Abstract
The osteogenic Runt-related (Runx2) transcription factor negatively regulates proliferation and ribosomal gene expression in normal diploid osteoblasts, but is up-regulated in metastatic breast and prostate cancer cells. Thus, Runx2 may function as a tumor suppressor or an oncogene depending on the cellular context. Here we show that Runx2-deficient primary osteoblasts fail to undergo senescence as indicated by the absence of β-gal activity and p16INK4a tumor suppressor expression. Primary Runx2-null osteoblasts have a growth advantage and exhibit loss of p21WAF1/CIP1 and p19ARF expression. Reintroduction of WT Runx2, but not a subnuclear targeting-defective mutant, induces both p21WAF/CIP1 and p19ARF mRNA and protein resulting in cell-cycle inhibition. Accumulation of spontaneous phospho-H2A.X foci, loss of telomere integrity and the Mre11/Rad50/Nbs1 DNA repair complex, and a delayed DNA repair response all indicate that Runx2 deficiency leads to genomic instability. We propose that Runx2 functions as a tumor suppressor in primary diploid osteoblasts and that subnuclear targeting contributes to Runx2-mediated tumor suppression.
Keywords: cancer, genomic instability, growth control, nuclear organization, osteoblast
When exposed to inappropriate growth signals, mammalian cells engage in elaborate mechanisms that provide alternative fates to prevent tumorigenesis. These defenses include apoptosis and senescence, which guard against unrestrained proliferation. Several tumor suppressors, including the retinoblastoma protein (pRb), p53, and the products of the INK4b–ARF–INK4a locus (p15–p19–p16), control telomere integrity, cellular senescence, and genomic stability (1–4). Tumors arise when cells override these intrinsic defenses. Cancer cells typically exhibit independence from extrinsic growth signals, lack of senescence or apoptosis, compromised telomeric integrity, and genomic instability, as well as changes in nuclear organization (2, 4–6). However, mice with genetic ablations of tumor suppressors are prone to tumorigenesis that is often lineage-restricted (7, 8). These observations suggest that lineage-specific mechanisms contribute to tumorigenesis.
Runt-related (Runx) transcription factors determine cell fate and regulate lineage-specific proliferation and differentiation (9–11). Genetic studies reveal that Runx1 is essential for definitive hematopoiesis, Runx2 is required for osteogenesis, and Runx3 is involved in gut development as well as neurogenesis (12–16). Runx proteins support epigenetic regulation at mitosis and control ribosomal gene expression during the cell cycle (9–11, 17–19). Runx transcription factors are nuclear-scaffolding proteins that integrate signaling pathways by organizing macromolecular complexes in nuclear microenvironments and facilitating chromatin remodeling (20). Mice with gene replacements expressing subnuclear targeting defective mutants (mSTDs) of Runx1 or Runx2 from the native loci exhibit phenotypes identical to mice in which the gene has been ablated (21, 22), suggesting that the biological activity of Runx proteins requires fidelity of subnuclear targeting.
Runx proteins are mutated, deleted, silenced, or ectopically expressed in a variety of solid tumors and leukemias (23–25). The Runx1 gene is a frequent target of chromosomal translocations in acute myeloid leukemia (AML) patients (24), whereas Runx3 is often deleted or silenced in gastric cancer cells (15, 25). These findings strongly suggest that Runx1 and Runx3 function as tumor suppressors. In contrast, Runx2 up-regulation in breast and prostate cancer cells, which metastasize to bone, correlates with their metastatic potential and coincides with higher expression of metastatic gene markers such as MMP9 (26–28). Furthermore, Runx2 up-regulation does not impede cell-cycle progression in cancer cells. In contrast, in normal osteoblasts, Runx2 ablation accelerates proliferation and increases ribosomal gene expression (18, 19, 29, 30). Together these observations indicate that Runx2 may function as a tumor suppressor in some cell types and have oncogenic potential in others. In this study, we show that Runx2 deficiency and defective subnuclear targeting in primary osteoblasts bypass senescence to promote immortalization and tumorigenic phenotype.
Results
Absence of Senescence in Runx2-Deficient Osteoblasts.
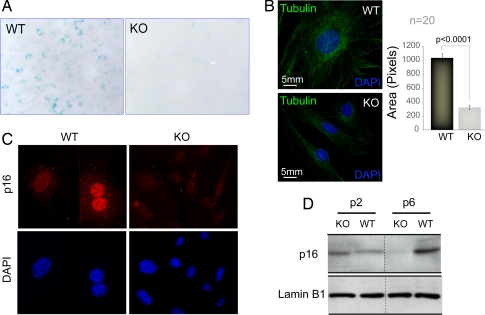
Cellular senescence is a physiological defense mechanism against unrestrained cell proliferation. As Runx2-null osteoblasts hyperproliferate, we investigated whether these cells exhibit disruption of pathways leading to senescence. Primary osteoblasts derived from calvaria of WT and Runx2-deficient mice at embryonic day 17.5 (E17.5) were initially examined for competency for senescence. Cells were cultured ex vivo following the standard 3T3 protocol and were stained for the activity of senescence-associated β-gal (SA β-gal) at passage 5. WT osteoblasts showed strong SA β-gal staining, whereas the null osteoblasts were essentially negative (Fig. 1A). Furthermore, WT osteoblasts are three to four times larger than null osteoblasts, another hallmark of senescence (Fig. 1B). We confirmed the loss of senescence in null osteoblasts by showing that, although the p16INK4a tumor suppressor is expressed at comparable levels in both cell types at passage 3, null osteoblasts lose the expression of p16 as a function of cell passage in culture (Fig. 1 C and D). Taken together, our findings show that disruption of the Runx2 gene in diploid osteoblasts results in a loss of cellular senescence.
Fig. 1.
Absence of senescence in Runx2-deficient osteoblasts. Primary calvarial osteoblasts isolated from WT and Runx2-deficient KO mice were cultured ex vivo according to the standard 3T3 protocol for cell passage. (A) WT and KO osteoblasts grown at passage 5 were stained for the activity of SA β-gal. WT osteoblasts showed significantly higher SA β-gal activity (blue), compared with KO osteoblasts. (B) At passage 5, both WT and KO osteoblasts were plated on gelatin-coated coverslips and immunostained for microtubules (tubulin, green) to assess the size of the cells 24 h after plating. Cells also were counterstained with DAPI to visualize the nucleus. The size of 20 WT and 20 KO cells was measured by using the image analysis software MetaMorph. The bar graph represents the average size of 20 WT and 20 KO cells in pixels. (C) WT and KO osteoblasts grown at passage 5 also were immunostained for p16INK4a, a tumor suppressor and marker for cellular senescence. The expression of p16INK4a was significantly reduced in KO osteoblasts. The results were further supported by Western blot analysis of cell lysates isolated from WT and KO osteoblasts at various passages. (D) The p16INK4a expression was gradually decreased in higher passage KO osteoblasts, whereas WT cells showed a high expression of p16INK4a.
Defective Subnuclear Targeting of Runx2 Prevents p21WAF1/CIP1-Mediated Cell-Cycle Inhibition in Primary Osteoblasts.
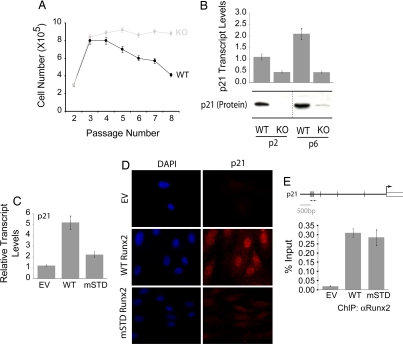
We investigated the mechanism by which loss of Runx2 provides a growth advantage to primary osteoblasts. Cell-cycle profiles of WT and null osteoblasts were indistinguishable at passage 3 (data not shown). However, WT osteoblasts showed a significantly decreased proliferative rate, compared with the null osteoblasts, when cultured ex vivo through eight passages (Fig. 2A). Because the cyclin-dependent kinase (CDK) inhibitor p21 is a direct target of Runx2 and attenuates cell-cycle progression in immature osteoblast progenitors (31), we monitored its expression in WT and null osteoblasts. Levels of p21 are significantly decreased in null osteoblasts at passages 2 and 6 (Fig. 2B). These results suggest that loss of p21 expression in the absence of Runx2 may be mechanistically related to escape from senescence. We next examined whether the subnuclear targeting function of Runx2 is required for its senescence-related activity. Reintroduction of WT Runx2, but not an mSTD that is incapable of targeting Runx2 to nuclear microenvironments (32), restores both transcript and protein levels of p21 (Fig. 2 C and D). However, the p21 gene promoter is occupied in vivo by both the WT and mSTD proteins (Fig. 2E). These results demonstrate that Runx2 deficiency and defective subnuclear targeting contribute to the ex vivo growth advantage of null osteoblasts through a p21-dependent mechanism.
Fig. 2.
Runx2-deficient osteoblasts exhibit growth advantage ex vivo owing to the loss of p21WAF1/CIP1. (A) Primary diploid WT and KO osteoblasts were cultured ex vivo according to the standard 3T3 protocol. Cells were counted at each passage and replated at the initial plating density (i.e., 3 × 105). The line graph represents the number of cells at the time of each passage. WT osteoblasts showed a considerable decrease in their rate of proliferation as a function of increasing passage. (B) Total cellular RNA was isolated from WT and KO osteoblasts grown at the indicated passages and was subjected to quantitative RT-PCR to assess the expression of p21WAF1/CIP1, a CDK2 inhibitor. KO osteoblasts showed a significant decrease in the expression of the p21WAF1/CIP1 transcript. KO osteoblasts grown on passage 3 were infected with retroviruses carrying the empty vector (EV) or cDNAs for the WT or an mSTD of Runx2. Cells were harvested 48 h after infection for total cellular RNA isolation, ChIP, or immunostaining for p21WAF1/CIP1. (C) As shown, the WT Runx2 restored the expression of p21WAF1/CIP1 in KO osteoblasts, whereas the mSTD Runx2 failed to enhance the p21WAF1/CIP1 expression. (D) Immunostaining revealed that the p21WAF1/CIP1 protein levels also were restored by the WT protein but not the mSTD Runx2 protein. (E) Both the WT and mSTD Runx2 proteins occupied the p21WAF1/CIP1 promoter in vivo as assessed by ChIP assay.
Runx2-Deficient Primary Osteoblasts Show Hallmarks of Genomic Instability.
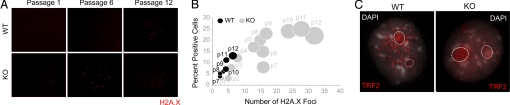
The loss of cellular senescence and concomitant growth advantage of Runx2-deficient cells may result in increased genomic instability to further promote immortalization. Therefore, we investigated whether null osteoblasts accumulate spontaneous DNA damage by immunostaining for phosphorylated histone H2A.X (γH2A.X) (Fig. 3A), a marker of double-strand DNA breaks. The number of γH2A.X foci per cell and the percentage of positive cells both increase as a function of cell passage in null osteoblasts (Fig. 3B). In contrast, WT osteoblasts exhibit lesser accumulation of γH2A.X foci and only at later passages. Telomere repeat factor-2 (TRF2) is a measure of telomeric integrity; loss of TRF2 is a key indicator of cellular immortalization and tumorigenic potential. Runx2-null osteoblasts show an absence of the punctate TRF2 staining present in WT cells (Fig. 3C), but cellular TERT-mediated telomerase activity is unaltered (data not shown). Together these findings show that genomic stability is compromised and suggest that DNA repair pathways are impaired in Runx2-deficient diploid osteoblasts.
Fig. 3.
Runx2-deficient osteoblasts exhibit hallmarks of genomic instability. (A) WT and KO osteoblasts grown at the indicated passages were subjected to immunofluorescence. Spontaneous DNA damage was assessed by staining cells with an antibody against the phosphorylated form of histone variant H2A.X (γH2A.X). (B) The increased incidence of spontaneous DNA damage in KO osteoblasts is quantitatively represented in a bubble graph. One hundred cells were counted for each of WT and KO osteoblasts. Cells that showed γH2A.X staining were scored as positive and are represented in the graph as percentage of positive cells (the y axis). In each positive cell, the number of γH2A.X foci also was counted and is represented on the x axis. Thus, the size of each bubble reflects the number of positive cells at each passage, as well as the number of γH2A.X foci in each positive cell. (C) WT and KO osteoblasts grown on gelatin-coated coverslips at passage 5 were immunostained for TRF2, a regulatory protein required for the maintenance and integrity of telomeric ends. WT osteoblasts exhibited the expected punctate staining of TRF2, whereas KO cells showed diffused TRF2 localization, indicating a loss of telomeric integrity. TRF2 is a known regulator of ribosomal gene expression and also is localized to nucleoli (demarcated here by dotted line circles).
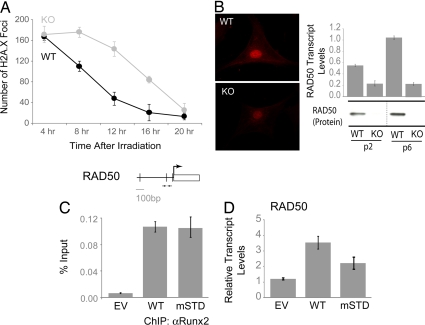
To investigate mechanisms contributing to the accumulation of spontaneous DNA damage in Runx2-null osteoblasts, we compared the DNA damage response of WT and null osteoblasts after a sublethal dose (5 Gy) of ionizing irradiation. Cells were immunostained for γH2A.X and counted (n = 50) at various time points after irradiation. Null osteoblasts display a pronounced delay in DNA repair (Fig. 4A). A comparison of WT and null osteoblasts for the expression of components of the Mre11/Rad50/Nbs1 (MRN) DNA repair complex shows that Rad50 expression is diminished in null osteoblasts at both the transcript (bar graphs) and protein (immunofluorescence and Western blots) levels (Fig. 4B). Reintroduction of either WT or mSTD Runx2 into null osteoblasts shows that both Runx proteins occupy the Rad50 promoter in vivo (Fig. 4C), although only the WT Runx2 restores expression of Rad50 (Fig. 4D). Accumulation of spontaneous DNA damage, absence of Rad50, and delayed DNA repair demonstrate that null osteoblasts exhibit intrinsic genomic instability. We propose that the enhanced propensity of Runx2-null osteoblasts to acquire chromosomal abnormalities may increase the potential to develop an immortalized phenotype ex vivo.
Fig. 4.
Runx2-deficient osteoblasts lack components of the MRN DNA repair complex and exhibit delayed DNA repair response. (A) WT and KO osteoblasts grown on coverslips at passage 3 were irradiated with 5 Gy ionizing radiation (IR). Cells were then subjected to immunofluorescence at the indicated time points after the IR. The appearance of γH2A.X foci was used as an assessment of DNA damage. The γH2A.X foci were counted in 50 WT and 50 KO osteoblasts. (B) Expression of Rad50, an integral component of the MRN complex involved in DNA damage response, was assessed in the WT and KO osteoblasts by immunofluorescence (Left) and qualitative RT-PCR (Right). KO osteoblasts grown at passage 3 were infected with retroviruses carrying the EV or cDNAs for the WT or a mSTD of Runx2. Cells were harvested 48 h after infection for total cellular RNA isolation and ChIP. (C) ChIP assay revealed that both WT and mSTD Runx2 proteins occupied Rad50 promoter in vivo. (D) However, only the WT protein was able to restore Rad50 transcript levels.
Runx2 Deficiency Results in Loss of the p19ARF Tumor Suppressor.
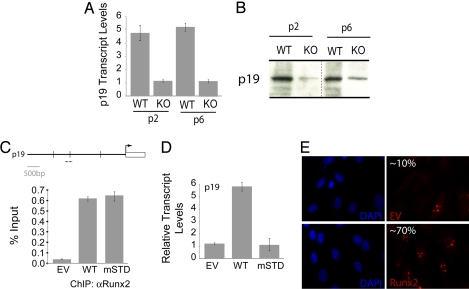
Senescence and genomic instability are controlled by the p53–p19ARF pathway. The p19ARF tumor suppressor is a known target of the hematopoietic factor Runx1 (33). Therefore, we directly addressed whether p19ARF is regulated by Runx2 in osteoblast progenitors. Although WT osteoblasts show robust expression, p19ARF expression is substantially decreased in Runx2-null osteoblasts at both the protein and mRNA levels (Fig. 5A). Forced expression of WT or mSTD Runx2 in Runx2-null osteoblasts shows that both proteins occupy the genomic p19ARF promoter in vivo as assessed by ChIP (Fig. 5C). However, only WT Runx2 restored p19ARF expression at mRNA and protein levels (Fig. 5 D and E). These data demonstrate that Runx2 directly regulates p19ARF tumor suppressor in a subnuclear targeting-dependent manner, and its deficiency may promote acquisition of tumorigenic properties.
Fig. 5.
Expression of p19ARF tumor suppressor is severely compromised in Runx2-null osteoblasts. (A) Total cellular RNA was isolated from WT and KO osteoblasts grown at the indicated passages and was subjected to quantitative RT-PCR to assess the expression of p19ARF, an effector of the p53 tumor suppressor pathway. KO osteoblasts showed a significant decrease in the expression of the p19ARF transcript. (B) Total cell lysates were resolved by gradient gel electrophoresis. Consistent with the p19ARF transcript levels, KO osteoblasts showed substantial down-regulation of p19ARF protein. KO osteoblasts grown on passage 3 were infected with adenoviruses carrying the EV or cDNAs for the WT or an mSTD of Runx2. (C–E) Cells were harvested 48 h after infection for ChIP (C), total cellular RNA isolation (D), or immunostaining (E) for p19ARF. (C) As shown, both the WT and mSTD Runx2 proteins occupied the p19ARF promoter in vivo as assessed by ChIP assay. (D) However, only WT Runx2 restored the expression of p19ARF in KO osteoblasts, whereas the mSTD Runx2 failed to enhance the p21WAF1/CIP1 expression. (E) In addition, immunostaining revealed that the p19ARF protein levels also were restored by the WT protein but not the mSTD Runx2 protein.
Discussion
In this study, we examined the biological function of Runx2 during osteoblast progenitor proliferation. Our results show that Runx2 directly controls the expression of key cell-cycle attenuators and checkpoint regulators, including p16INK4a, p21CIP1/WAF1, and p19ARF, whereas a subnuclear targeting-defective mutant of Runx2 does not. The combined loss of these principal factors upon ablation of Runx2 function sensitizes osteoprogenitors to mitogenic growth factor signaling, decreases options for cell-cycle arrest in response to genomic instability, and impairs activation of the proapoptotic p53 pathway. Concomitant with these molecular aberrations, Runx2 deficiency bypasses senescence, increases proliferative potential and immortalization, and causes impaired DNA damage responses and genomic instability. Taken together, our results provide converging evidence that Runx2 functions as a tumor suppressor in osteoblast progenitors.
Our findings that Runx2 has tumor suppressor properties in the osteogenic lineage are consistent with results reported for the other two Runx-related transcription factors, Runx1 and Runx3. Runx1/AML1 is frequently mutated or translocated in proliferative disorders of the hematopoietic lineage, including AML and myelodysplastic syndrome (24). Runx3 is deleted or silenced in cancers of the gastrointestinal tract, including gastric,colorectal, and pancreatic carcinomas (25). Thus, our data support the emerging concept that all three Runx proteins share inherent growth-suppressive and cancer-linked properties.
Notwithstanding the evident roles of Runx proteins in limiting cell proliferation, Runx2 also has been identified as an oncogene that cooperates with Myc in the etiology of T cell lymphomas (Til-1) (34). Furthermore, Runx2 is elevated and deregulated in osteosarcomas and is ectopically expressed in metastatic breast and prostate tumor cells (26–28). Although activation of Runx2 in cancer cells does not impede growth, it may reflect a gain of function that contributes to stages of tumorigenesis. We suggest that Runx2 biologically acts as a tumor suppressor or oncoprotein depending on cellular context, developmental stage, and/or tissue microenvironment.
The ability of Runx2 to influence major parameters of cell cycle and growth control may directly reflect its molecular activity as a scaffolding regulator that supports subnuclear integration of multiple proliferation-related signaling pathways (35, 36). Our findings demonstrate that subnuclear targeting of Runx2 is important for the expression of both p21CIP1/WAF1 and p19ARF, which mediate antiproliferative mechanisms in osteoblasts where Runx2 is endogenously expressed.
As we have observed for Runx2, loss of classical tumor suppressors (e.g., p53 and pRb) leads to increased proliferation, genomic instability, and acquisition of chromosomal abnormalities (7, 8). Runx2, p53, and pRb all control cell proliferation by regulating cell-cycle progression and mediating cell growth through effects on ribosomal biogenesis (18, 19, 30, 37, 38). Abrogation of tumor suppressor pathways controlled by p53 and pRb, which are ubiquitously expressed, results in the development of diverse tumors that are confined to specific tissues (7, 8), indicating that lineage-specific mechanisms are involved. Our results suggest that Runx2 belongs to an expanding class of proliferation regulators that are lineage-restricted and together may account for tissue-specificity of tumor formation.
Materials and Methods
Cell Culture.
Primary calvarial osteoblasts were isolated from mouse embryos (17.5 days postcoitum) of WT or Runx2-null mice. Cells were frozen at passage 2 in BamBanker freezing medium (Wako Chemical) for subsequent experiments. For experiments, cells were maintained in α-MEM containing 10% FCS (HyClon), penicillin, streptomycin, and l-glutamine.
Adenoviral Transduction of Cells.
Adenovirus expressing WT and mSTDs were generated by Adenovator system (BIOgene) and have been reported previously (39). Runx2-null cells were plated at a density of 1 × 105 cells per well of a six-well plate or 1 × 106 cells per 100-mm plate. Cultures at 70% confluence were infected with the Runx2 adenovirus in α-MEM without serum. After 90 min, cells were washed with serum-free α-MEM and fed with complete α-MEM until harvest.
SA β-Gal Staining.
SA β-gal staining was performed to detect the extent of senescent cells in the culture (40). Briefly, cells at 70% confluence were fixed in 2% formaldehyde/0.2% glutaraldehyde and incubated with staining solution [1 mg/ml X-gal, 5 mM K3[Fe(CN)6], 5 mM K4[Fe(CN)6]·3H2O, 2 mM MgCl2, 150 mM NaCl in 40 mM citric acid/sodium phosphate (pH 6.0)] for 4–6 h at 37°C. Cells were then observed under microscope and photographed.
In Situ Immunofluorescence.
WT and Runx2-null cells were plated at a density of 0.6 × 105 cells per well on gelatin-coated coverslips in six-well plates. Cells were processed in situ for whole cell, and digital microscopic analyses were carried out as described previously (41). Runx2, p21, p16, p19, and Rad50 were detected by rabbit polyclonal antibodies at a dilution of 1:200 (Santa Cruz Biotechnology) and tubulin with a mouse monoclonal antibody against β-tubulin (Sigma–Aldrich) at a dilution of 1:2,000. Secondary antibodies used were Alexa Fluor 488 anti-rabbit or Alexa Fluor 568 anti-mouse (Molecular Probes) at a dilution of 1:1,000. Slides were examined on a Zeiss Axioplan 2 microscope fitted with epifluorescence attached to a CCD camera. Images were saved and processed by using MetaMorph imaging software, version 6.1 (Universal Imaging).
Western Blot Analysis.
Whole-cell lysates from WT and Runx2-null cells infected with various adenoviruses were resolved by gradient gel electrophoresis, and proteins were transferred to an Immobilon (Millipore) membrane. The blots were blocked in PBS containing 5% nonfat dry milk for 1 h. Blots were incubated for 1 h at room temperature with 1:1,000 dilution of the primary antibody in PBS solution containing 1% milk. The blots were washed four times with the PBS-T solution and incubated for 1 h at room temperature with 1:5,000 dilution of appropriate horseradish peroxidase-conjugated goat secondary antibodies (Santa Cruz Biotechnology). After four washes with PBS-T solution, the immunoreactive bands were detected with ECL (Amersham Pharmacia Biotech) by exposing blots to XAR-5 film (Kodak).
RT-PCR and Quantitative Real-Time PCR.
First, 1 μg of total RNA was reverse-transcribed by using SuperScript first strand synthesis kit (Invitrogen) according to the manufacturer's protocol. Then, 1 μl of freshly reverse-transcribed cDNA and the fluorescent SYBR Green I dye was used to monitor DNA synthesis (SYBR Green PCR master mix; Applied Biosystems) in a real-time PCR assay.
ChIP Assay.
ChIP assays were performed as previously described (42). Briefly, formaldehyde cross-linking was performed for 10 min, and samples were sonicated to obtain DNA fragments with an average size of 400–500 bp. Protein–DNA complexes were immunoprecipitated by using Runx2 antibody (M-70; Santa Cruz Biotechnology). DNAs were purified and subjected to quantitative real-time PCR amplification. Graphs were represented as percentage of input.
Acknowledgments
This work was supported by National Institutes of Health Grants CA082834 and AR048818.
Footnotes
The authors declare no conflict of interest.
References
- 1.Gil J, Peters G. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 2.Stewart SA, Weinberg RA. Annu Rev Cell Dev Biol. 2006;22:531–557. doi: 10.1146/annurev.cellbio.22.010305.104518. [DOI] [PubMed] [Google Scholar]
- 3.Collado M, Blasco MA, Serrano M. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Sieber O, Heinimann K, Tomlinson I. Semin Cancer Biol. 2005;15:61–66. doi: 10.1016/j.semcancer.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Zink D, Fischer AH, Nickerson JA. Nat Rev Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- 6.Penman S. Proc Natl Acad Sci USA. 1995;92:5251–5257. doi: 10.1073/pnas.92.12.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Attardi LD, Donehower LA. Mutat Res. 2005;576:4–21. doi: 10.1016/j.mrfmmm.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 8.Macleod K. Curr Opin Genet Dev. 1999;9:31–39. doi: 10.1016/s0959-437x(99)80005-7. [DOI] [PubMed] [Google Scholar]
- 9.de Bruijn MF, Speck NA. Oncogene. 2004;23:4238–4248. doi: 10.1038/sj.onc.1207763. [DOI] [PubMed] [Google Scholar]
- 10.Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, Stein JL, Stein GS. Crit Rev Eukaryot Gene Expr. 2004;14:1–41. [PubMed] [Google Scholar]
- 11.Fukamachi H, Ito K. Oncogene. 2004;23:4330–4335. doi: 10.1038/sj.onc.1207121. [DOI] [PubMed] [Google Scholar]
- 12.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GWH, Beddington RSP, Mundlos S, Olsen BR, et al. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 14.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao Y-H, Inada M, et al. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 15.Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, et al. Cell. 2002;109:113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 16.Levanon D, Bettoun D, Harris-Cerruti C, Woolf E, Negreanu V, Eilam R, Bernstein Y, Goldenberg D, Xiao C, Fliegauf M, et al. EMBO J. 2002;21:3454–3463. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaidi SK, Young DW, Pockwinse SH, Javed A, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Proc Natl Acad Sci USA. 2003;100:14852–14857. doi: 10.1073/pnas.2533076100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young DW, Hassan MQ, Yang X-Q, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, et al. Proc Natl Acad Sci USA. 2007;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee S, Yang X, Xie R, Underwood J, Furcinitti P, et al. Nature. 2007;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- 20.Zaidi SK, Young DW, Javed A, Pratap J, Montecino M, van WA, Lian JB, Stein JL, Stein GS. Nat Rev Cancer. 2007;7:454–463. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- 21.North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, Marin-Padilla M, Speck NA. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- 22.Choi J-Y, Pratap J, Javed A, Zaidi SK, Xing L, Balint E, Dalamangas S, Boyce B, van Wijnen AJ, Lian JB, et al. Proc Natl Acad Sci USA. 2001;98:8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blyth K, Cameron ER, Neil JC. Nat Rev Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 24.Speck NA, Gilliland DG. Nat Rev Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 25.Bae SC, Choi JK. Oncogene. 2004;23:4336–4340. doi: 10.1038/sj.onc.1207286. [DOI] [PubMed] [Google Scholar]
- 26.Pratap J, Lian JB, Javed A, Barnes GL, van Wijnen AJ, Stein JL, Stein GS. Cancer Metastasis Rev. 2006;25:589–600. doi: 10.1007/s10555-006-9032-0. [DOI] [PubMed] [Google Scholar]
- 27.Pratap J, Javed A, Languino LR, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Mol Cell Biol. 2005;25:8581–8591. doi: 10.1128/MCB.25.19.8581-8591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Javed A, Barnes GL, Pratap J, Antkowiak T, Gerstenfeld LC, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Proc Natl Acad Sci USA. 2005;102:1454–1459. doi: 10.1073/pnas.0409121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pratap J, Galindo M, Zaidi SK, Vradii D, Bhat BM, Robinson JA, Choi J-Y, Komori T, Stein JL, Lian JB, et al. Cancer Res. 2003;63:5357–5362. [PubMed] [Google Scholar]
- 30.Galindo M, Pratap J, Young DW, Hovhannisyan H, Im HJ, Choi JY, Lian JB, Stein JL, Stein GS, van Wijnen AJ. J Biol Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westendorf JJ, Zaidi SK, Cascino JE, Kahler R, van Wijnen AJ, Lian JB, Yoshida M, Stein GS, Li X. Mol Cell Biol. 2002;22:7982–7992. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaidi SK, Javed A, Pratap J, Schroeder TM, Westendorf J, Lian JB, van Wijnen AJ, Stein GS, Stein JL. J Cell Physiol. 2006;209:935–942. doi: 10.1002/jcp.20791. [DOI] [PubMed] [Google Scholar]
- 33.Linggi B, Muller-Tidow C, van de LL, Hu M, Nip J, Serve H, Berdel WE, van der RB, Quelle DE, Rowley JD, et al. Nat Med. 2002;8:743–750. doi: 10.1038/nm726. [DOI] [PubMed] [Google Scholar]
- 34.Blyth K, Vaillant F, Hanlon L, Mackay N, Bell M, Jenkins A, Neil JC, Cameron ER. Cancer Res. 2006;66:2195–2201. doi: 10.1158/0008-5472.CAN-05-3558. [DOI] [PubMed] [Google Scholar]
- 35.Zaidi SK, Sullivan AJ, Medina R, Ito Y, van Wijnen AJ, Stein JL, Lian JB, Stein GS. EMBO J. 2004;23:790–799. doi: 10.1038/sj.emboj.7600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaidi SK, Sullivan AJ, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Proc Natl Acad Sci USA. 2002;99:8048–8053. doi: 10.1073/pnas.112664499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Budde A, Grummt I. Oncogene. 1999;18:1119–1124. doi: 10.1038/sj.onc.1202402. [DOI] [PubMed] [Google Scholar]
- 38.Voit R, Schafer K, Grummt I. Mol Cell Biol. 1997;17:4230–4237. doi: 10.1128/mcb.17.8.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Afzal F, Pratap J, Ito K, Ito Y, Stein JL, van Wijnen AJ, Stein GS, Lian JB, Javed A. J Cell Physiol. 2005;204:63–72. doi: 10.1002/jcp.20258. [DOI] [PubMed] [Google Scholar]
- 40.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Javed A, Guo B, Hiebert S, Choi J-Y, Green J, Zhao S-C, Osborne MA, Stifani S, Stein JL, Lian JB, et al. J Cell Sci. 2000;113:2221–2231. doi: 10.1242/jcs.113.12.2221. [DOI] [PubMed] [Google Scholar]
- 42.Gutierrez S, Liu J, Javed A, Montecino M, Stein GS, Lian JB, Stein JL. J Biol Chem. 2004;279:43581–43588. doi: 10.1074/jbc.M408335200. [DOI] [PubMed] [Google Scholar]