Abstract
In the vertebrate embryo, the cephalic neural crest cells (CNCCs) produce cells belonging to two main lineages: the neural [including neurons, glial cells of the peripheral nervous system (PNS), and melanocytes] and the mesenchymal (chondrocytes, osteoblasts, smooth muscle cells, and connective tissue cells), whereas the trunk NCCs (TNCCs) in amniotes yield only neural derivatives. Although multipotent cells have previously been evidenced by in vitro clonal analysis, the issue as to whether all of the mesenchymal and neural phenotypes can be derived from a unique NC stem cell has remained elusive. In the present work, we devised culture conditions that led us to identify a highly multipotent NCC endowed with both neural and mesenchymal potentials, which lies upstream of all the other NC progenitors known so far. We found that addition of recombinant Sonic Hedgehog (Shh) increased the number of CNCC progenitors yielding both mesenchymal and neural lineages and promoted the development of such precursors from the TNCC. Shh decreased the neural-restricted precursors without affecting the overall CNCC survival and proliferation. By showing a differential positive effect of Shh on the expression of mesenchymal phenotypes (i.e., chondrocytes and smooth muscle cells) by multipotent CNCCs, these results shed insights on the in vivo requirement of Shh for craniofacial morphogenesis. Together with evolutionary considerations, these data also suggest that the mesenchymal-neural precursor represents the ancestral form of the NC stem cell, which in extinct forms of vertebrates (the ostracoderms) was able to yield both the PNS and superficial skeleton.
Keywords: chondrogenesis, clonal culture, embryo, quail, stem cell
In the vertebrate body, a large diversity of cell types arises from the neural crest (NC), which forms from the dorsolateral ridges of the neural primordium and yields migratory cells that colonize a variety of tissues in the embryo. In trunk NC cells (TNCCs), the developmental repertoire is limited to neurons and glial cells of the peripheral nervous system (PNS), skin melanocytes, and endocrine (adrenomedullary) cells. In addition to these neural types of derivatives, the cephalic NCCs (CNCCs) generate a large amount of mesenchymal cells forming cartilages, bones, smooth muscles, adipose, and connective tissues in the head and neck (1, 2). In vitro culture and in vivo labeling of single NCCs have provided compelling evidence that some avian and mammalian NCCs are multipotent (3–15). At migration time, CNCCs and TNCCs are heterogeneous, including fully committed cells together with bi- and multipotent progenitors. Moreover, some of these progenitors exhibited the capacity to self-renew (11, 14–17). From previous in vitro clonal analysis of quail CNCCs, we proposed a model of NC lineage segregation in which neurons, glia, melanocytes, myofibroblasts/smooth muscle cells, and chondrocytes arise from progenitors that underwent progressive restrictions in the potentialities of a putative multipotent NC stem cell able to give rise to all of the NC cell types (1, 18). Although these studies revealed the presence of a few common progenitors to mesenchymal and neural cells in early migratory CNCCs, the existence of a unique precursor for all NC-derived lineages has so far remained hypothetical. In addition, the environmental factors that would support the development of such mesenchymal-neural NCCs are still awaiting identification.
Until now, most of the studies aimed at elucidating the influence of environmental factors on NCC clonal progeny have concerned growth factors acting on neuronal, glial, and melanocytic phenotypes (15, 16, 19–27). Transforming growth factor β was shown to drive TNCCs along the myofibroblast phenotype (19, 28). However, the signaling molecules able to promote mesenchymal (particularly chondrocytic) differentiation of multipotent NC progenitors remained to be identified. One candidate is the morphogen Sonic Hedgehog (Shh), which, among pleiotropic effects during embryogenesis, is crucial for the development of the brain, face, and cranial skeleton (29, 30). Previous studies have pointed to different roles of Shh signal on CNCCs in promoting the survival of migratory CNCCs, growth, and patterning of NC-derived mesenchyme and chondrogenesis in the branchial arch (30–34). However, whether Shh acts on a particular subset of CNCC progenitors was so far unknown. In the present work, we took advantage of the possibility to isolate pure populations of CNCCs from the quail embryo, and we used an efficient in vitro clonal culture system to investigate the effect of exogenous Shh on the NCC developmental repertoire. We show that Shh promotes CNCC and TNCC differentiation into chondrocytes. Moreover, Shh differentially favors the development of NC progenitors endowed with both mesenchymal skeletogenic and neural potentials, including a previously undescribed, highly multipotent CNCC lying upstream of all the other NC progenitors described so far.
Results
Shh Treatment Promotes Mesenchymal, Not Neural, Fates of CNCCs in Mass Cultures.
With the aim of analyzing the whole set of CNCC potentialities, we devised culture conditions that improve chondrocyte differentiation and explored the possible effect of the morphogen Shh on NC phenotypes (i.e., chondrocytes, myofibroblasts, melanocytes, neurons, and glial cells). For this purpose, mes-rhombencephalon CNCCs harvested after 24 h of primary culture were grown in the absence or presence of recombinant Shh at doses of 1, 10, and 100 ng/ml.
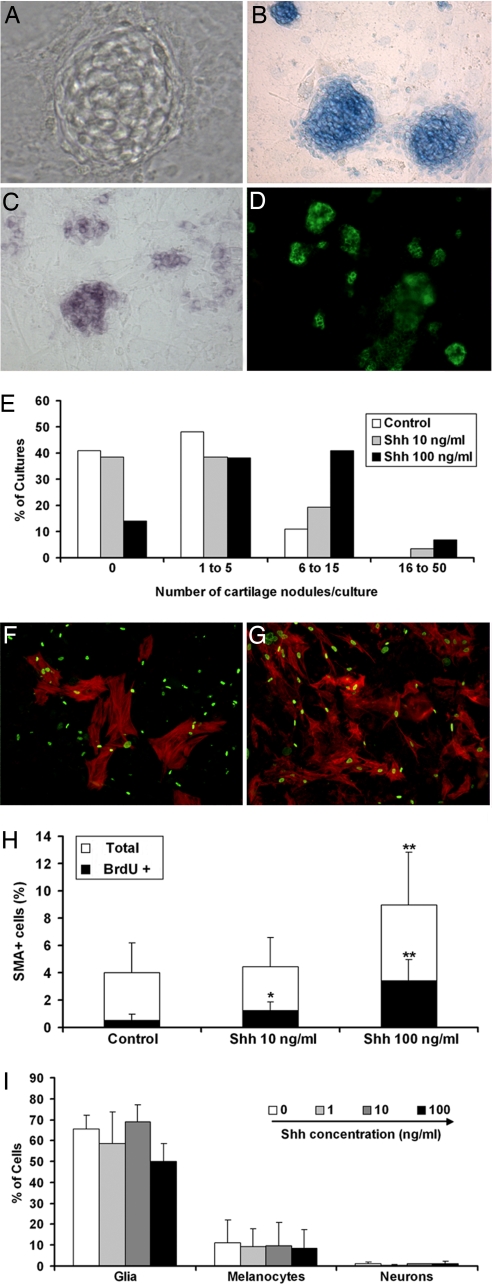
Treatment with Shh stimulated differentiation of chondrocytes in day 6 (d6) cultures as assessed by Alcian blue staining and expression of the early chondrocyte marker Sox9 (35) and chondroitin sulfate (Fig. 1A–D). Cartilage nodules differentiated in 58% (n = 119) of control cultures, whereas exposure to Shh at 10 and 100 ng/ml increased this frequency up to 63% (n = 135) and 88% (n = 138), respectively (P < 0.001). Although only 1 to 5 cartilage nodules developed in control cultures, up to 50 nodules per culture formed in the presence of 10 and 100 ng/ml Shh (Fig. 1E). Treatment with 1 ng/ml Shh did not alter chondrocyte differentiation, compared with controls (data not shown).
Fig. 1.
Influence of Shh treatment in CNCC mass cultures. Phenotype analysis in d6 cultures of mes-rhombencephalic NCCs grown in the absence and presence of Shh. (A–E) Chondrocytes are identified by phase-contrast microscopy (A), Alcian blue staining (B), expression of Sox9 transcripts (C), and immunoreactivity to chondroitin sulfate (D). (Magnification: A, ×200; B–D, ×55.) (E) Quantification of chondrogenesis is given by the number (%) of cultures containing indicated numbers of cartilage nodules per culture (n = 119, 135, and 138 cultures in control, 10 ng/ml Shh, and 100 ng/ml Shh, respectively). (F and G) Labeling of myofibroblasts with αSMA (red) and BrdU nuclear incorporation (green) in control (F) and 100 ng/ml Shh-treated (G) cultures. (H) The total number of αSMA+ cells and the number of BrdU+ αSMA+ cells are given as mean % (± SEM) of total CNCCs in control and Shh-containing media (n = 25; *, P < 0.05; **, P < 0.001). (I) Quantification of glia, melanocytes, and neurons (see Materials and Methods) is shown as mean % (± SEM) of CNCCs for each phenotype in Shh-treated and untreated cultures.
The total number of αSMA+ myofibroblasts as well as the proportion of SMA+ cells that incorporated BrdU after 1-h pulse was significantly higher after Shh treatment than in controls (Fig. 1 F–H), indicating that Shh enhances both differentiation and proliferation of myofibroblasts/smooth muscle cells. The number of glial cells, melanocytic cells, and neurons was not modified upon Shh treatment (Fig. 1I). Noticeably, Shh did not change the total number of CNCCs as determined by counting quail cell nuclei after Hoechst staining (data not shown). The rate of cell death, as determined by TUNEL staining, was low in both treated and untreated cultures (data not shown).
Therefore, continuous exposure of CNCCs to Shh differentially promoted generation of cells belonging to mesenchymal lineages (chondrocytes and myofibroblasts) and had no detectable effect on neural (neuronal, glial, and melanocytic) phenotypes.
Early Treatment of CNCCs with Shh Increases the Frequency of Chondrogenesis in d6 Mass Cultures.
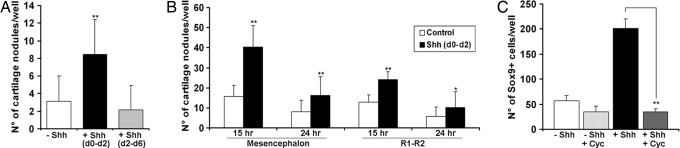
To further characterize the effect of Shh, we first studied CNCC response to different time windows of exposure to Shh. Addition of Shh during the first 48 h of culture (d0–d2) turned out to be necessary and sufficient for inducing the maximal increase in the number of cartilage nodules in d6 cultures, compared with untreated cells, whereas cultures treated with Shh from d2 to d6 were similar to untreated ones (Fig. 2A). We next investigated whether the chondrogenic ability of CNCCs varied according to the time at which migratory CNCCs were harvested from primary cultures, and we compared the chondrogenesis rate between CNCCs taken from either the mesencephalon or anterior rhombencephalon (rhombomere 1 R1 and R2). Consistent with previous results on CNCCs isolated from whole mes-rhombencephalon, d0 to d2 treatment with Shh increased chondrogenesis by mesencephalic and R1–R2 NCCs whenever harvested after 15 or 24 h of primary culture (Fig. 2B). However, mesencephalic NCCs provided a significantly higher rate of chondrogenesis than R1–R2 NCCs. Shortening the duration of primary culture to 15 h (instead of 24 h) resulted in a marked enhancement of chondrogenic differentiation by both mesencephalic and R1–R2 NCCs (Fig. 2B).
Fig. 2.
Early exposure to Shh is critical to increase chondrogenesis by CNCCs in mass cultures. (A) The mean number (± SEM) of cartilage nodules per d6 culture of mes-rhombencephalic NCCs is shown in the absence and presence of 100 ng/ml Shh added during the first 48 h (d0–d2) or from d2 to d6 (n = 28; **, P < 0.001). (B) Chondrogenesis by CNCCs isolated after 15- or 24-h migration from explanted mesencephalon, and anterior rhombencephalon (R1–R2) is given by mean number (± SEM) of cartilage nodules per d6 culture grown with and without 100 ng/ml d0–d2 Shh addition (n = 10; **, P < 0.001; *, P < 0.05). (C) Quantification of Sox9+ cells (mean number ± SEM) in d2 cultures of mesencephalic NCCs (harvested after 15 h) grown in control medium alone (−Shh) or with addition of either 100 ng/ml Shh (+Shh) or 5 μM cyclopamine (−Shh + Cyc) or both (+Shh + Cyc) (n = 4; **, P < 0.001). As determined by counting Hoechst-stained quail nuclei, the mean total number of cells did not significantly differ between culture conditions (data not shown).
To analyze the early response of mesencephalic NCCs to Shh, prechondrogenic cells were identified at d2 by the expression of Sox9 transcripts (36). Shh caused a 4-fold increase in the number of Sox9+ cells over controls while not altering the total cell number in d2 cultures (Fig. 2C). This effect was blocked by cyclopamine, a specific inhibitor of the Shh pathway (37). Treatment with both cyclopamine and Shh reduced the Sox9+ cell number to that observed in untreated cultures, whereas cyclopamine addition in the absence of Shh had no effect (Fig. 2C). To determine whether Shh influenced the proliferation rate of prechondrocytes, we analyzed BrdU incorporation by Sox9+ cells in d2 cultures. The proportion of BrdU+ Sox9+ cells did not differ in the absence (10.4 ± 5.4%) and presence (6.4 ± 2.1%; P = 0.2) of 100 ng/ml Shh. Moreover, neither labeling of collagen2a1+ chondrocytes with phosphohistone H3 in d4 cultures nor incorporation of BrdU by chondroitin sulfate+ cells in d6 cultures revealed a significant difference between Shh-treated and untreated cells (data not shown). Taken together, these results show that Shh acts during the first 48 h of culture to promote survival and/or differentiation of Sox9+ prechondrogenic cells, which are particularly numerous in the early mesencephalic NCCs.
Shh Increases Multipotent Mesenchymal Cells While Decreasing the Number of Only-Neural Progenitors in CNCC Clonal Cultures.
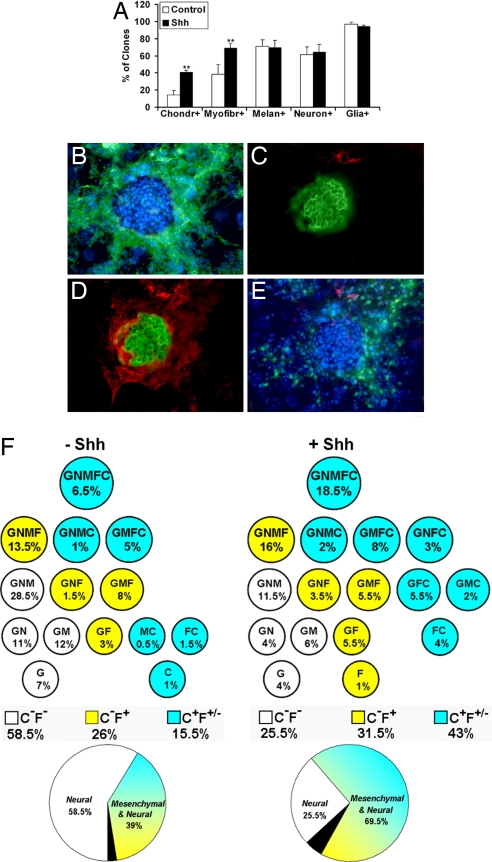
To identify the types of progenitors present in CNCCs, we performed clonal cultures by using the CNCC population that exhibited the highest chondrogenesis rate (i.e., NCCs that had migrated from explanted mesencephalon for 15 h). After 10 days in the absence or presence of 100 ng/ml Shh added during d0 to d2, the proportion of plated CNCCs that were clonogenic was similar in untreated and Shh-treated cultures [with a clonal efficiency of 47% (n = 126) and 55% (n = 150), respectively] (P = 0.2). The clones comprised from 50 to ≈10,000 cells in both conditions (data not shown). Phenotype analysis indicated that exposure to Shh significantly increased the overall frequency of the clones containing chondrocytes and those containing myofibroblasts without changing the proportion of those including glia, neurons, or melanocytes (Fig. 3A). Eighteen different clone types were recorded according to the cell types they contained (Fig. 3F). Although unipotent [i.e., chondrocytic (C), myofibroblastic (F), glial (G)] or bipotent [i.e., glial melanocytic (GM), glial myofibroblastic (GF), glial chondrocytic (GC), myofibroblastic chondrocytic (FC), and melanocytic chondrocytic (MC)] clonogenic cells were found, the vast majority of progenitors yielded three to five different cell types in both media. Noticeably, we identified a highly multipotent GNMFC progenitor able to give rise to all of the expected phenotypes [glial, neuronal, melanoblastic, myofibroblastic, and chondrocytic (Fig. 3 B–E)]. GNMFC progenitors showed a significantly higher frequency in the presence than in the absence of Shh (18.5% vs. 6.5%; P = 0.02) (Fig. 3F). The size of GNMFC progeny ranged from 80 to ≈5,000 cells and did not differ in both medium conditions, where the majority of these clones (44% vs. 57% in the absence and presence of Shh) included ≈1,000 to 5,000 cells. In cultures exposed to Shh, we identified two other, unreported so far, multipotent progenitors endowed with both mesenchymal and neural potentials: GNFC yielding glia, neurons, myofibroblasts, and chondrocytes and GMC progenitors generating glia, myofibroblasts, and chondrocytes. Together the chondrogenic progenitors reached 43% of clonogenic cells after Shh addition, compared with 15.5% (P = 0.002) in the absence of Shh (Fig. 3F).
Fig. 3.
Shh treatment of mesencephalic NCC clonal cultures promotes the development of mesenchymal-neural progenitors at the expense of neural ones. Phenotypic analysis of d10 clonal cultures of early harvested mesencephalic NCCs grown with and without 100 ng/ml Shh added during d0–d2 (data from 126 control and 150 Shh-treated colonies in three independent experiments). (A) Overall frequency (mean % ± SEM) of the clones containing chondrocytes, myofibroblasts, melanocytes, neurons, and glial cells (see Material and Methods) (**, P < 0.001). (B–E) Microscopic views (×140) of a GNMFC colony in Shh-treated cultures showing glial cells (HNK1+ in green, Hoechst-stained nuclei in blue) (B), a chondrocyte nodule (chondroitin sulfate+ in green) and neurons (TH/β-TubIII+ in red) (C), myofibroblasts (αSMA+ in red) surrounding the cartilage nodule (D), and melanocytic cells (MelEM+ in green) next to the neurons (in red) and the cartilage aggregate (Hoechst nuclear staining in blue) (E). (F) Scheme of the various types of progenitors recorded in the absence and presence of Shh according to the combination of cell types in their progeny (G, glial cells; N, neurons; M, melanocytes; F, myofibroblasts; C, chondrocytes). The frequency (% of total clones) of each progenitor type, as well as the total frequency of chondrogenic (C+F+/− in blue), myofibroblastic nonchondrogenic (C−F+ in yellow), and neural (C−F− in white) progenitors are indicated. Data are summarized by the respective contribution of mesenchymal-neural (in blue-yellow), only-neural (in white), and only-mesenchymal (i.e., C, F, and FC; in black) progenitors.
Taken together, the mesenchymal-neural progenitors accounted for 69.5% of clonogenic cells after exposure to Shh, compared with 39% in control medium (P < 0.001), because the frequency of purely mesenchymal precursors (i.e., C, F, and FC precursors) was low in both conditions. Conversely, only-neural progenitors developed with significantly higher frequency in the absence than in the presence of Shh (58.5% vs. 25.5%, respectively; P < 0.001) (Fig. 3F). Because the survival of clonogenic cells did not differ in the absence and presence of Shh, data argue that Shh promotes the development of multipotent progenitors belonging to both mesenchymal and neural lineages at the expense of CNCCs whose fate is restricted to only-neural cell types.
TNCCs Include Shh-Responsive, Multipotent Chondrogenic Progenitors.
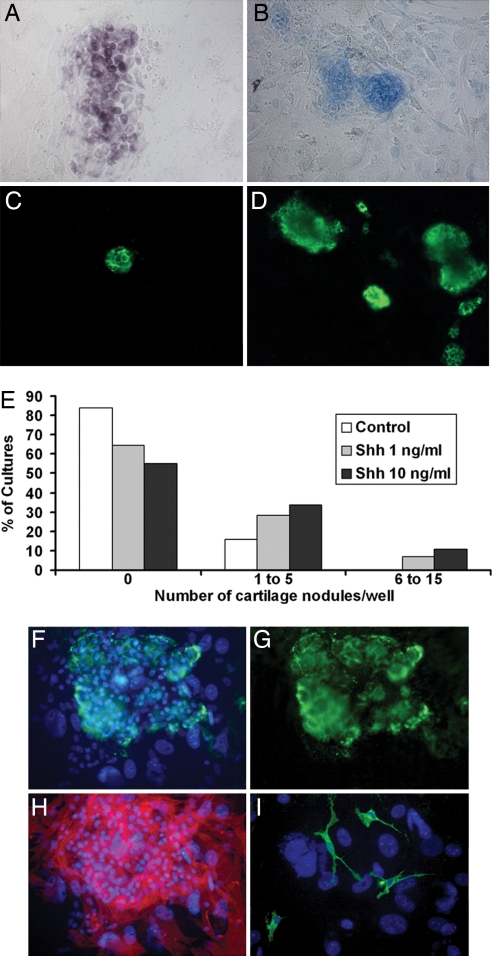
Because Shh promotes in vitro chondrogenesis by CNCCs, we investigated whether treatment with 1, 10, and 100 ng/ml Shh also was able to trigger chondrocyte differentiation of TNCCs. In d8 mass cultures prepared as for CNCCs, we found that TNCCs give rise to chondrocytic cells (Fig. 4A–D). Differentiation of chondrocytes occurred in 16% (n = 56) of control cultures versus 43% (n = 56; P < 0.0003) and 36% (n = 56; P = 0.02) of cultures exposed to 1 and 10 ng/ml Shh, respectively. Moreover, although TNCCs produced only 1–3 chondrocyte nodules in the absence of Shh, 6–15 nodules developed in the presence of 1 and 10 ng/ml Shh (Fig. 4E). TNCCs did not show enhancement of chondrogenesis after treatment with 100 ng/ml Shh (data not shown). Exposure to Shh did not influence TNCC differentiation along nonchondrocytic phenotypes, as assessed by counting the total number of glial, neuronal, melanocytic, and myofibroblastic cells in Shh-treated and untreated d8 cultures (data not shown).
Fig. 4.
Shh treatment of TNCC cultures promotes chondrogenesis. (A–D) Microscopic views of chondrocytes in d8 mass cultures of TNCCs in the absence (B and C) and presence (A and D) of 10 ng/ml Shh as identified by Sox9 expression (A), Alcian blue staining (B), and immunoreactivity to chondroitin sulfate (C and D). (Magnification: ×55.) (E) Quantification (%) of the cultures according to indicated increasing numbers of cartilage nodules in control medium and after exposure to 1 and 10 ng/ml Shh (n = 56). (F–I) Microscopic views show a glial, myofibroblastic, and chondrocytic (GFC) colony in the presence of 10 ng/ml Shh, which contains chondrocyte aggregates (G, chondroitin sulfate labeling in green; F, merged with Hoechst staining in blue), αSMA+ myofibroblasts (H, in red), and HNK1+ glial cells (I, in green). (Magnification: F–H, ×100; I, ×330.)
To investigate the developmental potential of chondrogenic TNCCs, we searched for the presence of chondrocytes in TNCC clonal cultures grown in the absence and presence of Shh. None of the TNCCs plated in control medium (n = 225) yielded a clonal progeny that included chondroitin sulfate-positive chondrocytes. In the presence of 10 ng/ml Shh, we identified one clone (of 225 plated cells) that contained chondrocyte nodules, glial cells, and myofibroblasts (Fig. 4 F–I).
Discussion
CNCCs contribute widely to vertebrate head development by yielding the cranial PNS and melanocytes, smooth muscle components, and most craniofacial skeletal structures (1, 2). Despite previous evidence that a rare CNCC subset has the potential to develop in vitro along some representatives of both mesenchymal (chondrogenic and myofibroblastic) and neural (neuronal, glial, and melanocytic) sublineages (4, 8, 15), the issue of whether the multiple CNCC-derived cell types arise from a common stem cell remained unanswered, and the growth factors regulating the differentiation of CNCC mesenchymal progenitors were still to be defined.
Taking advantage of the possibility of isolating CNCC populations from quail embryos and analyzing their developmental repertoire in single-cell culture, we identified a highly multipotent progenitor capable of giving rise to glia, neurons, melanoblasts, myofibroblasts, and chondrocytes in mesencephalic NCCs. This multipotent mesenchymal-neural progenitor lies upstream of the various progenitors that have been identified in the NC so far (18), thus supporting a hierarchical model of CNCC diversification whereby mesenchymal and neural cell types originate from a common NC stem cell. At present, specific markers to identify multipotent GNMFC cells are lacking, which allows isolating these cells and testing their ability to self-renew.
We challenged CNCC cultures with the signaling molecule Shh, which is required for craniofacial development and is expressed in ventral pharyngeal endoderm and branchial arch ectoderm during CNCC migration (29, 30). Highly multipotent mesenchymal-neural CNCC progenitors are responsive to Shh, which markedly increases their frequency in clonogenic cells. The GNMFC cell thus represents the most widespread progenitor (18.5%) recorded in the presence of Shh. These findings are in accordance with the positive role of Shh on stem cells in other systems such as the adult mammalian brain (38–40). In CNCCs, Shh increases the mesenchymal-neural progenitors (from 39% to 69.5%) while decreasing the only-neural ones (from 58.5% to 25.5%) and not altering the overall number of clonogenic CNCCs. This finding is consistent with Shh role in expanding the mesenchymal-neural progenitors at the expense of those restricted to a neural fate (Fig. 3F). The alternative possibility that Shh may act on survival and/or proliferation of mesenchymal-neural CNCCs is unlikely because the latter scenario would have involved an increase in the total number of progenitors in the presence of Shh. The present findings, however, are in line with the possibility that Shh may regulate NCC survival, adhesion, and neuronal differentiation at various developmental stages, as shown in other experimental systems (30, 32, 33, 41–43).
Stimulation of the development of mesenchymal-neural progenitors by Shh consists mainly of enhancing chondrogenic ability of CNCCs because the overall frequency of chondrogenic progenitors (C+F+/−) (Fig. 3F) highly increases in Shh-treated clones. Shh also increases the overall frequency of myofibroblastic progenitors. However, such an effect is only significant for those that possess chondrogenic potential (C+F+) (Fig. 3F). Our results argue that Shh, although favoring expression of mesenchymal phenotypes, is particularly efficient in driving CNCCs along the chondrogenic differentiation program. A proskeletogenic effect of Shh also has been shown in mesenchymal stem cells and somitic tissue (44–47).
The analysis of CNCC mass cultures supports the conclusions drawn from single-cell cultures that only the mesenchymal fates of CNCCs are promoted by Shh. Shh treatment enhances chondrogenesis only if applied within a d0–d2 time window, indicating an early action of Shh on chondrocyte progenitors (Fig. 2C). Shh increases by 4-fold the number of Sox9+ prechondrogenic cells with no detectable effect on their proliferation and the total CNCC number. Shh signaling on Sox9+ CNCCs is mediated through the receptor Smoothened because it is blocked by cyclopamine (37). Because in vivo Sox9 is expressed in all chondrocyte progenitors and differentiated chondrocytes (35, 48) and is required for the determination of the chondrogenic lineage in CNCCs (36), we hypothesize that Shh prochondrogenic action on multipotent CNCCs could be exerted through the induction and/or maintenance of Sox9 activity. Further investigations are required to know whether Shh can up-regulate Sox9 gene activity directly, as shown in mesoderm-derived chondrocytes (45), or whether it cooperates with other growth factors known to enhance chondrogenesis by CNCCs, such as FGF2 and FGF8 (33, 49).
Compared with mesencephalic NCCs, the response to Shh decreases in R1–R2 (Fig. 2B) and is low in R3–R8 (data not shown) mass cultures. Such graded potential to undergo Shh-dependent chondrogenesis by cultured CNCCs is in accordance with the different fates of rostrocaudal CNCCs in vivo, which show decreased generation of skeletal elements by rhombencephalic, compared with mesencephalic NCCs, and the absence of such derivatives in the TNC of amniotes. The skeletogenic ability of anteriormost CNCCs has been functionally related to the absence of Hox gene expression (50–52). Whether Hox gene status and Shh responsiveness of mesenchymal NC progenitors are linked thus remains to be explored.
The present work shows in vitro chondrocyte differentiation of TNCCs, a finding described previously (53–55). Similarly to CNCCs, chondrogenesis by TNCCs is markedly increased in the presence of Shh. However, compared with CNCCs, lower doses (1 and 10 ng/ml) of Shh are efficient, suggesting that Shh signal transduction or NCC behavior (or both) is differently regulated at cephalic and trunk levels. The rate of chondrogenesis is much lower in TNCCs than in CNCCs, as shown by the proportion of chondrogenic cultures and the number of cartilage nodules generated. Assuming that every nodule in mass cultures originates from a single chondrogenic cell, the proportion of plated cells that are chondrogenic in the absence of Shh can be evaluated to ≈0.04% in TNCCs (corresponding to a mean number of nodules of 0.3 per 800 cells) and 4% (16 nodules per 400 cells) in mesencephalic NCCs, respectively. In the presence of Shh, these estimated proportions of chondrogenic cells increase to 0.25% and 10% in TNCCs and mesencephalic NCCs, respectively. Such a low frequency of chondrogenic TNCCs likely explains that the TNCC clonal assay led to identifying a single chondrogenic progenitor of 225 cells plated in the presence of Shh. This progenitor yields glia, myofibroblasts, and chondrocytes, revealing that TNCCs include rare oligopotent cells endowed with both mesenchymal and neural potentials. Avian TNCCs are unable to form cartilage in vivo even when challenged by transplantations to cranial levels (56, 57). However, together with the recent evidence that cultured TNCCs differentiate into adipocytes (58), the present findings suggest that NCC potentialities, whether mesenchymal or neural, are widespread, but differentially distributed along the axial level in amniotes.
It is interesting to underline that the most primitive skeletal structures of the early vertebrates were superficial dermal calcified tissues bearing tooth-like structures, which have been assumed to be of NC origin (59). The craniofacial skeleton in higher vertebrates and the dorsal fin in teleosts are the only remnants of this more extended armor of extinct vertebrates. Therefore, we consider the mesenchymal-neural progenitors found in the avian NC as more primitive than the only-neural ones. According to this view, the latter could be derived from the former. In some of these neural progenitors, Shh signal is able to reveal the potentiality, which they have not completely lost, of giving rise to mesenchymal derivatives. The epigenetic processes controlling gene expression allow dormant mesenchymal potentialities to emerge when NC progenitor cells are submitted during a definite time window to the appropriate environmental signals such as Shh. Thus, one can put forward the hypothesis that most, if not all, of the NCCs were originally of the mesenchymal-neural type. During evolution, regression of the superficial bony structures substituted for by the internal skeleton resulted in strong reduction of mesenchymal potencies in the NCCs, which vanished, but did not completely disappear, in truncal region of amniotes.
Materials and Methods
Cell Cultures.
CNCCs were isolated from the neural primordium of six to seven somite-stage quail embryos at three different rostrocaudal levels: (i) whole mesencephalon-rhombencephalon, (ii) mesencephalon, and (iii) anterior rhombencephalon (i.e., R1–R2). TNCCs were obtained from thoracic neural tubes of 20–25 somite-stage quail embryos (15). After 15 or 24 h of primary culture, NCCs that had migrated around explanted neural tubes were harvested for secondary plating in mass cultures (400 cells per well for CNCCs and 800 cells per well for TNCCs) or in clonal cultures and grown on a feeder layer of growth-inhibited 3T3 fibroblasts (15) at a density of 24,000 3T3 cells per well in 96-well plates. Culture medium was DMEM 10% FCS (control medium) alone or supplemented with 1, 10, or 100 ng/ml mouse N-Shh (R&D Systems, Minneapolis, MN). In clonal cultures, 2% chicken embryo extract (15) was added to the control medium. Cultures were maintained at 37°C in a humidified 5%CO2/95% air atmosphere. Inhibition of Shh signaling was performed by treatment for 2 days with 5 μM cyclopamine (37) (Toronto Research Chemicals, Toronto).
Phenotype Analysis.
Cultures were fixed with 4% paraformaldehyde at times ranging from d2 to d10. Quail NCCs were distinguished from mouse 3T3 fibroblasts by nuclear staining with Hoechst 33342 (Sigma–Aldrich, St. Louis). Phenotypes derived from CNCCs and TNCCs were identified by using lineage-specific markers essentially as described previously (15): melanoblast/melanocyte early marker and melanin for melanocytic cells, Schwann Cell Myelin Protein and HNK1 for glial cells, and α smooth muscle actin (SMA) (clone 1A4; Sigma–Aldrich) for myofibroblasts/smooth muscle cells. Neurons and adrenergic cells were detected by using β-Tubulin III (Promega, Madison, WI) and tyrosine hydroxylase antibodies, respectively. Immunoreactivity to collagen 2a1 (Chemicon International, Temecula, CA) and chondroitin sulfate (Sigma–Aldrich) and Alcian blue staining identified chondrocytes. Secondary antibodies were from Southern Biotechnology (Birmingham, AL). Fluorescence was observed under an inverted ×70 microscope (Olympus, Tokyo, Japan). In situ hybridization for chick Sox9 was performed as described (17). Proliferation assays consisted of immunodetection of phosphohistone H3 (Sigma–Aldrich) and BrdU incorporation after 1-h pulse (Cell Proliferation Kit; Roche Diagnostics, Indianapolis). Differences in cell and colony numbers were analyzed by using Student's two-tailed t test or χ2 test and were considered statistically significant at P < 0.05.
Acknowledgments
We thank José M. Brito for helpful discussions, Andrea Gonçalves Trentin for continuous support to G.W.C., and Paul Scotting (University of Nottingham, Nottingham, U.K.) for the chicken Sox9 probe. This work was supported by the Centre National de la Recherche Scientifique, Association Pour la Recherche Contre le Cancer Grant N3929, and a Coordenação de Aperfeiçoamento de Pessoal de Nível Superior Fellowship (to G.W.C.).
Abbreviations
- d6
day 6
- GNMFC
glial, neuronal, melanoblastic, myofibroblastic, and chondrocytic
- NC
neural crest
- NCC
NC cell
- CNCC
cephalic NCC
- TNCC
trunk NCC
- PNS
peripheral nervous system
- Shh
Sonic Hedgehog.
Footnotes
The authors declare no conflict of interest.
References
- 1.Dupin E, Creuzet S, Le Douarin NM. Adv Exp Med Biol. 2006;589:96–119. doi: 10.1007/978-0-387-46954-6_6. [DOI] [PubMed] [Google Scholar]
- 2.Le Douarin N, Kalcheim C. The Neural Crest. Cambridge, UK: Cambridge Univ Press; 1999. [Google Scholar]
- 3.Sieber-Blum M, Cohen AM. Dev Biol. 1980;80:96–106. doi: 10.1016/0012-1606(80)90501-1. [DOI] [PubMed] [Google Scholar]
- 4.Baroffio A, Dupin E, Le Douarin NM. Proc Natl Acad Sci USA. 1988;85:5325–5329. doi: 10.1073/pnas.85.14.5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronner-Fraser M, Fraser SE. Nature. 1988;335:161–164. doi: 10.1038/335161a0. [DOI] [PubMed] [Google Scholar]
- 6.Bronner-Fraser M, Fraser SE. Neuron. 1989;3:755–766. doi: 10.1016/0896-6273(89)90244-4. [DOI] [PubMed] [Google Scholar]
- 7.Dupin E, Baroffio A, Dulac C, Cameron-Curry P, Le Douarin NM. Proc Natl Acad Sci USA. 1990;87:1119–1123. doi: 10.1073/pnas.87.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baroffio A, Dupin E, Le Douarin NM. Development. 1991;112:301–305. doi: 10.1242/dev.112.1.301. [DOI] [PubMed] [Google Scholar]
- 9.Ito K, Sieber-Blum M. Dev Biol. 1991;148:95–106. doi: 10.1016/0012-1606(91)90320-3. [DOI] [PubMed] [Google Scholar]
- 10.Sieber-Blum M. Neuron. 1991;6:949–955. doi: 10.1016/0896-6273(91)90235-r. [DOI] [PubMed] [Google Scholar]
- 11.Stemple DL, Anderson DJ. Cell. 1992;71:973–985. doi: 10.1016/0092-8674(92)90393-q. [DOI] [PubMed] [Google Scholar]
- 12.Ito K, Morita T, Sieber-Blum M. Dev Biol. 1993;157:517–525. doi: 10.1006/dbio.1993.1154. [DOI] [PubMed] [Google Scholar]
- 13.Hagedorn L, Suter U, Sommer L. Development. 1999;126:3781–3794. doi: 10.1242/dev.126.17.3781. [DOI] [PubMed] [Google Scholar]
- 14.Morrison SJ, White PM, Zock C, Anderson DJ. Cell. 1999;96:737–749. doi: 10.1016/s0092-8674(00)80583-8. [DOI] [PubMed] [Google Scholar]
- 15.Trentin A, Glavieux-Pardanaud C, Le Douarin NM, Dupin E. Proc Natl Acad Sci USA. 2004;101:4495–4500. doi: 10.1073/pnas.0400629101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee HY, Kleber M, Hari L, Brault V, Suter U, Taketo MM, Kemler R, Sommer L. Science. 2004;303:1020–1023. doi: 10.1126/science.1091611. [DOI] [PubMed] [Google Scholar]
- 17.Real C, Glavieux-Pardanaud C, Le Douarin NM, Dupin E. Dev Biol. 2006;300:656–669. doi: 10.1016/j.ydbio.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 18.Le Douarin NM, Creuzet S, Couly G, Dupin E. Development. 2004;131:4637–4650. doi: 10.1242/dev.01350. [DOI] [PubMed] [Google Scholar]
- 19.Shah NM, Marchionni MA, Isaacs I, Stroobant P, Anderson DJ. Cell. 1994;77:349–360. doi: 10.1016/0092-8674(94)90150-3. [DOI] [PubMed] [Google Scholar]
- 20.Dupin E, Le Douarin NM. Dev Biol. 1995;168:529–548. doi: 10.1006/dbio.1995.1100. [DOI] [PubMed] [Google Scholar]
- 21.Shah NM, Groves AK, Anderson DJ. Cell. 1996;85:331–343. doi: 10.1016/s0092-8674(00)81112-5. [DOI] [PubMed] [Google Scholar]
- 22.Shah NM, Anderson DJ. Proc Natl Acad Sci USA. 1997;94:11369–11374. doi: 10.1073/pnas.94.21.11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stone JG, Spirling LI, Richardson MK. J Cell Sci. 1997;110:1673–1682. doi: 10.1242/jcs.110.14.1673. [DOI] [PubMed] [Google Scholar]
- 24.Lahav R, Dupin E, Lecoin L, Glavieux C, Champeval D, Ziller C, Le Douarin NM. Proc Natl Acad Sci USA. 1998;95:14214–14219. doi: 10.1073/pnas.95.24.14214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagedorn L, Floris J, Suter U, Sommer L. Dev Biol. 2000;228:57–72. doi: 10.1006/dbio.2000.9936. [DOI] [PubMed] [Google Scholar]
- 26.Morrison SJ, Perez SE, Qiao Z, Verdi JM, Hicks C, Weinmaster G, Anderson DJ. Cell. 2000;101:499–510. doi: 10.1016/s0092-8674(00)80860-0. [DOI] [PubMed] [Google Scholar]
- 27.Kleber M, Lee HY, Wurdak H, Buchstaller J, Riccomagno MM, Ittner LM, Suter U, Epstein DJ, Sommer L. J Cell Biol. 2005;169:309–320. doi: 10.1083/jcb.200411095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen S, Lechleider RJ. Circ Res. 2004;94:1195–1202. doi: 10.1161/01.RES.0000126897.41658.81. [DOI] [PubMed] [Google Scholar]
- 29.Tapadia MD, Cordero DR, Helms JA. J Anat. 2005;207:461–477. doi: 10.1111/j.1469-7580.2005.00484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brito JM, Teillet MA, Le Douarin NM. Proc Natl Acad Sci USA. 2006;103:11607–11612. doi: 10.1073/pnas.0604751103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahlgren SC, Bronner-Fraser M. Curr Biol. 1999;9:1304–1314. doi: 10.1016/s0960-9822(00)80052-4. [DOI] [PubMed] [Google Scholar]
- 32.Ahlgren SC, Thakur V, Bronner-Fraser M. Proc Natl Acad Sci USA. 2002;99:10476–10481. doi: 10.1073/pnas.162356199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abzhanov A, Tabin CJ. Dev Biol. 2004;273:134–148. doi: 10.1016/j.ydbio.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 34.Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP. Genes Dev. 2004;18:937–951. doi: 10.1101/gad.1190304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Q, Eberspaecher H, Lefebvre V, De Crombrugghe B. Dev Dyn. 1997;209:377–386. doi: 10.1002/(SICI)1097-0177(199708)209:4<377::AID-AJA5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 36.Mori-Akiyama Y, Akiyama H, Rowitch DH, de Crombrugghe B. Proc Natl Acad Sci USA. 2003;100:9360–9365. doi: 10.1073/pnas.1631288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 38.Machold R, Hayashi S, Rutlin M, Muzumdar MD, Nery S, Corbin JG, Gritli-Linde A, Dellovade T, Porter JA, Rubin LL, et al. Neuron. 2003;39:937–950. doi: 10.1016/s0896-6273(03)00561-0. [DOI] [PubMed] [Google Scholar]
- 39.Lewis PM, Gritli-Linde A, Smeyne R, Kottmann A, McMahon AP. Dev Biol. 2004;270:393–410. doi: 10.1016/j.ydbio.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 40.Palma V, Lim DA, Dahmane N, Sanchez P, Brionne TC, Herzberg CD, Gitton Y, Carleton A, Alvarez-Buylla A, Altaba AR. Development. 2005;132:335–344. doi: 10.1242/dev.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jarov A, Williams KP, Ling LE, Koteliansky VE, Duband JL, Fournier-Thibault C. Dev Biol. 2003;261:520–536. doi: 10.1016/s0012-1606(03)00351-8. [DOI] [PubMed] [Google Scholar]
- 42.Ota M, Ito K. Dev Dyn. 2003;227:544–551. doi: 10.1002/dvdy.10336. [DOI] [PubMed] [Google Scholar]
- 43.Fu M, Lui VC, Sham MH, Pachnis V, Tam PK. J Cell Biol. 2004;166:673–684. doi: 10.1083/jcb.200401077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teillet M, Watanabe Y, Jeffs P, Duprez D, Lapointe F, Le Douarin NM. Development. 1998;125:2019–2030. doi: 10.1242/dev.125.11.2019. [DOI] [PubMed] [Google Scholar]
- 45.Zeng L, Kempf H, Murtaugh LC, Sato ME, Lassar AB. Genes Dev. 2002;16:1990–2005. doi: 10.1101/gad.1008002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dwyer JR, Sever N, Carlson M, Nelson SF, Beachy PA, Parhami F. J Biol Chem. 2007;282:8959–8968. doi: 10.1074/jbc.M611741200. [DOI] [PubMed] [Google Scholar]
- 47.Kempf H, Ionescu A, Udager AM, Lassar AB. Dev Dyn. 2007;236:1954–1962. doi: 10.1002/dvdy.21205. [DOI] [PubMed] [Google Scholar]
- 48.Ng LJ, Wheatley S, Muscat GE, Conway-Campbell J, Bowles J, Wright E, Bell DM, Tam PP, Cheah KS, Koopman P. Dev Biol. 1997;183:108–121. doi: 10.1006/dbio.1996.8487. [DOI] [PubMed] [Google Scholar]
- 49.Sarkar S, Petiot A, Copp A, Ferretti P, Thorogood P. Development. 2001;128:2143–2152. doi: 10.1242/dev.128.11.2143. [DOI] [PubMed] [Google Scholar]
- 50.Couly G, Creuzet S, Bennaceur S, Vincent C, Le Douarin NM. Development. 2002;129:1061–1073. doi: 10.1242/dev.129.4.1061. [DOI] [PubMed] [Google Scholar]
- 51.Couly G, Grapin-Botton A, Coltey P, Ruhin B, Le Douarin NM. Development. 1998;125:3445–3459. doi: 10.1242/dev.125.17.3445. [DOI] [PubMed] [Google Scholar]
- 52.Creuzet S, Couly G, Vincent C, Le Douarin NM. Development. 2002;129:4301–4313. doi: 10.1242/dev.129.18.4301. [DOI] [PubMed] [Google Scholar]
- 53.Abzhanov A, Tzahor E, Lassar AB, Tabin CJ. Development. 2003;130:4567–4579. doi: 10.1242/dev.00673. [DOI] [PubMed] [Google Scholar]
- 54.Ido A, Ito K. Dev Dyn. 2006;235:361–367. doi: 10.1002/dvdy.20635. [DOI] [PubMed] [Google Scholar]
- 55.McGonnell IM, Graham A. Curr Biol. 2002;12:767–771. doi: 10.1016/s0960-9822(02)00818-7. [DOI] [PubMed] [Google Scholar]
- 56.Nakamura H, Ayer-le Lievre CS. J Embryol Exp Morphol. 1982;70:1–18. [PubMed] [Google Scholar]
- 57.Lwigale PY, Conrad GW, Bronner-Fraser M. Development. 2004;131:1979–1991. doi: 10.1242/dev.01106. [DOI] [PubMed] [Google Scholar]
- 58.Billon N, Iannarelli P, Monteiro MC, Glavieux-Pardanaud C, Richardson WD, Kessaris N, Dani C, Dupin E. Development. 2007;134:2283–2292. doi: 10.1242/dev.002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith MM, Hall BK. Biol Rev Camb Philos Soc. 1990;65:277–373. doi: 10.1111/j.1469-185x.1990.tb01427.x. [DOI] [PubMed] [Google Scholar]