Abstract
Cleavage and release (shedding) of membrane proteins is a critical regulatory step in many normal and pathological processes. Evidence suggests that the antiaging transmembrane protein Klotho (KL) is shed from the cell surface by proteolytic cleavage. In this study, we attempted to identify the enzymes responsible for the shedding of KL by treating KL-transfected COS-7 cells with a panel of proteinase inhibitors and measuring cleavage products by Western blot. We report that metalloproteinase inhibitors, including EDTA, EGTA, and TAPI-1, inhibit the shedding of KL, whereas insulin increases shedding. The effects of the inhibitors in KL-transfected COS-7 cells were repeated in studies on rat kidney slices ex vivo, which validates the use of COS-7 cells as our model system. Tissue inhibitor of metalloproteinase (Timp)-3 effectively inhibits KL cleavage, whereas Timp-1 and Timp-2 do not, a profile that indicates the involvement of members of the A Desintegrin and Metalloproteinase (ADAM) family. Cotransfection of KL with either ADAM10 or ADAM17 enhances KL cleavage, whereas cotransfection of KL with small interference RNAs specific to ADAM10 and ADAM17 inhibits KL secretion. These results indicate that KL shedding is mediated mainly by ADAM10 and ADAM17 in KL-transfected COS-7 cells. The effect of insulin is abolished when ADAM10 or ADAM17 are silenced. Furthermore, we demonstrate that the effect of insulin on KL shedding is inhibited by wortmannin, showing that insulin acts through a PI3K-dependent pathway. Insulin enhances KL shedding without increasing ADAM10 and ADAM17 mRNA and protein levels, suggesting that it acts by stimulating their proteolytic activities.
Keywords: antiaging protein, insulin signaling, sheddase, metalloproteinase, amyloid precursor protein
Klotho (KL), an antiaging protein, was named after the goddess who spins the threads of life (1). Mice lacking KL exhibit many changes that occur during aging, including atherosclerosis, osteoporosis, infertility, and cognitive decline. They also have a short life span (1). In contrast, mice overexpressing KL live 30% longer than wild-type mice and are more resistant to oxidative stress (2). Another correlation linking KL and aging comes from a microarray analysis comparing young and aged brains. As part of our studies of age-associated cognitive decline in a rhesus monkey model, we found that the expression of KL was decreased in the aged brain (3). The gene for the mammalian KL has two transcripts that encode a long type I transmembrane protein and a short secreted protein. The long isoform of KL, originating from the transmembrane isoform, is found in serum and cerebrospinal fluid (CSF), suggesting that the extracellular domain of KL is cleaved and released from the cell membrane (4). Identifying the proteases responsible for KL shedding may lead to research designed to regulate the aging process.
The extracellular domains of numerous integral membrane proteins such as KL are released from the cell surface by enzymes called sheddases (5). Protein ectodomain shedding plays a crucial role in development, inflammation, and disease. The sheddases include members of various proteinase families, such as metalloproteinases, A Disintegrin and Metalloproteinases (ADAMs) (6, 7), and serine proteases (8). ADAMs are particularly important for ectodomain shedding of proteins such as Notch, the amyloid-β precursor protein (APP), and TGF-α, which, as a result of shedding, transactivate surface receptors in autocrine or paracrine mechanisms (9–11).
In this study, we searched for the sheddase responsible for KL ectodomain shedding from the cell membrane in KL-transfected COS-7 cells by examining a panel of putative inhibitors, including insulin. We selected insulin because it is a substrate and a competitive inhibitor of insulin degrading enzyme (IDE), and KL has been reported to be involved in the inhibition of the insulin signaling pathway (2). We show that ADAM10 and ADAM17 are capable of shedding KL from the plasma membrane, and that insulin can stimulate KL shedding.
Results
COS-7 Cells Have KL Sheddase Activities.
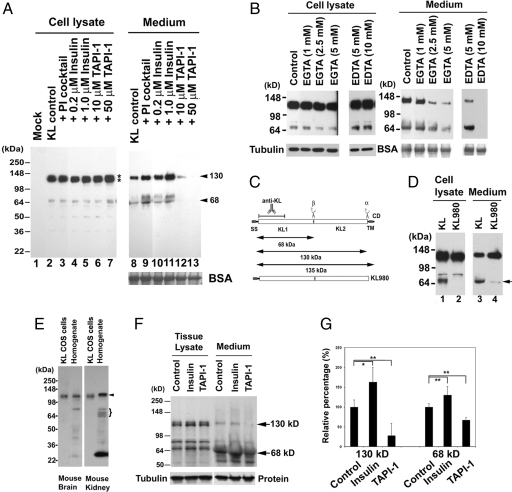
Mock-transfected COS-7 cells do not express a detectable level of endogenous KL (Fig. 1A, lane 1). However, on transfection of KL, we could detect two major KL fragments in the medium: a 130-kDa band and a 68-kDa doublet (Fig. 1A, lane 8). This finding indicates that COS-7 cells have KL sheddase activity. The 68-kDa doublet consists of a distinct band ≈64 kDa and a broad band ≈68 kDa, which is likely the glycosylated form of the lower band. In the cell lysate, KL appears as a doublet: The secreted 130-kDa fragment runs in between the 135- and 120-kDa doublets seen in the lysate [Fig. 1A, lanes 2–7 (lysate) and 8–13 (medium)]. This molecular mass of secreted KL is consistent with the results describing the secreted KL in serum and CSF (12). The major KL fragment found in the COS-7 cells medium and in the CSF was the 130-kDa fragment (Fig. 1A, lane 8) (12). Therefore, we named the cleavage that produced the 130-kDa fragment α-cut and the cleavage that produced the 68-kDa fragment β-cut (Fig. 1C). We used pulse-chase to study the kinetics of KL processing and observed KL fragments in the medium after a 30-min chase [supporting information (SI) Text and SI Fig. 6]. Because the KL antibody recognizes the N terminus of KL (Fig. 1C), a fragment corresponding to KL2 is likely produced, but cannot be recognized by this antibody. When we used KL C-terminally tagged with V5 or GFP, we were able to show that KL2 is released as well (SI Fig. 7).
Fig. 1.
Searching for the regulators affecting KL shedding in COS-7 cells and ex vivo. (A) Western blots from COS-7 cells transiently transfected with no DNA control (Mock, lane 1) or with the KL plasmid (lanes 2–13). After exposure to various inhibitors in serum-free DMEM (shown above blot), protein samples were collected from either the cell lysate or medium. BSA is shown as the TCA precipitation loading control. COS-7 cells do not express detectable levels of endogenous KL (lane 1). Exogenous KL expressed as a doublet of 135- and 120-kDa bands (lanes 2–7, asterisks). The blot was slightly overexposed to detect all of the KL processing fragments in the cells. At lower exposure of the blot, we could clearly see the doublet, similarly to that seen in G. The estimated molecular masses of the KL fragments in the medium are indicated (130 and 68 kDa, arrowheads). (B) After KL transfection, COS-7 cells were treated without (Control) or with various concentrations of EGTA and EDTA, as indicated, in serum-free DMEM. BSA and tubulin were used as loading controls for protein precipitation from the medium and total protein concentration in the cell lysates. (C) Schematic diagram of the KL protein structure and its processing in COS-7 cells. KL protein contains a signal sequence (SS), two homologous domains (KL1 and KL2), a transmembrane domain (TM), and a short cytoplasmic domain (CD). The anti-KL antibody recognition region is indicated. The α- and β-cleavage sites and the estimated molecular mass of the KL fragments are illustrated. KL980 indicates the truncated form of KL with amino acids 1–980, in which the TM and CDs of KL have been removed. (D) Comparison of the processing pattern of KL and KL980 lacking the TM region. The arrow indicates that the 68-kDa fragment is not detectable in the cell lysate (lane 2) and barely detectable in the medium (lane 4). (E) Expression pattern of KL in mouse brain and kidney tissues. Both full-length KL (arrowhead) and KL fragments (brackets) similar to the processing pattern in COS-7 cells were found in mouse kidney tissue. The ≈30-kDa band is a breakdown product of KL, and we also see it reproducibly in human CSF (data not shown). (F) Rat kidney slices were treated without (Control) or with 5 μM insulin or 100 μM TAPI-1 in serum-free DMEM for 4 h. The KL in the medium and the tissue was analyzed by Western blotting. Note that full-length KL appears as a doublet in the tissue (135 and 120 kDa), and a single band of a 130-kDa KL fragment was detected in the medium. The 130- and 68-kDa KL fragments are indicated (arrows). Tubulin and a protein in the medium were used as loading controls. (G) Statistical analysis of the results from F. The intensities of the 130- and 68-kDa bands were analyzed and normalized to the KL bands from the tissue lysate by using the average intensity of the controls as 100% from three to four independent experiments. The SD is calculated by using the nonbiased or n − 1 method. Significance of results was found by using Student's t test. *, P < 0.05; **, P < 0.005.
Regulation of KL Ectodomain Shedding by Metalloproteinase Inhibitors.
The amount of KL expressed in each experimental condition was similar (Fig. 1A, lanes 2–7), indicating similar efficiency levels for each KL transfection. The protease inhibitor mixture containing inhibitors for all classes of proteinases (serine, cysteine, metallo, and aspartic), 1 mM EDTA, and 1 mM EGTA failed to inhibit KL shedding for both 130- and 68-kDa fragments (Fig. 1A, lanes 8 and 9). It has been reported that low calcium concentrations could increase KL secretion (12). We similarly observed a slight increase in KL shedding when cells were treated with a protease inhibitor mixture, which contains 1 mM EDTA and 1 mM EGTA (Fig. 1A, compare lane 9 with lane 8). Insulin increased KL shedding in a dose-dependent manner without changing the KL protein expression in the cell lysate (Fig. 1A, compare lanes 10 and 11 with lane 8 for medium; compare lanes 4 and 5 with lane 2 for the cell lysate). TAPI-1, a general ADAM and matrix metalloproteinase (MMP) inhibitor, inhibited both the α- and β-cut of KL shedding in a dose-dependent manner (Fig. 1A, lanes 12 and 13). These results suggested that the sheddase could be a member of the ADAM or MMP families.
ADAM and MMP family members are Zn-dependent proteinases. Thus, their activities should be inhibited by the metal ion chelators EDTA and EGTA. However, the proteinase inhibitor (PI) mixture we used (containing 1 mM EDTA and 1 mM EGTA) did not reduce the sheddase activity. Because the concentration of Ca in DMEM is 1.3 mM, the EDTA and EGTA in the mixture may not have enough chelating power to remove all of the zinc ions from the sheddase. To test this hypothesis, we examined the sheddase activity with increasing amounts of these chelators. We found that both EGTA and EDTA inhibit KL shedding in a dose-dependent manner (Fig. 1B). With 10 mM EDTA in the medium, we were able to completely block both α- and β-cleavages (Fig. 1B, lane 6), whereas EGTA inhibited the secretion in a concentration-dependent manner (Fig. 1B, lanes 8–10). These data clearly demonstrate the involvement of one or more metalloproteinases in KL shedding.
Membrane Anchoring Is Critical for the β-Cleavage of KL.
To test whether membrane anchoring is required for KL β-cleavage, we constructed a truncated form of KL (KL980), in which the TM and cytoplasmic regions of KL after amino acid 980 were removed (Fig. 1C). We transfected KL980 into COS-7 cells and examined the KL shedding pattern. The 68-kDa band disappeared in the cell lysate and was barely detectable in the medium (Fig. 1D, arrow). This result shows that membrane anchoring is critical for KL β-cleavage, and it is likely that a transmembrane form of sheddase is involved in the KL cleavage.
Regulation of KL Shedding ex Vivo.
Western blot analysis of mouse brain and mouse kidney homogenates demonstrates comparable KL immunoreactive bands to those seen in KL-transfected COS-7 cells (Fig. 1 A and E). These results suggest that the transfected KL is processed in COS-7 cells similarly to endogenous KL in tissues of two separate organs.
To confirm the physiological relevance of the KL-transfected COS-7 cells, we investigated the effect of insulin and TAPI-1 on KL shedding in rat kidney slices. We found that insulin slightly, but significantly, increased KL shedding, and TAPI-1 reduced KL shedding significantly in kidney slices (Fig. 1 F and G). The effects of insulin and TAPI-1 were more apparent in cell culture (Fig. 1A) most likely because of the limited diffusion/accessibility for insulin and TAPI-1 to only the cells on the surface of the kidney tissue slices. These results suggest that KL shedding is regulated in the kidney similarly to what we observed in the COS-7 cells, and thus we decided to use COS-7 cells as a model system to search for KL sheddases.
Insulin Increases KL Shedding.
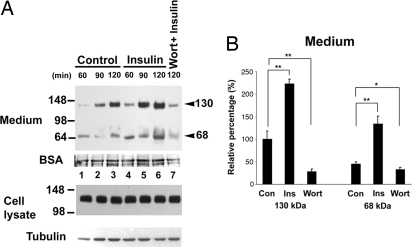
Unexpectedly, the addition of insulin increased KL shedding. We tested whether wortmannin, a fungal metabolite that specifically inhibits PI3K, could inhibit/reduce KL shedding in the presence of insulin. Fig. 2 shows the kinetics of KL shedding with or without insulin in the medium. Insulin increased KL shedding at all time points (Fig. 2A). The average increase of KL shedding by insulin is 2- to 3-fold (Fig. 2B). Pretreatment of the cells with wortmannin for 15 min before the 2-h serum-free DMEM incubation with insulin reduced KL shedding (Fig. 2). The insulin-induced KL shedding was blocked by the ADAM and MMP inhibitor, TAPI-1 (data not shown). These results suggest that the insulin pathway is involved in KL shedding by influencing an ADAM or MMP.
Fig. 2.
Regulation of KL shedding by insulin signaling pathway. (A) KL-transfected COS-7 cells were treated without (Control, lanes 1–3) or with (lanes 4–7) 1 μM insulin in serum-free DMEM for the times indicated. In lane 7, the cells were treated with 100 nM wortmannin, the insulin pathway inhibitor, for 15 min before insulin treatment for 120 min (Wort + Insulin). The proteins in the medium and cells were analyzed as described in Fig. 1. BSA and tubulin were used as loading controls. (B) Densitometric analysis of the intensity of the 130- and 68-kDa bands obtained from Western blotting from control (Con), insulin-treated (Ins), and wortmaninn-plus-insulin-treated (Wort) for 120 min from three independent transfections (for typical results, see lanes 3, 6, and 7 in A). The intensity of each band was normalized to the intensities of BSA or tubulin from each sample. The relative percentage using the average intensity of the 130-kDa band in control as 100% was analyzed and plotted. Error bars represent SD of the relative percentage (n = 3). *, P < 0.05; **, P < 0.005.
Characterization of Metalloproteinases Participating in Shedding of KL.
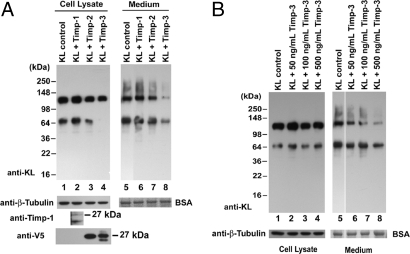
Tissue inhibitors of metalloproteinases (Timps) are important endogenous regulators of metalloproteinase activity. To provide more insight into the identity of the KL sheddase, we examined the effects of three Timps (Timp-1, Timp-2, and Timp-3) on KL shedding. Cotransfection of Timp-1 and Timp-2 did not affect KL shedding (Fig. 3A, compare lanes 2 and 3 with lane 1 and lanes 6 and 7 with lane 5), whereas cotransfection of Timp-3 inhibited β-cleavage in the cells and both α- and β-cleavage on plasma membrane, with resulting reduced 130-and 68-kDa KL fragments in the medium (Fig. 3A, compare lanes 4 and 8 with lanes 1 and 5).
Fig. 3.
Effects of Timp-1, Timp-2, and Timp-3 on KL shedding. (A) COS-7 cells were transfected with KL alone (KL control) or cotransfected with KL and Timp1, Timp-2, or Timp-3 as indicated. Forty-eight hours after transfection, the cells were washed and incubated in serum-free medium as described before. The protein samples in the cell lysates and medium were collected and analyzed by Western blotting. BSA and tubulin were used as loading controls. The expression of Timp-1 was analyzed by Western blotting with anti-Timp-1 antibody, and the expressions of Timp-2 and Timp-3 were analyzed with an anti-V5 antibody. The apparent molecular masses of the bands are indicated. Statistical analysis of the densitometric results is shown in SI Text. (B) Forty-eight hours after transfection, KL-transfected COS-7 cells were incubated in serum-free DMEM without (KL control) or with various amounts of the Timp-3 peptide, as indicated. The proteins in the cell lysates and medium were analyzed as described in Fig. 1.
We then applied increasing amounts of Timp-3 peptide to the medium and tested whether Timp-3 can inhibit KL shedding from outside the cells. Indeed, the addition of the Timp-3 peptide to the medium inhibited KL shedding in a dose-dependent manner (Fig. 3B). In comparing the results of cotransfection of Timp-3 versus applying Timp-3 to the medium, it appeared that cotransfection of Timp-3 inhibited β-cleavage as indicated by a significant reduction in the 68-kDa band (Fig. 3A, lane 4), which was not observed when applying Timp-3 peptide from outside (Fig. 3B, lanes 2–4). However, we cannot exclude the possibility that α-cut also is inhibited intracellularly. SI Fig. 8 shows the statistical analysis of Timp-3 inhibition of KL shedding on both α- and β-cleavages. These results suggest that the cleavage of KL in COS-7 cells is carried out by metalloproteinases that are specifically inhibited by Timp-3, and that this cleavage occurs both intracellularly and at the cell surface.
ADAM10 and ADAM17 Are Involved in both the α- and β-Cuts.
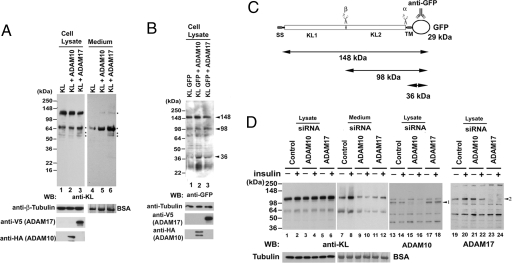
A recent publication revealed that the sheddase ADAM17 had an inhibitory profile similar to what we had found (13). Thus, we decided to determine whether ADAM17 and ADAM10 also are responsible for KL ectodomain shedding. We cotransfected ADAM10 or ADAM17, together with KL, and examined the effects on KL shedding. Cotransfection of KL with either ADAM10 or ADAM17 increased the generation of KL fragments (Fig. 4A, compare lanes 5 and 6 with lane 4), and the effect of the ADAM17 transfection was more profound than that of the ADAM10 transfection on KL shedding in both the cell lysate and medium (Fig. 4A compare lanes 3 and 6 with lanes 2 and 5). As expected, we saw a more significant effect of the cotransfection from the medium samples compared with the cell lysates (Fig. 4A, compare lanes 2 and 3 with lane 1 and lanes 5 and 6 with lane 4) because the cell lysates represent the steady state of KL expression in 48 h, whereas the KL fragments from the medium resulted from 2 h incubation of the cells in serum-free DMEM. The anti-KL antibody was generated by using a recombinant protein corresponding to amino acids 55–261 in the KL1 domain of KL (Fig. 1C) (14). Because the secreted 130-kDa fragment migrates on SDS/PAGE gels in between the 135-kDa doublet seen in the cell lysate, we sought to produce a fusion protein in which the full-length and cleaved fragments are better separated. We produced a KL construct, KL GFP, with a GFP tag at the C terminus and cotransfected it with ADAM10 or ADAM17. We were able to detect with the anti-GFP antibody KL fragments of 148, 98, and 36 kDa in the cells (Fig. 4 B and C), although the estimated molecular mass for full-length KL GFP is 164 kDa. Cotransfection of KL GFP with ADAM10 and ADAM17 increased KL GFP processing, as indicated by increased intensity of the 36- and 98-kDa KL fragments (Fig. 4B, compare lanes 2 and 3 with lane 1). We noticed that ADAM10 and ADAM17 may have different preferences for the α- and β-cuts because we observed a stronger 36-kDa band after cotransfection with ADAM10 and a stronger 98-kDa band for cotransfection with ADAM17 (Fig. 4B). These results suggest that ADAM10 and ADAM17 are involved in both the α- and β-cleavages of KL ectodomain shedding.
Fig. 4.
ADAM10 and ADAM17 are responsible for KL shedding in COS-7 cells. (A) COS-7 cells were cotransfected with KL, with empty vector, or with either ADAM10 or ADAM17 as indicated. Forty-eight hours after transfection, the cells were washed and incubated in serum-free medium as described before. The protein samples in the cell lysates and medium were collected and analyzed by Western blotting. BSA and tubulin were used as loading controls. The expression of ADAM17 was analyzed by Western blotting with anti-V5 antibody, and the expression of ADAM10 was analyzed with an anti-HA antibody. Note that the cotransfection of KL with ADAM10 and ADAM17 resulted in increased KL fragments in cells and medium (lanes 3, 5, and 6; asterisks). This particular exposure was chosen to emphasize the difference in KL fragments intensities with cotransfection of KL with either ADAM10 or ADAM17. (B) Comparison of the processing pattern of KL and KL GFP construct cotransfected with ADAM10 or ADAM17. The blots were analyzed with anti-GFP antibody as indicated. The estimated molecular masses of the KL fragments are shown. (C) Schematic diagram of the KL GFP construct. The anti-GFP antibody recognition site is indicated. The α- and β-cleavage sites and the estimated molecular masses of the KL GFP fragments are illustrated. (D) Western blots of samples from COS-7 cells transfected with KL with control siRNA (Control) or with KL and siRNA specific to either ADAM10 or ADAM17 were analyzed as described before using antibodies to KL, ADAM10, and ADAM17. Tubulin and BSA were used as loading controls. ADAM10 (arrowhead 1) and ADAM17 (arrowhead 2) protein are indicated. Statistical analysis of the results from D is shown in SI Text.
Silencing of ADAM10 and ADAM17 with siRNA.
Silencing endogenous ADAM10 or ADAM17 resulted in a significant reduction of both α- and β-cleavages (Fig. 4D, lanes 9–12, compared with lanes 7 and 8). Western blotting by using anti-ADAM10 antibody revealed that the ADAM10 protein was largely reduced with treatment of ADAM10 siRNA (Fig. 4D, compare lane 15 with lane 13, arrowhead 1). A similar effect was observed with anti-ADAM17 antibody (i.e., ADAM17 siRNA reduced ADAM17 protein) (Fig. 4D, compare lane 23 with lane 19). The statistical analysis of the results from Fig. 4D is shown in SI Fig. 9.
Regulation of KL Shedding by Insulin, ADAM10, and ADAM17.
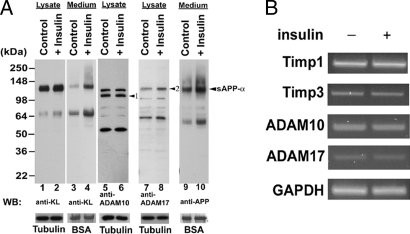
We demonstrated that insulin can increase KL shedding, and that both ADAM10 and ADAM17 are involved in KL cleavage. To investigate whether the effect of insulin on KL shedding has a direct effect on either ADAM10 or ADAM17, we examined the effects of ADAM10 and ADAM17 on KL shedding with siRNA specific to either ADAM10 or ADAM17 with or without insulin treatment. The results showed that silencing either ADAM10 or ADAM17 could significantly reduce the effects of insulin on increasing KL shedding (Fig. 4D, compare lanes 10 and 12 with lanes 9 and 11). Next, we examined ADAM10 and ADAM17 protein levels and activity with or without insulin treatment. Because the APP also is a substrate for both ADAM10 and ADAM17, which act on APP as α-secretase to release secreted APPα (sAPPα) (15, 16), we tested the levels of endogenous sAPPα as an indicator of α-secretase activity. As expected, insulin induces an increase in the levels of sAPPα in the medium (Fig. 5A, lanes 9 and 10). However, we did not find a significant change in either ADAM10 or ADAM17 protein levels (Figs. 4D and 5A and SI Fig. 10). In addition, we did not detect changes in Timp-1, Timp-3, ADAM10, or ADAM17 in mRNA levels by using RT-PCR (Fig. 5B). The statistical analysis of the results from Fig. 5 A and B is shown in SI Fig. 10. These results suggest that insulin increases KL shedding through regulation of both ADAM10 and ADAM17 proteolytic activity without affecting their expression levels.
Fig. 5.
Effect of Insulin on ADAM10 and ADAM17 activities, mRNA level, and KL shedding in COS cells. (A) KL-transfected COS-7 cells were treated without (Control) or with (+Insulin) 1 μM insulin for 2 h by using the same sample preparation and analysis procedures as described in Fig. 1. The blots were analyzed with the indicated antibodies. BSA and tubulin were used as loading controls. The ADAM10 and ADAM17 proteins (arrowheads 1 and 2, respectively), and a secreted derivative of APP (arrowhead sAPPα) are indicated. (B) RT-PCR of specific genes. Total RNA was purified from transfected cells with no insulin treatment (−) or with insulin treatment (+) as described in A for the genes indicated. Statistical analysis of the results are shown in SI Text.
Discussion
In this study, we show that members of the ADAMs family, ADAM10 and ADAM17, can cleave the extracellular domain of KL to generate 130- and 68-kDa KL fragments. Furthermore, insulin considerably enhances KL shedding in COS-7 cells. Notably, we found that TAPI-1 and insulin exhibit the same effects on KL secretion ex vivo in rat kidney slices. We further demonstrate that overexpression of either ADAM10 or ADAM17 leads to an increase in both the 130- and 68-kDa KL fragments, whereas silencing of either ADAM10 or ADAM17 with siRNA leads to a decrease of both fragments (Fig. 4 A, B, and D). These results indicate that ADAM10 and ADAM17 are involved in both α- and β-cleavages of KL in COS-7 cells. There are at least 23 human ADAMs from the 40 ADAMs identified thus far (http://people.virginia.edu/∼jw7g/Table_of_the_ADAMs.html). At this time, we cannot rule out the involvement of other sheddases in KL cleavage in COS cells or other cells or tissues.
Although our results suggest that ADAM10 and ADAM17 are responsible for both the α- and β-cleavages of KL, leading to its shedding, the efficiency of the α- and β-cuts is not the same. In most cases, the 130-kDa fragment is the major product of KL shedding. This result is consistent with the previously reported study where the 130-kDa fragment was the dominant form of KL in kidney, serum, CSF, and choroid plexus, whereas only insignificant other fragments of KL were detected (12, 14). It would be interesting to determine whether the biological properties of the KL1 and KL2 fragments differ from those of the major 130-kDa KL fragment.
Most interestingly, the finding that sheddase activity can be enhanced by insulin suggests the involvement of the insulin signaling pathway in the release of KL from cell membranes. We propose a possible negative feedback loop of insulin regulation by KL, in which insulin initiates a signaling cascade and/or gene expression that results in the trafficking and/or activation of ADAM10 and/or ADAM17. This result, in turn, increases the release of the KL protein (the 130-kDa KL1 and KL2 fragments) and other ADAM10 and ADAM17 substrates (i.e., TNF-α and sAPPα) into the medium. KL has been shown to block insulin and insulin-like growth factor 1 receptor phosphorylation of the insulin receptor substrate (IRS) and also subsequent downstream activation of PI3K and Akt-1 (17, 18). The KL fragments can then feedback through an as-yet-unknown process to turn off insulin signaling. TNF-α also has been shown to contribute to the inhibition of the insulin signaling pathway (19).
The mechanism of insulin-induced shedding is unknown, but there are several possibilities that are not mutually exclusive. It has been reported that, in CHO cells, insulin stimulates a 2- to 3-fold increase in the endocytic recycling pathway, implicating that the vesicle-associated proteins have an increased chance to be at the cell surface (20). In further support for the role of insulin in vesicle trafficking is the recent report that in adipocytes insulin causes the fodrin/spectrin remodeling, leading to the translocation of GLUT4 to the membrane (21). Because shedding of transmembrane proteins requires their trafficking by the secretory pathway to the plasma membrane, a faster recycling induced by insulin would cause an increase of KL sheddases, KL, or both at the cell surface, resulting in increased KL shedding. Another possible mechanism of ADAM17 activation by insulin is by the down-regulation of Timp-3, an ADAM17 inhibitor. In support of this mechanism are findings from the insulin receptor (IR) heterozygous mice (Insr+/−) that develop diabetes with more than five times the increased insulin level in the serum. These mice have reduced Timp-3 and increased ADAM17 activity (22). However, we did not detect any change in the Timp-3 mRNA level in our system (Fig. 5B and SI Fig. 10) in favor of the hypothesis that insulin enhances KL shedding through protein translocation or trafficking.
Insulin can increase shedding of transmembrane proteins, including KL and APP. The up-regulation of the nonamyloidogenic processing of APP by ADAM17 is of particular interest because it results in reduced Aβ formation due to a lower amount of APP available for β-secretase cleavage. Insulin has been previously shown to regulate sAPPα release by the activity of PI3K. Because of the physiological role of PI3K in the translocation of glucose transporter-containing vesicles, the authors speculate that PI3K involvement in APP metabolism is at the level of vesicular trafficking of APP or secretase-containing vesicles (23). However, here we posit that insulin enhances sAPPα release by the same mechanism as KL release: the activation of ADAM10 and ADAM17 by insulin's effects on the intracellular trafficking of the ADAMs.
The KL transgenic mice are excellent models to explain the interaction between KL and insulin as described in the elegant work of Kurosu et al. (2) and reviewed by Unger (18). Mice overexpressing KL are insulin-resistant. In these mice, increased KL levels lead to increased repression of the autophosphorylation of the IR. As a result, the IRS is less phosphorylated, reducing its association with PIK3 p85. This finding leads to less phosphorylation of FoxO transcription factors, their subsequent entry into the nucleus, and the up-regulation of SOD and catalase. Thus, although these mice are insulin-resistant because their IR signaling is blocked and less GLUT4 is available at the membrane, they are more resistant to oxidative stress and live longer and healthier lives. In contrast, in KL-mutant mice with loss-of function mutation, in response to insulin signaling, GLUT4 is translocated to the membrane, but FoxO is phosphorylated and cannot enter the nucleus to activate transcription of antioxidant genes, resulting in major organ failure. Our results are consistent with the findings in transgenic KL mice, but add insight into the mechanism of KL-insulin relationship (i.e., the involvement of ADAM10 and ADAM17). The findings presented here provide an additional molecular link between KL and insulin resistance. It is still unclear whether KL works by binding to its yet-to-be-identified receptor or whether it has a more direct effect on the phosphorylation of the IR.
The delineation of the complexity of the multiple factors that participate in KL shedding provides many targets for pharmacologic intervention.
Materials and Methods
Materials.
The proteinase inhibitor mixture, including 3 μg/ml aprotinin, 5 μg/ml leupeptin, 0.7 μg/ml pepstatin, 1 mM Pefabloc, 0.5 mM EDTA, and 1 mM EGTA, was from Roche. TAPI-1 was purchased from Peptides International. Insulin was a gift from Konstantin Kandror (Boston University School of Medicine, Boston). Wortmannin was purchased from Calbiochem. Timp-3 peptide was purchased from Oncogene. All other chemicals were from Sigma–Aldrich unless otherwise specified.
Plasmid Construction.
Details of plasmid construction are provided in SI Text.
Cell Culture, Transfection, and Protein Sample Collection.
Details of cell culture, transfection, and protein sample collection are provided in SI Text.
Tissue Preparation.
Details of tissue preparation are provided in SI Text.
Western Blotting.
Details of Western blotting are provided in SI Text. The rat anti-KL antibody KM2076 (1:2,000) is described in ref. 14 and was kindly provided by Kyowa Hakko Kogy (Tokyo) o. Other primary antibodies used are described in SI Text. The identity of the fragments visualized by Western blot was confirmed by trypsin digestion and analysis of the digestion fragments was done by mass spectrometry (see SI Fig. 7).
Protease Inhibition Studies.
The reagents used included a proteinase inhibitor mixture (antipain, leupeptin, pepstatin, PMSF, aprotinin, EDTA, and EGTA), TAPI-1, and insulin. Forty-eight hours after transfection, the growth medium was replaced with serum-free DMEM containing the proteinase inhibitors and incubated for 2 h. The medium was collected and TCA-precipitated together with 10 μg/ml (final concentration) BSA as a carrier. Both the cell lysates and TCA-precipitated samples from the medium were analyzed by SDS/PAGE, followed by Western blotting with the anti-KL antibody.
siRNA Transfection.
Control, ADAM10, and ADAM17 siRNA were purchased from Dharmacon. Transfection of siRNA was performed by using Lipofectamine plus transfection reagent (Invitrogen). A final concentration of 100 nM or 200 nM siRNA was used in each transfection according to the manufacturer's protocol.
RT-PCR.
Details of RT-PCR are provided in SI Text.
Supplementary Material
Acknowledgments
We thank K. Kandror for suggestions during the preparation of the manuscript and the gift of insulin; R. Yamin for carefully reading the manuscript; S. Russek (Boston University School of Medicine, Boston) for the rat tissues; J. Luebke for assistance in preparing kidney tissue slices; Kyowa Hakko Kogyo for the KL monoclonal antibody KM2076; F. Fahrenholz (Johannes Gutenberg-Universität, Mainz, Germany) for the ADAM10 cDNA clone; H. Dietz (Johns Hopkins School of Medicine, Baltimore) for the KL cDNA clones; and B. Slack (Boston University School of Medicine) for the TAPI-1 inhibitor. This work was supported by National Institutes of Health/National Institute on Aging Grant AG-00001.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709805104/DC1.
References
- 1.Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi H, Suga T, Utsugi T, Ohyama Y, Kurabayashi M, Kaname T, Kume E, et al. Nature. 1997;390:45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- 2.Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, McGuinness OP, Chikuda H, Yamaguchi M, Kawaguchi H, et al. Science. 2005;309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duce JA, Podvin S, Hollander W, Kipling D, Rosene DL, Abraham CR. Glia. 2008;56:106–117. doi: 10.1002/glia.20593. [DOI] [PubMed] [Google Scholar]
- 4.Matsumura Y, Aizawa H, Shiraki-Iida T, Nagai R., Kuro-o M, Nabeshima Y. Biochem Biophys Res Commun. 1998;242:626–630. doi: 10.1006/bbrc.1997.8019. [DOI] [PubMed] [Google Scholar]
- 5.Ehlers MR, Riordan JF. Biochemistry. 1991;30:10065–10074. doi: 10.1021/bi00106a001. [DOI] [PubMed] [Google Scholar]
- 6.Moss ML, Lambert MH. Essays Biochem. 2002;38:141–153. doi: 10.1042/bse0380141. [DOI] [PubMed] [Google Scholar]
- 7.Seals DF, Courtneidge SA. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 8.Urban S, Lee JR, Freeman M. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 9.Blobel CP. Cell. 1997;90:589–592. doi: 10.1016/s0092-8674(00)80519-x. [DOI] [PubMed] [Google Scholar]
- 10.Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN, et al. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 11.Borrell-Pages M, Rojo F, Albanell J, Baselga J, Arribas J. EMBO J. 2003;22:1114–1124. doi: 10.1093/emboj/cdg111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imura A, Iwano A, Tohyama O, Tsuji Y, Nozaki K, Hashimoto N, Fujimori T, Nabeshima Y. FEBS Lett. 2004;565:143–147. doi: 10.1016/j.febslet.2004.03.090. [DOI] [PubMed] [Google Scholar]
- 13.Tsakadze NL, Sithu SD, Sen U, English WR, Murphy G, D'Souza SE. J Biol Chem. 2006;281:3157–3164. doi: 10.1074/jbc.M510797200. [DOI] [PubMed] [Google Scholar]
- 14.Kato Y, Arakawa E, Kinoshita S, Shirai A, Furuya A, Yamano K, Nakamura K, Iida A, Anazawa H, Koh N, et al. Biochem Biophys Res Commun. 2000;267:597–602. doi: 10.1006/bbrc.1999.2009. [DOI] [PubMed] [Google Scholar]
- 15.Buxbaum JD, Liu KN, Luo Y, Slack JL, Stocking KL, Peschon JJ, Johnson RS, Castner BJ, Cerretti DP, Black RA. J Biol Chem. 1998;273:27765–27767. doi: 10.1074/jbc.273.43.27765. [DOI] [PubMed] [Google Scholar]
- 16.Slack BE, Ma LK, Seah CC. Biochem J. 2001;357:787–794. doi: 10.1042/0264-6021:3570787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres PU, Prie D, Molina-Bletry V, Beck L, Silve C, Friedlander G. Kidney Int. 2007;71:730–737. doi: 10.1038/sj.ki.5002163. [DOI] [PubMed] [Google Scholar]
- 18.Unger RH. Nat Med. 2006;12:56–57. doi: 10.1038/nm0106-56. [DOI] [PubMed] [Google Scholar]
- 19.Pedersen BK. Essays Biochem. 2006;42:105–117. doi: 10.1042/bse0420105. [DOI] [PubMed] [Google Scholar]
- 20.Johnson AO, Subtil A, Petrush R, Kobylarz K, Keller SR, McGraw TE. J Biol Chem. 1998;273:17968–17977. doi: 10.1074/jbc.273.28.17968. [DOI] [PubMed] [Google Scholar]
- 21.Liu L, Jedrychowski MP, Gygi SP, Pilch PF. Mol Biol Cell. 2006;17:4249–4256. doi: 10.1091/mbc.E06-04-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Federici M, Hribal ML, Menghini R, Kanno H, Marchetti V, Porzio O, Sunnarborg SW, Rizza S, Serino M, Cunsolo V, et al. J Clin Invest. 2005;115:3494–3505. doi: 10.1172/JCI26052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solano DC, Sironi M, Bonfini C, Solerte SB, Govoni S, Racchi M. FASEB J. 2000;14:1015–1022. doi: 10.1096/fasebj.14.7.1015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.