Abstract
Competition between random genetic drift and natural selection play a central role in evolution: Whereas nonbeneficial mutations often prevail in small populations by chance, mutations that sweep through large populations typically confer a selective advantage. Here, however, we observe chance effects during range expansions that dramatically alter the gene pool even in large microbial populations. Initially well mixed populations of two fluorescently labeled strains of Escherichia coli develop well defined, sector-like regions with fractal boundaries in expanding colonies. The formation of these regions is driven by random fluctuations that originate in a thin band of pioneers at the expanding frontier. A comparison of bacterial and yeast colonies (Saccharomyces cerevisiae) suggests that this large-scale genetic sectoring is a generic phenomenon that may provide a detectable footprint of past range expansions.
Keywords: genetic distance, founder effect, microorganisms, range expansion
A principal tenet of modern evolutionary biology is that Darwinian selection and random genetic drift compete in driving evolutionary change (1). It is widely accepted that genetic drift can have significant effects on small populations (2) that may even lead to speciation (3). In large populations, however, random sampling effects are generally considered weak compared with selection (4–6) (law of large numbers). A major departure from this paradigmatic behavior occurs when large populations undergo range expansions. The descendents of individuals first settling in a new territory are most likely to dominate the gene pool as the expansion progresses (7–9). Random sampling effects among these pioneers results in genetic drift that can have profound consequences on the diversity of the expanding population. Indeed, spatially varying levels of genetic diversity and colonization patterns appear to be correlated in many species (10–13). For example, the often observed south–north gradient in neutral genetic diversity [“southern richness to northern purity” (14)] on the northern hemisphere is thought to reflect past range expansions induced by glacial cycles (12). Although these trends indicate that genetic drift during range expansions has shaped the gene pool of many species, the underlying spatial mechanism remains obscure: Diversity gradients are often difficult to interpret and potentially interfere with the signal of spreading beneficial mutations (15, 16). In fact, a major challenge of present-day population genetics is to decide whether natural selection or a past demographic process is responsible for the prevalence of common mutations (17).
Here, we use simple microbial systems to study the nature of random genetic drift in range expansions of large populations. We observe chance effects that segregate the gene pool into well defined, sector-like regions of reduced genetic diversity. The genetic segregation on the population level is the consequence of number fluctuations on a much smaller scale, within a thin region of reproducing pioneers at the expanding frontier. We expect these patterns to be a general signature of continuous range expansions in populations exhibiting moderate rates of turnover and migration. Our results have significant implications for the identification and demographic interpretation of neutral mutations that swept through a population merely by chance.
Results and Discussion
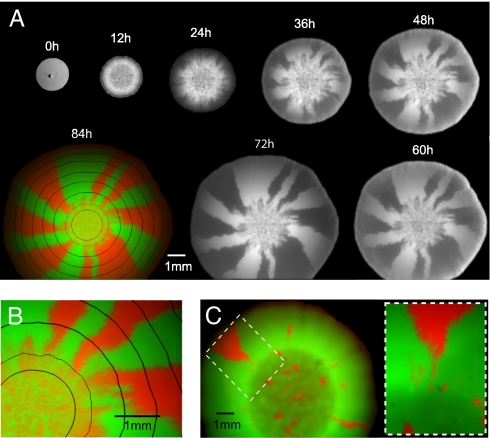
We used two strains of fluorescently labeled bacteria (Escherichia coli) to track both the neutral gene dynamics and the population growth during a range expansion. The strains were genetically identical except for expressing either cyan fluorescent protein (CFP) or yellow fluorescent protein (YFP), which differ only by a single point mutation, from the same constitutively active promoter (see Materials and Methods). We mixed both strains to obtain cultures with different proportions and placed a droplet containing cells at the center of an agar plate containing rich growth medium (LB). Population growth and mutant distribution were monitored for 4 days by means of a stereo microscope. Fig. 1 summarizes the colonial development of a founder population of CFP- to YFP-tagged strains at mixing ratios of 50:50 and 95:5. The time series of fluorescence images in Fig. 1A shows that, in our experiments with immotile E. coli on hard agar, there is no noticeable temporal change in the mutant distribution behind the population front. The mutant distribution evolves only at the leading edge, much like a (dichromatic) carpet that is knitted from the inside to the outside. The final state of this “genetic” carpet (Fig. 1 B and C) thus can be taken as a frozen record of the colonization process.
Fig. 1.
Fluorescent images of bacterial colonies grown from a mixture of CFP- and YFP-labeled cells reveal spatial segregation of neutral genetic markers. (A) Images, taken at 12-h intervals, of a growing colony founded by a 50:50 mixture of bacteria (≈106 E. coli cells) carrying plasmids with either CFP or YFP. Even though the bacteria were otherwise genetically identical, the growing colony shows complete segregation of the two neutral markers (CFP and YFP) over time. The progression of fluorescent images suggests that the dynamics of segregation is restricted to the edges of the colony, whereas, except for a gradual thickening, the interior distribution of CFP and YFP is essentially frozen. The bright-field image at 0 h indicates the initial extent of the founding population, which is mostly lined up on a circle (shown enlarged in SI Fig. 5; the central dark spot is a surface irregularity caused by the delivering pipette). Within 12 and 72 h, the YFP signal is shown; at 84 h (final time point), YFP (green) and CFP (red) images are overlaid on each other with the physical boundary (black lines) of the growing colony at earlier times obtained from bright-light images. (B) Enlarged view at 84 h of the founding region (innermost black circle) and its immediate surroundings. (C) Fluorescence image of a colony produced by a founder population (≈103 cells) with 95% YFP- and 5% CFP-labeled mutants taken 62 h after inoculation. The magnified region shows that, by chance, a sector of the minority population (CFP) can successfully surf (7–9) even in this imbalanced case.
The observed mutant distributions can be partitioned into areas of two very different patterns. The central region of Fig. 1B, which we refer to as the homeland, exhibits a dense speckle pattern reminiscent of the homogeneous and well mixed initial population. This homeland is bounded by a ring of higher speckle density, which was formed at the edge of the initial droplet (innermost black line in Fig. 1B) during evaporation of its water content [most likely due to flows generated by the evaporation process (18), see supporting information (SI) Fig. 5]. From this ring toward the boundary of the mature colony, the population segregates into single-colored domains. A large number of differently labeled flares radiate and gradually coalesce into a few large sectors that reach the edge of the mature colony. Fig. 1C, which corresponds to the unbalanced mixture 95:5 of YFP to CFP, also shows a subdivision into an inner and outer region. The main difference is that the spots and flares are so dilute that their evolution is effectively “noninteracting” and can be individually identified. In particular, only two spots of the minority population evolve into sectors that actually reach the edge of the mature colony.
The formation of morphological sectors, often visible to the naked eye, is a well known phenomenon in microbial colonies harboring cells in different epigenetic states or carrying nonneutral mutations that occur during growth (19, 20). In cases in which the incipient microbial population consists of very few individuals [representing a bottleneck (2)], sector angles are frequently used to infer the initial composition [e.g., in a colony-sectoring assay (21–23)]. In contrast, here, sectoring arises neither because of a small founder population (the initial droplet of Fig. 1A contains ≈106 cells) nor because of mutations and/or epigenetic changes; the genetically segregated domains are nonmorphological, i.e., indiscernible under white light. Furthermore, when populations isolated from sectors of one type were back-competed against ancestral strains of the other type, no significant fitness difference could be detected (SI Fig. 6). We argue, instead, that Fig. 1 is a visible manifestation of random genetic drift acting at the leading edge of a range expansion. Genetic drift refers to the fluctuations of genotype frequencies because of the randomness in the reproduction process, as opposed to persistent differences in their phenotype. These fluctuations can ultimately lead to the fixation of neutral, or even deleterious, genes if they are strong compared with mutations and selection. In a permanently well mixed population grown in a well shaken test tube, genetic drift is usually weak, because the large number of individuals guarantees an almost deterministic growth of the population (law of large numbers). In the present case of a continual range expansion, however, chance effects are strongly enhanced, because the effective population of reproducing individuals is a small fraction of the total population. Only a very thin layer of growing cells (the “pioneers”) at the boundary of the colony is able to pass on their genes to the next layer of outwardly growing cells. Apparently, the reduction in effective population size due to this continual bottleneck is so strong that the population quickly segregates at the wave front into different domains fixed for one of the two mutant strains. The thickness of this boundary layer of growing pioneers is ≈30 μm, which can be estimated from the product of the measured growth velocity of 1.3 × 10−2 μm/s and the cell doubling time of 38 min at the edge of the population. The latter has been extracted by means of time-lapse microscopy of the advancing population front at cellular resolution (see SI Materials and Methods, SI Fig. 7, and SI Movie 1).
After the wave front is segregated into monochromatic flares very early in our experiments, a coarsening process occurs: As the colony grows further, some of the flares are squeezed out of the colonization front and others dominate. Very few domains “survive” and reach the edge of the mature colony. From the view point of population genetics, the significance of this coarsening process is that it leads to a gradual decrease in the genetic diversity at the wave front (9, 13) and generates a characteristic sectoring pattern in the neutral genetic makeup of a population. Understanding this process is also of practical importance, because it is a major source of randomness in the relation between the final sector angles and the composition of the incipient microbial population (23).
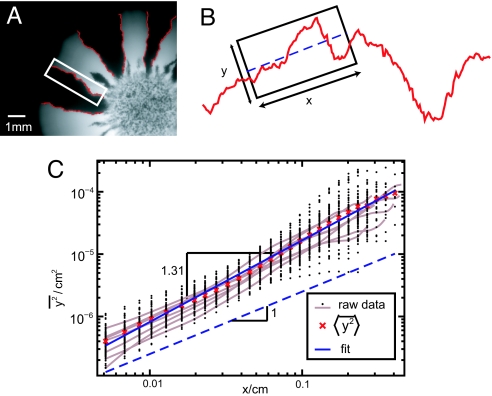
The coarsening dynamics is intimately related with the rugged path of the boundaries separating neighboring domains. A single-colored domain looses contact with the wave front when the leading ends of the flanking boundaries meet and annihilate each other (for an illustration, see SI Fig. 8). Such an unsuccessful sector is, henceforth, trapped in the bulk of the colony without any chance to participate in the further colonization of the agar plate. The sectors surviving this annihilation process, on the other hand, grow in size at the cost of those left behind and appear to surf (7–9) on the wave front. The irregular shapes of domain boundaries drive the decimation of sectors at the colonization front: Domain boundaries would not collide (and annihilate) if they were straight radial lines. To dissect the coarsening mechanism, we studied the wandering of individual domain boundaries quantitatively. Suppose, as a null model, that the leading tip of these lines carry out diffusive random walks around their average directions (24). Then, the mean square transverse displacement from a straight line should increase linearly with the length L of the random walk. To test this hypothesis, we have plotted the mean square displacement from the best linear fit, extracted from a set of 92 domain walls, as a function of length in Fig. 2C on a double-logarithmic scale. Our data indicate that the mean square displacement is indeed well described by a power law, over more than two decades. However, its increase with length L is stronger than linear, i.e., significantly faster than expected for a diffusive random walk; we find the scaling L2ζ with a wandering exponent ζ = 0.65 ± 0.05 > 1/2.
Fig. 2.
Random meandering of domain boundaries is superdiffusive. (A) From fluorescent images of E. coli colonies, domain boundaries were extracted (red lines) by using an edge-detection algorithm. (B) To test whether these domain boundaries resemble time traces of diffusive random walks, we quantified their contour fluctuations as a function of contour length. To this end, we chose a sliding window of size x, parameterized the contour by the transverse displacements y from the linear best fit, and determined the mean square displacement y2 (see Materials and Methods). (C) The mean square transverse displacement is shown as a function of the size x of the sliding window. Ninety-two domain boundaries sampled from 12 different E. coli colonies were analyzed (black points). Even though the data for individual domain boundaries (gray lines) are quite noisy, the ensemble average (red crosses) very closely follows a straight line in this double logarithmic plot. The exponent 2ζ = 1.31 ± 0.1 of the fitted power law (blue line) indicates that boundaries carry out superdiffusive random walks around their average growth direction, i.e., they wander more vigorously than conventional random walks, which have a wandering exponent given by 2ζ = 1.
The superdiffusive character (ζ > 1/2) of the domain wall motion can be explained by the observation that the shape of the colonial edge locally biases the course of the domain boundary. Indeed, in our experiments we found that despite vigorous zigzag motion, there is a clear correlation between the course of the domain wall and the shape of the colony. Notice from the colored mutant distribution in Fig. 1 A and B, in which a series of black lines documents the advance of the population front, that the average local orientation of domain boundaries seems to be nearly perpendicular to the colonial edge (see SI Fig. 9 for a histogram of these local orientations). When this locally preferred direction does not coincide with the overall (i.e., time-averaged) growth direction, the leading tip of the domain wall drifts transverse to the growth direction, as illustrated in Fig. 3. This drift fluctuates in time, just as the shape of the colonial front does, and has zero mean. By providing a fluctuating bias, a time-dependent interface roughness is thus expected to amplify the random wandering of the domain boundaries. The following scaling argument suggests that this effect can even change the character of the random walk: Suppose the interface kinetics of the colony is characterized (25) by a roughness exponent (25) χ and a dynamical exponent ζ; i.e., an interface segment of linear dimension L exhibits displacements of the order Lχ from a straight line and relaxes on a time scale Lζ. We then argue that, during the time t, a transverse drift ∝ L(t)χ − 1 ∼ t(χ − 1)/z acts on the leading tip of a domain wall that is proportional to the tilting angle of a typical undulation on the scale L. This drift entails transverse displacements of the random walk scaling like t1+(χ − 1)/z, suggesting a wandering exponent of ζ = 1 + (χ − 1)/z. Our deterministic analysis is valid only in the case of superdiffusion, such that any intrinsic regular diffusion is negligible on sufficiently large times. A simple bacterial growth model, the Eden model‖ (26), is known to be characterized by χ = 1/2 and z = 3/2 in one dimension, leading to a wandering exponent of ζ = 2/3 [verified by simulations (27)], which is consistent with the exponent ζ = 0.65 ± 0.05 found in our experiments (Fig. 2C).
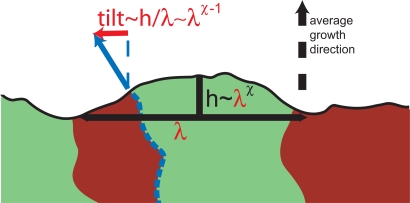
Fig. 3.
The roughness of the colonization front (black line) influences the wandering of locally perpendicular domain boundaries (red lines). In this sketch, the middle domain boundary (dashed blue) tends to follow the blue arrow indicating the local growth direction of the colony. Because of the stochastic surface growth, the local growth direction deviates from the average growth direction (black dashed arrow). Consequently, the domain boundary is subject to a drift (red arrow) transverse to the average growth direction, which is proportional to the local tilt of the interface. This tilt depends on the roughness of the edge of the colony, which we assume to be characterized by a roughness exponent χ: On a length scale λ, the edge of the colony exhibits stochastic shape undulations of amplitude h ∼ λχ. The corresponding tilt, h/λ ∼ λχ − 1, biases the path of the considered domain boundary until the surface irregularity causing it has relaxed. Because of the roughness of the colonial edge, the domain boundaries thus experience a fluctuating bias, which may significantly change their wandering statistics, as explained in the text.
Because the endpoints of domain walls carry out random walks (with the radial axis representing time) and annihilate when they collide, their gradual decimation may be described by a one-dimensional model of annihilating random walkers (28) (with periodic boundary conditions). These random walkers, which come in pairs, annihilate completely for large enough times, if their available space is finite. This scenario applies to a colony that advances approximately as a straight line, as can be induced experimentally through a line inoculation (compare Fig. 4 C and D). For describing the case of circular microbial colony, such as Fig. 1, this analysis has to be modified to account for the fact that the circumference grows linearly in time. Because of this inflation, two domain walls tend to drift apart at a rate proportional to the velocity of the expanding population front. Incorporating this feature into an annihilating walker model with inflation reveals a cross-over time beyond which the increase in the size of the circumference is faster then the random walk of the domain walls. As a consequence, domain walls stop colliding, and their mutual annihilation ends. Thus, domain walls have a nonzero survival probability in circular colonies. A simple scaling argument balancing the opposing effects of inflation and domain-wall wandering indicates that the expected genetic diversity, as reflected by the sectoring pattern, is reminiscent of the shape of the precolonial habitat rather than the initial number of founder cells. In particular, we expect the relationship Ns ∼ r01 − ζ between the average number Ns of surviving sectors and the radius r0 of the homeland (see SI Text). The enhanced wandering (ζ ≈ 0.65 > 1/2) of domain walls amplifies the effects of genetic drift and leads to a more rapid decrease in genetic diversity than expected for a purely diffusive wandering.
Fig. 4.
Segregation patterns of two different microbial species are qualitatively similar but different in detail. (A and B) Both bacteria (E. coli) and yeast (S. cerevisiae) colonies exhibit spatial gene segregation when they grow colonies on agar plates. However, the number of (surviving) sectors is much larger in yeast colonies. For the linear inoculations (C and D), the Petri dish was gently touched with a sterile razor blade that was previously wetted by a liquid culture of a binary mixture of mutants. (E and G) Continuous patches of boundary regions and homeland (bounded by dashed line) at a magnification of ×51 for E. coli (E) and yeast (G). (F and H) Images at single-cell resolution (×100). (F) Tip of a sector dies out (E. coli). (H) Section boundary at the frontier (yeast).
Our neutral model for the coarsening dynamics in expanding E. coli colonies suggests that the sectoring phenomenon is not unique to the considered microbial species but caused by the nature of chance effects during colonization. Indeed, our experiments on growing colonies of haploid Saccharomyces cerevisiae give qualitatively similar results (Fig. 4). Again, we observed an initial coarsening phase, in which domain boundaries annihilate, followed by a stationary phase, in which the sectoring pattern is stable. Compared with E. coli, however, yeast colonies have a much larger number of surviving sectors and much straighter domain boundaries. Both observations are consistent with our model of annihilating domain boundaries, because the frequency of annihilation events is expected to decrease for straighter domain boundaries. The disparate domain wall wandering could be due to differences in the mode of replication, the shape of the cells, or the thickness of the colonies. However, even though the local genetic patterns in colonies of different microbial species might be quite different, the genetic segregation on larger scales seems to be a more general consequence of a microbial range expansion.
Conclusions
The observations presented here strongly suggest that enhanced genetic drift at expanding frontiers can dramatically alter the genetic makeup of large microbial populations. Neutral mutants quickly segregate into monoclonal domains that further coarsen as the colony grows. This coarsening process is driven by the rugged, fractal path of the domain boundaries. The final sectoring of circular colonies is controlled by a balance between the deterministic inflation of the colonial perimeter, dominant at large times, and the stochastic meandering of domain boundaries causing them to annihilate on short times.
More generally, our experiments demonstrate that mutations at expanding frontiers can sweep through a population without any selective advantage. This observation supports theoretical arguments and genetic evidence (15, 29) that common alleles in a population might not necessarily reflect positive selection but, instead, recent range expansions, which have probably occurred in many species (11, 12). Our characterization of the resulting sector-like regions of reduced genetic diversity may serve as a basis for discriminating these surfing mutations from sweeping beneficial mutations. The possibility of superdiffusive wandering (identified here for E. coli) would have to be considered in such a discrimination.
In light of inferring past migrations from spatially resolved genetic data, our results suggest that, apart from a much anticipated general reduction of genetic diversity by a range expansion, a fragmentation of the colonized regions into sectors has to be considered, which is stabilized during expansion by an inflationary effect. Although demonstrated here only for nonmotile microbial species, we believe that the underlying segregation mechanism is quite generic for populations growing continuously and isotropically in two dimensions. As long as these genetic patterns are not blurred by subsequent dispersal and population turnover, they provide a record of the colonization dynamics and information about the precolonial habitat.
Materials and Methods
Bacterial Strains and Plasmids.
Our strains of immotile E. coli are based on a DH5α background and contain plasmids expressing enhanced (e)CFP (A206K) and Venus YFP (A206K), respectively. These strains are identical except for the fluorescent protein structures: Genes for those proteins are cloned between the SacI and XbaI sites of the vector pTrc99A, which is Ampicillin resistant and isopropyl β-d-thiogalactoside (IPTG) inducible. The A206K mutation eliminates protein–protein aggregation. DH5α showed sufficient background level expression even in the absence of IPTG. This expression allowed us to visualize the cells in the absence of any IPTG in the medium. Measurements of the exponential growth rate of both ancestral strains, and of isolates derived from sectors, indicate that the fitness differences are <5% (see SI Fig. 6).
Bacterial Growth Conditions.
The two strains of E. coli were grown overnight in separate liquid cultures at 37°C in rich growth medium (LB) supplemented with Ampicillin. After tenfold dilution in fresh medium, we mixed them to obtain cultures with different proportions of the two kinds of cells. Droplets of different sizes were then pipetted on LB-agar plates (1.5% Agar) with Ampicillin, and grown at 37°C for up to 84 h. Population growth and mutant distribution were monitored at regular time intervals by means of a stereo microscope. No noticeable dependence of the final mutant distribution on the growth phase of the cells at inoculation time could be detected. The cell density used for the inoculation influenced the patchiness of the homeland, but had no apparent effect on the sectoring pattern.
Yeast Strains and Plasmids.
The haploid strains (mating type) of S. cerevisiae, obtained from the E. O'Shea laboratory (Harvard University), are derived from a K699 background and have eCFP or Venus (YFP) driven by the ADH1 promoter integrated into the leu2 locus.
Yeast Growth Conditions.
The two strains were grown overnight in separate liquid cultures at 30°C in yeast extract/peptone/dextrose (YPD) medium. After tenfold dilution in fresh medium, we mixed them to obtain cultures with different proportions of the two kinds of cells. Droplets of different sizes (1–8 μl) were then pipetted on YPD-agar plates, and grown at 30°C for up to 5 days before microscopy.
Microscopy and Image Processing.
The cells were observed by using a Zeiss fluorescent binocular microscope (Zeiss) and with a Zeiss Axiovert 200M (Zeiss), both with a Hamamatsu ORCA-AG camera (Hamamatsu). Emission from YFP was visualized at 535 nm (30-nm bandwidth) upon excitation at 500 nm (20-nm bandwidth), and emission of CFP was visualized at 470 nm (30-nm bandwidth) upon excitation at 430 nm (25-nm bandwidth). The image overlays in Figs. 1 B and C and 4 were created by using Adobe Photoshop (Adobe Systems). The fluorescence channels complement each other without any noticeable gaps (i.e., no loss of plasmids). For Fig. 2C, the contours of 92 domain boundaries (of typical length 0.5 cm) from 18 repeated experiments were extracted on a resolution of 5 μm by using the edge-detection algorithm of the software ImageJ (http://rsb.info.nih.gov/ij/). A C++ code was used to quantify the wandering of a given section (length L) of a domain boundary in the following way. First, a longitudinal axis was defined by fitting a straight line to the considered contour (best linear fit). Second, the contour was parameterized in terms of its displacement transverse to the longitudinal axis y(x) as a function of the longitudinal coordinate x ∈ (0, L) (see Fig. 2B). Third, we computed the mean square displacement y2(L) = ∫0Ldx y2(x)/L. To improve statistics, we averaged this quantity over the possible ways to define a section of length L within a domain boundary of larger length (running average). There are various other ways to quantify the wandering, which should all give the same asymptotic scaling. Because, however, the range of our data are limited to two orders of magnitude (see Fig. 2C), the fitted exponent somewhat depends on such a definition. We estimated the error in the exponent ζ ≈ 0.65 ± 0.05 by probing its dependence on the quantification method. This source of error is, by far, larger than the standard deviation of the ensemble average in Fig. 2C (red crosses) from the fitted line (blue).
Supplementary Material
Acknowledgments
We thank J. Shapiro and T. Shimizu for helpful discussions; T. Shimizu (Harvard University) together with H. Berg (Harvard University) and V. Sourjik (Center for Molecular Biology Heidelberg, ZMBH, Heidelberg, Germany) for the E. coli strains and plasmids; the O'Shea laboratory for S. cerevisiae strains; and M. Bathe, K. Foster, M. Nowak, B. Stern, J. Wakeley, and J. Xavier for providing comments on the manuscript. This work was supported by German Research Foundation Grant Ha 5163/1 (to O.H.), National Science Foundation through Grant DMR-0654191, the Harvard Materials Research Science and Engineering Center through Grant DMR-D2138D5 (to D.R.N.), a Human Frontiers Grant (to S.R.), a Keck Futures Initiative Grant (to S.R.), and a National Institute of General Medical Sciences Center grant (to P.H. and S.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710150104/DC1.
Eden M (1961) Proceedings of the Fourth Berkeley Symposium on Mathematical Statistics and Probability, June 20–30, 1960, Berkeley, CA, 4:233.
References
- 1.Nei M. Mol Biol Evol. 2005;22:2318–2342. doi: 10.1093/molbev/msi242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nei M, Maruyama T, Chakraborty R. Evolution (Lawrence, Kans) 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- 3.Mayr E. Animal Species and Evolution. Cambridge, MA: Belknap; 1963. [Google Scholar]
- 4.Elena SF, Lenski RE. Nat Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 5.Hegreness M, Shoresh N, Hartl D, Kishony R. Science. 2006;311:1615–1617. doi: 10.1126/science.1122469. [DOI] [PubMed] [Google Scholar]
- 6.Hartl DL, Clark AG. Principles of Population Genetics. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 7.Edmonds CA, Lillie AS, Cavalli-Sforza LL. Proc Natl Acad Sci USA. 2004;101:975–979. doi: 10.1073/pnas.0308064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klopfstein S, Currat M, Excoffier L. Mol Biol Evol. 2006;23:482–490. doi: 10.1093/molbev/msj057. [DOI] [PubMed] [Google Scholar]
- 9.Hallatschek O, Nelson DR. Theor Popul Biol. 2007 doi: 10.1016/j.tpb.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Cavalli-Sforza LL, Menozzi P, Piazza A. Science. 1993;259:639–646. doi: 10.1126/science.8430313. [DOI] [PubMed] [Google Scholar]
- 11.Cavalli-Sforza LL, Feldman MW. Nat Genet. 2003;33:266–275. doi: 10.1038/ng1113. [DOI] [PubMed] [Google Scholar]
- 12.Hewitt G. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 13.Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Proc Natl Acad Sci USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hewitt GM. Biol J Linnean Soc. 1996;58:247–276. [Google Scholar]
- 15.Currat M, Excoffier L, Maddison W, Otto SP, Ray N, Whitlock MC, Yeaman S. Science. 2006;313:172a. doi: 10.1126/science.1122822. [DOI] [PubMed] [Google Scholar]
- 16.Schroeder KB, Schurr TG, Long JC, Rosenberg NA, Crawford MH, Tarskaia LA, Osipova LP, Zhadanov SI, Smith DG. Biol Lett. 2007;3:218–223. doi: 10.1098/rsbl.2006.0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pennisi E. Science. 2007;316:1690–1692. doi: 10.1126/science.316.5832.1690. [DOI] [PubMed] [Google Scholar]
- 18.Deegan RD, Bakajin O, Dupont TF, Huber G, Nagel SR, Witten TA. Nature. 1997;389:827–829. doi: 10.1103/physreve.62.756. [DOI] [PubMed] [Google Scholar]
- 19.Shapiro JA. BioEssays. 1995;17:597–607. doi: 10.1002/bies.950170706. [DOI] [PubMed] [Google Scholar]
- 20.Ben-Jacob E. Philos Trans R Soc London Ser A. 2003;361:1283–1312. doi: 10.1098/rsta.2003.1199. [DOI] [PubMed] [Google Scholar]
- 21.Koshland D, Kent JC, Hartwell LH. Cell. 1985;40:393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- 22.Murray AW, Szostak JW. Cell. 1983;34:961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- 23.McMurray MA, Gottschling DE. Science. 2003;301:1908–1911. doi: 10.1126/science.1087706. [DOI] [PubMed] [Google Scholar]
- 24.Berg HC. Random Walks in Biology. Princeton: Princeton Univ Press; 1993. [Google Scholar]
- 25.Halpin-Healy T, Zhang Y. Phys Rep. 1995;254:215–414. [Google Scholar]
- 26.Kardar M, Parisi G, Zhang YC. Phys Rev Lett. 1986;56:889–892. doi: 10.1103/PhysRevLett.56.889. [DOI] [PubMed] [Google Scholar]
- 27.Saito Y, Mueller-Krumbhaar H. Phys Rev Lett. 1995;74:4325–4328. doi: 10.1103/PhysRevLett.74.4325. [DOI] [PubMed] [Google Scholar]
- 28.Toussaint D, Wilczek F. J Chem Phys. 1983;78:2642–2647. [Google Scholar]
- 29.Biek R, Henderson JC, Waller LA, Rupprecht CE, Real LA. Proc Natl Acad Sci USA. 2007;104:7993–7998. doi: 10.1073/pnas.0700741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.