Abstract
Single-cell resolution lineage information is a critical key to understanding how the states of gene regulatory networks respond to cell interactions and thereby establish distinct cell fates. Here, we identify a single pair of neural stem cells (neuroblasts) as progenitors of the brain insulin-producing neurosecretory cells of Drosophila, which are homologous to islet β cells. Likewise, we identify a second pair of neuroblasts as progenitors of the neurosecretory Corpora cardiaca cells, which are homologous to the glucagon-secreting islet α cells. We find that both progenitors originate as neighboring cells from anterior neuroectoderm, which expresses genes orthologous to those expressed in the vertebrate adenohypophyseal placode, the source of endocrine anterior pituitary and neurosecretory hypothalamic cells [Whitlock KE (2005) Trends Endocrinol Metab 16:145–151]. This ontogenic-molecular concordance suggests that a rudimentary brain endocrine axis was present in the common ancestor of humans and flies, where it orchestrated the islet-like endocrine functions of insulin and glucagon biology.
Keywords: glucagon, insulin, neuroblast, pituitary, hypothalamus
The principal insulin producing-cells (IPCs) in higher metazoans, such as flies and mammals, direct organismal growth, metabolism, aging, and reproduction via a conserved signal transduction pathway (1). Gut- or pancreas-based IPCs, with endodermal origin, emerged as the principal IPC locus with the evolution of lower vertebrates such as the jawless fish (2). In contrast, the principal IPCs of invertebrates are found in the nervous system and are likely of ectodermal origin. Despite this difference, the possibility that gene regulatory modules may be conserved for cell fate programming the principal IPCs of all higher animals, irrespective of germ layer origin, has led us to address the development of islet-like cells in Drosophila.
Results and Discussion
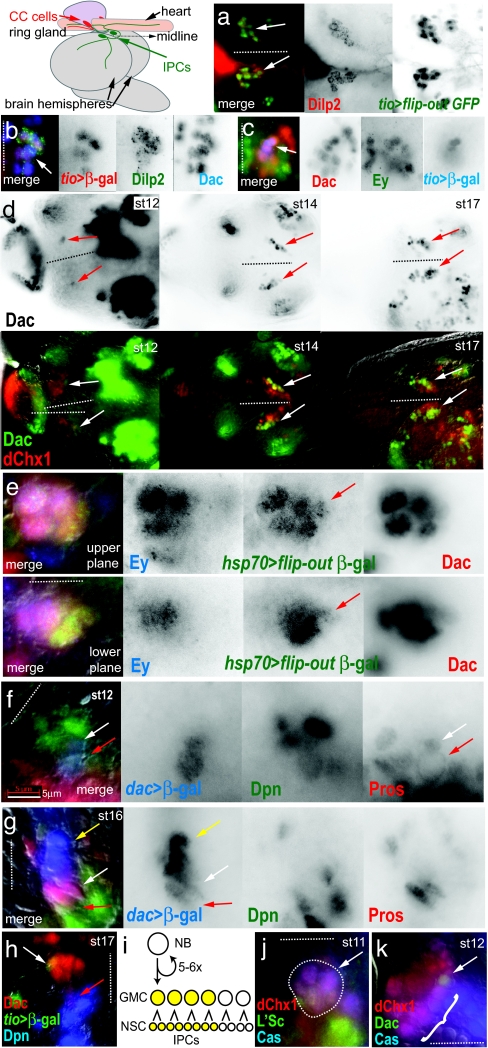
Brain IPCs in Drosophila were first recognized by their expression of insulin (Drosophila insulin-like peptide, Dilp2) at the end of embryonic development (3). Our goal is to understand the developmental origin of these cells. The absence of morphological and vital markers for identifying brain neuroblasts for dye-labeled lineage tracing necessitated the combined use of mosaic analysis to demonstrate lineage relationships and immunohistology to follow cell identities. In this study, we used 16 molecular lineage markers corresponding to conserved genes to follow cells in fixed embryos. To identify genes involved in early IPC lineage development, before the differentiation of IPCs, we screened 650 transposable GAL4-transgene insertions, obtained from public collections, that reported gene enhancer activity (GAL4 enhancer traps) in the CNS. Enhancer-driven GAL4 activity was used to trigger heritable and irreversible lineage labeling (4), which was assayed for coexpression with Dilp2 in late larval brains, thereby identifying lineage markers and potential developmental determinants. We found that enhancers near the genes dachshund (dac), eyeless (ey), optix, and tiptop (tio) (5) each triggered IPC lineage labeling by the time of Dilp2 expression onset just before hatching (late-stage 17). tio enhancer-triggered labeling was highly specific to the IPCs within the pars intercerebrallis (PI), the dorsomedial brain region harboring the IPCs and other neurosecretory cells (coexpression of Dilp2 and tio enhancer lineage labeling in late larval stage IPCs shown in Fig. 1a, arrows in merged image). Antibody staining of Dac, Ey, and Optix proteins recapitulated enhancer reporter labeling and revealed expression in the tio+ cell cluster in late-stage embryos just after IPC differentiation (coexpression of tio, Dilp2, and Dac shown in Fig. 1b, arrow in merged image), and before IPC differentiation at early-stage 17 (coexpression of Dac, Ey, and tio shown in Fig. 1c, arrow in merged image). Thus, we molecularly identified a bilateral cluster of 10–12 Dac+ Ey+ cells, 6–8 of which expressed tio before continuing on to express insulin (Dilp2) slightly later in development.
Fig. 1.
IPC lineage development in the Drosophila brain. The diagram, at top left, shows the positions of the IPCs, within in the pars intercerebralis, and the CC cells within the ring gland, relative to the embryonic and larval protocerebrum. All brains labeled by antibodies are as indicated with the text color corresponding to color channels in merged images. The position of the midline is indicated by dotted lines with anterior to the left when midline is horizontal, or anterior to the top when midline is vertical. Embryonic stages of labeled brains are as indicated. For an indication of scale, note that the individual cells of the IPC lineage are typically 3–8 μm at all stages. (a) Dorsal view of IPCs in third-instar larval brain shows the specificity of the tio enhancer for IPCs (arrows). (b) IPCs in the pharate larval brain express tio and Dac (arrow). (c) Before onset of Dilp2 expression, tio+ cells comprise the anterior part of a 10- to 12-cell cluster of Dac+ Ey+ cells (arrow). (d) The expansion of the Dac+ IPC lineage from one cell at stage 12 to 10–12 cells by stage 17 (arrows). (Upper) Series shows Dac expression only. (Lower) Series shows the Dac+ cluster expands on the posterior-lateral side of the anterior PI (6), marked by dChx1. (e) The Dac+ IPC lineage is marked by a single marked clone at early stage 17 (red arrow). The two focal planes show the entire Dac+ group. Ey expression appears in the older anterior cells of the Dac+ group at this stage. (f and g) The IPC lineage is produced from a dividing Dpn+ Dac+ neuroblast with membrane-localized Pros (red arrow). At stage 12 (f), neuroblast has divided once and the GMC daughter cell shows nuclear localized Pros (white arrow). At stage 16 (g), the neuroblast is still present (red arrow), as are the GMCs (white arrow). Cells to the anterior of the Dac+ group are early IPCs, lacking both Dpn and Pros expression (yellow arrow). (h) IPCs arise from the first-born GMCs. tio expression (white arrow), which labels the IPCs, is in the anterior of the Dac+ group, whereas the IPC neuroblast is at the posterior (red arrow). (i) Lineage model for IPCs. (j) The dChx1+ Cas+ L'Sc+ proneural group, which gives rise to the IPC neuroblast, in the anterior neuroectoderm (area in dotted outline, arrow). (k) The dChx1+ Cas+ cells delaminate as a chain of neuroblasts (bracket). The first to delaminate is the Dac+ IPC neuroblast (arrow). (Scale bar: 5 μm.)
We tested the hypothesis that the Dac+ Ey+ cluster was generated by the proliferation of a single neuroblast. The pre-Dilp2 Dac+ Ey+ cluster comprised 10–12 cells at stage 17, but only a single Dac+ cell at stage 12, suggesting that a lineage expanded from a single progenitor beginning at stage 12 (Dac expression in the IPC cluster at stages 12, 14, and 17 shown in Fig. 1d Upper, arrows). The Dac+ cluster maintains a posterior and lateral position within the anterior PI, identified by dChx1 expression (6), which allows following it during the morphogenetic changes in the developing brain (dChx1 and Dac expression within growing IPC cluster shown in Fig. 1d Lower, arrows). To mark progenitors and their lineage descendants, stage 11–12 embryos harboring both a heat-shock promoter-flip recombinase (hsp70-flp) transgene and an FRT-mediated flip-out Actin promoter-LacZ reporter were heat-shocked to induce random clone marking events in cell lineages (7). After aging embryos for 6 h at 25°C to reach stage 16–17, we occasionally recovered marked clusters of clonally related cells that comprised the 10–12 cell Dac+ Ey+ cluster (colabeling of the Dac+ Ey+ IPC cluster by a β-gal+ clone shown in Fig. 1e, arrow; Ey expression is limited to the more anterior, more differentiated cells of the cluster). Clones that partly labeled the Dac+ Ey+ cluster, which were posterior in the cluster, were interpreted as being labeled by a lineage marking event induced after the neuroblast had divided one or more times. It was unlikely that multiple marking events accounted for the apparent clonal labeling of IPCs because the frequency of marked clone induction was extremely low (tens per brain). We also found clones that labeled neighboring cells, but did not label Dac+ Ey+ cells, suggesting there was a lineage restriction that defined the Dac+ Ey+ cluster. Thus, all data were consistent with a lineage model whereby one neuroblast produced 10–12 Dac+ Ey+ cells, 6–8 of which were IPCs.
We further tested whether the single Dac+ cell progenitor of IPCs seen at stage 12 was indeed a neuroblast by using markers of neuroblast lineage development. Asymmetrically dividing neuroblasts can be identified by nuclear expression of the pan-neuroblast marker Deadpan (Dpn) (8) and Prospero (Pros) localization to the plasma membrane (9). We found that the single Dac+ cell expressed Dpn and also showed Pros localization at the plasma membrane, which indicated that it was a neuroblast (Fig. 1f, red arrow). As the Dac+ cluster increased in cell number with age, we found that Pros was present in the nucleus of Dac+ cells anterior to the Dac+ neuroblast, which indicated that these were the neuroblast daughter cells, or ganglion mother cells (GMCs) generated by asymmetric neuroblast divisions (nuclear-localized Pros in Dac+ GMCs shown in Fig. 1 f and g, red arrows) (9). By stage 14, the most anterior Dac+ cells in the cluster lacked Dpn and Pros, suggesting that they were early, undifferentiated neurons or neurosecretory cells generated by GMC cell divisions (Dac+ Dpn− Pros− early, undifferentiated IPCs shown in Fig. 1g, yellow arrow). We also found that tio expression occurs in the most anterior Dac+ cells of the lineage group (Fig. 1h, white arrow), furthest from the posterior-located Dac+ neuroblast (Fig. 1h, red arrow), suggesting that the six to eight IPCs are the products of the first three to four GMCs to be generated by asymmetric neuroblast division. This observation confirmed our interpretation of the marked clone data that showed partial labeling by a clone occupies the posterior, more recently formed region of the Dac+ Ey+ cluster, near the IPC neuroblast. Thus, we observed a histological pattern of cell identities and divisions within the Dac+ IPC lineage group that was consistent with the generic lineage development of a single neuroblast, with the IPCs being produced from the first three to four GMCs formed (diagram of IPC lineage model shown in Fig. 1i).
We further sought to identify the precise origin of the IPC neuroblast within the neuroectoderm epithelium and the blastoderm embryo to place this lineage in the context of early axial patterning. The IPC neuroblast was first recognized by Dac expression only after neuroblast formation, but before its first division. However, preceding the formation of the IPC neuroblast, the markers Castor (Cas) and dChx1 (6) and the proneural factor Lethal of Scute (L'Sc) (10) showed coexpression in eight nearby cells of the neuroectoderm epithelium (Fig. 1j, dotted outline and arrow). Cas and dChx1 were maintained in all neuroblast lineages that delaminated from this group, as indicated by coexpression of Dpn (data not shown). The IPC neuroblast was the only neuroblast from this group to express Dac, and it was always the first Dpn+ neuroblast to delaminate, becoming the most posterior in a chain of delaminating Cas+ dChx1+ neuroblasts (Fig. 1k, arrow shows Dac+ Cas+ dChx1+ neuroblast and bracket shows other Dac− Cas+ dChx+ neuroblasts). We also found that the Cas+ dChx1+ L'Sc+ proneural group lies within a “gap gene” head stripe corresponding to the Bicoid responsive giant head stripe 1 (gt1) (11), which suggested that the IPC neuroblast, or its earliest progenitor, arose from this pattern element of the precellular blastoderm (see Fig. 3f).
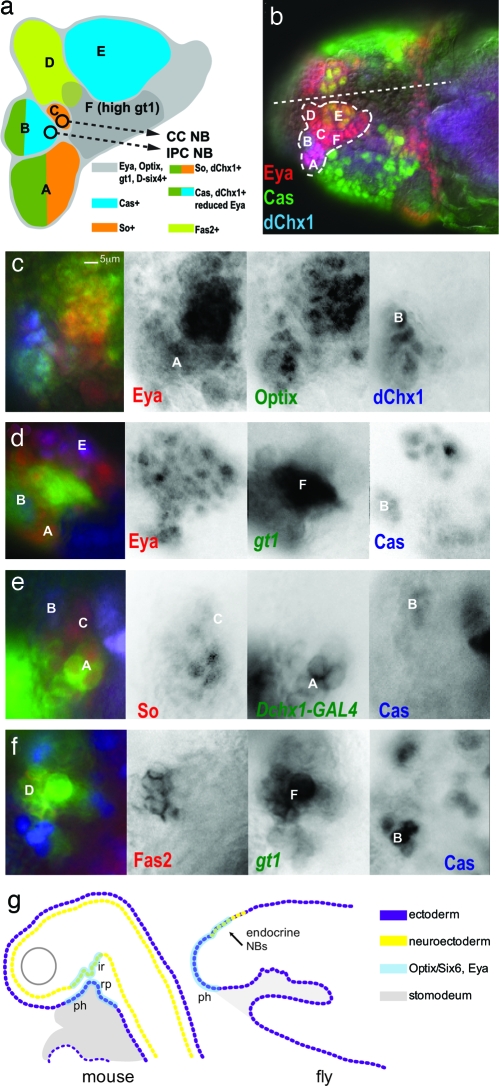
Fig. 3.
The gt head stripe 1 in stage 10–11 embryos has molecular similarity to the vertebrate hypohyseal placode. (a) A summary diagram of expression labeling data from b–f, which shows the subdivision of neuroectoderm and underlying neuroblast lineages into 2–12 cell groups with common gene expression. The color-coding for gene expression is given in the diagram. The CC cell and IPC neuroblast progenitors delaminate from the surface neuroectoderm at stages 10 and 12, respectively. At stage 13, regions B (anterior PI) and D (Pars lateralis) invaginate to form neurogenic placodes underneath the surface neuroectoderm and continue to generate neuroblasts (6). The CC cell and IPC neuroblasts are not included in the brain neuroblast map of Urbach and Technau (35) (data not shown). (b–f) All brains labeled by antibodies are as indicated with the text color corresponding to color channels in merged images. Letter labels correspond to those in the diagram (a). Anterior is to the left. (b) Embryo head with dotted outline showing region of placode gene expression (So, D-six4, Optix, and Eya). The position of the midline is indicated by dotted line. Expression patterns of Eya, Cas, and dChx1. (c) Expression patterns of Eya, Optix, and dChx1. Region B shows low-level Eya expression. (d) Expression patterns of Eya, Cas, and gt1. Region F shows high-level gt1 expression. (e) Expression patterns of So, Cas, and dChx1. The Mz-VUM enhancer of the Dchx1 gene (shown) labels the dChx1+ cells of region A at this stage. (f) Expression patterns of Fas2, gt1, and Cas. Fas2 labeling of region D corresponds to the developing Pars lateralis (6). (g) Comparison of hypophyseal gene expression (Optix/Six6 and Eya) in the fate maps of mouse and fly during the early development of the brain endocrine axis and pharynx. The neurohypohyseal diverticulum of the infundibuar region (ir) will give rise to neurosecretory hypothalamic cells (neuhypophysis). Rathke's pouch (rp) is an invagination of the oral ectoderm that gives rise to the anterior pituitary (adenohypohysis). In both mammals and flies, Optix/Six6+ Eya cells are fated to become either pharynx (ph) or endocrine progenitors. (Scale bar: 5 μm.)
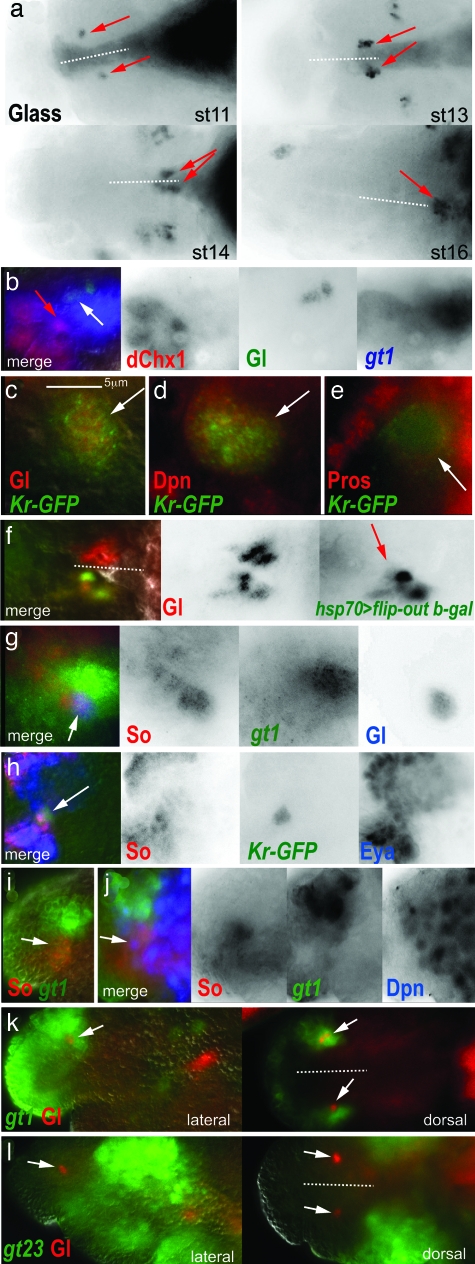
β Cell and α cell development in mammals shares a largely common pathway (12), thus we also sought to study the origin of the α-like cells in Drosophila and their development relative to the IPC lineage. Corpora cardiaca (CC) cells are analogous in function to islet α cells (13). These neuroendocrine cells reside in the endocrine ring gland, just dorsal to the brain (Fig. 1 diagram). CC cells produce and secrete a glucagon-like peptide, adipokinetic hormone, in response to circulating glucose levels, via a conserved Katp sensor. The gene glass (gl) is a marker of CC cells and their precursors that specifically labels the CC lineage beginning at stage 10 (Fig. 2a, arrows show Gl+ CC cell precursors at various stages) (14). We found that the Gl+ group of cells expanded in number to form a bilateral pair of six to eight cell clusters, aligned at the border of the brain and the developing foregut (stage 13). The Gl+ clusters then migrated out of the protocerebrum (stage 14), and posterior along the roof of the pharynx, to ultimately coalesce at the midline within the prospective ring gland (stage 16). Remarkably, the first Gl+ cells (Fig. 2b, white arrow) appeared a single cell diameter apart from the dChx1+ cluster containing the IPC neuroblast (Fig. 2b, red arrow), also within the gt1 stripe.
Fig. 2.
CC cell lineage development in the Drosophila brain. All brains labeled by antibodies are as indicated with the text color corresponding to color channels in merged images. The position of the midline is indicated by dotted lines with anterior to the left when midline is horizontal, or anterior to the top when midline is vertical. Embryonic stages of labeled brain are as indicated. For an indication of scale, note that the individual cells of the CC lineage are typically 3–8 μm at all stages. (a) Gl expression labels the CC cell precursor group. The CC cell cluster (red arrows) expands from a single cell (stage 11) to a group of six to eight (stage 16). The cell cluster migrates out of the developing brain (stage 13), along the developing foregut (stage 14), to coalesce in the presumptive ring gland (stage 16). (b) The first Gl+ cells to appear (white arrow) are gt1+ and are within a cell diameter of the dChx1+ Cas+ neuroectoderm (red arrow). (c–e) In stage 10–11 embryos, the CC cell precursor group is associated with a Kr-GFP+ neuroblast. (c) The Gl+ cells are labeled by the Kr-GFP reporter (arrow), as is an adjacent neuroblast that is Dpn+ (d, arrow) and that has membrane localized Pros (e, arrow). (f) The Gl+ CC cell lineage is marked by a single marked clone at early stage 17 (red arrow). The CC cell group from the contralateral brain hemisphere is not labeled by the clone. (g–l) Early gene expression within the CC cell group in stage 10–11 embryos. (g) The first Gl+ cell to appear expresses So and gt1 (arrow). (h) The Kr-GFP+ CC cell group expresses So and Eya. (i) So and gt1 coexpression in the surface neuroectoderm epithelium (arrow). (j) 1–2 Dpn+ neuroblasts arise from the So+ gt1 neuroectoderm (arrow). (k and l) Gl+ CC cell precursors are a component of the gt head stripe 1 (k, arrows), but not the gt head stripes 2 and 3 (l, arrows). (Scale bar: 5 μm.)
These results suggested that the CC cell lineage, like the IPC lineage, is also generated from a progenitor within the gt1+ dorsal neuroectoderm. Indeed, a neuroblast progenitor for CC cells was suggested by expression of a Kruppel reporter (Kr-GFP) (15) found to specifically label the Gl+ cells (Fig. 2c, arrow) and an adjacent cell that both was Dpn+ (Fig. 2d, arrow) and showed membrane localized Pros (Fig. 2e, arrow), indicating that it was a neuroblast. We tested, as for IPCs, if CC cells were derived from a single progenitor, perhaps the Kr-GFP+ neuroblast. We recovered Gl+ β-gal+-marked clones that comprised all or part of a CC cell cluster, after their migration to the prospective ring gland at stage 16 (Fig. 2f, arrow; contralateral CC cell cluster is not labeled by clone). Because labeled CC cells had moved from their point of origin in the developing PI, we could not determine whether a progenitor also produced other cells besides the CC cells, which did not similarly migrate. Together, these observations suggest that the CC cells are related by lineage to a neuroblast progenitor.
Typically, neuroblasts inherit the expression of cell specification factors from their point of origin in the patterned neuroectoderm before the neuroblast forms (16). We found that this was the case with the IPC neuroblast, which retains dChx1 and Cas expression from the neuroectoderm. We therefore hypothesized that this may also be the case for the CC cell neuroblast. CC cell specification was shown to require the function of gt, sine oculis (so), twist (twi), and snail (sna) (14). Indeed, we found that all of these factors were expressed in the Gl+ CC cell lineage (gt1 and So coexpression in the first Gl+ cells shown in Fig. 2g, arrow; Twi and Sna, data not shown). Moreover, the Kr-GFP+ cell group, containing the neuroblast and CC cell precursors, also expressed Eyes absent (Eya), the cognate protein tyrosine phosphatase of So (Fig. 2h, arrow). We subsequently found that at stage 10, the time that Gl+ cells are first detected, a region of gt1+ neurectoderm shows expression of So (corresponding to regions A and C in Fig. 3a). We also found that one to two So+ gt1+ neuroblasts can be detected by labeling with Dpn at this stage. Thus, we propose that the So+ Eya+ gt1+ neuroectoderm gives rise to the Kr-GFP+ So+ Eya+ gt1+ neuroblast, which is the single progenitor of the CC cells.
Our model of a dorsal neurectoderm origin for CC cells is in disagreement with another extant model. The anterior ventral furrow (AVF) epithelium was suggested to be the CC cell origin based on gene expression and function studies implicating So, Gt, Twi, and Sna in CC cell formation (14, 17). To distinguish between the AVF and dorsal neuroectoderm as possible origins of CC cells, we have used two newly available gt promoter fragment reporters whose expression persists late enough in development, beyond endogenous protein and transcript expression, to serve as a coarse-grain lineage marker of CC cells (11). The AVF is marked by the gt23 reporter, whose expression is limited to the two gt head stripes posterior to gt1 at the blastoderm stage (11). This reporter does not label the Gl+ cells (Fig. 2l). However, as we have shown, the Gl+ cells arise in the context of the most anterior gt head stripe, gt1 (compare Fig. 2 k and l), which reaffirms their proposed origin from the gt1+ neuroectoderm.
We further investigated the organization of this gt1+ segment-derived proendocrine neuroectoderm with respect to the conserved factors Optix, So, Eya, and dChx1. Optix and Eya expression aligned with the gt1 reporter expression domain (Optix+ Eya+ gt1+ region diagramed as gray background field within Fig. 3a, corresponding to the region in the dotted outline of Fig. 3b; Eya and Optix coexpression shown in Fig. 3c; Eya and gt1 coexpression shown in Fig. 3d). The D-six4 gene also shows expression specific to this domain (18, 19). Our labeling studies showed that this domain is subdivided into several small compartments of 2–12 cells with discrete gene expression profiles (see Fig. 3a for summary and Fig. 3 b–f for labeling data (A-F label domains in microscopy images that correspond with the diagram). Our data indicate that the IPC neuroblast was derived from compartment B (Optix+, dChx1+, Cas+, So−, low-level Eya) and the CC cell neuroblast arose from the adjacent compartment C (Optix+, So+, Eya+, dChx1−). This somewhat surprising finding suggests that the largely common developmental pathway of β and α cells may be partly conserved in Drosophila, perhaps with respect to a domain of Sine oculis/Six family and Eya gene expression.
The early expression of the mouse ortholog of the Drosophila homeodomain gene optix, Six6, demarcates the hypophyseal placode and infundibular region, which give rise to the anterior pituitary and neurosecretory hypothalamus, respectively (20–22). Mutation of the Six6 gene leads to reduction of the pituitary in mice (20) and humans (23). The hypophyseal placode and adjacent ectoderm also expresses the other so-called “placode genes,” Six1, Six4, and Eya, and this coexpression pattern is conserved in amphibians (24), fish (25, 26), and lower chordates such as ascidians (27, 28). In mice, the anterior pituitary is reduced in size in the double mutant of Eya1 and Six1 (29), and in zebrafish, Eya1 is essential for differentiation of all pituitary cell types except for prolactin-expressing cells (25). In Drosophila, So (14) and Eya (data not shown) are essential for CC cell formation. Thus, there is a striking conservation of the molecular signature of tissues that give rise to elements of the brain endocrine axis in flies, mammals, lower vertebrates, and lower chordates.
There are also parallels between vertebrate and fly with respect to tissue morphogenesis within the developing brain endocrine system and adjacent oral ectoderm, although there appears to be considerable variation on a general theme. For example, in mouse, the progenitors of the anterior pituitary and neurosecretory hypothalamus appear to arise respectively from Rathke's pouch, an invagination of the oral ectoderm, and the neurectoderm, which do not start as neighboring regions, but come into direct contact only after neurulation. However, in the zebrafish, which does not form a Rathke's pouch (30), the progenitors of the anterior pituitary and neurosecretory hypothalamic cells (GnRH1+) arise from neighboring regions of the hypohyseal placode, which is situated directly dorsal to the stomodeal ectoderm (31). In Drosophila, the ventral cells of the gt1+ Optix+ Eya+ ectoderm invaginate to form the roof of the pharynx, the fly's oral ectoderm, whereas the dorsal cells contribute to the endocrine axis. Therefore, there is considerable evidence for evolutionarily conservation of the close relationship between the oral ectoderm and the developing compartments of the endocrine axis, all of which express the hypophyseal placode genes (Fig. 3g). The gene expression profile and specification of endocrine cell functions from the anterior ectoderm appears to be more “fixed” across the bilateria, whereas the pattern of accompanying tissue morphogenesis and diversity of cell types is more variable, just as has been demonstrated for the specification of the bilaterian CNS, eye, gut, and heart.
Our model contrasts with the prior suggestion, based on the proximity of developing CC cells to the posterior foregut in the moth, Manduca, that CC cells originate from neurogenic placodes of the foregut that engender the stomatogastric nervous system (32, 33). Because CC cell progenitors were not identified in those studies, and subsequent mutational analysis in Drosophila demonstrated that the CC cells develop independently of the stomatogastric nervous system and posterior foregut (14), we suggest that our model of CC cell origin is the most strongly supported.
We propose that the brain endocrine systems of invertebrates and vertebrates are derived from a common ancestry because they both develop from a domain of Eya and sine oculis/Six family gene expression that comprises the anterior neuroectoderm and adjacent oral ectoderm. Indeed, these results extend prior observations that the neurosecretory cells of the PI and ring gland show other aspects of homology to the hypothalamic-pituitary axis (33). The specification of islet-like cells within a conserved brain endocrine axis raises the intriguing possibility that islet organogenesis, which is a derived feature of vertebrates, may have coopted brain endocrine cis-regulatory modules for specification of islet fates in endoderm. Indeed, the ectopic expression of the nominal rat insulin promoter reporter in anterior pituitary and hypothalamus underscores the similar gene regulatory state of these endocrine tissues (34). We expect that further genetic analysis of endocrine cell fate specification within the gt1 domain of Drosophila will lead to insights into the patterning and organogenesis of endocrine compartments and provide the basis for identifying conserved pan-IPC regulatory modules with relevance to mammalian systems.
Materials and Methods
Antibody Dilution.
Antibodies used were: mouse anti-Eya diluted 1:250 (mAB10H6; Developmental Studies Hybridoma Bank), rabbit anti-Optix diluted 1:50 (gift of F. Pignoni, Harvard University, Boston), guinea pig anti-So diluted 1:5,000 (gift of I. Rebay, Whitehead Institute, Boston), rabbit anti-Giant diluted 1:800 (gift of J. Reinitz, Stony Brook University, Stony Brook, NY), rabbit anti-Twist diluted 1:5,000 (gift of S. Roth, University of Cologne, Cologne, Germany), mouse anti-Glass undiluted (mAB 9B2.1; Developmental Studies Hybridoma Bank), mouse anti-Dac diluted 1:100 (mABdac2-3; Developmental Studies Hybridoma Bank), rabbit anti-Ey 1:200 (gift of U. Walldorf, University of Saarlandes, Saarbrücken, Germany; P. Callaerts, University of Leuven, Leuven, Belgium), guinea pig anti-Dpn diluted 1:500 (gift of Y. N. Jan, University of California, San Francisco), mouse anti-Pros diluted 1:20 (mABMR1A; Developmental Studies Hybridoma Bank), guinea pig anti-dChx1 diluted 1:50 (gift of T. Erclik, University of Toronto, Toronto), rat anti-L'Sc diluted 1:500 (gift of G. Boekhoff-Falk, University of Wisconsin, Madison), anti-Fas2 diluted 1:10 (mAB1D4; Developmental Studies Hybridoma Bank), chicken anti-β-Gal diluted 1:100 (Abcam), and rabbit anti-Cas diluted 1:1,000 (gift of W. Odenwald, National Institutes of Health, Bethesda).
Transgenes and Fly Strains.
optix (NP2631), tiptop (NP707) and dachshund (NP2446)-related GAL4 insertions were obtained from R. Ueda (National Institute of Genetics, Mishima, Japan). eyeless (ey) (OK107) and sna-GAL4 (T. Ip), dac-LZ (L. Fasano, Centre National de la Recherche Scientifique, Marseille, France), gt1-LacZ and gt23-LacZ (S. Small, University of New York, New York), Kr-GFP (15), Mz-VUM GAL4 (T. Erclik). Lineage marking line was comprised of Actin5c<CD2 >GAL4 (F. Pignoni), UAS-flp, and UAS-LacZ or UAS-GFP. Heat shock clones have been described (9).
Images.
Images were collected with a Zeiss Axioplan microscope equipped with the Zeiss HRm camera, the 100X Alpha-Plan Fluar, and 20X Plan-Apo objectives and Axiovision acquisition software. Images were prepared with Adobe Photoshop.
Acknowledgments
We thank C. Doe (University of Oregon, Eugene), J. Skeath (Washington University, St. Louis), H. Broihier (Case Western Reserve University, Cleveland), V. Hartenstein (University of California, Los Angeles), J. Reinitz, S. Roth, Z.-C. Lai (Pennsylvania State University, University Park), R. Renkawitz-Pohl (Philipps-Universität, Marburg, Germany), F. Pignoni, L. Fasano, P. Callerts, J. Kumar (University of Indiana, Bloomington), T. Erclik, S. Small, W. Odenwald, U. Walldorf, E. Bier (University of California at San Diego, La Jolla), Y. N. Jan, I. Rebay, G. Boekhoff-Falk, N. Bonini (University of Pennsylvania, Philadelphia), R. Ueda, P. Taghert (Washington University, St. Louis), T. Preat (Centre National de la Recherche Scientifique, Paris), and A. Ghysen (University of Montpellier, Montpellier, France) for generously providing reagents and V. Hartenstein, S. DiNardo, M. Ramalho-Santos, and R. Derynck for helpful discussions. S.W. was supported by a University of Pennsylvania Diabetes and Endocrinology Research Center pilot and feasibility grant and National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases Grant DK19525. N.T. and E.J.R. were supported by National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases Grant R01 DK069492. E.J.R. was supported by a Juvenile Diabetes Research Foundation transition award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Tatar M, Bartke A, Antebi A. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- 2.Falkmer S. Endocrinol Metab Clinics North Am. 1993;22:731–752. [PubMed] [Google Scholar]
- 3.Rulifson EJ, Kim SK, Nusse R. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- 4.Weigmann K, Cohen SM. Development (Cambridge, UK) 1999;126:3823–3830. doi: 10.1242/dev.126.17.3823. [DOI] [PubMed] [Google Scholar]
- 5.Laugier E, Yang Z, Fasano L, Kerridge S, Vola C. Dev Biol. 2005;283:446–458. doi: 10.1016/j.ydbio.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 6.de Velasco B, Erclik T, Shy D, Sclafani J, Lipshitz H, McInnes R, Hartenstein V. Dev Biol. 2007;302:309–323. doi: 10.1016/j.ydbio.2006.09.035. [DOI] [PubMed] [Google Scholar]
- 7.Rulifson EJ, Wu CH, Nusse R. Mol Cell. 2000;6:117–126. [PubMed] [Google Scholar]
- 8.Urbach R, Schnabel R, Technau GM. Development (Cambridge, UK) 2003;130:3589–3606. doi: 10.1242/dev.00528. [DOI] [PubMed] [Google Scholar]
- 9.Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F. Nature. 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- 10.Martin-Bermudo MD, Martinez C, Rodriguez A, Jimenez F. Development (Cambridge, UK) 1991;113:445–454. doi: 10.1242/dev.113.2.445. [DOI] [PubMed] [Google Scholar]
- 11.Ochoa-Espinosa A, Yucel G, Kaplan L, Pare A, Pura N, Oberstein A, Papatsenko D, Small S. Proc Natl Acad Sci USA. 2005;102:4960–4965. doi: 10.1073/pnas.0500373102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sander M, German MS. J Mol Med. 1997;75:327–340. doi: 10.1007/s001090050118. [DOI] [PubMed] [Google Scholar]
- 13.Kim SK, Rulifson EJ. Nature. 2004;431:316–320. doi: 10.1038/nature02897. [DOI] [PubMed] [Google Scholar]
- 14.De Velasco B, Shen J, Go S, Hartenstein V. Dev Biol. 2004;274:280–294. doi: 10.1016/j.ydbio.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 15.Casso D, Ramirez-Weber F, Kornberg TB. Mech Dev. 2000;91:451–454. doi: 10.1016/s0925-4773(00)00248-3. [DOI] [PubMed] [Google Scholar]
- 16.Skeath JB, Thor S. Curr Opin Neurobiol. 2003;13:8–15. doi: 10.1016/s0959-4388(03)00007-2. [DOI] [PubMed] [Google Scholar]
- 17.de Velasco B, Mandal L, Mkrtchyan M, Hartenstein V. Dev Genes Evol. 2006;216:39–51. doi: 10.1007/s00427-005-0029-4. [DOI] [PubMed] [Google Scholar]
- 18.Clark IB, Boyd J, Hamilton G, Finnegan DJ, Jarman AP. Dev Biol. 2006;294:220–231. doi: 10.1016/j.ydbio.2006.02.044. [DOI] [PubMed] [Google Scholar]
- 19.Seo HC, Curtiss J, Mlodzik M, Fjose A. Mech Dev. 1999;83:127–139. doi: 10.1016/s0925-4773(99)00045-3. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Perissi V, Liu F, Rose DW, Rosenfeld MG. Science. 2002;297:1180–1183. doi: 10.1126/science.1073263. [DOI] [PubMed] [Google Scholar]
- 21.Whitlock KE. Trends Endocrinol Metab. 2005;6:145–151. doi: 10.1016/j.tem.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Rios J, Gallardo ME, Rodriguez de Cordoba S, Bovolenta P. Mech Dev. 1999;83:155–159. doi: 10.1016/s0925-4773(99)00017-9. [DOI] [PubMed] [Google Scholar]
- 23.Gallardo ME, Lopez-Rios J, Fernaud-Espinosa I, Granadino B, Sanz R, Ramos C, Ayuso C, Seller MJ, Brunner HG, Bovolenta P, et al. Genomics. 1999;61:82–91. doi: 10.1006/geno.1999.5916. [DOI] [PubMed] [Google Scholar]
- 24.Schlosser G, Ahrens K. Dev Biol. 2004;271:439–466. doi: 10.1016/j.ydbio.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Nica G, Herzog W, Sonntag C, Nowak M, Schwarz H, Zapata AG, Hammerschmidt M. Dev Biol. 2006;292:189–204. doi: 10.1016/j.ydbio.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 26.Seo HC, Drivenes, Ellingsen S, Fjose A. Mech Dev. 1998;73:45–57. doi: 10.1016/s0925-4773(98)00028-8. [DOI] [PubMed] [Google Scholar]
- 27.Schlosser G. J Exp Zool. 2005;304:347–399. doi: 10.1002/jez.b.21055. [DOI] [PubMed] [Google Scholar]
- 28.Brugmann SA, Moody SA. Biol Cell. 2005;97:303–319. doi: 10.1042/BC20040515. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, et al. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 30.Chapman SC, Sawitzke AL, Campbell DS, Schoenwolf GC. J Comp Neurol. 2005;487:428–440. doi: 10.1002/cne.20568. [DOI] [PubMed] [Google Scholar]
- 31.Whitlock KE. Brain Behav Evol. 2004;64:126–140. doi: 10.1159/000079742. [DOI] [PubMed] [Google Scholar]
- 32.Copenhaver PF, Taghert PH. Development (Cambridge, UK) 1991;113:1115–1132. doi: 10.1242/dev.113.4.1115. [DOI] [PubMed] [Google Scholar]
- 33.Hartenstein V. J Endocrinol. 2006;190:555–570. doi: 10.1677/joe.1.06964. [DOI] [PubMed] [Google Scholar]
- 34.Gannon M, Shiota C, Postic C, Wright CV, Magnuson M. Genesis. 2000;26:139–142. doi: 10.1002/(sici)1526-968x(200002)26:2<139::aid-gene12>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 35.Urbach R, Technau GM. Development (Cambridge, UK) 2003;130:3621–3637. doi: 10.1242/dev.00533. [DOI] [PubMed] [Google Scholar]