foundation Grants NI 1110/1-1 and FOR855 (to A.H., M.M., and D.N.), JA696/4–3 and SFB646 TP A5 (to T.G.-D., and R.-P.J.), and SFB 594 (to J.B. and K.R.); by the Helmholtz Association (VH-NG-142 to D.N.); and by the Boehringer-Ingelheim-Fonds (M.M).
Note Added in Proof.
While the manuscript was under review, a related study was published by Dunn et al. (39). Both independent studies provide complementing data showing that Myo4p is monomeric.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706780104/DC1.
References
- 1.Soldati T, Schliwa M. Nat Rev Mol Cell Biol. 2006;7:897–908. doi: 10.1038/nrm2060. [DOI] [PubMed] [Google Scholar]
- 2.St. Johnston D. Nat Rev Mol Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- 3.Vale RD. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 4.Sellers JR, Veigel C. Curr Opin Cell Biol. 2006;18:68–73. doi: 10.1016/j.ceb.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Sweeney HL, Houdusse A. Curr Opin Cell Biol. 2007;19:57–66. doi: 10.1016/j.ceb.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Richards TA, Cavalier-Smith T. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- 7.Li XD, Jung HS, Mabuchi K, Craig R, Ikebe M. J Biol Chem. 2006;281:21789–21798. doi: 10.1074/jbc.M602957200. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Taylor DW, Krementsova EB, Trybus KM, Taylor KA. Nature. 2006;442:208–211. doi: 10.1038/nature04719. [DOI] [PubMed] [Google Scholar]
- 9.Thirumurugan K, Sakamoto T, Hammer JA, III, Sellers JR, Knight PJ. Nature. 2006;442:212–215. doi: 10.1038/nature04865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shav-Tal Y, Singer RH. J Cell Sci. 2005;118:4077–4081. doi: 10.1242/jcs.02543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller M, Heuck A, Niessing D. Cell Mol Life Sci. 2007;64:171–180. doi: 10.1007/s00018-006-6286-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshimura A, Fujii R, Watanabe Y, Okabe S, Fukui K, Takumi T. Curr Biol. 2006;16:2345–2351. doi: 10.1016/j.cub.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 13.Olkkonen VM, Ikonen E. J Cell Sci. 2006;119:5031–5045. doi: 10.1242/jcs.03303. [DOI] [PubMed] [Google Scholar]
- 14.Karcher RL, Roland JT, Zappacosta F, Huddleston MJ, Annan RS, Carr SA, Gelfand VI. Science. 2001;293:1317–1320. doi: 10.1126/science.1061086. [DOI] [PubMed] [Google Scholar]
- 15.Jansen RP, Dowzer C, Michaelis C, Galova M, Nasmyth K. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- 16.Böhl F, Kruse C, Frank A, Ferring D, Jansen RP. EMBO J. 2000;19:5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estrada P, Kim J, Coleman J, Walker L, Dunn B, Takizawa P, Novick P, Ferro-Novick S. J Cell Biol. 2003;163:1255–1266. doi: 10.1083/jcb.200304030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruse C, Jaedicke A, Beaudouin J, Böhl F, Ferring D, Güttler T, Ellenberg J, Jansen RP. J Cell Biol. 2002;159:971–982. doi: 10.1083/jcb.200207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Münchow S, Sauter C, Jansen RP. J Cell Sci. 1999;112:1511–1518. doi: 10.1242/jcs.112.10.1511. [DOI] [PubMed] [Google Scholar]
- 20.Schmid M, Jaedicke A, Du T-D, Jansen R-P. Curr Biol. 2006;16:1538–1543. doi: 10.1016/j.cub.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 21.Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD. Proc Natl Acad Sci USA. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takizawa PA, Vale RD. Proc Natl Acad Sci USA. 2000;97:5273–5278. doi: 10.1073/pnas.080585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long RM, Gu W, Lorimer E, Singer RH, Chartrand P. EMBO J. 2000;19:6592–6601. doi: 10.1093/emboj/19.23.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andoh T, Oshiro Y, Hayashi S, Takeo H, Tani T. Biochem Biophys Res Commun. 2006;351:999–1004. doi: 10.1016/j.bbrc.2006.10.139. [DOI] [PubMed] [Google Scholar]
- 25.Olivier C, Poirier G, Gendron P, Boisgontier A, Major F, Chartrand P. Mol Cell Biol. 2005;25:4752–4766. doi: 10.1128/MCB.25.11.4752-4766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jambhekar A, McDermott K, Sorber K, Shepard KA, Vale RD, Takizawa PA, DeRisi JL. Proc Natl Acad Sci USA. 2005;102:18005–18010. doi: 10.1073/pnas.0509229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chartrand P, Meng XH, Hüttelmaier S, Donato D, Singer RH. Mol Cell. 2002;10:1319–1330. doi: 10.1016/s1097-2765(02)00694-9. [DOI] [PubMed] [Google Scholar]
- 28.Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tyson CB, Lord PG, Wheals AE. J Bacteriol. 1979;138:92–98. doi: 10.1128/jb.138.1.92-98.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 31.Trybus KM, Freyzon Y, Faust LZ, Sweeney HL. Proc Natl Acad Sci USA. 1997;94:48–52. doi: 10.1073/pnas.94.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krementsova EB, Hodges AR, Lu H, Trybus KM. J Biol Chem. 2006;281:6079–6086. doi: 10.1074/jbc.M510041200. [DOI] [PubMed] [Google Scholar]
- 33.Reck-Peterson SL, Tyska MJ, Novick PJ, Mooseker MS. J Cell Biol. 2001;153:1121–1126. doi: 10.1083/jcb.153.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 35.Gonsalvez GB, Urbinati CR, Long RM. Biol Cell. 2005;97:75–86. doi: 10.1042/BC20040066. [DOI] [PubMed] [Google Scholar]
- 36.Jüschke C, Ferring D, Jansen RP, Seedorf M. Curr Biol. 2004;14:406–411. doi: 10.1016/j.cub.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 37.Li JF, Nebenfuhr A. J Biol Chem. 2007;282:20593–20602. doi: 10.1074/jbc.M700645200. [DOI] [PubMed] [Google Scholar]
- 38.Reisen D, Hanson MR. BMC Plant Biol. 2007;7:6. doi: 10.1186/1471-2229-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunn BD, Sakamoto T, Hong MS, Sellers JR, Takizawa PA. J Cell Biol. 2007;178:1193–1206. doi: 10.1083/jcb.200707080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niessing D, Hüttelmaier S, Zenklusen D, Singer RH, Burley SK. Cell. 2004;119:491–502. doi: 10.1016/j.cell.2004.10.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.