Abstract
Maintaining connectivity for broad-scale ecological processes like dispersal and gene flow is essential for conserving endangered species in fragmented landscapes. However, determining which habitats should be set aside to promote connectivity has been difficult because existing models cannot incorporate effects of multiple pathways linking populations. Here, we test an ecological connectivity model that overcomes this obstacle by borrowing from electrical circuit theory. The model vastly improves gene flow predictions because it simultaneously integrates all possible pathways connecting populations. When applied to data from threatened mammal and tree species, the model consistently outperformed conventional gene flow models, revealing that barriers were less important in structuring populations than previously thought. Circuit theory now provides the best-justified method to bridge landscape and genetic data, and holds much promise in ecology, evolution, and conservation planning.
Keywords: Gulo gulo, isolation by resistance, landscape connectivity, Swietenia macrophylla, landscape genetics
Preserving and restoring connectivity for broad-scale ecological processes, such as dispersal and gene flow, has become a major conservation priority (1, 2). Conservation organizations are investing considerable resources—and asking governments to do the same—to set aside land to promote connectivity (3). A major impediment to this goal is the difficulty in predicting how different land use, climate change, or reserve design scenarios will affect connectivity, and conservation planning decisions are often made without quantifying benefits for the ecological processes they are meant to conserve. If scarce conservation dollars are to be spent effectively, conservation biologists need clear, efficient, and reliable tools relating landscape composition and pattern to important ecological processes (4).
Gene flow is a critical ecological process with conservation benefits ranging from promoting the persistence of small populations to spreading adaptive traits in changing environments (5–8). Because of these important ecological and evolutionary roles, a new and rapidly growing field—landscape genetics—is primarily dedicated to understanding and predicting how landscape characteristics affect gene flow (5). By combining genetic, computational, and spatial analytic tools unavailable a decade ago, the interdisciplinary field has yielded insights relevant not only to conservation (e.g., refs. 8 and 9), but to fields such as evolution (10), infectious disease ecology (11), and population ecology (12).
Yet progress in all of these fields has been hampered by a lack of models capable of predicting gene flow from landscape structure. As a result, typical landscape genetic analyses simply detect genetic discontinuities and propose ad hoc explanations based on coincident landscape features, rather than testing a priori predictions of how such features are expected to influence genetic structure. This hinders hypothesis testing and leaves conservation planners without validated metrics of how landscape change will affect genetic connectivity. Although a growing number of studies are incorporating landscape data into genetic predictions and conservation plans using least-cost path (LCP) modeling (9, 13–16), gene flow among real populations is not restricted to single, optimal pathways, as LCP models assume; rather, it occurs over multiple pathways and often involves indirect allele movements spanning many generations (17). Thus, to support conservation planning and to enable hypothesis testing in landscape genetics, theoretically justified models of how landscape features facilitate or impede gene flow over multiple, direct and indirect pathways are needed.
Here, we use electrical circuit theory to overcome these limitations and test its performance using data from threatened plant and animal species. The isolation-by-resistance (IBR) model improves over previous gene flow models by using the resistance distance, a graph-theoretic distance metric based in circuit theory (18), to simultaneously consider all possible pathways connecting population pairs (19). The analogy between electrical and genetic connectivity is simple: as multiple or wider conductors connecting two electrical nodes allow greater current flow than would a single, narrow conductor, multiple or wider habitat swaths connecting populations allow greater gene flow. The model is further grounded in rigorous connections between effective resistances in electrical networks and gene coalescence times in population networks (19), giving it a stronger theoretical foundation than either conventional isolation-by-distance (IBD) models (17, 20) or popular LCP models. The IBR model has been shown to reliably predict the effects of landscape heterogeneity on gene flow and genetic structure in artificial population networks (19) but has thus far not been tested with real landscape or genetic data.
Results
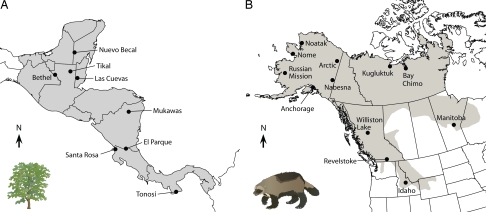
To test the IBR model, we used landscape and genetic data from two threatened species. Both are vulnerable to extinction because of habitat loss and overexploitation (21), and conservation efforts rely on our ability to identify important landscape connections and barriers. The plant data set consisted of eight Central American populations of big-leaf mahogany (Swietenia macrophylla), an insect-pollinated tropical tree, with 287 individuals typed at seven microsatellite loci by Novick et al. (22) (Fig. 1A). The animal data set consisted of 12 western north American populations of the wolverine (Gulo gulo), a large circumpolar mustelid, with 461 animals typed at 12 microsatellite loci by Kyle and Strobeck (23) (Fig. 1B). These previous studies had used conventional IBD analyses to draw conclusions regarding the evolutionary history and future conservation needs of the species. However, such analyses assume habitat to be unbounded and were unable to incorporate effects of spatial heterogeneity, such as range limits, into predictions of genetic differentiation.
Fig. 1.
Sample locations and modeled ranges. (A) Eight big-leaf mahogany populations (22), and modeled range following the Central American landmass from the Isthmus of Tehuantepec south through Panama (shading); extension of modeled range further north or south did not alter results. Sample sizes ranged from 16 to 55 individuals per population. (B) Twelve wolverine populations (23), and contiguous wolverine range in the western United States and Canada. Sample sizes ranged from 14 to 67 individuals per population. All results were consistent using alternative map projections, raster cell sizes, and genetic distance measures.
We reanalyzed each data set using IBR to determine whether incorporating known spatial heterogeneity could improve predictions of genetic differentiation among the sampled populations. Although IBR can accommodate multiple habitat types and landscape features, each with different effects on gene flow (19), for this initial empirical test we used simple, readily obtainable spatial data representing coarse-scale limits to each species' range (Fig. 1). For comparison we repeated analyses with LCP models using the same spatial data. We then evaluated the performance of IBR, LCP, and IBD models as predictors of observed differentiation between population pairs using Mantel tests (24) and causal modeling (25).
Model Performance.
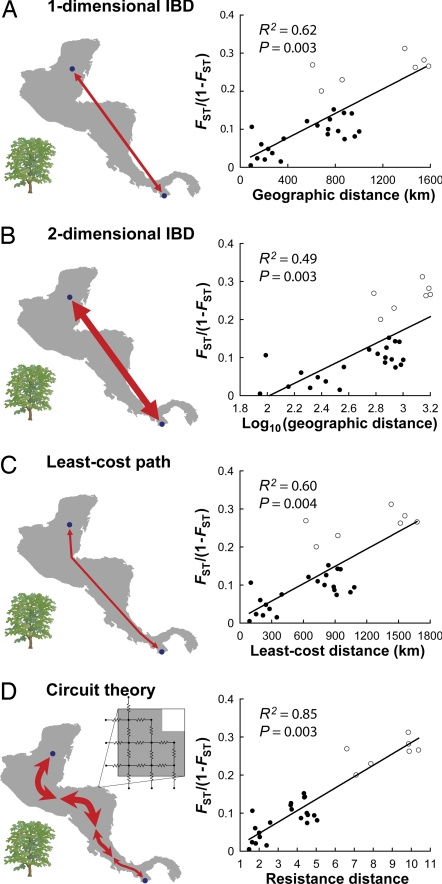
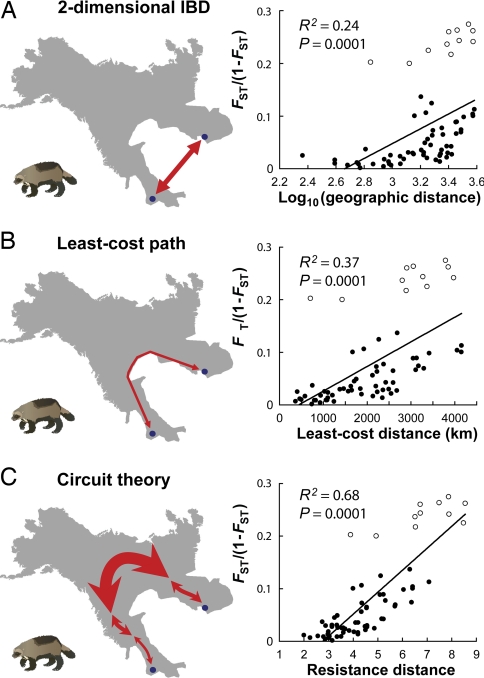
Circuit theory consistently outperformed IBD models for both species. Model fit improved 37–73% for mahogany (Fig. 2) and 188% for wolverines (Fig. 3). By contrast, LCP models yielded more modest improvements or none at all (Figs. 2C and 3B). Causal modeling also strongly supported the IBR hypothesis. For both species, a strong positive relationship between IBR predictions and genetic distance remained after controlling for all competing models (mahogany, r > 0.82, P < 0.0005; wolverines, r > 0.79, P < 0.0001). Conversely, partial correlations between all other models and genetic distances were either nonsignificant or negative (r < 0) after controlling for IBR.
Fig. 2.
Circuit theory outperforms standard models of genetic differentiation among mahogany populations. Range maps illustrate how pairwise isolation is measured under the competing models between two example populations (Nuevo Becal and Tonosí), and scatterplots show performance of the models for all population pairs. Models compared are one-dimensional IBD (A), which assumes habitat to be arranged in an infinite linear array; two-dimensional IBD (B), which assumes habitat to be arranged in an infinite plane; LCP (C), which incorporates spatial heterogeneity but assumes differentiation to vary with distance measured along a single, optimal path; and IBR (D), which predicts greater gene flow and less genetic differentiation with multiple pathways and wider habitat swaths connecting populations. (D Inset) Habitat modeled as a conductive surface. Open circles indicate pairwise comparisons including Tonosí; filled circles indicate comparisons among the remaining seven populations. Linear regression lines include all populations.
Fig. 3.
Circuit theory outperforms standard models of genetic differentiation among wolverine populations. Range maps illustrate how pairwise isolation is measured under the competing models between two example wolverine populations (Idaho and Manitoba). Models compared are two-dimensional IBD (A), the better justified IBD model (see IBD Predictions under Methods), LCP (B), and IBR (C). Open circles indicate pairwise comparisons including Idaho. Linear regression lines include all populations.
Barrier Effects.
Incorporating range shape using circuit theory not only improved gene flow predictions, but also illuminated the effects of other important factors driving population structure. Both of the original studies had found residual variation unexplained by distance and had attributed that variation to migration barriers or other factors driving differentiation within each species' range. For example, Novick et al. (22) noted that the southernmost mahogany population [Tonosí (Fig. 1A)] appeared to be more differentiated from other populations than would be expected from distance alone, and speculated that the increased differentiation may result from geographic barriers and/or historical events. We confirmed that the population was more differentiated than expected from either its distance [r = 0.82, P = 0.0002 (Fig. 2A)] or log-transformed distance [r = 0.87, P = 0.0001 (Fig. 2B)] from the other populations. But evidence for increased differentiation was considerably weakened after controlling for IBR [r = 0.40, P = 0.025 (Fig. 2D)], and the additional differentiation, as measured by partial regression coefficients, was 51–55% smaller.
Correcting for range shape also diminished the inferred importance of barriers among the wolverine populations. As noted by Kyle and Strobeck (23), the Idaho population (Fig. 1B) was more differentiated from others than expected under IBD [r = 0.92, P = 0.0001 (Fig. 3A)]. The authors concluded that this likely resulted from low effective population sizes and restricted gene flow in the southern portion of the species' range, possibly due to barriers or population fragmentation (23). We found that Idaho was still more differentiated than expected from IBR predictions [r = 0.87, P = 0.0001 (Fig. 3C)], but the increased differentiation, as measured by partial regression coefficients, was 23% less than under IBD.
Robustness to Alternative Model Parameterizations.
To examine the effects of different modeling choices on model performance, we repeated analyses with alternative genetic distance measures, map projections, node connection schemes, and extents and resolutions of habitat maps. The improved performance of IBR was consistent across all alternative model parameterizations, as shown in Tables 1 and 2. The use of different cell sizes and genetic distance measures had only modest effects on model fit, and the IBR model consistently outperformed IBD and LCP models in each case. Resistance distances calculated using alternative map projections, with diagonal connections between nodes, or with the modeled mahogany range extended to the north or south were highly correlated with original calculations (r > 0.99) and had no effect on correlations with any of the genetic distance measures.
Table 1.
Results of mahogany analyses using different spatial resolutions and genetic distance metrics
| Spatial distance metric | Genetic distance metric |
|
|---|---|---|
| FST /(1 − FST) | RST /(1 − RST) | |
| Resistance distance (5-km cell size) | 0.85 | 0.77 |
| Resistance distance (50-km cell size) | 0.83 | 0.73 |
| Geographic distance | 0.62 | 0.46 |
| Log10 (geographic distance) | 0.49 | 0.41 |
| Least-cost distance | 0.59 | 0.44 |
Table entries are R2 values. The resistance distance was superior to geographic and least-cost distances in all cases.
Table 2.
Results of wolverine analyses using different spatial resolutions and genetic distance metrics
| Spatial distance metric | Genetic distance metric |
||
|---|---|---|---|
| FST /(1 − FST) | DLR | DS | |
| Resistance distance (5-km cell size) | 0.68 | 0.70 | 0.75 |
| Resistance distance (50-km cell size) | 0.71 | 0.73 | 0.77 |
| Log10 (geographic distance) | 0.24 | 0.24 | 0.28 |
| Least-cost distance | 0.37 | 0.37 | 0.43 |
Table entries are R2 values. The resistance distance was superior to geographic and least-cost distances in all cases.
Discussion
Three main results from our analyses have relevance for landscape genetics and for conservation. First, circuit theory consistently outperformed competing models for both the plant and animal data sets. Combined with its stronger theoretical foundation and better performance with simulated landscape and genetic data (19), this real-world validation means that IBR is better justified for use in landscape genetics and conservation planning than commonly applied IBD or LCP models. Second, we found that range shape—a factor rarely considered in landscape genetic studies—can dramatically affect genetic structure. Thus, applying models that ignore range shape could distort genetic data analyses or predictions of how landscape change will affect genetic connectivity. Finally, much of landscape genetics is concerned with detecting barriers (5), but conventional analyses may exaggerate their effects. In both of the cases we examined, traditional tests for barrier effects could only demonstrate that something other than geographic distance was at work, where “something” might simply reflect nuisance factors like oceans or range limits. By correcting for range shape, the IBR model allowed stronger inference about genuine impediments to gene flow.
Circuit theory gives a better picture of gene flow in these landscapes for several reasons. Unlike IBD models (Fig. 2 A and B and Fig. 3A), IBR explicitly incorporates effects of limited and irregular habitat extent, and obviates the need to choose between one- and two-dimensional habitat models. Unlike LCP models (Figs. 2C and 3B), IBR also accounts for multiple pathways and wider habitat swaths connecting populations (Figs. 2D and 3C). Such features strongly affect patterns of genetic differentiation by increasing migration rates, decreasing rates of genetic drift, and lengthening within-population coalescence times relative to between-population coalescence times (17, 19). For the mahogany data, IBR correctly predicted greater gene flow in the north, where habitat is broadly distributed (and more potential pathways exist), and less in the south, where habitat becomes more constricted. By contrast, within-habitat LCP distances differed little from Euclidean distances and offered no improvement over IBD predictions.
Improvement in model fit was greatest for the wolverine data (Fig. 3), where a more irregular range shape had larger effects on patterns of genetic differentiation. Northern populations are well connected by broad swaths of habitat, but southern populations such as Revelstoke, Manitoba, and especially Idaho (Fig. 1B) are in peninsular portions of the range, where we would expect genetic differentiation to increase more rapidly with distance. Here, LCP predictions improved over IBD by measuring within-habitat distances (Fig. 3B), but IBR also considered habitat width and availability of additional pathways (Fig. 3C), greatly improving connectivity estimates.
Explicitly incorporating the effects of range shape can alter conclusions regarding the roles that barriers and/or historical events have had in generating observed patterns of genetic structure. For mahogany, doing so called into question whether the southernmost population was more differentiated than would be expected in the absence of barriers. Although habitat gaps may currently separate northern and southern mahogany populations (26, 27), they apparently have not yet resulted in substantial divergence (Fig. 2D). Conversely, although correcting for range shape diminished the inferred importance of barriers among the wolverine populations, doing so also increased confidence in their effects. We found that much of Idaho's isolation can be explained by the shape of the wolverine's range, particularly its peninsular southern limit where habitat becomes more constricted (Fig. 3C). Still, because we have accounted for range shape, the substantial remaining variance can now be more confidently attributed to the factors cited by Kyle and Strobeck (23), namely barriers to dispersal or lower population densities in the south.
Importantly, the improved performance of the IBR model was evident when using even coarse-scale, binary habitat data. Moreover, model performance was relatively insensitive to choice of cell size (Tables 1 and 2). These results indicate that process-based connectivity models can still be informative in the many cases for which only poor-quality spatial data are available.
When spatial and genetic data permit, the IBR model can also incorporate more subtle effects of spatial heterogeneity within species' ranges. Model formulations can include fine-scaled variation in habitat quality, landscape resistances derived from detailed movement models, and/or connections between nonadjacent grid cells to accommodate long-distance dispersal (19). Such refinements will necessitate careful choice of appropriate scales of analysis. Even in complex landscapes, however, preliminary simulations indicate that as long as raster grids capture relevant landscape elements (e.g., corridors and barriers), resistance distances measured using different cell sizes are highly correlated (B.H.M., unpublished data).
Of course, genetic structuring is affected not only by spatial heterogeneity, but also by population history, and many factors other than recurrent processes of drift and migration likely affect structuring in these (and nearly all) natural populations. Indeed, both species are undoubtedly undergoing rapid demographic changes due to anthropogenic pressure. However, if equilibrium processes are to be invoked at all (as they are in IBD analyses), circuit theory better predicts their effects than do traditional approaches.
Still, the utility of circuit theory is not limited to equilibrium situations. As with IBD analyses, scatterplots of predicted vs. observed genetic differentiation can be examined to test for equilibrium and evaluate the historical influences of gene flow and genetic drift on population structure (28). IBR can improve upon this method because it accounts for spatial heterogeneity in null model predictions, rather than assuming habitat to be unbounded or simply divided by discrete barriers. Applying these diagnostics to our IBR plots (Figs. 2D and 3C) indicates no evidence for the nonequilibrium effects detectable by the method (28).
We also see potential for directly modeling nonequilibrium processes by drawing on well established methods for analyzing dynamic circuit behavior. The methods incorporate circuit elements such as capacitors and inductors that store and release energy over time and resist instantaneous changes in currents and voltages. Because they can predict transient behavior resulting from initial conditions, perturbations, or changes in network configurations over time, these methods may be useful in predicting lasting effects of historical events in complex population networks. Recent work relating dynamic circuit behavior to random walks (29) is particularly encouraging. Just as the IBR model is based on links between static circuit analyses and random walk theory (19), these findings may pave the way for modeling nonequilibrium processes as well.
More immediately, circuit theory should be of broad use in other areas of connectivity research relevant to ecology, evolution, and conservation. Such applications appear overdue. For example, it has long been known that patterns of current flow across electrical networks have precise relationships with movement patterns and fates of random walkers in analogous graphs (30); this means that circuit theory can be applied to predict not only emergent patterns of gene flow over large distances, but also patterns of individual movement, mortality, and dispersal success at finer scales. As with gene flow predictions, such analyses can better assess the importance of multiple pathways for dispersal than popular LCP models, while well established connections with random walk theory give them a stronger theoretical foundation. We hope that these new models will be used to identify and map critical linkages connecting threatened populations in landscapes under development pressure. Coupled with the present work, such efforts should make increasingly clear how spatial heterogeneity affects connectivity for important ecological and evolutionary processes.
Methods
We chose plant and animal data sets from the literature according to the following criteria. We required that there be published pairwise FST values, large numbers of sampled individuals, published geographic location data, and readily definable geographic ranges. We used the spatial data to generate predictions of relative differentiation between population pairs expected under IBD, LCP, and IBR models, and we compared model predictions with actual genetic distances measured between populations. These analyses involved several choices regarding how each species' range was mapped and discretized to calculate resistance distances, as well as how genetic differentiation among populations was measured. We evaluated the consequences of the choices we made at each step of the modeling process by repeating analyses under different conditions.
Genetic Data.
We obtained pairwise FST values from refs. 22 and 23. These were linearized by using the formula FST/(1 − FST), as advocated by Rousset (20).
Range Maps.
The IBR model can be applied with landscape data varying in complexity from binary range maps to fine-scaled land-cover maps with habitats of differing qualities and permeabilities to movement (19). We used simple binary habitat/nonhabitat maps reflecting objectively definable limits to each species' range for two reasons. First, applying more detailed representations of habitat for either species would have required subjectively choosing among a large number of possible multivariate habitat models. Second, if the IBR model is to be widely applicable, it must make useful predictions of genetic structure using simple, readily obtainable data on habitat quality and distribution.
For wolverines, such a range map was already available (31). For big-leaf mahogany, existing range maps differ markedly from one another (26, 27) and may not adequately capture the species' long-term regional distribution because mahogany is long-lived and likely experienced range expansions during glacial periods (22). Currently, the species is widespread throughout Central America, occurring in every country and in every Mexican state from the Isthmus of Tehuantepec through Panama (26, 27, 32). We therefore considered the Central American landmass to be a suitable and parsimonious compromise between fine-grained models of the species' present distribution and homogeneous, unbounded habitat arrays assumed in IBD analyses. The resulting range model included all Mexican states and Central American countries from the Isthmus of Tehuantepec in the north through Panama in the south (Fig. 1A).
We mapped each species' range using the UTM coordinate system. We then converted each map to a raster grid using 5-km cell sizes, a resolution that was coarse enough to calculate resistance distances using a standard desktop computer, yet fine enough to capture the salient features of each species' range and minimize discretization errors. At this scale, the mahogany and wolverine range maps contained 31,426 and 249,606 habitat cells, respectively.
IBD Predictions.
When conducting IBD analyses, two competing models must be considered: the first assumes habitat to be arrayed in an infinite one-dimensional lattice, whereas the second assumes an infinite two-dimensional lattice. The two models predict genetic differentiation to increase with raw and log-transformed Euclidean distances, respectively (20). Neither model was entirely appropriate for the mahogany range. A two-dimensional model was clearly more appropriate in the wider, northern part of the range (Fig. 1A), but a one-dimensional model was more appropriate in the south, where habitat becomes more elongated (20). We therefore tested both raw and log-transformed geographic distances as predictors of genetic differentiation between mahogany populations. For wolverines, only the two-dimensional IBD model was justified given the broad shape of their range (Fig. 1B).
IBR Predictions.
The IBR model predicts a linear relationship between genetic distance and the resistance distance, a distance metric that takes into account habitat configuration between samples. We calculated resistance distances among sample population locations using Circuitscape version 2.2, a Java software package that takes raster habitat data as input and produces a matrix of resistance distances among population pairs (19). The program represented the two range maps as graphs by replacing habitat cells with nodes and connecting adjacent nodes with unit resistors; non-unit resistors would be used to incorporate data on variation in habitat quality within a species' range, when available (19). Resistors connected first-order neighbors (habitat nodes occurring in the four cardinal directions), as shown in Fig. 2D Inset. The program then calculated resistance distances by using nodal analysis, an algorithm that is widely applied in circuit analysis and described in detail in ref. 19.
LCP Predictions.
Landscape genetic analyses using LCP models assume that genetic distances between population pairs increase with cost-weighted distances measured along the optimal (least-cost) routes connecting them (9, 13–16). We calculated LCP distances (reflecting the shortest within-habitat routes connecting sample pairs; Figs. 2C and 3B), using the PATHMATRIX (33) software package.
Model Comparisons.
To determine which model (one-dimensional IBD, two-dimensional IBD, LCP, or IBR) had the greatest support as a predictor of genetic differentiation, we calculated correlations between genetic distance matrices and matrices of geographic distances, log-transformed geographic distances, LCP distances, and resistance distances. We further compared competing models using causal modeling (25) as applied by Cushman et al. (16); the model with the best support should exhibit not only the highest simple correlation with genetic distance, but also a significant, positive partial correlation with genetic distance after controlling for each of the competing models. We evaluated the significance of correlations and partial correlations using Mantel (24) and partial Mantel (34) tests, computed using the R Package (35) with 10,000 randomizations.
Barrier Effects.
Both of the original studies had found evidence that one population was more differentiated from others than could be explained by distance. We used partial Mantel tests to determine whether increased differentiation remained significant after controlling for IBD, LCP, and IBR predictions.
Robustness to Modeling Choices.
To ensure that model performance was not sensitive to arbitrary choices made regarding range maps or genetic distance measures, we repeated analyses under different conditions. First, to determine whether results were sensitive to choice of cell size, node connection scheme, or distortions introduced by any particular map projection, we calculated resistance distances using cell sizes that were 10-fold larger (50 × 50 km), with nodes connected to both first- and second-order (diagonal) neighbors, and with equidistant conic and Albers equal area conic map projections. Second, in addition to the modeled range shown in Fig. 1, big-leaf mahogany also occurs in the Mexican state of Vera Cruz and in South America. Although including these areas was not expected to affect model predictions because it did not provide new, independent pathways connecting samples, we repeated calculations with modeled habitat extending further to the north (Vera Cruz) and south (Venezuela). Finally, in addition to FST, we calculated correlations using other genetic distance measures reported in the original studies (RST for mahogany, DS and DLR for wolverines).
Acknowledgments
We thank M. Slatkin, P. Flikkema, E. Damschen, J. Novembre, R. Fredrickson, P. Keim, C. Kyle, R. Novick, L. DeWald, T. Theimer, J. Gregg, N. Schumaker, and the Sisk and Waits laboratory groups for discussions. We also thank the Integration and Application Network for supplying wolverine and mahogany symbols. This work was supported in part by graduate fellowships from the Environmental Protection Agency Science to Achieve Results program and the Merriam–Powell Center for Environmental Research. B.H.M. was supported as a Postdoctoral Associate at the National Center for Ecological Analysis and Synthesis, a center funded by National Science Foundation Grant DEB-0553768, the University of California at Santa Barbara, and the State of California.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Rosenberg DK, Noon BR, Meslow EC. Bioscience. 1998;47:677–687. [Google Scholar]
- 2.Crooks KR, Sanjayan M, editors. Connectivity Conservation. Cambridge, UK: Cambridge Univ Press; 2006. [Google Scholar]
- 3.Kareiva P. In: Connectivity Conservation: Maintaining Connections for Nature. Crooks K, Sanjayan MA, editors. Cambridge, UK: Cambridge Univ Press; 2006. pp. 293–295. [Google Scholar]
- 4.Calabrese JM, Fagan WF. Front Ecol Environ. 2004;2:529–536. [Google Scholar]
- 5.Manel S, Schwartz MK, Luikart G, Taberlet P. Trends Ecol Evol. 2003;18:189–197. [Google Scholar]
- 6.Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I. Nature. 1998;392:491–494. [Google Scholar]
- 7.Slatkin M. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- 8.Templeton AR, Robertson RJ, Brisson J, Strasburg J. Proc Natl Acad Sci USA. 2001;98:5426–5432. doi: 10.1073/pnas.091093098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Epps CW, Wehausen JD, Bleich VC, Torres SG, Brashares JS. J Appl Ecol. 2007;44:714–724. [Google Scholar]
- 10.Petren K, Grant PR, Grant BR, Keller LF. Mol Ecol. 2005;14:2943–2957. doi: 10.1111/j.1365-294X.2005.02632.x. [DOI] [PubMed] [Google Scholar]
- 11.Real LA, Henderson JC, Biek R, Snaman J, Jack TL, Childs JE, Stahl E, Waller L, Tinline R, Nadin-Davis S. Proc Natl Acad Sci USA. 2005;102:12107–12111. doi: 10.1073/pnas.0500057102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rueness EK, Stenseth NC, O'Donoghue M, Boutin S, Ellegren H, Jakobsen KS. Nature. 2003;425:69–71. doi: 10.1038/nature01942. [DOI] [PubMed] [Google Scholar]
- 13.Adriaensen F, Chardon JP, Blust GD, Swinnen E, Villalba S, Gulinck H, Matthysen E. Landsc Urban Plann. 2003;64:233–247. [Google Scholar]
- 14.Coulon A, Cosson JF, Angibault JM, Cargnelutti B, Galan M, Morellet N, Petit E, Aulagnier S, Hewison AJM. Mol Ecol. 2004;13:2841–2850. doi: 10.1111/j.1365-294X.2004.02253.x. [DOI] [PubMed] [Google Scholar]
- 15.Vignieri SN. Mol Ecol. 2005;14:1925–1937. doi: 10.1111/j.1365-294X.2005.02568.x. [DOI] [PubMed] [Google Scholar]
- 16.Cushman SA, McKelvey KS, Hayden J, Schwartz MK. Am Nat. 2006;168:486–499. doi: 10.1086/506976. [DOI] [PubMed] [Google Scholar]
- 17.Slatkin M. Evolution (Lawrence, Kans) 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- 18.Klein DJ, Randic M. J Math Chem. 1993;12:81–85. [Google Scholar]
- 19.McRae BH. Evolution (Lawrence, Kans) 2006;60:1551–1561. [PubMed] [Google Scholar]
- 20.Rousset F. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.World Conservation Union. IUCN Red List of Threatened Species. Gland, Switzerland: World Conservation Union; 2006. [Google Scholar]
- 22.Novick RR, Dick CW, Lemes MR, Navarro C, Caccone A, Bermingham E. Mol Ecol. 2003;12:2885–2893. doi: 10.1046/j.1365-294x.2003.01951.x. [DOI] [PubMed] [Google Scholar]
- 23.Kyle JC, Strobeck C. Mol Ecol. 2001;10:337–347. doi: 10.1046/j.1365-294x.2001.01222.x. [DOI] [PubMed] [Google Scholar]
- 24.Mantel N. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 25.Legendre P, Legendre L. Numerical Ecology. Amsterdam: Elsevier; 1998. [Google Scholar]
- 26.Lamb FB. Mahogany of Tropical America: Its Ecology and Management. Ann Arbor, MI: Univ of Michigan Press; 1966. [Google Scholar]
- 27.Calvo J. Evaluation of Mahogany (Swietenia macrophylla King) in Mesoamerica. Costa Rica: PROARCA/CAPAS, Tropical Science Center; 2000. [Google Scholar]
- 28.Hutchison DW, Templeton AR. Evolution (Lawrence, Kans) 1999;53:1898–1914. doi: 10.1111/j.1558-5646.1999.tb04571.x. [DOI] [PubMed] [Google Scholar]
- 29.Qian H, Nassif SR, Sapatnekar SS. IEEE Trans Comput Aided Des Integr Circuits Syst. 2005;24:1204–1224. [Google Scholar]
- 30.Doyle PG, Snell JL. Random Walks and Electric Networks. Washington, DC: Math Assoc of Am; 1984. [Google Scholar]
- 31.Wilson DE. In: Wild Mammals of North America. Chapman JA, Feldhamer GA, editors. Baltimore: Johns Hopkins Univ Press; 1982. pp. 644–652. [Google Scholar]
- 32.Pennington TD, Styles BT, Taylor DAH. Flora Neotrop. 1981;28:1–449. [Google Scholar]
- 33.Ray N. Mol Ecol Notes. 2005;5:177–180. [Google Scholar]
- 34.Smouse PE, Long JC, Sokal RR. Syst Zool. 1986;35:627–632. [Google Scholar]
- 35.Legendre P, Vaudor A. The R Package: Multidimensional Analysis, Spatial Analysis. Montréal: Département de Sciences Biologiques, Université de Montréal; 1991. [Google Scholar]