Abstract
Cilia project from cells as membranous extensions, with microtubule structural cores assembling from basal bodies by intraflagellar transport (IFT). Here, we report a ciliary role for the inositol 1,3,4,5,6-pentakisphosphate 2-kinase (Ipk1) that generates inositol hexakisphosphate. In zebrafish embryos, reducing Ipk1 levels inhibited ciliary beating in Kupffer's vesicle and decreased ciliary length in the spinal canal, pronephric ducts, and Kupffer's vesicle. Electron microscopy showed that ciliary axonemal structures were not grossly altered. However, coincident knockdown of Ipk1 and IFT88 or IFT57 had synergistic perturbations. With GFP-Ipk1 enriched in centrosomes and basal bodies, we propose that Ipk1 plays a previously uncharacterized role in ciliary function.
Keywords: cilia, inositol hexakisphosphate, kidney development, left–right asymmetry, zebrafish
Inositol signaling pathways constitute intracellular communication networks that coordinate cell growth, differentiation, death, and metabolism. Following the classic signaling paradigm, activation of cell surface receptors by extracellular stimuli results in altered lipid and soluble inositol polyphosphate (IP) levels (1). Lipid-anchored phosphatidylinositol 4,5-bisphosphate is the effective starting point for soluble IP production, with regulated hydrolysis by phospholipase C producing two essential signaling molecules, diacylglycerol and soluble inositol 1,4,5-triphosphate (IP3) (2). IP3 is sequentially phosphorylated to produce inositol tetrakisphosphate isomers, inositol 1,3,4,5,6-pentakisphosphate (IP5), inositol hexakisphosphate (IP6), and inositol pyrophosphates (3). The functions, targets, and roles in development and disease for many of the IPs are unknown.
Recently, we reported that zebrafish embryos depleted of the IP5 2-kinase (Ipk1), which catalyzes IP6 synthesis, fail to execute proper left–right (LR) body placement of organs like the liver, pancreas, and heart (4). Strikingly, a reduction in the left-biased intracellular Ca2+ flux was coincidentally observed in cells surrounding the ciliated Kupffer's vesicle (KV), a structure orthologous to the mouse node. Others have documented roles for motile cilia in initial events establishing the LR body axis and in generating counterclockwise fluid flow in the node (5). In zebrafish embryos, alterations in proper KV morphogenesis, ciliary motility, and directional fluid flow all perturb LR asymmetry (6, 7). Two mechanisms have been proposed for the initiating event that results in asymmetric gene expression. One, cilia-driven fluid flow results in a morphogen concentrating toward the nodal left side to influence intracellular signaling (5). Two, fluid flow results in directional bending of mechanosensor immotile cilia in the node that triggers a left-sided asymmetric Ca2+ flux (8). Both mechanisms are based on a crucial role for nodal ciliary function.
Cilia project from many eukaryotic cell types as membranous extensions with a microtubule-based central core termed the axoneme. Microtubule nucleation and organizing centers for the ciliary axonemes are basal body structures, derived from centrioles (9, 10). Motile cilia have an axoneme with nine microtubule doublets, two central microtubule singlets, radial spokes linked to the microtubules, and inner and outer dynein arm motors (10), termed canonical “9 + 2” structures. Ciliary beating results from the sliding of outer microtubule doublets relative to one another, powered by the motor activity of axonemal dynein (11). In contrast, nonmotile, sensory cilia do not have two central microtubule singlets or dyneins, termed a “9 + 0” axonemal type. Mouse nodal cilia have been observed with both 9 + 0 and 9 + 2 axonemes (5, 12, 13), reflecting proposed populations of motile and immotile cilia (8). Zebrafish KV cilia have a reported 9 + 2 axoneme (6). Ciliary assembly, motility, and maintenance depend on bidirectional intraflagellar transport (IFT) along axonemal microtubules (9, 14) and involve anterograde kinesin-2 motors and retrograde dynein motors.
Here, we have analyzed further the mechanism for LR asymmetry defects in zebrafish embryos with altered Ipk1 levels and perturbed IP production. We report an unidentified role for Ipk1 in ciliary beating and length maintenance and propose that Ipk1 is an important component of the basal body with IP production mediating ciliary function.
Results and Discussion
ipk1 Knockdown Reduces Ciliary Beating and Length.
In our previous studies of ipk1 function, we analyzed loss-of-function phenotypes by injecting cleavage-stage zebrafish embryos with an antisense morpholino oligonucleotide (MO) that blocks ipk1 mRNA translation (ipk1MO1 embryos) (4). The ipk1MO1 effects were specific and directly altered IP6 production in vivo. We did not detect gross changes in KV morphogenesis or global KV cilia number in the ipk1MO1 embryos (4). However, we did not directly assay ciliary beating or cilia function. Because Ca2+ flux in cells surrounding the KV was altered and LR asymmetry randomized (4), it was critical to more fully analyze ciliary functions and structures.
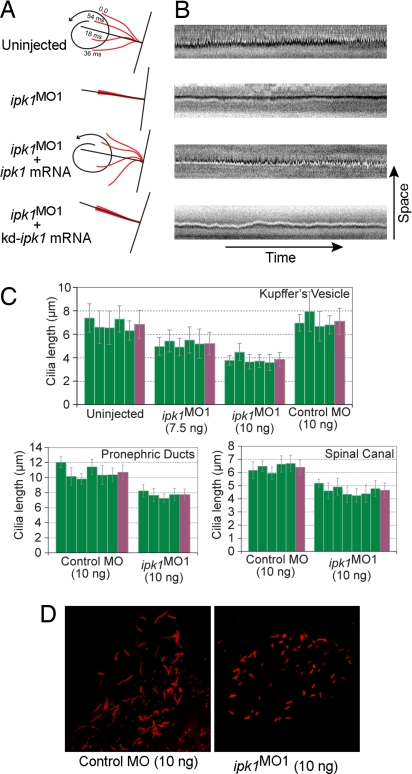
First, we compared KV ciliary beating in uninjected and ipk1MO1 embryos. In wild-type embryos, cilia beat with a vortical motion in a counterclockwise orientation at a frequency of 12.95 ± 2.21 Hz (n = 6) [Fig. 1A and supporting information (SI) Movies 1 and 2]. In contrast, in ipk1MO1 embryos, cilia quivered in a vibration-like motion devoid of counterclockwise rotation (Fig. 1A and SI Movies 3 and 4). In kymographs (Fig. 1B), the time–space trajectories for a cilium in an uninjected embryo showed deflections at regular intervals. However, the cilium in ipk1MO1 embryos had a straight-line trajectory with no significant deflection (Fig. 1B). Thus, in contrast to our prior speculation (4), the cilia in ipk1MO1 embryos are functionally perturbed and do not show ciliary beating.
Fig. 1.
ipk1 knockdown reduces normal ciliary beating and length. (A and B) Schematic diagrams (A) and kymographs (B) showing trajectory of a cilium in uninjected, ipk1MO1-injected, ipk1MO1 plus wild-type ipk1 mRNA-injected, or ipk1MO1 plus kinase-dead (kd) ipk1 mRNA-injected embryos. (C) Graphs for measurements of ciliary length from individual embryos (green), with the mean ciliary length (far right, purple) determined from multiple independent measurements for KV (n > 26), pronephric duct (n > 13), and spinal canal (n > 14) cilia. (D) Representative images of KV cilia (red) from embryos injected with 10 ng of either control MO or ipk1MO1. n, number of cilia measured per embryo.
To directly test whether the kinase activity of Ipk1 is required for ciliary beating, we performed a series of complementation experiments. Synthetic mRNA encoding wild-type Ipk1 or a kinase-dead (kd) Ipk1 was engineered that lacked recognition by the ipk1MO1. Ciliary beating was measured in the KV of embryos coinjected with the ipk1MO1 and wild type ipk1 or kd-ipk1 mRNA. Significantly, the ciliary beating defect in ipk1MO1 embyros was rescued by coinjection of wild-type ipk1 mRNA (Fig. 1 A and B and SI Movies 5 and 6). However, coinjection of mRNA encoding the kd-Ipk1 failed to complement (Fig. 1 A and B and SI Movies 7 and 8). As controls, synthetic mRNAs were coinjected with a five-mismatch control MO, and no apparent change in ciliary beating was observed (SI Movies 9 and 10). Thus, Ipk1 kinase activity is critical for ciliary beating.
Second, we quantitatively analyzed ciliary length in the KV and other ciliated organs. To do this, we performed whole-mount anti-acetylated tubulin immunocytochemistry on 12-h-after-fertilization (hpf) [eight-somite stage (SS)] (for KV) and 30 hpf (for pronephric duct and spinal canal) embryos (Fig. 1D and SI Fig. 5). In ipk1MO1 embryos, we observed a dose-dependent, statistically significant decrease in KV ciliary length (Fig. 1C and SI Table 1). KV ciliary length decreased ≈46% when 10 ng of ipk1MO1 was injected per embryo. In contrast, 10 ng injection of a five-mismatch control MO had no effect. This length defect was not apparent in our previous studies (4) wherein no more than 7.5 ng of the ipk1MO1 was injected per embryo. With 10 ng of the ipk1MO1, we also detected ≈27% length decrease in pronephric duct and spinal canal cilia (Fig. 1C). We conclude that Ipk1 plays a key role in ciliary length maintenance in multiple organs. A role for an enzyme in the soluble IP production pathway in ciliary beating and length control has not been previously reported.
Decreased ciliary length in Ipk1-depleted embryos could result from a delay in embryonic growth and development. As controls, we examined expression of three different genes required during zebrafish segmentation that have no role in LR asymmetry: krox20, a Zn-finger transcription factor expressed during segmentation (15), myoD, a member of myogenic basic helix–loop–helix (bHLH) family of transcription factors expressed in developing somites (16), and deltaC, a cyclic gene encoding a Notch ligand expressed in presomitic mesoderm and the posterior half of each somite (17). Expression patterns of these genes were indistinguishable between control MO and ipk1MO1 embryos (SI Fig. 6). These results correlate directly with our prior analysis of expression for the nodal-related gene southpaw (4). The ipk1MO1 injection does not alter early bilateral southpaw expression in paraxial mesoderm precursors. However, ipk1MO1 injection perturbs the asymmetric expression of southpaw during 15-16 SS, when it is expressed in the left LPM, preceding fully randomized expression at 22 SS (4). Altogether, Ipk1 depletion altered early LR-specific expression of southpaw without perturbing expression of krox20, myoD, and deltaC. These results indicate that Ipk1 depletion did not indirectly alter the developmental timing. We conclude that the ciliary defect observed in ipk1MO1 embryos is specific to Ipk1 function in cilia.
Normal Ciliary Axonemal Organization Is Not Perturbed in Ipk1-Deficient Embryos.
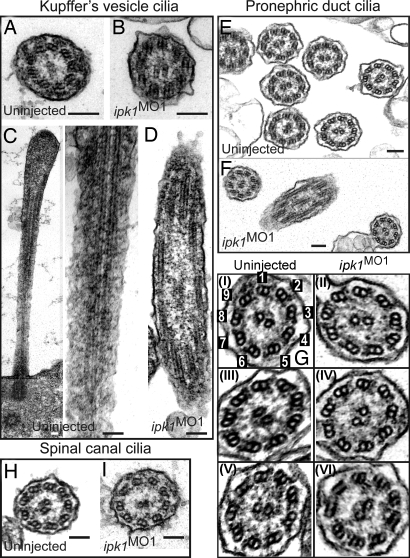
To further investigate the ciliary defects, we examined cilia ultrastructure in ipk1MO1 embryos by thin-section transmission electron microscopy (TEM). For KV cilia, TEM was performed on 11.5-hpf embryos, whereas 24 hpf embryos were used for pronephric duct and spinal canal cilia. No difference in the ultrastructural organization of KV cilia was noted between the wild-type and ipk1MO1 embryos (Fig. 2 A and B). Microtubule tracks in the KV cilia were visible in the longitudinal sections in both sets of embryos (Fig. 2 C and D). The ciliary ultrastructures in the pronephric duct (Fig. 2 E–G) and spinal canal (Fig. 2 H and I) were also not markedly altered in ipk1MO1 embryos. There were inner and outer dynein arms visible on the microtubule doublets in both sets of embryos (Fig. 2G). For example, outer dynein arms are present in the ipk1MO1 sections of Fig. 2G-II (off doublet 6) and Fig. 2G-IV (off doublet 5) that are similar to those in the uninjected of Fig. 2G-I (off doublet 5) and Fig. 2G-III (off doublet 4). Inner dynein arms are noted in the ipk1MO1 sections of Fig. 2G-II (off doublet 7), Fig. 2G-IV (off doublet 4), and Fig. 2G-VI (off doublet 1) as in the uninjected Fig. 2G-I (off doublet 9), Fig. 2G-III (off doublet 3), and Fig. 2G-V (off doublet 3). Although the ipk1MO1 embryos might harbor undetectable subtle perturbations of ciliary structure and spacing of dynein arms, the loss of Ipk1 activity did not grossly alter the overall framework for ciliary axonemal organization.
Fig. 2.
ipk1 knockdown does not affect ciliary axoneme organization. (A and B) TEM cross-sections of cilia in KV for uninjected (A) and ipk1MO1 (B) embryos. (C and D) Longitudinal TEM sections showing axonemal microtubules in KV cilia in uninjected (C) and ipk1MO1 (D) embryos. (E–I) TEM sections showing pronephric duct cilia (E, F, and G) and spinal canal cilia (H and I). Enlarged images of independent pronephric duct cilia TEM sections are shown in G. Cilia sections in uninjected (E, G-I, G-III, G-V, and H) and ipk1MO1 (F, G-II, G-IV, G-VI, and I) embryos are shown. In G–I, numerical tags are assigned to each of the nine outer microtubule doublets for reference to dynein arms in the text. (Scale bars, 0.1 μm.)
Codepletion of Ipk1 and Either IFT88 or IFT57 Has Synergistic Effects on LR Asymmetry Establishment.
Several possibilities existed by which Ipk1 and IP production might affect ciliary beating and length regulation, including reduced ciliary IFT and/or axonemal dynein motor activity. LR defects result from mutations in genes encoding microtubule motor proteins involved in either ciliogenesis, such as Kif3a (18, 19), Kif3b (12) and Tg737/Polaris (IFT88) (20), or ciliary motility, such as LR dynein (Lrd) (21). Blocking IFT inhibits tubulin incorporation at the tip of cilia and results in cilia shortening (14, 22). MO-mediated knockdowns of either the ift88 or ift57 genes perturb normal LR asymmetry and ciliary length in multiple organs (including KV and the pronephric duct) without altering normal ciliary 9 + 2 organization (6, 23). Thus, the phenotypes associated with IFT defects correlated with our observations in the ipk1MO1 embryos.
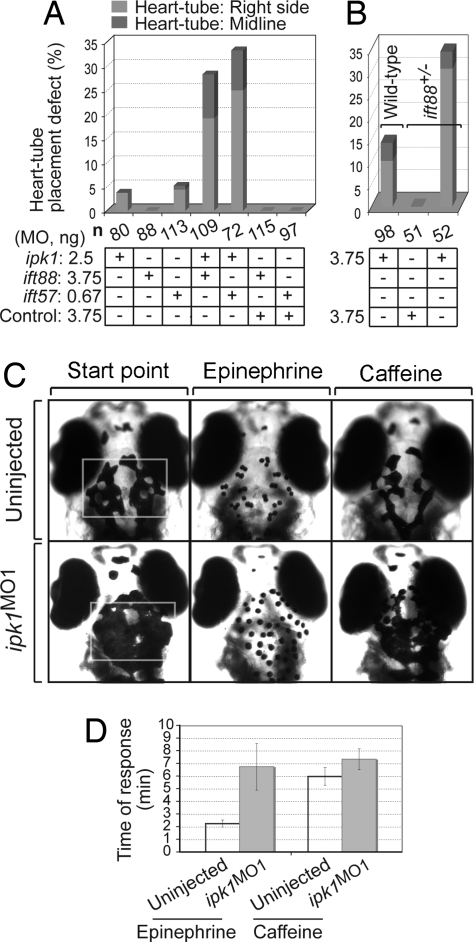
To test for connections between Ipk1 function and IFT, we examined the effect of codepleting Ipk1 and IFT proteins. If Ipk1 function and IFT are linked, combining suboptimal levels of the ipk1MO1 and ift88MO or ift57MO should result in synergistic perturbations on LR asymmetry establishment. First, we determined the maximal concentrations of ipk1MO1, ift88MO, and ift57MO that individually had negligible effects on LR asymmetry (SI Fig. 7). Interestingly, combining the suboptimal concentration of ipk1MO1 with that for ift88MO or ift57MO resulted in a highly significant percentage of randomized heart laterality (Fig. 3A). In contrast, coinjecting the control MO with ift88MO or ift57MO did not perturb normal heart laterality (Fig. 3A). We also tested the effect of injecting suboptimal concentrations of ipk1MO1 into embryos obtained from a cross of heterozygous ift88 mutant fish (previously known as polaris or oval) (24). The ift88+/− embryos injected with ipk1MO1 had a significantly greater percentage of laterality defects than either the ift88+/− embryos injected with the control MO or wild-type embryos injected with ipk1MO1 (Fig. 3B). These findings suggest that Ipk1 and IFT88/IFT57 act on the same or parallel pathways underling LR asymmetry establishment.
Fig. 3.
Ipk1 links to IFT and microtubule-mediated organelle transport. (A and B) Codepletion of Ipk1 and either IFT88 or IFT57 synergistically perturbs LR asymmetry. (A) Embryos were injected with varying amounts of either a single MO (ipk1MO1, ift88MO, ift57MO, control MO) or a MO combination (ipk1MO1+ift88MO, ipk1MO1+ift57MO, control MO+ift88MO, control MO+ift57MO). (B) Embryos derived from crossing either heterozygous ift88+/− mutant or wild-type fish were injected with ipk1MO1 or control MO. Scores for heart-tube placement defect in embryos at 28 hpf are graphed. (C and D) ipk1 knockdown perturbs microtubule-mediated organelle transport. (C) Five days after fertilization embryos [uninjected and ipk1MO1-injected (Left)] were treated with 0.5 mg/ml epinephrine, and time (min) required for all melanosomes in the head and trunk (within the white rectangle) to become perinuclear (Center) was determined. Time required for retracted melanosomes in epinephrine-treated endpoint embryos to fully disperse on exposure to 1 mg/ml caffeine (Right) was also determined. (D) Graph of the response time for epinephrine and caffeine treatments in uninjected and ipk1MO1 embryos.
ipk1 Knockdown Perturbs Microtubule-Dependent Cellular Transport.
To test whether Ipk1 plays a role in general microtubule-based motor protein transport, we used a model system for monitoring organelle dynamics and assayed microtubule-dependent transport of melanosomes in zebrafish larvae (25). Melanosomes are pigment-containing vesicles that travel bidirectionally along microtubule tracks. Their anterograde movement (toward microtubule plus-ends with dispersion to the cell periphery) is accomplished by proteins of the kinesin superfamily, with their retrograde movement (toward microtubule minus-ends and retraction to the cell center) achieved by dynein and dynein-associated proteins (25). Retraction of melanosomes to the cell center is stimulated by epinephrine, whereas melanosome dispersion to the cell periphery is induced by caffeine treatment (26, 27).
We measured the rates of melanosome retraction and dispersion in wild-type and ipk1MO1 embryos. The epinephrine-induced retraction was 3-fold delayed in ipk1MO1 embryos (Fig. 3 C and D and SI Table 2), reflecting perturbation of dynein-dependent retrograde transport. Kinesin-2 dependent anterograde transport was less altered, with a modest 1.2-fold delay in caffeine-induced dispersion (Fig. 3 C and D and SI Table 2). We conclude that Ipk1 is also required for microtubule-dependent organelle transport. This mechanism might be independent of the Ipk1 role in ciliary function; however, other proteins with roles in both ciliary IFT and such organelle transport have been reported (see below) (26).
Ipk1 Is Enriched in Centrosomes and Basal Bodies.
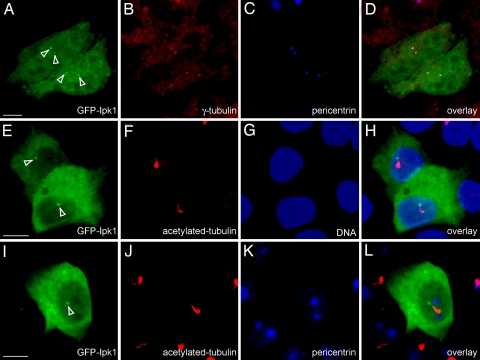
We next examined the cellular distribution of an ectopically expressed GFP-tagged Ipk1 in different model cell-culture systems. In HeLa cells, GFP-Ipk1 was ubiquitously distributed throughout the cell and enriched in distinct perinuclear foci (Fig. 4A). Colocalization experiments showed that GFP-Ipk1 foci overlapped with the staining for two established centrosomal proteins, γ-tubulin and pericentrin (28, 29) (Fig. 4 A–D).
Fig. 4.
GFP-Ipk1 is enriched in centrosomes and at basal bodies of cilia. Localization of GFP-Ipk1 in HeLa (A–D) and LLC-PK1 Cl4 (E–L) cells is shown. Cells transfected with pEGFPc1-Ipk1 were fixed in 3% paraformaldehyde and imaged for GFP fluorescence. Coimmunostaining with anti-γ-tubulin (B) and anti-pericentrin (C and K) antibodies detected centrosomes. Anti-acetylated-tubulin antibodies detected cilia (F and J), and TO-PRO3 staining was for nuclear DNA (G). Overlays show colocalization of GFP Ipk1 with γ-tubulin/pericentrin in HeLa cells (D), and with pericentrin and/or at the base of the cilium in LLC-PK1-Cl4 cells (H and L). White arrowheads in A, E, and I indicate GFP-Ipk1 foci. (Scale bars, 5 μm.)
In two ciliated cell types (LLC-PK1-C14 and MDCK cells), the GFP-Ipk1 signal was enriched at the cilium base, with the foci localized at only one end of the ciliary axoneme (as detected with acetylated tubulin) (Fig. 4 E–H and SI Fig. 8). The GFP-Ipk1 did not colocalize in the ciliary axoneme itself. Instead, GFP-Ipk1 localization signal overlapped with that of antipericentrin (Fig. 4 I–L). We also examined GFP-tagged human Ipk1 and found that the localization was identical to that for the zebrafish GFP-Ipk1 (data not shown). We concluded that GFP-Ipk1 is concentrated in both centrosomes and the ciliary basal body structure.
To examine whether Ipk1 has a centrosomal structural role, we monitored the localization of the bona fide centrosomal component γ-tubulin (29) in Ipk1-depleted embryos (12 hpf). We did not observe any difference in the centrosomal localization of γ-tubulin between ipk1MO1 and control MO-injected embryos (SI Fig. 9). The difference between the mean fluorescence of the anti-γ-tubulin signals from these two sets of embryos was statistically insignificant (P = 0.7835; SI Fig. 9). This suggests that Ipk1 depletion does not grossly perturb centrosomal structure/organization.
Conclusions and Implications
Here, we document defective ciliary beating and length maintenance in the KV of ipk1MO1 embryos. We also find that GFP-Ipk1 localizes to the base of the cilium at basal bodies, and codepletion of Ipk1 and IFT proteins synergistically perturbs LR asymmetry. This defines a previously uncharacterized role for Ipk1 in ciliary function. There are several potential mechanisms by which Ipk1 could influence ciliary function. We speculate that Ipk1 regulates factors directly related to ciliary motor function in the KV cells. Because Ipk1 knockdown results in perturbed IP production (4), we conclude that the Ipk1 kinase activity is required in this mechanism. This is further supported by our results showing that the kinase-dead ipk1 mRNA did not complement the ipk1MO1 ciliary beating defect. It is also possible that the Ipk1 protein itself is independently necessary. Because GFP-Ipk1 is not enriched in the axoneme, it is likely not an IFT or structural axonemal protein. However, soluble IPs might access the axoneme. IPs and IP6 itself are known to directly modulate protein activity through IP binding (3, 30–32). We predict that Ipk1, IP6, and/or soluble IPs contribute to IFT by acting as a cofactor for an IFT, cilia, or basal body protein and allowing for their optimal activity.
There is some evidence that centrosomes contain specific RNAs (33). Ipk1 was first discovered in a Saccharomyces cerevisiae genetic screen for factors required in mRNA export and nuclear pore complex (NPC) function (34). The NPC is a large macromolecular assembly embedded in the nuclear envelope (35) of which multiple protein subunits share structural folds in common with proteins of clathrin-coated vesicles (36–38). Intriguingly, proteins of the IFT complex are also homologous to these same coated vesicle components (38, 39). Thus, Ipk1 and IP6 production might play similar roles at both the NPC and in cilia. At the NPC, IP6 stimulates the RNA-dependent ATPase activity of the DEAD-box protein Dbp5 in a Gle1-dependent manner (30, 32). This activation of Dbp5 is required for remodeling of the RNA-binding protein composition of exported mRNA-particles (40). Our localization of GFP-Ipk1 to centrosomes and basal bodies makes it tantalizing to consider that IP6 production by Ipk1 might regulate a putative centrosomal RNA component. Identifying the Ipk1 or IP target in basal bodies or cilia is a future goal.
Localization of GFP-Ipk1 to the basal body suggests it either participates in ciliogenesis or mediates communication between the cilium and cell interior. A role for centrosomal proteins in ciliogenesis has been reported (41). Of note, aside from shorter length and lack of KV ciliary beating, cilia develop normally in the ipk1MO1 embryos, with their overall density and abundance not affected. It is possible that Ipk1 regulates initial cilia assembly but that either the maternal Ipk1 transmission or a hypomorphic MO-mediated gene knockdown provides sufficient Ipk1 for initial cilia assembly but not for ciliary function.
The lack of KV ciliary beating in the ipk1MO1 embryos correlates directly with our prior studies showing the lack of left-biased Ca2+ flux at the KV in the ipk1MO1 embryos (4). To date, all reports of nonbeating nodal cilia mutants show defects in LR asymmetry establishment (reviewed in ref. 5). In addition to regulating ciliary beating, Ipk1 could independently contribute to Ca2+ signaling and cell–cell communication across multicellular fields in the embryo. Interestingly, ipk1 expression is not detected in the KV, but only in cells surrounding the KV (4). The ipk1 expression levels in the KV could be below the detection limit of in situ hybridization assays, and Ipk1 is present in the KV cells. Alternatively, if Ipk1 is only in the surrounding cells, this indicates potential nonautonomous regulation of KV ciliary function.
Recent studies have shown that the knockdowns of zebrafish Bardet–Biedl syndrome (BBS) genes disrupt LR asymmetry, KV formation and ciliary maintenance as well as retrograde transport of melanosomes (26). These BBS phenotypes have clear parallels with the ipk1 knockdown phenotype. Interestingly, GFP-Ipk1 localization at centrosomes and basal bodies is also similar to that reported for the BBS4 and BBS8 proteins (42). We propose that the functions of Ipk1 and of the protein(s) encoded by the BBS gene(s) might be linked to a common mechanism in ciliary function. A wealth of prior studies suggest that the pathologies observed in LR asymmetry defects and polycystic kidney disease are linked to perturbations in the formation or function of cilia (11, 14). Our findings connecting Ipk1 and soluble IP production to such an important cellular organelle might facilitate our understanding of the mechanisms underlying ciliary function and cell signaling, and impact therapeutics for these diseases.
Methods
Morpholino Antisense Oligonucleotides and Complementing mRNA.
One-cell-stage embryos were injected with either 7.5 ng or 10 ng of the published ipk1MO1 (5′-GTCCATTTTATCCAGTTCCATAACC-3′), or a control antisense MO with five mismatches (5′-GTGCAATTTATGCACTTGCATAACC-3′) (4). Published MOs (ift88MO: CTGGGACAAGATGCACATTCTCCAT and ift57MO: CCTCCGCCATCCCTCTCTCTTTCT) were used to block translation of polaris/ift88 and hippi/ift57 transcripts (6). MOs were purchased from Gene Tools (ipk1MO1, control MO), or Open Biosystems (ift88MO, ift57MO). Synthetic capped mRNA encoding wild-type Ipk1 was prepared from linearized ipk1/pCDNA3 plasmid (pSW3012) harboring silent mutations (C to T and G to A at nucleotide positions 7 and 9, respectively, of the ORF) in the ipk1MO1-binding site. For mRNA encoding kinase-dead (kd)-Ipk1, site-directed mutagenesis of pSW3012 changed the sequence encoding the Cys at position 168 to Tyr (pSW3369). mRNA was generated by using a mMassage mMachine kit (Ambion) and purified with a Sephadex G50 (fine) spin column (Roche). Embryos at the one-cell stage were coinjected with 7.5 ng of MO (ipk1MO1 or control MO) and 500 pg of synthetic mRNA.
Zebrafish Immunohistochemistry and Whole-Mount in Situ Hybridization.
Embryos were fixed overnight at 4°C in 4% paraformaldehyde, 0.15 mM CaCl2, 4% sucrose in 0.1 M phosphate buffer (pH 7.3) and treated with acetone at −20°C for 7 min, and indirect immunofluorescence was performed with 1:200 anti-acetylated tubulin monoclonal antibody (Sigma) or 1:250 monoclonal anti-γ-tubulin antibody (Sigma) and 1:500 Alexa Fluor 568 goat anti-mouse conjugate (Molecular Probes). Images were obtained by using a Zeiss LSM510 Meta laser scanning confocal microscope. Fluorescence intensities of the anti-γ-tubulin signals were measured by using Volocity software (Improvision). Whole-mount in situ hybridizations were performed as described (43). Images were obtained by using a Retiga 1300 CCD camera (Qimaging) mounted on a stereomicroscope (Leica MZFLIII) and OPENLAB software.
Ciliary Length Measurement and Videomicroscopy.
Ciliary length measurements were made with Volocity software. Base and ends of immunolabeled individual cilia were marked in the confocal z-stack images and outlined, and the calculated length was recorded. For visualizing KV ciliary beating, live embryos (12 hpf) were removed from their chorions, mounted in SeaPlaque low-melting agarose (Biowhittaker Molecular Applications) (1.0% in embryo medium) in microwells of glass-bottom culture dishes (MatTek), and covered with embryo medium. Movies were acquired for 30 sec by using OPENLAB software (Improvision) at 55 frames per second with a 63× DIC objective on a Zeiss Axiovert 200 inverted fluorescence microscope equipped with a Retiga Exi Fast camera (Qimaging). The movies were played frame by frame to determine the number of frames (n) for a complete ciliary beat cycle, and frequency was calculated (1/n × 0.018 Hz). Kymographs were obtained by drawing a line across a ciliary trajectory by using ImageJ software (National Institutes of Health, developed by W. Rasband) and MultipleKymograph plugin (developed by J. Rietdorf and A. Seitz).
Electron Microscopy.
Zebrafish embryos were fixed in 4% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, postfixed in 1% OsO4 in cacodylate buffer, dehydrated through an ethanol series, equilibrated in propylene oxide, and then embedded in epoxy resin. Thin sections were stained with uranyl acetate and lead citrate and examined in a Hitachi H-800 electron microscope.
Melanosome Transport Assay.
Zebrafish day 5 larvae were dark-adapted for maximal melanophore dispersion, and the assay was performed in embryo medium as described (26). Larvae were exposed to epinephrine (0.5 mg/ml) for melanosome retraction, washed with excess embryo medium, and then treated with caffeine (1 mg/ml, pH 7.0).
Cell Culture and Immunofluorescence.
HeLa, LLC-PK1 Cl4, and MDCK cells were grown to 70–80% confluency on 22-mm-diameter glass coverslips in DMEM with 10% FBS (GIBCO) and 2 mM glutamine without antibiotics. A sequence encoding zebrafish ipk1 was cloned in pEGFPc1 and pEGFPn3 (Clontech) for amino (GFP-Ipk1) (pSW3235) and carboxyl-terminal (Ipk1-GFP) (pSW3236) fusions. Both fusion proteins localized similarly, with GFP-Ipk1 used in all studies shown. Sequence encoding the gene for human Ipk1 was cloned into the pEGFPc1 vector for GFP-hIpk1 (pSW3351). Plasmids were transfected by using Lipofectamine 2000 (Invitrogen), and cells were grown for 24–36 h, fixed for 30 min in 3% paraformaldehyde at room temperature (RT), and analyzed for direct GFP fluorescence. Fixed cells were permeabilized with PBS/0.2% Triton X-100 for 10 min, blocked in 3% milk for 10 min, incubated for 1 h with primary antibody (diluted in PBS/3% milk) at RT and then for 1 h with secondary antibody [Alexa Fluor-conjugated secondary antibodies (1:500; Invitrogen) in PBS/3% milk]. Primary antibodies included mouse monoclonal anti-acetylated tubulin antibody (1:200; Sigma), rabbit polyclonal anti-pericentrin antibody (1:500; Abcam), and mouse monoclonal anti-γ tubulin antibody (1:250; Sigma). TO-PRO-3 (Invitrogen) was used for nuclear counterstaining.
Supplementary Material
Acknowledgments
We thank M. J. Tyska for critical input and the LLC-PK1 Cl4 cells, J. R. Goldenring for the MDCK cells, I. N. Kaverina and B. J. Cha for primary antibodies, and L. Solnica-Krezel, the B.A. and S.R.W. laboratory members for discussions. Confocal microscopy equipment was made available by the Vanderbilt University Medical Center Cell Imaging Core Resource, supported by National Institutes of Health (NIH) Grants 1S10RR15682, CA68485, and DK20593. This work was supported by the American Heart Association Postdoctoral Fellowship Grant (to B.S.), NIH Grants NS46668 (to B.A.) and GM51912 (to S.R.W.), and the Vanderbilt University Academic Venture Capital Fund and a Vanderbilt University Zebrafish Pilot Project Grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706934104/DC1.
References
- 1.Berridge MJ. Nature. 1993;361:315–325. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 2.Irvine RF. Nat Rev Mol Cell Biol. 2003;4:586–590. doi: 10.1038/nrm1152. [DOI] [PubMed] [Google Scholar]
- 3.York JD. Biochim Biophys Acta. 2006;1761:552–559. doi: 10.1016/j.bbalip.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Sarmah B, Latimer AJ, Appel B, Wente SR. Dev Cell. 2005;9:133–145. doi: 10.1016/j.devcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Hirokawa N, Tanaka Y, Okada Y, Takeda S. Cell. 2006;125:33–45. doi: 10.1016/j.cell.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Kramer-Zucker AG, Olale F, Haycraft CJ, Yoder BK, Schier AF, Drummond IA. Development (Cambridge, UK) 2005;132:1907–1921. doi: 10.1242/dev.01772. [DOI] [PubMed] [Google Scholar]
- 7.Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Development (Cambridge, UK) 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- 8.McGrath J, Somlo S, Makova S, Tian X, Brueckner M. Cell. 2003;114:61–73. doi: 10.1016/s0092-8674(03)00511-7. [DOI] [PubMed] [Google Scholar]
- 9.Scholey JM. Annu Rev Cell Dev Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- 10.Praetorius HA, Spring KR. Annu Rev Physiol. 2005;67:515–529. doi: 10.1146/annurev.physiol.67.040403.101353. [DOI] [PubMed] [Google Scholar]
- 11.Ibanez-Tallon I, Heintz N, Omran H. Hum Mol Genet. 2003;12:R27–R35. doi: 10.1093/hmg/ddg061. [DOI] [PubMed] [Google Scholar]
- 12.Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 13.Caspary T, Larkins CE, Anderson KV. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Rosenbaum JL, Witman GB. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 15.Oxtoby E, Jowett T. Nucleic Acids Res. 1993;21:1087–1095. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Grunwald DJ, Riggleman B. Development (Cambridge, UK) 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- 17.Smithers L, Haddon C, Jiang YJ, Lewis J. Mech Dev. 2000;90:119–123. doi: 10.1016/s0925-4773(99)00231-2. [DOI] [PubMed] [Google Scholar]
- 18.Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LS. Proc Natl Acad Sci USA. 1999;96:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeda S, Yonekawa Y, Tanaka Y, Okada Y, Nonaka S, Hirokawa N. J Cell Biol. 1999;145:825–836. doi: 10.1083/jcb.145.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murcia NS, Richards WG, Yoder BK, Mucenski ML, Dunlap JR, Woychik RP. Development (Cambridge, UK) 2000;127:2347–2355. doi: 10.1242/dev.127.11.2347. [DOI] [PubMed] [Google Scholar]
- 21.Supp DM, Brueckner M, Kuehn MR, Witte DP, Lowe LA, McGrath J, Corrales J, Potter SS. Development (Cambridge, UK) 1999;126:5495–5504. doi: 10.1242/dev.126.23.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall WF, Rosenbaum JL. J Cell Biol. 2001;155:405–414. doi: 10.1083/jcb.200106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bisgrove BW, Snarr BS, Emrazian A, Yost HJ. Dev Biol. 2005;287:274–288. doi: 10.1016/j.ydbio.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 24.Tsujikawa M, Malicki J. Neuron. 2004;42:703–716. doi: 10.1016/s0896-6273(04)00268-5. [DOI] [PubMed] [Google Scholar]
- 25.Marks MS, Seabra MC. Nat Rev Mol Cell Biol. 2001;2:738–748. doi: 10.1038/35096009. [DOI] [PubMed] [Google Scholar]
- 26.Yen HJ, Tayeh MK, Mullins RF, Stone EM, Sheffield VC, Slusarski DC. Hum Mol Genet. 2006;15:667–677. doi: 10.1093/hmg/ddi468. [DOI] [PubMed] [Google Scholar]
- 27.Nascimento AA, Roland JT, Gelfand VI. Annu Rev Cell Dev Biol. 2003;19:469–491. doi: 10.1146/annurev.cellbio.19.111401.092937. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerman WC, Sillibourne J, Rosa J, Doxsey SJ. Mol Biol Cell. 2004;15:3642–3657. doi: 10.1091/mbc.E03-11-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moudjou M, Bordes N, Paintrand M, Bornens M. J Cell Sci. 1996;109:875–887. doi: 10.1242/jcs.109.4.875. [DOI] [PubMed] [Google Scholar]
- 30.Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. Nat Cell Biol. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- 31.Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. Science. 2005;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Nat Cell Biol. 2006;8:711–716. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- 33.Alliegro MC, Alliegro MA, Palazzo RE. Proc Natl Acad Sci USA. 2006;103:9034–9038. doi: 10.1073/pnas.0602859103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.York JD, Odom AR, Murphy R, Ives EB, Wente SR. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- 35.Tran EJ, Wente SR. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 36.Mans BJ, Anantharaman V, Aravind L, Koonin EV. Cell Cycle. 2004;3:1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- 37.Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A. Proc Natl Acad Sci USA. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jekely G, Arendt D. BioEssays. 2006;28:191–198. doi: 10.1002/bies.20369. [DOI] [PubMed] [Google Scholar]
- 40.Tran EJ, Zhou Y, Corbett AH, Wente SR. Mol Cell. 2007 doi: 10.1016/j.molcel.2007.09.019. in press. [DOI] [PubMed] [Google Scholar]
- 41.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 42.Ansley SJ, Badano JL, Blacque OE, Hill J, Hoskins BE, Leitch CC, Kim JC, Ross AJ, Eichers ER, Teslovich TM, et al. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- 43.Hauptmann G, Gerster T. Methods Mol Biol. 2000;137:139–148. doi: 10.1385/1-59259-066-7:139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.