Abstract
Evolutionarily significant selective sweeps may result in long stretches of homozygous polymorphisms in individuals from outbred populations. We developed whole-genome homozygosity association (WGHA) methodology to characterize this phenomenon in healthy individuals and to use this genomic feature to identify genetic risk loci for schizophrenia (SCZ). Applying WGHA to 178 SCZ cases and 144 healthy controls genotyped at 500,000 markers, we found that runs of homozygosity (ROHs), ranging in size from 200 kb to 15 mb, were common in unrelated Caucasians. Properties of common ROHs in healthy subjects, including chromosomal location and presence of nonancestral haplotypes, converged with prior reports identifying regions under selective pressure. This interpretation was further supported by analysis of multiethnic HapMap samples genotyped with the same markers. ROHs were significantly more common in SCZ cases, and a set of nine ROHs significantly differentiated cases from controls. Four of these 9 “risk ROHs” contained or neighbored genes associated with SCZ (NOS1AP, ATF2, NSF, and PIK3C3). Several of these risk ROHs were very rare in healthy subjects, suggesting that recessive effects of relatively high penetrance may explain a proportion of the genetic liability for SCZ. Other risk ROHs feature haplotypes that are also common in healthy individuals, possibly indicating a source of balancing selection.
Keywords: genomewide, selection, haplotype, HapMap, susceptibility
The recent development of microarray platforms, capable of genotyping hundreds of thousands of SNPs, has provided an opportunity to rapidly identify novel susceptibility genes for complex phenotypes. Studies employing genotyping microarrays have typically used a whole-genome association (WGA) approach, in which each SNP is examined individually for association with disease (1); multiple testing requires that statistical thresholds for WGA approach 10−7 or lower (2). Given the presumably polygenic nature of complex illness, this conservative strategy inevitably results in false negatives in the search for susceptibility genes (3). At the same time, structural properties of WGA datasets, including patterns of linkage disequilibrium (LD), have not yet been exploited in these analyses. Consequently, we developed an analytic approach, termed whole-genome homozygosity association (WGHA), which first identifies patterned clusters of SNPs demonstrating extended homozygosity and then employs both genomewide and regionally specific statistical tests for association to disease. In the present study, we used WGHA in a case-control dataset of patients with schizophrenia (SCZ) and healthy volunteers, genotyped at ≈500,000 SNPs, to detect novel susceptibility loci for SCZ.
SCZ is a disease with estimated lifetime morbid risk approaching 1% worldwide. Although genetic epidemiologic studies have revealed high heritability estimates (70–80%) for SCZ, identification of susceptibility genes remains challenging. As with other complex diseases, linkage studies have revealed multiple candidate regions with modest LOD scores (4), whereas studies of individual candidate genes are inherently limited in scope. By contrast, WGHA (described in detail below) presents an opportunity for rapidly identifying susceptibility loci broadly across the genome, yet with resolution sufficient to implicate a circumscribed set of candidate genes. WGHA is designed to be sensitive for detecting loci under selective pressure, and recent data suggest that signatures of evolutionary selection may be strongly observed in genes regulating neurodevelopment (5, 6). Thus, WGHA may be particularly effective for SCZ, which is thought to have a primary pathophysiological basis in abnormal neurodevelopmental processes (7).
Regions of extended homozygosity across large numbers of consecutive SNPs form the basis of WGHA analysis. In general, extent of homozygosity is a function of LD within a chromosomal region, which in turn is a function of recombination rates and population history (8–10). Size and structure of LD blocks vary widely across the genome and across populations (11), and regions of extensive long-range LD may be indicative of partially complete selective sweeps of functional significance (12). For example, variants of the extended haplotype homozygosity test (13) have been used to examine identity-by-descent across unrelated chromosomes in HapMap (14) and other population samples, identifying known loci under selection (e.g., LCT in Europeans, see refs. 15 and 16). A logical consequence of such identity across unrelated chromosomes is that long stretches of homozygosity may be observed in healthy individuals from outbred populations lacking any known consanguineous parentage (17, 18). However, the relative commonality of this phenomenon has not been systematically documented in large datasets at high resolution. Moreover, although homozygosity mapping has successfully identified disease loci in pedigrees marked by Mendelian illness (19), the ability of such a method to detect susceptibility loci in common disease has not been examined in a case-control study. We present data addressing both normal patterns of homozygosity and use of these patterns in WGHA mapping of SCZ.
Results
The sample of 178 unrelated Caucasian SCZ cases and 144 unrelated Caucasian, sex-matched controls were ascertained and psychiatrically diagnosed at a single geographic site (the Zucker Hillside Hospital, ZHH), as described in ref. 20. DNA extracted from whole blood was assayed at 500,568 SNPs (mean spacing = 5.8 kb; mean heterozygosity = 27%). After performance of quality control procedures (see Methods for details), 444,763 autosomal (and pseudoautosomal) SNPs demonstrating genotype call replicability >99.4% were available for WGHA analysis.
Identification of Common ROHs in ZHH Subjects.
The first step of WGHA analysis is the identification of runs of homozygosity (ROHs) in each subject, defined in the present study as any window of 100 or more consecutive SNPs on a single chromosome not receiving a heterozygous call (see Methods for details). Because WGHA seeks to identify frequently observed variants that can be statistically compared in a case-control design, only those ROHs in which 10 or more subjects share ≥100 identical homozygous calls were retained for further analysis. Each common ROH was then scored “present” or “absent” for each subject.
A total of 339 common ROHs were thus identified [supporting information (SI) Table 4], encompassing ≈12–13% of the genome as measured both by number of included SNPs and total chromosomal length. The six longest ROHs, ranging from 6 to 15.6 mb, encompass the centromeres of chromosomes 3, 5, 8, 11, 16, and 19. In part, this is a function of long regions with no SNPs ascertained; nevertheless, in each case, these centromeric gene deserts are flanked by homozygous regions containing hundreds of SNPs. This phenomenon may reflect meiotic drive, the selective bias toward transmission of a single meiotic product that has been observed at centromeric DNA across species (5, 21). The greatest number of consecutive SNPs (852) is found in roh172, spanning the centromere of chromosome 8; this region, which contains the gene encoding syntrophin γ1 (SNTG1), has been highlighted in several genomewide studies of selective sweeps (5, 14–16), providing a positive control for our method.
There are nine ROHs that were very common (>25% frequency) in ZHH healthy controls. As displayed in Table 1, publicly available data indicate that these regions are not marked by excessive copy number variation or segmental duplication, nor do they appear to have abnormally low recombination rates (14). However, examination of Haplotter data (ref. 15, accessed at http://hg-wen.uchicago.edu/selection/haplotter.htm) indicates high scores for each of these regions on one or more measures of positive selection in Caucasian samples. Several gene categories identified in studies of selective pressure (5, 14–16) are evident in these regions, including genes involved in the immune system (on chromosomes 6p, 12q, and 5q), olfactory receptors (6p and 11p), members of the dystrophin protein complex (SNTG1 and DGKZ), and many other CNS-expressed genes (e.g., GPHN, UNC5D, and ATXN2). Across all 339 regions, ROH frequency in controls was significantly correlated with maximal integrated haplotype score (iHS, ref. 15; r = 0.33, P = 3.4 × 10−10) and Tajima's D (r = 0.30, P = 2.8 × 10−8); these correlations are comparable to the intercorrelation of maximal iHS and D for the same regions (r = 0.30, P = 1.3 × 10−8).
Table 1.
List of nine ROHs with frequency ≥25% in the healthy cohort (n = 144)
| ROH | Chr | Start (B35) | End (B35) | Length, bp | No. of SNPs | Control, n (%) | iHS max | Tajima D max | Fst max | No. of deletions/duplications/hotspots | Core SNP | Allele |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| roh172 | 8 | 42588087 | 53273605 | 10,685,518 | 852 | 77 (53.5) | 3.90 | 2.00 | 0.90 | 0/6/32 | rs11136221 | C |
| roh134 | 6 | 26216147 | 29800284 | 3,584,137 | 489 | 52 (36.1) | 2.40 | 1.50 | 0.52 | 0/3/13 | rs2859365 | A |
| roh89 | 4 | 32428277 | 34888919 | 2,460,642 | 252 | 52 (36.1) | 3.50 | 2.40 | 0.62 | 9/2/17 | rs7340793 | G |
| roh241 | 11 | 46212732 | 49874378 | 3,661,646 | 323 | 48 (33.3) | 2.30 | 1.75 | 0.70 | 11/1/3 | rs10838852 | T |
| roh291 | 14 | 65475183 | 67065410 | 1,590,227 | 197 | 47 (32.5) | 2.50 | 2.75 | 0.99 | 0/0/9 | rs2053149 | C |
| roh171 | 8 | 33590815 | 36749387 | 3,158,572 | 443 | 41 (28.5) | 0.60 | 2.80 | 0.42 | 0/0/19 | rs2719307 | A |
| roh238 | 11 | 37492528 | 40090659 | 2,598,131 | 386 | 39 (27.1) | 1.10 | 5.10 | 0.45 | 0/0/25 | rs7938730 | T |
| roh275 | 12 | 109752647 | 111733445 | 1,980,798 | 205 | 37 (25.7) | 1.10 | 1.80 | 0.22 | 0/0/8 | rs17696736 | G |
| roh125 | 5 | 129481382 | 132022568 | 2,541,186 | 286 | 36 (25.0) | 1.20 | 1.25 | 0.65 | 1/1/12 | rs31251 | A |
Chromosomal coordinates listed from National Center for Biotechnology Information (NCBI) build 35. Columns 8–10 represent maximal values for alternate metrics of positive selection, derived from Haplotter (ref. 15, http://hg-wen.uchicago.edu/selection/haplotter.htm). Number of deletions, duplications, and recombination hotspots derived from HapMap version 21a (ref. 14, http://hapmap.org). Alleles are listed consistent with strand as displayed in HapMap v21a. Note that all alleles are designated as derived alleles according to dbSNP build 127 (except for rs7340793, for which ancestral/derived alleles are not available).
The final columns of Table 1 indicate the “core” SNP and allele demonstrating the maximal degree of overlap across all carriers of a given ROH. Extent of allelic/haplotypic sharing is variable because ROHs can vary in length across individual subjects with differing degrees of overlap and extension (see Methods); however, in general, common ROHs represent carriers of the same alleles. Core SNPs were determined by the most strongly significant genotypic χ2 comparison (within the ZHH healthy control cohort) of carriers vs. noncarriers of each ROH (10−4 < P values <10−21). Notably, for each core SNP, ROH carriers bear the derived (nonancestral) allele, consistent with an incomplete selective sweep.
Validation of ROH Methodology in HapMap Samples.
Using publicly available data (www.affymetrix.com), we applied analogous methods to Affymetrix 500K data derived from all unrelated individuals in each of the three major HapMap populations (Caucasian, African, and Asian). For each population, we identified all ROHs of length ≥100 SNPs that were present in at least 20% of subjects using all available Affymetrix SNPs (no filtering applied). As described below, we tested a series of hypotheses, to support our interpretation that common ROHs indicate loci under selective pressure as well as to eliminate the possibility that biased SNP selection on the Affymetrix array might have served to confound this interpretation. Specifically, we predicted considerable overlap between ROHs identified in our control cohort and Caucasian HapMap samples and considerable disjunction with African and Asian samples. Moreover, we predicted that the African cohort would possess fewer ROHs, whereas the Asian cohort would demonstrate a greater frequency of ROHs, based on relative age and homogeneity of the respective lineages (9, 22).
Of the 32 ROHs that were found in the HapMap CEU (CEPH Utah residents with ancestry from Northern and Western Europe) sample (n = 60 founders), all but one overlapped with a ROH that was common (>5%) in our Caucasian controls. Moreover, the four most common ROHs in the CEU sample coincided with four of the five most common ROHs in our control sample (roh172, roh134, roh89, and roh291). By contrast, no common ROHs were identified in the YRI (Yoruba from Ibadan, Nigeria) founder sample (n = 60) using the same 20% threshold, consistent with their much more ancient lineage and resultant increase in recombination events. Results from the YRI sample also provided an important test of a potential artifact. We examined the heterozygosity (in YRI founders) of the 1,673 SNPs that were constituents of the most commonly found in ROHs in our Caucasian control sample. In YRI founders, the heterozygosity of these 1,673 SNPs (31.2%) was higher than the mean heterozygosity across the remainder of the array (28.8%). This result demonstrates that the identification of ROHs is not driven by artifactual properties of specific SNPs on the array (i.e., these are not SNPs that always lead to low heterozygosity calls due to poor signal/noise characteristics or absolute rarity).
Consistent with very recent data on allele frequency spectra (22), the Asian HapMap samples show greater long-range LD relative to the CEU samples, despite the fact that the Asian samples combine two distinct subgroups [CHB and JPT (Han Chinese from Beijing and Japanese from Tokyo)]. By using the same 20% frequency threshold, more than three times as many ROHs were identified as in the CEU sample. Moreover, the most common ROH in the Asian sample (53.3% frequency) was not among the common ROHs identified in the CEU sample. Located in the centromeric region of chromosome 16, this region overlapped with roh306 (SI Table 4), which was only the 40th most common ROH in the Caucasian ZHH control sample.
Comparison of ROH Frequency in ZHH Patients and Controls.
The total number of common ROHs marked “present” was summed for each ZHH subject to permit genomewide comparison across diagnostic groups. Of a total possible sum of 339, patients with schizophrenia demonstrated a significantly greater number of common ROHs (mean = 31.7, SD = 12.3) relative to healthy volunteers (mean = 28.0, SD = 12.8; t320 = 2.62, P = 0.009). Nine individual ROHs significantly (P < 0.01) differed in frequency between cases and controls (Table 2); each was more common in SCZ cases.
Table 2.
List of nine risk ROHs significantly overrepresented in SCZ cases (P < 0.01)
| ROH | Chr | Start (B35) | End (B35) | Length, bp | No. of SNPs | Cases, n (%) | Control, n (%) | χ2 | P | Genes | Core SNP | Risk allele |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| roh250 | 11 | 102488778 | 102947117 | 458,339 | 103 | 14 (7.9) | 0 (0.0) | Fisher | 0.0004 | DYNC2H1 | rs11225703 | T |
| roh321 | 18 | 37022928 | 37619977 | 597,049 | 119 | 15 (8.4) | 1 (0.7) | Fisher | 0.0012 | (PIK3C3) | rs2848745 | C |
| roh314 | 17 | 41169023 | 42622984 | 1,453,961 | 211 | 40 (22.5) | 14 (9.7) | 9.271 | 0.0023 | CRHR1, IMP5, MAPT, STH, KIAA1267, LRRC37A, ARL17, LRRC37A2, NSF; WNT3, WNT9B, GOSR2, RPRML, CDC27 | rs17651507 | T |
| roh52 | 2 | 175671012 | 176445047 | 774,035 | 115 | 17 (9.6) | 2 (1.4) | Fisher | 0.0032 | CHN1, ATF2, ATP5G3 | rs2437896 | T |
| roh15 | 1 | 158440777 | 159015569 | 574,792 | 173 | 20 (11.2) | 4 (2.8) | 8.256 | 0.0041 | DUSP12, ATF6, OLFML2B, NOS1AP | rs2499846 | T |
| roh129 | 5 | 154592379 | 155033077 | 440,698 | 116 | 13 (7.3) | 1 (0.7) | Fisher | 0.0042 | (SGCD, MRPL22) | rs4958803 | G |
| roh291 | 14 | 65475183 | 67065410 | 1,590,227 | 197 | 86 (48.3) | 47 (32.6) | 8.068 | 0.0045 | GPHN, C14orf54, MPP5, ATP6V1D, EIF2S1, PLEK2 | rs2053149 | C |
| roh55 | 2 | 188489676 | 190772106 | 2,282,430 | 274 | 31 (17.4) | 10 (6.9) | 7.855 | 0.0051 | GULP1, DIRC1, COL3A1, COL5A2, WDR75, SLC40A1, NS3TP1, ASNSD1, ANKAR, OSGEPL1, ORMDL1, PMS1, GDF8 | rs7582658 | G |
| roh173 | 8 | 57989122 | 58616467 | 627,345 | 120 | 20 (11.2) | 5 (3.5) | 6.7 | 0.0096 | IMPAD1 | rs2119783 | G |
Genes previously associated with SCZ listed in bold. Alleles listed consistent with strand as displayed in HapMap v21a.
Several features of these nine “risk ROHs” are notable. First, greater than half (54.9%) of healthy controls, but only 19.1% of SCZ subjects, did not have any risk ROHs present in their WGHA data (χ2 = 44.7, df = 1, P = 2.3 × 10−11; permuted P = 0.0022; odds ratio = 5.15, 95% CI = 3.13–8.46). Moreover, as the number of risk ROHs increases, risk of illness increases dramatically. Using logistic regression, the total number of risk ROHs significantly predicted group status (χ2 = 62.6, df = 1, P = 2.51 × 10−15; permuted P = 0.00095), with each additional risk ROH imparting a hazard ratio of 2.83 (95% CI = 2.10–3.81; see also Table 3).
Table 3.
Odds of SCZ as a function of number of risk ROHs present in a given individual
| #Risk ROHs (sum) | Cases, n | Cases, % | Control, n | Control, % | OR* | 95% CI |
|---|---|---|---|---|---|---|
| 0 | 34 | 19.1 | 79 | 54.9% | ||
| 1 | 70 | 39.3 | 49 | 34.0% | 3.3 | 1.9–5.7 |
| 2 | 43 | 24.2 | 13 | 9.0% | 5.4 | 3.7–16.1 |
| 3 | 25 | 14.0 | 3 | 2.1% | 24† | 6.9–83.9 |
| 4 | 5 | 2.8 | 0 | 0.0% | ||
| 5 | 1 | 0.6 | 0 | 0.0% |
Odds ratios (OR) computed by using sum = 0 as reference category.
*OR compared to Sum = 0.
†Sum ≥ 3 compared to Sum = 0.
Six of the nine risk ROHs listed in Table 2 range from uncommon to very rare in healthy controls. One ROH (roh250), containing the gene encoding the dynein cytoplasmic 2, heavy-chain 1 protein (DYNC2H1 on chromosome 11q), was exclusively observed in SCZ; in other words, this genetic variant demonstrated 100% penetrance for illness in our sample. However, one very common ROH in healthy subjects (roh291, see Table 1) also conferred risk for SCZ (χ2 = 8.1, df = 1, P = 0.0045). The odds ratio for this ROH was moderate (1.93; 95% CI = 1.22–3.04), although population attributable risk was 12% because of its commonality. This ROH is centered on the very large (≈675 kb) gene GPHN, which codes for gephyrin, a protein scaffold that serves to anchor GABA receptors in the postsynaptic membrane. Patients with schizophrenia who exhibited this ROH tended to carry the same derived allele as was noted in those controls carrying the ROH (rs2053149 C). Comparison of CC genotype frequency for this core allele in patients carrying the ROH to control non-ROH carriers was strongly significant (P = 1.37 × 10−18). Core SNP for other risk ROHs in Table 2 was determined by the homozygous allele that was most common to patients carrying the ROH yet least common among controls not carrying the ROH. For six of the nine risk ROHs, all or nearly all patients (0–2 exceptions) carried the same core allele, which was the derived allele; however, a sizable fraction of patients carrying roh55, roh314, and roh321 demonstrated homozygosity at the alternate alleles.
Genes Within Risk ROHs.
Four of the nine ROHs contain or immediately neighbor genes that have been linked to schizophrenia, a result that is significantly unlikely by chance (binomial distribution P < 0.01) even if a 10% prior probability is assigned to each region (4, 23). Specifically, roh15 on chromosome 1q contains NOS1AP (formerly CAPON), which has been related to schizophrenia in both genetic linkage and association studies, as well as in postmortem gene-expression studies (24). This protein competes with PSD95 for binding to neuronal nitric oxide synthase (nNOS), thereby disrupting neuronal NMDA receptor transmission at the postsynaptic density. Similarly, roh52 contains ATF2, a downstream target of the mitogen-activated protein kinase/extracellular signal-regulated kinase signaling pathway triggered by nNOS; protein levels of activating transcription factor 2 have been reported to be elevated in postmortem SCZ brain tissue (25). Further, roh314 contains NSF (encoding N-ethylmaleimide sensitive fusion), which regulates dissociation of the SNARE complex and binds to the GluR2 subunit of AMPA glutamate receptors. Abnormalities in this gene have been also linked with schizophrenia in both gene-expression and genetic-association studies (23, 26). In addition to NSF, roh314 (at chromosome 17q21) contains MAPT (microtubule-associated protein tau). MAPT has been reported to contain a common inversion under selective pressure, resulting in a distinctive haplotypic genealogy that has been associated with multiple neurological disorders (27).
Two ROHs that were significantly overrepresented in patients with SCZ contained no known genes (roh321 on chromosome 18q and roh129 on 5q). Although both regions include one or more ESTs and may harbor as-yet-unknown regulatory elements, it is also possible that the extent of allelic hitchhiking is not fully captured by our ROH methodology and may impact genes immediately neighboring these regions (8). Consequently, the first gene located within 500 kb in either direction of these ROHs is listed in parentheses in Table 2. PIK3C3 (adjacent to roh129) encodes phosphoinositide-3-kinase, class 3. A promoter region variant in this gene has been associated with SCZ in three studies to date (23).
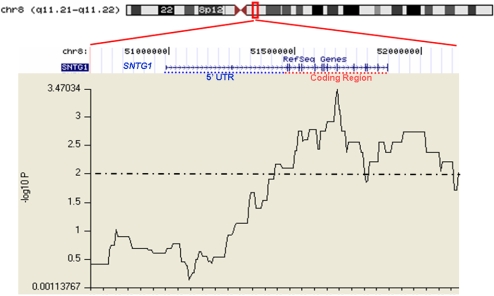
Finally, exploratory analyses examining binarized individual SNP data revealed subregions of two additional ROHs that were significantly overrepresented in SCZ cases relative to controls (SI Table 5). Segments of the very large ROH on chromosome 8 (roh172), demonstrated a strong differentiation between cases and controls (maximal χ2 = 12.9, df = 1, P = 3.28 × 10−4) occurring directly in the coding region of SNTG1 (Fig. 1). Notably, SNTG1 is expressed exclusively in neurons, including hippocampal pyramidal cells, cerebellar Purkinje cells, and multiple cortical regions, where it binds to dystrophin, the dystrobrevins, and diacylglycerol kinase, ζ(DGKZ) in the postsynaptic density.
Fig. 1.
Graphical depiction of statistical comparisons (SCZ vs. control) at individual SNPs within roh172 on chromosome 8q. Chromosomal context is depicted in ideogram at the top. Gene location (build 35 coordinates) for SNTG1 is depicted immediately below the ideogram. Coding region of SNTG1 is indicated by a red dotted line; exons are indicated by horizontal lines. The jagged line depicts −log10 P values for case-control comparisons at each binarized SNP.
Discussion
Using dense, whole-genome microarray SNP data, we observed that ROHs ranging in size from 200 kb to >15 mb were common even in healthy individuals from an outbred population (U.S. Caucasians residing in New York City/Long Island). These homozygous regions are both too common and too small to suggest recent consanguinity (28). Our data are consistent with the possibility that common ROHs mark regions under selective pressure for several reasons; specifically, the most common ROHs in the present study (i) have been implicated in prior studies using varying coalescent models and statistical assumptions and are strongly correlated with other reported measures of selection (5, 14–16); (ii) are characterized by the derived allele; (iii) contain genes recognized by other methods as under strong selective pressure in Caucasians, such as SNTG1 (included in roh172) and ALDH2 (roh275); (iv) are replicated in Caucasian HapMap samples; (v) are not replicated in non-Caucasian HapMap samples. Because the SNP selection of the current generation of whole-genome microarrays is still limited and does not permit uniform coverage across the genome, the presence of SNP ascertainment bias limits formal statistical testing of the evidence for selection (29). In addition, it is important to note that effects of population bottlenecks and neutral drift can sometimes mimic results deriving from positive selection.
Nevertheless, ROH frequency is a readily available measure for statistical comparisons in a case-control design. In case-control comparison, we observed that ROHs were overrepresented in SCZ at a genomewide level. The effect size (Cohen's d) was ≈0.30, a small to moderate effect comparable with the effect size seen across many biological studies of schizophrenia. Although subtle differences in ascertainment between groups cannot be fully ruled out, the finding of increased homozygosity associated with heightened disease risk is predicted by classical genetic models (30) and is supported by empirical data from Drosophila and other organisms (31). Intriguingly, studies of population isolates and consanguineous families demonstrate elevated rates of schizophrenia (32, 33).
The presence of nine specific ROHs was associated with illness susceptibility both individually and cumulatively. Four of these regions implicated genes related to postsynaptic (largely glutamatergic) receptor complexes implicated in SCZ pathophysiology. These genes include NOS1AP and NSF, each of which has been associated with schizophrenia, as well as GPHN and SGCD, which have not been previously examined in SCZ association studies. A fifth region spanning the coding region of SNTG1 was associated with SCZ in exploratory analyses; syntrophin abnormalities in SCZ are consistent with the accumulating evidence associating DTNBP1 haplotypic variation with SCZ susceptibility (23).
It should be noted that results for at least one risk region (roh314) may be influenced by the frequent presence of copy number variation at chromosome 17q21 (34); however, it is unlikely that results of the present study are primarily reflective of copy number variation, for four reasons. First, HapMap data suggest that duplications in this region are far more common than deletions (34), whereas deletions are more likely to create a spurious pattern of homozygous calls (35). Second, deletions in this region have been associated with mental retardation (36), which is not observed in our study. Third, chromosomal locations containing highly common ROHs (Table 1) are not generally marked by frequent copy number variation in publicly available databases (34). Fourth, inspection of raw intensity plots from microarrays analyzed for the present study are not consistent with frequent, large regions of copy number variation in the neighborhood of common ROHs (data not shown). Further research is needed to carefully examine the role of copy number variation in SCZ.
It is noteworthy that most of the risk ROHs demonstrated low frequencies in the general population. Future studies may determine whether these rare variants, conferring high risk ratios in small subpopulations, demarcate dissociable subtypes of illness at the genetic level. It is possible that this form of genetic heterogeneity coexists with the multifactorial, common-disease/common-variant mode of inheritance that is generally studied in whole-genome association. Twin studies of heritability of schizophrenia demonstrate considerable heterogeneity in MZ/DZ concordance rates (37), which may be consistent with a disease that can follow either multifactoral polygenicity or oligogenic heterogeneity modes of transmission in different families (38). As a simplified example, a single allele with 10% frequency (1% homozygosity) in the general population, conveying 10-fold increased risk under a recessive model, could account for a large portion of the sibling recurrent risk (estimated at 10%) in a small number of families with schizophrenia (10%). Such an allele would likely be missed by other methodologies, including standard WGA and linkage designs.
Finally, at least two risk ROHs were relatively common in controls, possibly reflecting positive selection. It is perhaps counterintuitive that such ROHs would be commonly observed in patients with schizophrenia. However, results are consistent with a model of rare, deleterious recessive effects associated with an allele or haplotype with overdominant properties (15, 39). These balancing effects may either be the result of the same allele, as in HBB and malaria, or from distal alleles that have hitchhiked on a region undergoing selection (5). Although WGHA currently lacks the spatial resolution to identify the causative allele(s), regions reported in the present study provide fairly narrow windows containing highly plausible candidates for further investigation.
Methods
Participants.
As described in ref. 20, patients with SCZ spectrum disorders (total n = 178, including 158 patients with schizophrenia, 13 patients with schizoaffective disorder, and 7 with schizophreniform disorder) were recruited from the inpatient and outpatient clinical services of The Zucker Hillside Hospital, a division of the North Shore–Long Island Jewish Health System. After written informed consent was provided, the Structured Clinical Interview for DSM-IV Axis I disorders (SCID, version 2.0) was administered by trained raters from a single site and team. Information obtained from the SCID was supplemented by a review of medical records and interviews with family informants when possible; all diagnostic information was compiled into a narrative case summary and presented to a consensus diagnostic committee, consisting of a minimum of three senior faculty. Subjects were representative of the ZHH patient sample and were unselected for any particular features other than ethnicity (Caucasian) and availability of DNA.
Healthy controls (n = 144) were recruited by use of local newspaper advertisements, flyers, and community internet resources. After written informed consent was provided, the nonpatient SCID (SCID-NP) was administered to rule out the presence of an Axis I psychiatric disorder; a urine toxicology screen for drug use and an assessment of the subject's family history of psychiatric disorders were also performed. Exclusion criteria included (current or past) Axis I psychiatric disorder, psychotropic drug treatment, substance abuse, a first-degree family member with an Axis I psychiatric disorder, or the inability to provide written informed consent. Patients (65 female/113 male) and controls (63 female/81 male) did not significantly differ in sex distribution (P > 0.05).
All subjects self-identified as Caucasian, non-Hispanic. As described in ref. 20, population structure was tested by examination of 210 ancestry informative markers (AIMs). AIMs included all SNPs on the array that passed initial quality control procedures and demonstrated a frequency difference of ≥0.5 in comparisons between Caucasian individuals and Asians or African-Americans in data made publicly available by Shriver and colleagues (40) (http://146.186.95.23/biolab/voyage/psa.html). Two tests of structure were performed, both of which indicated no significant stratification. First, analysis with the STRUCTURE (41) program (using multiple levels of K) confirmed that all subjects were drawn from a single population; second, comparison of cases and controls on allelic frequency across the 210 AIMs revealed no differences beyond those expected by chance.
Genotyping.
Genomic DNA extracted from whole blood was hybridized to two oligonucleotide microarrays (42) containing ≈262,000 and ≈238,000 SNPs (mean spacing = 5.8 kb; mean heterozygosity = 27%) as per manufacturer's specifications (Affymetrix). Patients and controls were proportionally distributed on each plate and were processed together to minimize confounding plate artifacts. Genotype calls were obtained by using the Bayesian Robust Linear Model with Mahalanobis distance classifier (BRLMM) algorithm thresholded at 0.5 applied to batches of 100 samples. Quality control procedures followed several steps (20). First, samples that obtained mean call rates <90% across both chips (or <85% for a single chip) were rejected. Mean call rate of remaining samples (total n = 322) was 97%. Twenty two of these cases were successfully repeated, and concordance of the two calls (reliability) for each SNP was evaluated. SNPs with more than one discrepancy were excluded from further analyses. Concordance across the remaining 454,699 SNPs exceeded 99.4%. Additionally, 9,936 SNPs in the sex-linked (i.e., nonpseudoautosomal) portion of the X chromosome were deleted, yielding 444,763 SNPs available for WGHA analysis. For WGHA, individual SNPs with low call rates even in valid cases were included, as were SNPs not in Hardy–Weinberg equilibrium in the control sample, because SNPs with these properties may be indicative of structural genomic variation of interest (35). It should be noted that the major results reported in Tables 1 and 2 were not substantively changed when analyses were performed on only the 439,511 SNPs that met strict QC criteria (Hardy–Weinberg equilibrium P > 0.001 in controls, and call rate >85). Specifically, patients still exhibited an average of four more ROHs compared with controls (P = 0.006). Each of the nine “risk ROHs” described in Table 2 remained significant at the P < 0.01 level. Additionally, each of the nine most common ROHs in healthy controls (Table 1) remained prevalent at a frequency ≥24%. All statistical analyses described above were conducted by using HelixTree software (Golden Helix).
WGHA: Construction of ROHs.
WGHA analysis entails several within-subject and across-subject analytic steps, each performed with customized python scripting in the HelixTree environment: First, SNP data from each chromosome of each subject were interrogated for runs of homozygosity (ROHs), which are long series of consecutive SNPs that are homozygous (uncalled SNPs are permitted within a run, because these may indicate genomic phenomena of interest). A fixed threshold of 100 consecutive SNPs was selected in a manner analogous to a recently published study that used the Affymetrix 10K chip to study recessive effects in known consanguineous families (28). It is acknowledged that patterns of LD are quite variable across the genome and that the threshold could be dynamically adjusted to account for this regional variability. However, dynamically adjusting ROH requirements for regional LD would confound the primary goal of the ROH approach, which is the identification of regions of strong, extended LD.
Additionally, it is possible that variable SNP properties on the Affymetrix array can result in a nonuniform distribution of heterozygosity; for example, locally dense marker spacing or lower minor allele frequency could result in ROHs that do not reflect meaningful biological phenomena. We addressed this potential confound to the interpretability of ROHs in two ways: (i) Mean SNP density across all 339 ROHs in SI Table 4 (≈7.1 kb) was lower than SNP density than the average across the entire 500K array (≈5.8 kb). Even excluding seven ROHs that span centromeres, which might artificially inflate the SNP spacing, the average marker spacing in the remaining ROHs is 6.0 kb, which is still slightly greater than mean spacing across the array. (ii) Minor allele frequency (and thus, heterozygosity) of SNPs in common ROHs (as identified in ZHH controls) was higher than the array average when measured in HapMap YRI samples (see Results).
Our criterion of 100 consecutive SNPs was selected to be more than an order of magnitude larger than the mean haploblock size in the human genome, without being so large as to be very rare, which would prohibit meaningful group comparisons. As an approximation, putting aside regional variability in LD and heterozygosity, the likelihood of observing 100 consecutive chance events can be described as follows: Because mean heterozygosity across all SNPs in the ZHH was observed to be 27%, any given SNP has, on average, a 0.73 chance of being called homozygous. Given 444,763 reliable SNPs and 322 subjects, a minimum run length of 70 would be required to produce <5% randomly generated ROHs across all subjects (0.7370 × 444,763 × 322 = 0.04), assuming complete independence of all SNPs. Because of linkage disequilibrium, SNP calls are not fully independent, thereby inflating the likelihood of chance occurrence of biologically meaningless ROHs. Genomewide identification of tag SNPs within windows of 70 markers by using the Carlson method (2) as implemented in HelixTree revealed 314,869 separable tag groups, representing a 29.3% reduction of information compared with the total number of original SNPs. Thus, run size of 100 SNPs was selected to approximate the degrees of freedom of 70 independent SNP calls.
Each subject's SNP data were then converted to binary calls (0 or 1) at each position indicating whether that SNP is a member of a ROH for that individual. Next, at each position, data from all subjects were examined to determine whether a minimum number of individuals share a ROH call at a given position. Because the purpose of this investigation was the identification of statistical differences between biologically meaningful ROHs in a case-control design, SNPs with <10 ROH calls across the entire sample were eliminated, resulting in 65,422 SNPs with 10 or more ROH calls, an 85% reduction from the original pool of SNPs. Taking this strategy a step further, “common” ROHs were identified that contained a minimum of 100 consecutive ROH calls across 10 or more subjects. A total of 339 such ROHs were identified across the genome, ranging in size from 100 to 852 SNPs in length (mean = 161, SD = 82, median = 133, see SI Table 4). A subject whose individual ROH calls overlapped with a common ROH was called present for that common ROH. Thus, each subject could have a total (sum) score ranging from 0 to 339.
WGHA Statistical Plan.
Based on these definitions, the statistical plan followed several steps for the identification of differences between cases and controls. First, this total score for common ROHs was compared between cases and controls by using Student's t test; this constituted a single genomewide test for difference in ROH frequency, with α set to 0.05. Next, as a planned post hoc examination of any significant genomewide difference, case-control comparisons of frequency of presence for each common ROH were examined by using χ2 tests (or Fisher's exact test when expected values <10 were found for any cell); although α would be protected by the preceding genomewide comparison, the threshold for significance for this analysis was set to P < 0.01 to further reduce the risk of false positives. Third, the cumulative effect of these risk-imparting ROHs (i.e., the dose-dependence of the presence of “risk ROHs”) was tested with logistic regression. Because the predictor variables for these logistic regression analyses were the ROHs already identified as significantly differentiating cases and controls, the raw P values for these regressions should be considered as strongly anticonservative. Therefore, empirical P values were calculated by using 100,000 permutations of the full ROH dataset for each regression analysis.
Finally, as an exploratory analysis to potentially identify smaller regions of difference between cases and controls, χ2 tests were performed on the 54,600 binarized SNP calls within common ROHs. Analogous to the dual-thresholding procedures commonly used in voxelwise brain imaging studies (43), statistical significance for these exploratory analyses was defined as 50 or more consecutive SNPs significantly differing between cases and controls at the P < 0.01 level (see SI Text for WGHA methods summary).
Supplementary Material
Acknowledgments
We thank Dr. Irving I. Gottesman for helpful suggestions regarding interpretation of the data. This work was supported by the Donald and Barbara Zucker Foundation, internal funding from the North Shore–Long Island Jewish Health System, a Keyspan Fellowship Award (to T.L.), and grants from the Stanley Foundation (to A.K.M.), the National Alliance for Research on Schizophrenia and Depression (to A.K.M.), and the National Institutes of Health (MH065580 to T.L., MH074543 to J.M.K., and MH001760 to A.K.M.).
Footnotes
Conflict of interest statement: C.L. is employed with Golden Helix and holds >5% equity in the company. Golden Helix subsequently developed a commercial version of the data analysis methodologies described in this paper.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710021104/DC1.
References
- 1.Hirschhorn JN, Daly MJ. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 2.Carlson CS, Eberle MA, Kruglyak L, Nickerson DA. Nature. 2004;429:446–452. doi: 10.1038/nature02623. [DOI] [PubMed] [Google Scholar]
- 3.Storey JD, Tibshirani R. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, Schwab SG, Pulver AE, Faraone SV, et al. Am J Hum Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans PD, Gilbert SL, Mekel-Bobrov N, Vallender EJ, Anderson JR, Vaez-Azizi LM, Tishkoff SA, Hudson RR, Lahn BT. Science. 2005;309:1717–1720. doi: 10.1126/science.1113722. [DOI] [PubMed] [Google Scholar]
- 7.Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, Sawamura N, Park U, Kudo C, Okawa M, et al. Nat Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- 8.McVean GA, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 9.Reich DE, Schaffner SF, Daly MJ, McVean G, Mullikin JC, Higgins JM, Richter DJ, Lander ES, Altshuler D. Nat Genet. 2002;32:135–142. doi: 10.1038/ng947. [DOI] [PubMed] [Google Scholar]
- 10.Coop G, Przeworski M. Nat Rev Genet. 2007;8:23–34. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- 11.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 12.Kim Y, Nielsen R. Genetics. 2004;167:1513–1524. doi: 10.1534/genetics.103.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 14.International HapMap Consortium. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voight BF, Kudaravalli S, Wen X, Pritchard JK. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang ET, Kodama G, Baldi P, Moyzis RK. Proc Natl Acad Sci USA. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson J, Morton NE, Collins A. Hum Mol Genet. 2006;15:789–795. doi: 10.1093/hmg/ddi493. [DOI] [PubMed] [Google Scholar]
- 18.Simon-Sanchez J, Scholz S, Fung HC, Matarin M, Hernandez D, Gibbs JR, Britton A, de Vrieze FW, Peckham E, Gwinn-Hardy K, et al. Hum Mol Genet. 2007;16:1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- 19.Miyazawa H, Kato M, Awata T, Kohda M, Iwasa H, Koyama N, Tanaka T, Kyo S, Okazaki Y, Hagiwara K. Am J Hum Genet. 2007;80:1090–1102. doi: 10.1086/518176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lencz T, Morgan TV, Athanasiou M, Dain B, Reed CR, Kane JM, Kucherlapati R, Malhotra AK. Mol Psychiatry. 2007;12:572–580. doi: 10.1038/sj.mp.4001983. [DOI] [PubMed] [Google Scholar]
- 21.Talbert PB, Bryson TD, Henikoff S. J Biol. 2004;3:18. doi: 10.1186/jbiol11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keinan A, Mullikin JC, Patterson N, Reich D. Nat Genet. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen NC, Bagade S, Tanzi R, Bertram L. [Accessed May 2, 2007];The SchizophreniaGene Database. Schizophrenia Research Forum. Available at www.schizophreniaforum.org/res/sczgene/default.asp.
- 24.Xu B, Wratten N, Charych EI, Buyske S, Firestein BL, Brzustowicz LM. PLoS Med. 2005;2:e263. doi: 10.1371/journal.pmed.0020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyosseva SV, Elbein AD, Hutton TL, Griffin ST, Mrak RE, Sturner WQ, Karson CN. Arch Gen Psychiatry. 2000;57:685–691. doi: 10.1001/archpsyc.57.7.685. [DOI] [PubMed] [Google Scholar]
- 26.Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 27.Hardy J, Pittman A, Myers A, Fung HC, de Silva R, Duckworth J. Alzheimer Dis Assoc Disord. 2006;20:60–62. doi: 10.1097/01.wad.0000201853.54493.d8. [DOI] [PubMed] [Google Scholar]
- 28.Woods CG, Cox J, Springell K, Hampshire DJ, Mohamed MD, McKibbin M, Stern R, Raymond FL, Sanford R, Malik Sharif S, et al. Am J Hum Genet. 2006;78:889–896. doi: 10.1086/503875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R. Genome Res. 2005;15:1496–1502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markow TA, Gottesman II. Genetica. 1993;89:297–305. [Google Scholar]
- 31.Wright A, Charlesworth B, Rudan I, Carothers A, Campbell H. Trends Genet. 2003;19:97–106. doi: 10.1016/s0168-9525(02)00033-1. [DOI] [PubMed] [Google Scholar]
- 32.Myles-Worsley M, Coon H, Tiobech J, Collier J, Dale P, Wender P, Reimherr F, Polloi A, Byerley W. Am J Med Genet B. 1999;88:4–10. doi: 10.1002/(sici)1096-8628(19990205)88:1<4::aid-ajmg2>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 33.Bulayeva KB. Croat Med J. 2006;47:641–648. [PMC free article] [PubMed] [Google Scholar]
- 34.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, Dallaire S, Gabriel SB, Lee C, Daly MJ, et al. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 36.Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, Hurst JA, Stewart H, Price SM, Blair E, Hennekam RC, et al. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 37.Sullivan PF, Kendler KS, Neale MC. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 38.Goldman D, Oroszi G, Ducci F. Nat Rev Genet. 2005;6:521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- 39.Crespi B, Summers K, Dorus S. Proc R Soc London Ser B. 2007;274:2801–2810. doi: 10.1098/rspb.2007.0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H, Jovel C, Pfaff C, Jones C, Massac A, Cameron N, et al. Hum Genet. 2003;112:387–399. doi: 10.1007/s00439-002-0896-y. [DOI] [PubMed] [Google Scholar]
- 41.Pritchard JK, Stephens M, Donnelly P. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kennedy GC, Matsuzaki H, Dong S, Liu WM, Huang J, Liu G, Su X, Cao M, Chen W, Zhang J, et al. Nat Biotechnol. 2003;21:1233–1237. doi: 10.1038/nbt869. [DOI] [PubMed] [Google Scholar]
- 43.Poline JB, Worsley KJ, Evans AC, Friston KJ. NeuroImage. 1997;5:83–96. doi: 10.1006/nimg.1996.0248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.